Abstract
Staphylococcus epidermidis is a commensal bacterium which widely colonizes in human skin and mucous membrane and rarely causes clinically manifested infections. S. epidermidis surface protein I (SesI) is considered to be the major virulence factor of S. epidermidis infection, but its pathogenesis is not clear. Here, we demonstrated that the prevalence of sesI among S. epidermidis invasive isolates (20.8%, 26/125) was significantly higher than that among colonizing isolates (3.8%, 4/106). The positive rates of biofilm-associated genes (aap, icaA, IS256) and resistance-associated genes mupA among the sesI-positive isolates were significantly higher than those among sesI-negative isolates (p < 0.05). And antimicrobial susceptibility testing showed that the resistance rates of sesI-positive isolates to ciprofloxacin, gentamicin and trimethoprim/sulfamethoxazole were significantly higher than those among sesI-negative isolates. Interestingly, 80.8% (21/26) of sesI-positive isolates belong to ST2 determined by MLST, while ST2 was not found among any of the 99 sesI-negative invasive isolates, indicating that there is a strong association between carriage of sesI and ST2 clone. In order to further study the role of sesI gene in pathogenesis, the sesI gene mutant (S. epidermidis RP62AΔsesI) and complementary expression strain (S. epidermidis RP62AΔsesI-C) were successfully constructed. All experimental data indicated that sesI may promote S. epidermidis to adhere and aggregate, but it had no obvious effect on the mature stage of biofilm formation. Taken together, these results suggest that sesI, along with antimicrobial and other biofilm-associated genes enables S. epidermidis easier for colonization and adhesion and contributes to the spread of S. epidermidis, especially ST2 clone.
Keywords: Staphylococcus epidermidis, sesI, biofilm, adhesion, aggregation, virulence-associated genes
Introduction
Staphylococcus epidermidis, a commensal bacterium which colonizes frequently the skin and mucous membranes of humans, is part of the human epithelial microflora and less frequently involved in clinically manifested infections (Becker et al., 2014). However, with the increasing use of indwelling or implanted medical devices including catheters, heart valves, vascular bypass grafts, nervous shunts and prosthetic implants, and the increase of multimorbid, elderly, and immunocompromised patients, S. epidermidis is becoming one of the major opportunistic pathogens responsible for hospital-acquired infections, especially foreign body-related bloodstream infections (Becker et al., 2014). Compared with Staphylococcus aureus, S. epidermidis possesses fewer virulence properties (Otto, 2004).
Biofilm formation is the major virulence factor of S. epidermidis isolates associated with device-related infections (Otto, 2004). Polysacchariden intercellular adhesin (PIA) encoded by icaADBC operon is the best-studied mediator for biofilm formation in S. epidermidis (Otto, 2004). In addition to PIA, many components have been found to be associated with biofilm formation, such as teichoic acids (TA), accumulation-associated protein (Aap), extracellular matrix-binding protein (Embp), biofilm-associated protein (Bap), phenol-soluble modulins (PSMs) (Otto, 2012). Arginine catabolic mobile element (ACME), a novel genomic island, which can enhance the capacity of this species to colonize human skin, mucosal surfaces and in-dwelling medical devices, is another important virulence-associated factor (Barbier et al., 2011; Onishi et al., 2013). The virulence-associated factors of S. epidermidis are not well characterized. Only a few cell-wall-associated proteins of S. epidermidis, including serine-aspartate repeat protein (SdrG), Embp, Ebps and AtlE, were found to have potential adhesin function binding to extracellular host matrix molecules, such as fibrinogen, fibronectin, collagen, vitronectin and elastin (Becker et al., 2014). These cell-wall-associated proteins usually display a common C-terminal cell wall sorting signal (CWS) with LPXTG [leucine–proline–variable amino acid (X)–threonine–glycine] motif. Bowden et al. reported that S. epidermidis surface (Ses) proteins had a similar structural organization to the previously described cell-wall-anchored proteins from S. aureus and other Gram-positive cocci and certain genes encoding Ses proteins were found more frequently in disease isolates relative to colonizing isolates (Bowden et al., 2005). Bowden et al. (2005) also found that the prevalence of sesI gene among invasive isolates causing bacterimia was higher than that among contaminant and colonizing isolates, but there was no significant difference between these groups. But Soderquist et al. (2009) found that sesI was not found among the normal S. epidermidis flora of healthy individuals without any healthcare association, but was found in approximately 50% of clinical isolates causing invasive infections. The association between the presence of sesI and invasive or colonizing isolates was not clear. The role of SesI in the pathogenicity of S. epidermidis is still unknown. In the present study, the aim is to investigate the prevalence of sesI among S. epidermidis clinical and colonizing isolates, microbiological and molecular characteristics of sesI-positive isolates and the role of sesI in the biofilm formation of S. epidermidis.
