To the Editor
ORMDL3, located on chromosome 17q21, is highly linked to asthma in genetic association studies1. The SNP linking ORMDL3 to asthma is associated with increased levels of ORMDL3 expression2. Mice overexpressing increased levels of human ORMDL have spontaneous increased levels of airway hyperreactivity (AHR) and increased airway smooth muscle (ASM) in the absence of airway inflammation3. ORMDL3 is localized to the endoplasmic reticulum (ER) and regulates several downstream pathways including the ER localized transcription factor ATF6α4–7. Prior studies of hORMDL3zp3−Cre transgenic mice which overexpress human ORMDL3 have demonstrated that these mice have increased lung expression of ATF6α3 suggesting that ATF6α may be one of the downstream pathways contributing to increased ASM and increased AHR in hORMDL3zp3−Cre transgenic mice.
In this study we have utilized ATF6α−/− mice to determine whether ATF6α contributes to AHR and could explain a downstream pathway from ORMDL3 to increased AHR. These studies (all methods described in the Supplementary On-Line Repository Methods) demonstrated that there was no difference in levels of ASM mass (Figure 1A–B) or AHR (Figure 1C) in naïve ATF6α−/− mice compared to naïve WT mice. However house dust mite (HDM) challenged ATF6α−/− mice had significantly less ASM mass compared to HDM challenged WT mice (Figure 1A–1B). This reduction in ASM mass in HDM challenged ATF6α−/− mice was associated with reduced AHR to Mch compared to HDM challenged WT mice (p<0.05 Mch 24 mg/ml; p<0.01 Mch 48 mg/ml) (Figure 1C). There was no significant difference in levels of peribronchial fibrosis (Fig 1D), mucus (Fig 1E), BAL inflammation, peribronchial inflammation, or selected BAL cytokines (IL-5, IL-13, TGF-β1) comparing HDM challenged WT and HDM challenged ATF6α−/− mice (Supplementary On-Line Repository Figure E1A–K).
Figure 1. Reduced peribronchial smooth muscle and AHR in HDM challenged ATF6α−/− mice.
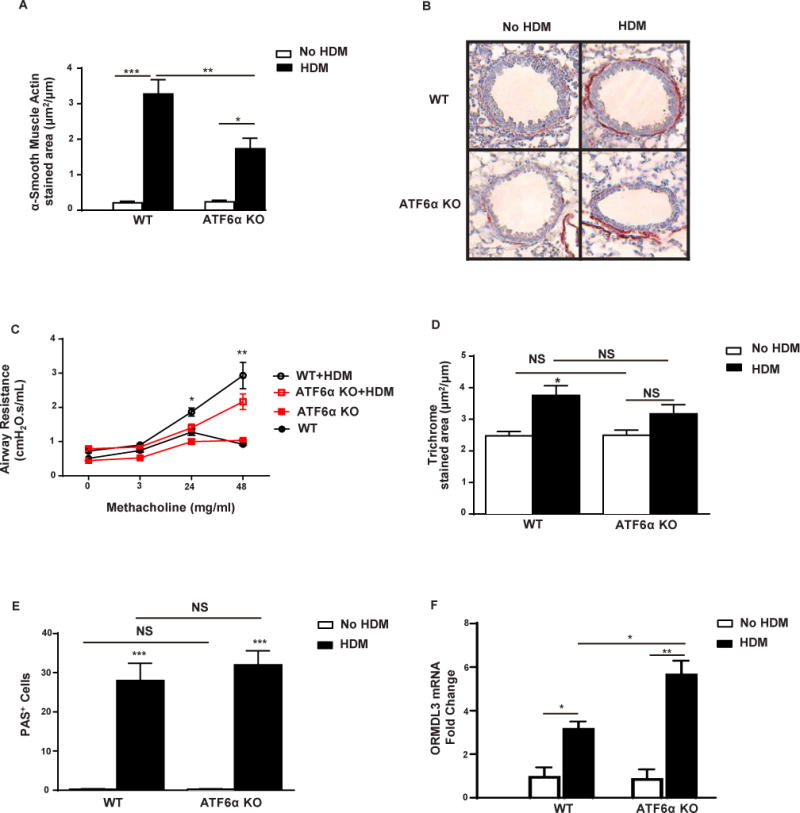
ATF6α−/− or WT mice (n = 8 mice/group) were challenged with HDM intranasally on day 0, 7, 14, 21. On day 24, AHR to methacholine was assessed, and lung tissues were processed for immunohistology and image analysis. A–B, Levels of peribronchial smooth muscle mass were quantitated by immunohistochemistry using an anti-α-smooth muscle actin Ab and image analysis (original