Materials and Methods
Bacterial Isolates
One hundred and twenty-five non-duplicate S. epidermidis isolates were collected from the blood and catheter of inpatients in 2002–2008 and 2012–2014 at the First Affiliated Hospital of Wenzhou Medical University, Wenzhou, China. 68 (54.4%) and 57 (45.6%) S. epidermidis isolates were isolated from blood and catheter respectively. The identification of S. epidermidis isolates was performed using Gram-staining, catalase test, coagulase test, and a VITEK-2 automated platform (bioMérieux, Marcy l’Etoile, France). S. epidermidis isolates from more than one set of blood culture were considered to be invasive isolates. For catheter samples, the isolates with more than 15 colonies of pure culture were regarded as invasive isolates (Liduma et al., 2012). One hundred and six S. epidermidis isolates obtained from the urethral orifices of healthy volunteers with no symptoms were considered to be colonizing isolates. These colonizing isolates were identified as S. epidermidis by 16S rRNA sequencing (Shang et al., 2005). The Ethics Committee of the First Affiliated Hospital of Wenzhou Medical University exempted this study from review because the present study focused on bacteria. Verbal informed consents were obtained from all participants.
Escherichia coli DH5α, Escherichia coli DC10B, S. epidermidis RP62A and pKOR1 were shown in Supplementary Files. The S. epidermidis RP62A is biofilm former and carries sesI, icaA, mecA and aap. And antimicrobial susceptibility testing showed that S. epidermidis RP62A was resistant to trimethoprim/sulfamethoxazole, clindamycin and erythromycin, while sensitive to tetracycline, penicillin, ciprofloxacin, linezolid and vancomycin. And the MLST analysis showed S. epidermidis RP62A belonged to ST128.
Antimicrobial Susceptibility Testing
In vitro antimicrobial susceptibility testing for S. epidermidis invasive isolates was performed using the disk diffusion method according to the guidelines recommended by the Clinical and Laboratory Standards Institute (CLSI, 2014). Antimicrobial agents tested included ciprofloxacin (5 μg), clindamycin (2 μg), erythromycin (15 μg), gentamicin (10 μg), chloramphenicol (30 μg), linezolid (30 μg), penicillin (10 U), tetracycline (30 μg), and trimethoprim/sulfamethoxazole (1.25/23.75 μg). All antimicrobial disks were obtained from Oxoid Ltd., and S. aureus ATCC25923 was used as the quality control strain for antimicrobial susceptibility testing. The minimum inhibitory concentration of vancomycin was determined by the agar dilution method according to CLSI guidelines (CLSI, 2014).
Multi-locus Sequence Typing
Multi-locus sequence typing for S. epidermidis invasive isolates was performed by amplifying internal fragments of seven housekeeping genes including arcC, aroE, gtr, mutS, pyrR, tpiA, and yqiL, as described previously (Thomas et al., 2007). Allele numbers and STs were assigned using the online database available at http://www.mlst.net.
PCR Detection of Virulence- and Resistance-Associated Genes
All S. epidermidis invasive isolates were tested for the presence of mecA gene by PCR amplification as described previously (Murakami et al., 1991). The presence of virulence-associated genes including aae, aap, ACME-arcA, atlE, bhp, gehD, icaA, IS256 and sesI was also detected by PCR amplification using previously described primers (Bowden et al., 2002; Heilmann et al., 2003; Catalanotti et al., 2005; Soderquist et al., 2009; Du et al., 2013; Cala et al., 2015). Detection of genes for mupirocin resistance (mupA) and chlorhexidine-based antiseptic resistance (qacA/B) was also performed using previously described PCR primers (Li et al., 2013).
Construction of S. epidermidis sesI Mutant Strain and Its Corresponding Complemented Expression Strains
According to the results of the whole genome sequence of S. epidermidis RP62A, two sets of primers (Supplementary Files) containing additionally a KpnI restriction site were designed to amplify the upstream and downstream fragments of sesI gene, respectively. Genomic DNA (gDNA) of S. epidermidis RP62A was used as a template. At first, the upstream and downstream fragments of sesI gene were connected each other via the specific restriction sites (KpnI) by T4 DNA ligase to yield the homologous arm fragment with the deletion of sesI gene. Then the temperature sensitivity shuttle plasmid pKOR1 and the homologous arm fragment generated recombination reaction under the action of the BP Clonase II Enzyme, then formatted the homologous recombinant plasmid (named pKsesI). The recombinant plasmid (pKsesI) was firstly transferred into E. coli DH5α, then transferred into E. coli DC10B to modify, ultimately electroporated into S. epidermidis RP62A competent cells. The allelic exchange procedure was performed as described previously (Bae and Schneewind, 2006). In order to eliminate other gene mutations that may arise during the process of construction of mutant strains effecting on the results of phenotypic studies, a complementary strain was constructed. Amplifying the whole sesI gene and its upstream promoter and ribosome binding region (primers listed in Supplemental Files), then the gene fragments were connected to the expression vector pRB473 by the specific restriction sites, forming the recombinant complementary expression plasmid (pRBsesI). After modified by E. coli DC10B, pRBsesI was electroporated into the sesI mutant strain (RP62AΔsesI). The mutant strain and complementary expression strain was verified by PCR, fluorescent quantitative PCR (RT-qPCR) and sequencing.
RNA Extraction, cDNA Synthesis, and Quantitative Reverse Transcription-PCR (qRT-PCR)
Real time PCR was used to verify whether sesI gene was knocked out and complementary expression strain was constructed successfully. For RNA isolation, the overnight cultures were diluted 1:100 in TSB and grown to post-exponential phase at 37°C with shaking at 200 rpm. Total RNA was isolated and purified using a PureLink RNA Mini Kit (Invitrogen, Carlsbad, CA, United States); then was reverse transcribed into cDNA using a PrimeScript RT reagent kit (TaKaRa, Japan), according to the manufacturer’s protocol. The real-time PCR was carried out using a SsoFas EvaGreen Supermix kit (Bio-Rad, United States) with the Bio-Rad CFX96 Manager software. S. epidermidis RP62A wild type strain was used as a control (relative expression = 1), and gyrB was used as a reference gene to investigate genes of interest. RNA transcript levels were calculated by the method of delta delta Ct (ΔΔCt) (Montgomery et al., 2012). Data analysis was carried out using Bio-Rad CFX software. Each reaction was performed in triplicate. All measurements were independently conducted three times. All primers used are listed in Supplementary Files.
Growth Curve
Staphylococcus epidermidis RP62A wild type strain, S. epidermidis RP62A, sesI mutant strain (S. epidermidis RP62AΔsesI) and S. epidermidis RP62A sesI complementary expression strain (S. epidermidis RP62AΔsesI-C) were cultured overnight in 5 ml TSB at 37°C with shaking at 200 rpm. The overnight cultures were diluted into 50 ml TSB to obtain the same starting optical density (OD) at 562 nm. The growth of each strain was monitored by Microplate Manager 6 (Bio-Rad, United States) software at 1 h intervals for a total of 12 h with sterile TSB medium as control, then tested again at 24 h. The growth curve test was repeated at least two times.