magnification ×200). C, Airway resistance in response to methacholine was measured by flexiVent. WT + HDM vs WT, p<0.001 (Mch 48 mg/ml); ATF6α−/− +HDM vs ATF6α−/−, p<0.01 (Mch 48 mg/ml). Asterisk p values depicted in Figure are for WT+HDM vs ATF6α−/− +HDM. D, Levels of peribronchial fibrosis were quantitated as the area of trichrome staining. E, Levels of lung mucus were quantitated by PAS staining. F, Levels of ORMDL3 mRNA were quantitated in the lungs of WT and ATF6α−/− mice before and after HDM challenge by RT-qPCR. *P <0.05, **P <0.01, ***P <0.001 and NS (not significant).
Levels of ORMDL3 were similar at baseline in the lungs of WT and ATF6α−/− mice (Fig 1F). ORMDL3 was significantly induced in the lungs of both WT and ATF6α−/− mice following HDM challenge. Interestingly, levels of lung ORMDL3 were induced to a higher degree in HDM challenged ATF6α−/− compared to HDM challenged WT mice. This suggests that there may be a feedback loop from ATF6α to ORMDL3 such that ATF6α inhibits allergen induced expression of ORMDL3 in the lung. When ATF6α is inhibited (i.e. ATF6α−/− mice), the brake on inhibition of ORMDL3 is removed resulting in increased levels of ORMDL3 under conditions of HDM challenge. Thus, ATF6α may be both downstream of ORMDL34, as well as mediate a novel feedback loop to inhibit ORMDL3 under conditions of allergen challenge.
As our studies demonstrated that ATF6α−/− mice had reduced levels of HDM induced ASM and AHR without reduced levels of airway inflammation, we examined whether an intrinsic effect of ATF6α in mouse ASM (mASM) cells could influence ASM contraction, proliferation, or apoptosis in vitro. These studies demonstrated that mASM cells from both WT and ATF6α−/− mice had similar low levels of spontaneous contraction as assessed in an ASM collagen gel contraction assay (Figure 2A). Incubation of WT ASM cells with either Mch (Figure 2A) or histamine (Figure 2B), resulted in a rapid increase in contraction by 15 minutes which persisted through 30 minutes. In contrast, incubation of ATF6−/− deficient mASM cells resulted in a significantly reduced contractile response to agonists at 15 minutes which persisted through 30 minutes (p<0.01 histamine; p<0.05 Mch; ATF6α−/− mice vs WT mice) (Figure 2A, Figure 2B).
Figure 2. Effect of ATF6α on mouse and human ASM contraction, proliferation, and apoptosis in vitro.