Initial Adhesion Assay in Vitro
The overnight cultures were inoculated to 30 ml TSB medium according to the ratio of 1:200, and incubated at 37°C with shaking at 220 rmp until OD562 = 0.7∼0.8. Then the cell suspension was transferred to a microcentrifuge tube to pellet the cells, and were washed three times with sterile PBS buffer. Then PBS buffer was used to resuspend bacteria and adjust the OD562 = 0.6. A total of 3 ml cell suspensions was pipetted into the wells of 6-well flat bottomed polystyrene plates, which each contained a glass disk. Plates were incubated for 1h at 37°C without shaking, after which the disks were washed three times with PBS to remove the bacteria which were not adhered to the disks. Afterward, the disks were put into a sterile 50 ml microcentrifuge tube and pounded to pieces. Placed on the vortex oscillator to mix for 1 min with 15 s interval for five times, in order to remove the bacteria of adhesion on the coverlips completely. The cell suspension was diluted by 104, 105 and 106, respectively. Hundred microliter cell suspension was cultured on blood agar plates using the spiral plating system, and detected the endpoint numbers of CFU after incubated overnight at 37°C. Assays were performed in triplicate.
Aggregation Assay
Staphylococcus epidermidis PR62A wild type strain, S. epidermidis RP62AΔsesI and S. epidermidis RP62AΔsesI-C were incubated overnight in 5 ml sterile TSB medium at 37°C with shaking at 200 rpm, then the overnight culture medium of three strains were coated on glass slides, respectively. After drying, the gram stain were immediately performed. The aggregation ability of different strains could be observed under the microscope. Assays were performed in triplicate.
Biofilm Formation Assay
The ability of biofilm formation of different strains was tested using a semi-quantitative adherence assay in 96-well polystyrene microtiter plates (BD Biosciences) as previously described (Shahrooei et al., 2009, 2012). Briefly, the overnight cultures were diluted 1:200 with fresh TSB medium whether or not supplemented with 4% NaCl, 1% glucose or 4% EtOH, subsequently, adjusted to obtain the same starting optical density (OD) at 562 nm. Two hundred microliter of the diluted cultures of different bacteria were pipetted into sterile 96-well polystyrene microtiter plates with three attached wells of each bacteria, then incubated overnight at 37°C without shaking. After incubation, the suspension cultures were removed from each well, a total of 200 μl sterile PBS buffer was added to the wells, and rinsed three times gently, then dried afterward. The adhered material was stained with 200 μl of a 1% (w/v) crystal violet (Sigma) solution for 10 min, and subsequently, the wells were washed three times with water and again dried. The OD562 of the biofilm was measured by Microplate Manager 6 (Bio-Rad, United States) software. The experiment was repeated twice.
Scanning Electron Microscopy
Biofilm formed on glass coverslips (10 mm in diameter) was observed by scanning electron microscopy (SEM) as previously described (Pintens et al., 2008). Briefly, an overnight bacterial culture was diluted in proportion to 1:200 in TSB medium to obtain the same starting optical density (OD) at 562 nm. A glass disk was put in advance into the bottom of the sterile flat bottomed polystyrene plates (Costar 3524; Corning, NY, United States) and 1 ml of the cell suspensions was added into the wells. Then, the plates were incubated overnight at 37°C without shaking, after which each well was rinsed three times with PBS buffer, each time for 10 min with slight shaking. Biofilms formed on the glasses were fixed with 2.5% glutaraldehyde in 0.1 M sodium cacodylate buffer (pH 7.4) by incubation for 2 h at room temperature. Next the coverslips were washed three times with 0.1 M sodium phosphate buffer, each time for 15 min. Thereafter, 1% osmiumtetroxide was used for a post-fixation step for 2 h at 4°C, followed by three times washed with distilled water for 10 min each, and then dehydrated in a series of ascending ethanol baths (25, 50, 75, 95 and 100%) for 10 min each. The samples were freeze-dried for 5 h and ultimately coated with gold and palladium in an evaporator. The observations were usually performed with a scanning electron microscope (FEI Quanta 200; United States). The experiment was repeated twice.
Triton X-100 Induced Autolysis
In order to test the potential role of sesI for autolysis in S. epidermidis, Triton X-100 induced autolysis was performed. The overnight cultures were diluted in 50 ml TSB containing 1 M NaCl, then bacterial cells were pelleted by centrifugation from early exponentially growing cultures (OD562 = 0.7). Afterward, the cells were washed twice with 50 ml of ice-cold water and re-suspended into 50 ml of Tris-HCl (pH 7.2) containing 0.05% (vol/vol) TritonX-100. Autolysis was monitored during incubation at 37°C by Microplate Manager 6 (Bio-Rad, United States) software at half an hour intervals for a total of 3 h. The experiment was repeated twice.
Statistical Analysis
Chi-square or Fisher’s exact tests were used to compare the rates of biofilm-associated genes and antimicrobial resistance genes among sesI-positive vs. sesI-negative S. epidermidis isolates using SPSS statistical software (version 19, IBM SPSS Statistics). Statistical significance was associated with a two-sided P-value of <0.05. Real-time RT-PCR results were performed using GraphPad prism 6 software. A P-value of <0.05 was considered a significant difference.
Results
Prevalence of sesI among S. epidermidis Invasive and Colonizing Isolates
Twenty-six (20.8%) of 125 S. epidermidis invasive isolates were positive for sesI, while only 4 (3.8%) of 106 colonizing isolates were positive for this gene. The prevalence of sesI among S. epidermidis invasive isolates was significantly higher than that among colonizing isolates.
Antimicrobial Susceptibility Characteristics
The resistance rates of sesI-positive and sesI-negative S. epidermidis invasive isolates to antimicrobial agents tested were shown in Table 1. All invasive isolates tested were susceptible to vancomycin. Interestingly, the resistance rates of sesI-positive isolates to ciprofloxacin, gentamicin and trimethoprim/sulfamethoxazole were significantly higher than those among sesI-negative isolates (Table 1).
Table 1.
Antimicrobial resistance profiles and virulence gene carriage among S. epidermidis clinical isolates.
|
sesI-positiveisolatesb(N = 26) |
sesI-negative isolates (N = 99) |
p-valuec | |||
|---|---|---|---|---|---|
| No. | % | No. | % | ||
| Antimicrobialsa | |||||
| Ciprofloxacin | 19 | 73.1 | 19 | 19.2 | 0.000 |
| Chloramphenicol | 5 | 19.2 | 38 | 38.4 | 0.067 |
| Clindamycin | 17 | 65.4 | 49 | 49.5 | 0.149 |
| Erythromycin | 21 | 80.8 | 81 | 81.8 | 0.902 |
| Gentamicin | 21 | 80.8 | 20 | 20.2 | 0.000 |
| Linezolid | 0 | 0.0 | 0 | 0.0 | N/A |
| Penicillin | 25 | 96.2 | 93 | 93.9 | 0.662 |
| Tetracycline | 5 | 19.2 | 31 | 31.3 | 0.996 |
| SXTb | 22 | 84.6 | 29 | 29.3 | 0.000 |
| Vancomycin | 0 | 0.0 | 0 | 0.0 | N/A |
| Virulence genes | |||||
| aae | 25 | 96.2 | 97 | 98.0 | 0.588 |
| aap | 25 | 96.2 | 56 | 56.6 | 0.000 |
| ACME-arcA | 9 | 34.6 | 49 | 49.5 | 0.176 |
| atlE | 26 | 100 | 96 | 97.0 | 0.369 |
| bhp | 1 | 3.9 | 23 | 23.2 | 0.026 |
| gehD | 13 | 50.0 | 34 | 34.3 | 0.142 |
| icaA | 20 | 76.9 | 28 | 28.3 | 0.000 |
| IS256 | 18 | 69.2 | 35 | 35.4 | 0.002 |
| Resistance genes | |||||
| mupA | 8 | 30.8 | 14 | 14.1 | 0.048 |
| qacA/B | 13 | 50.0 | 31 | 31.3 | 0.076 |
| mecA | 25 | 96.2 | 89 | 89.9 | 0.316 |
aSXT, trimethoprim/sulfamethoxazole; bN, number of isolates; cNA, not analyzed.
Prevalence of mupA and qacA/B
Although the positive rate of qacA/B among sesI-positive S. epidermidis invasive isolates were higher than that among sesI-negative isolates, there were no significant differences between the two groups. However, the prevalence of mupA among sesI-positive S. epidermidis invasive isolates was higher than that among sesI-negative isolates, which has significantly difference between the two groups (p < 0.05) (Table 1).