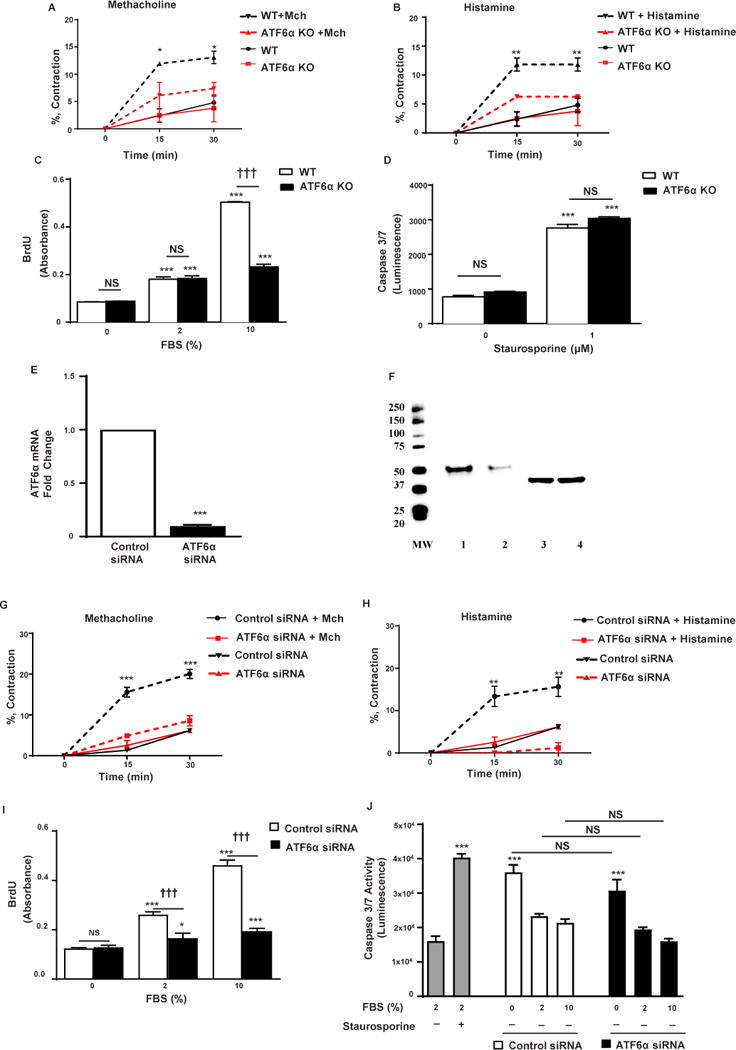
Levels of smooth muscle contraction in WT or ATF6α−/− mASM cells were assessed at baseline (time 0 min), as well as 15 and 30 min after incubation with either A, methacholine WT + Mch vs WT, p<0.05 (15 min), p<0.05 (30 min); ATF6α−/− + Mch vs ATF6α−/−, p=NS (15 min), p=NS (30 min); Asterisk p values depicted in Figure are for WT+ Mch vs ATF6α−/− + Mch, or B, histamine WT + histamine vs WT, p<0.01 (15 min), p<0.01 (30 min); ATF6α−/− + histamine vs ATF6α−/−, p<0.05 (15 min), p< NS (30 min); Asterisk p values depicted in Figure are for WT+ histamine vs ATF6α−/− + histamine. With agonist-induced smooth muscle contraction the area of the gel decreases significantly. C, Cell proliferation was assessed by BrdU incorporation in WT or ATF6−/− mASM cells incubated with either 0%, 2%, or 10% FBS. D, Caspase3/7 activity was measured by Caspase-Glo assay in WT or ATF6−/− mASM cells cultured in the presence of staurosporine (1μM) or absence of staurosporine (0 μM). E, The efficiency of siRNA knockdown of ATF6α mRNA (RT-qPCR), and F, ATF6α protein (western blot) in hASMS cells was assessed. Lane 1 and 2 are ATF6α. Lane 3 and 4 are GAPDH. MW is molecular weight marker, lane 1 hASM control siRNA, lane 2 hASM ATF6α siRNA, lane 3 hASM control siRNA, lane 4 hASM ATF6α siRNA. G–H, Levels of smooth muscle contraction in ATF6α siRNA or control siRNA transfected hASM cells were assessed at baseline (time 0 min), as well as 15 and 30 min after incubation with either G, methacholine, Control siRNA + Mch vs Control siRNA, p<0.001 (15 min), p< 0.001 (30 min); ATF6α siRNA+ Mch vs ATF6α siRNA, p=NS (15 min), p=NS (30 min); Asterisk p values depicted in Figure are for Control siRNA+ Mch vs ATF6α siRNA + Mch, or H, histamine, Control siRNA + histamine vs Control siRNA, p<0.01 (15 min), p< 0.01 (30 min); ATF6α siRNA+ histamine vs ATF6α siRNA, p=NS (15 min), p<0.05 (30 min); Asterisk p values depicted in Figure are for Control siRNA+ histamine vs ATF6α siRNA + histamine. I, Cell proliferation was assessed by BrdU incorporation in ATF6α siRNA or control siRNA transfected hASM cells incubated with either 0%, 2%, or 10% FBS. J, Caspase3/7 activity was measured by Caspase-Glo assay in hASM cells cultured in 2% FBS in the presence or absence of staurosporine (grey bars). In addition ATF6α siRNA or control siRNA transfected hASM cells were cultured in the presence or absence of FBS (0, 2, and 10%). *P <0.05, **P <0.01, ***P <0.001, †††P <0.001, and NS (not significant).
In addition, ATF6α−/− mASM cells had a minimal proliferative response to FBS 10% in vitro as assessed by BrdU incorporation, which was significantly less than that observed with WT mASM cells stimulated with FBS 10% (p<0.001; ATF6α−/− vs WT)(Figure 2C). Both WT and ATF6α−/− mASM cells had a similar minimal proliferative response to FBS 2% (Figure 2C). Thus, ATF6α−/− contributes to mASM cell proliferation under conditions of an increased mASM cell proliferative stimulus. Next, we examined whether the decrease in proliferation in ATF6α−/− mASM cells was due to apoptosis as assessed with a caspase 3/7 assay. Staurosporine, a known inducer of apoptosis8, induced a similar significant increase in mASM cell apoptosis in both WT mice (p<0.001) and ATF6α−/− mice (p<0.001) as detected by a significant increase in caspase3/7 activity (Figure 2D). Thus, ATF6α does not modulate levels of apoptosis in proliferating mASM cells in vitro.
We also examined whether ATF6α regulated human ASM (hASM) contraction, proliferation, and apoptosis in vitro. In these studies, we used siRNA to ATF6α as compared to scrambled control siRNA to effectively knockdown ATF6α mRNA (p<0.001)(Figure 2E), and ATF6α protein (Figure 2F) in primary hASM cells. Incubation of hASM cells transfected with ATF6α siRNA with either Mch (Figure 2G) or histamine (Figure 2H), resulted in a significantly reduced contractile response to agonists at 15 minutes which persisted through 30 minutes (p<0.01 histamine; p<0.001 Mch; ATF6α siRNA vs control siRNA). The addition of histamine to ATF6α siRNA treated cells further blocked contraction (p<0.05)(Figure 2H) through an at present unknown mechanism. There was no difference in contraction in non-transfected ASM cells compared to control siRNA treated ASM cells at baseline or in response to methacholine (Fig E2A), or histamine (Fig E2B). In addition, hASM cells knocked down with ATF6α siRNA had a reduced proliferative response to FBS 2% or FBS 10% which was significantly less than that observed with control scrambled siRNA transfected hASM cells stimulated with either FBS 2% (p<0.001)(Figure 2I), and FBS 10% (p<0.001)(Figure 2I). Levels of apoptosis were not increased in either hASM cells transfected with ATF6α siRNA or control siRNA and incubated with 0% FBS, 2% FBS or 10% FBS (Figure 2J). Levels of eotaxin-1 in hASM stimulated with IL-13 (Fig E1L) were no different in hASM cells in which ATF6α is knocked down with siRNA or control siRNA. Thus, in vitro in hASM cells in which ATF6α is knocked down with siRNA there is inhibition of contraction, and proliferation, but there is no effect on eotaxin-1 production.