Prevalence of Biofilm-Associated Genes
In the present study, the positive rate of icaA among sesI-positive S. epidermidis isolates (76.9%, 20/26) was significantly higher than that among sesI-negative isolates (28.3%, 28/99). The positive rate of aap among sesI-positive isolates was high to 96.2%, which was significantly higher than that among sesI-negative isolates (56.6%) (P < 0.05). All the sesI-positive S. epidermidis isolates were positive for atlE, and the prevalence of atlE in sesI-negative S. epidermidis isolates was 97.0%. And the prevalence of another multifunctional autolysin/adhesin gene aae, which encodes Aae with bacteriolytic activity that binds to fibrinogen, vitronectin and fibronectin was 96.2% vs. 98.0% in both sesI-positive and sesI-negative S. epidermidis invasive isolates. ACME-arcA was also found in both sesI-positive and sesI-negative S. epidermidis invasive isolates (34.6% vs. 49.5%). The prevalence of IS256 among sesI-positive isolates was significantly higher than that among sesI-negative isolates. However, bhp, a gene homologous to bap and assumed to promote biofilm formation, was found more frequent among sesI-negative isolates than sesI-positive isolates (Table 1).
Genetic Diversity of S. epidermidis Invasive Isolates
Distinct ST patterns were showed in Table 2. Among the 125 S. epidermidis invasive isolates, 80.8% (21/26) of sesI-positive isolates were belong to ST2 determined by MLST. A total of 45 STs were found among 99 sesI-negative isolates with ST466 being the most prevalent ST (11.1%, 11/99), followed by ST130 (8.1%, 8/99) and ST59 (7.1%, 7/99), indicating that sesI-negative S. epidermidis invasive isolates were more diverse genetically than sesI-positive isolates. Interestingly, ST2 was not found among the 99 sesI-negative invasive isolates.
Table 2.
Sequence type (ST) distribution among sesI-positive and –negative S. epidermidis invasive isolates.
| MLST |
sesI-positive isolates (N = 26) |
sesI-negative isolates (N = 99) |
p-valuec | ||
|---|---|---|---|---|---|
| No. | % | No. | % | ||
| ST2 | 21 | 80.8 | 0 | 0.0 | 0.000 |
| ST5 | 0 | 0.0 | 1 | 1.0 | 0.607 |
| ST14 | 0 | 0.0 | 1 | 1.0 | 0.607 |
| ST16 | 0 | 0.0 | 2 | 2.0 | 0.465 |
| ST218 | 0 | 0.0 | 2 | 2.0 | 0.465 |
| ST47 | 0 | 0.0 | 2 | 2.0 | 0.465 |
| ST20 | 0 | 0.0 | 5 | 5.1 | 0.242 |
| ST21 | 0 | 0.0 | 1 | 1.0 | 0.607 |
| ST57 | 0 | 0.0 | 2 | 2.0 | 0.465 |
| ST59 | 3 | 11.5 | 7 | 7.1 | 0.455 |
| ST89 | 1 | 3.9 | 1 | 1.1 | 0.305 |
| ST125 | 0 | 0.0 | 5 | 5.1 | 0.242 |
| ST130 | 0 | 0.0 | 8 | 8.1 | 0.134 |
| ST206 | 0 | 0.0 | 1 | 1.0 | 0.607 |
| ST210 | 0 | 0.0 | 5 | 5.1 | 0.242 |
| ST226 | 0 | 0.0 | 2 | 2.0 | 0.465 |
| ST227 | 0 | 0.0 | 2 | 2.0 | 0.465 |
| ST234 | 0 | 0.0 | 2 | 2.0 | 0.465 |
| ST235 | 0 | 0.0 | 2 | 2.0 | 0.465 |
| ST263 | 0 | 0.0 | 3 | 3.0 | 0.369 |
| ST267 | 0 | 0.0 | 1 | 1.0 | 0.607 |
| ST419 | 0 | 0.0 | 1 | 1.0 | 0.607 |
| ST466 | 0 | 0.0 | 11 | 11.1 | 0.075 |
Initial Adhesion Assay in Vitro
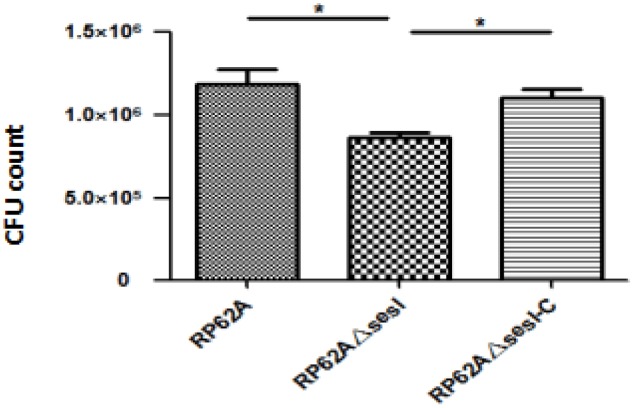
It has been confirmed that S. epidermidis surface proteins have the ability to adhere human epithelial cells and extracellular matrix proteins. The in vitro initial adhesion experiment was used to verify the role of sesI in the initial adhesion stage of bacterial biofilm formation. The ability of initial adhesion for S. epidermidis RP62AΔsesI was significantly lower than that for S. epidermidis RP62A and S. epidermidis RP62AΔsesI-C (Figure 1) (p < 0.05).