In summary, in this study we have demonstrated that ATF6α expression in mouse and human ASM cells in vitro plays an important role in ASM proliferation and contractility which are key features of asthma. Prior studies have also demonstrated that in epithelial cells ER resident protein 57 which is regulated by ATF6α, plays a role in fibrosis and AHRE8,E9. In vivo studies with ATF6α−/− mice demonstrate an important role for ATF6α in mediating increased levels of ASM and increased AHR in response to allergen challenge. In contrast, ATF6α did not play a significant role in mediating allergen induced airway inflammation. As ORMDL3 regulates ATF6α3,4 these studies suggest that ATF6α may be one of the important pathways downstream of ORMDL3 playing an important role in ASM and AHR changes characteristic of asthma. In this regard genes on chromosome 17q21 (ORMDL3, GSDMB)3,9 and the downstream pathways of ORMDL3 (ATF6α) may play an important part in the structural changes in the airway, which contrasts with chromosome 5q which harbors genes (IL-4, IL-5, IL-9, IL-13) which play an important role in the immune and inflammatory response in asthma.
Supplementary Material
Capsule Summary.
ORMDL3 on chromosome 17q21 is highly linked to asthma and regulates several downstream pathways including ATF6α. We demonstrated that ATF6α in airway smooth muscle (ASM) plays an important role in ASM contractility, proliferation, and AHR.
Abbreviations
- AHR
Airway hyperreactivity
- ASM
Airway smooth muscle
- ATF6α
Activating transcription factor 6α
- ATF6α−/− mice
Activating transcription factor 6α deficient mice
- BAL
Bronchoalveolar lavage
- ER
Endoplasmic reticulum
- HDM
House dust mite
- Mch
Methacholine
- ORMDL3
Orosomucoid like 3
- WT
Wild type
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Moffatt MF, Kabesch M, Liang L, Dixon AL, Strachan D, Heath S, et al. Genetic variants regulating ORMDL3 expression contribute to the risk of childhood asthma. Nature. 2007;448:470–3. doi: 10.1038/nature06014. [DOI] [PubMed] [Google Scholar]
- 2.Verlaan DJ, Berlivet S, Hunninghake GM, Madore AM, Larivière M, Moussette S, et al. Allele-specific chromatin remodeling in the ZPBP2/GSDMB/ORMDL3 locus associated with the risk of asthma and autoimmune disease. Am J Hum Genet. 2009;85:377–93. doi: 10.1016/j.ajhg.2009.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Miller M, Rosenthal P, Beppu A, Mueller JL, Hoffman HM, Tam AB, et al. ORMDL3 transgenic mice have increased airway remodeling and airway responsiveness characteristic of asthma. J Immunol. 2014;192:3475–87. doi: 10.4049/jimmunol.1303047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Miller M, Tam AB, Cho JY, Doherty TA, Pham A, Khorram N, et al. ORMDL3 is an inducible lung epithelial gene regulating metalloproteases, chemokines, OAS, and ATF6. Proc Natl Acad Sci U S A. 2012;109:16648–53. doi: 10.1073/pnas.1204151109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Adachi Y, Yamamoto K, Okada T, Yoshida H, Harada A, Mori K. ATF6 is a transcription factor specializing in the regulation of quality control proteins in the endoplasmic reticulum. Cell Struct Funct. 2008;33:75–89. doi: 10.1247/csf.07044. [DOI] [PubMed] [Google Scholar]
- 6.Haze K, Yoshida H, Yanagi H, Yura T, Mori K. Mammalian transcription factor ATF6 is synthesized as a transmembrane protein and activated by proteolysis in response to endoplasmic reticulum stress. Mol Biol Cell. 1999;10:3787–99. doi: 10.1091/mbc.10.11.3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Walter P, Ron D. The unfolded protein response: from stress pathway to homeostatic regulation. Science. 2011;334:1081–6. doi: 10.1126/science.1209038. [DOI] [PubMed] [Google Scholar]
- 8.Vagner T, Mouravlev A, Young D. A novel bicistronic sensor vector for detecting caspase-3 activation. J Pharmacol Toxicol Methods. 2015;72:11–18. doi: 10.1016/j.vascn.2014.11.006. [DOI] [PubMed] [Google Scholar]
- 9.Das S, Miller M, Beppu A, Mueller J, McGeough M, Vuong C, et al. GSDMB induces an asthma phenotype characterized by increased airway responsiveness and remodeling without lung inflammation. Proceedings National Academy Sciences USA. 2016;113:13132–13137. doi: 10.1073/pnas.1610433113. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