FIGURE 1.
Comparison of initial adhesion ability of wild type (RP62A), isogenic sesI mutant (RP62AΔsesI), and sesI-complemented (RP62AΔsesI-C) strains. ∗Indicate P < 0.05.
Aggregation Assay More Dispersed
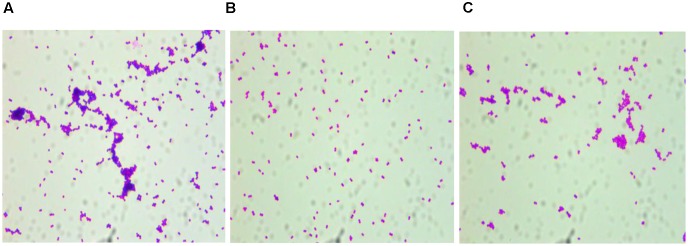
The overnight cultures of all three strains tested were coated on glass coverslips after Gram staining, and observed under the microscope, showing that the distribution of S. epidermidis RP62A and S. epidermidis RP62AΔsesI-C could be aggregated into blocks. However, the distribution of S. epidermidis RP62AΔsesI was more dispersed. And the distribution density of S. epidermidis RP62A was significantly higher than that of S. epidermidis RP62AΔsesI (Figure 2).
FIGURE 2.
Comparison of aggregation ability of different strains. Photograph were taken by oil immersion lens. (A) wild type (RP62A), (B) the sesI mutant strain, (C) the complement strain.
Biofilm Formation Assay
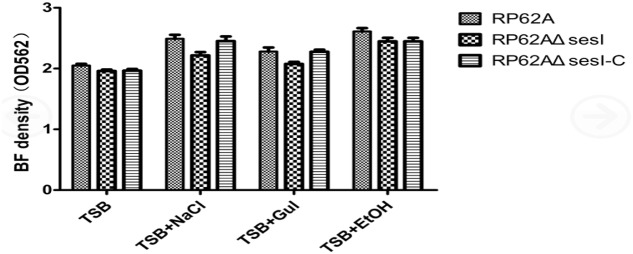
The biofilm formation ability of different strains was tested using a semi-quantitative adherence assay in 96-well polystyrene microtiter plates. The results showed that there was no significant difference among S. epidermidis RP62A, S. epidermidis RP62AΔsesI and S. epidermidis RP62AΔsesI-C in the mature stage of biofilm formation whether or not under the defined predisposing conditions (added 0.5%Glu, 4%NaCl and 4% ethanol) (P > 0.05) (Figure 3).
FIGURE 3.
Comparison of biofilm formation in mature stage under different conditions for different strains.
Scanning Electron Microscopy
Scanning electron microscopy can be used to directly and effectively observe the formation of biofilm. After a series of fixed, washed, dewatered, dried and sprayed gold for biofilm. S. epidermidis RP62A, S. epidermidis RP62AΔsesI and S. epidermidis RP62AΔsesI-C could be seen to accumulate and form microcolonies by scanning electron microscope. The microcolonies produced by all the three strains tested were overlapped with each other, forming dense clumps. It can be seen that there was no difference in the distribution and the sparsity degree of biofilm in the mature stage (Figure 5).
FIGURE 5.
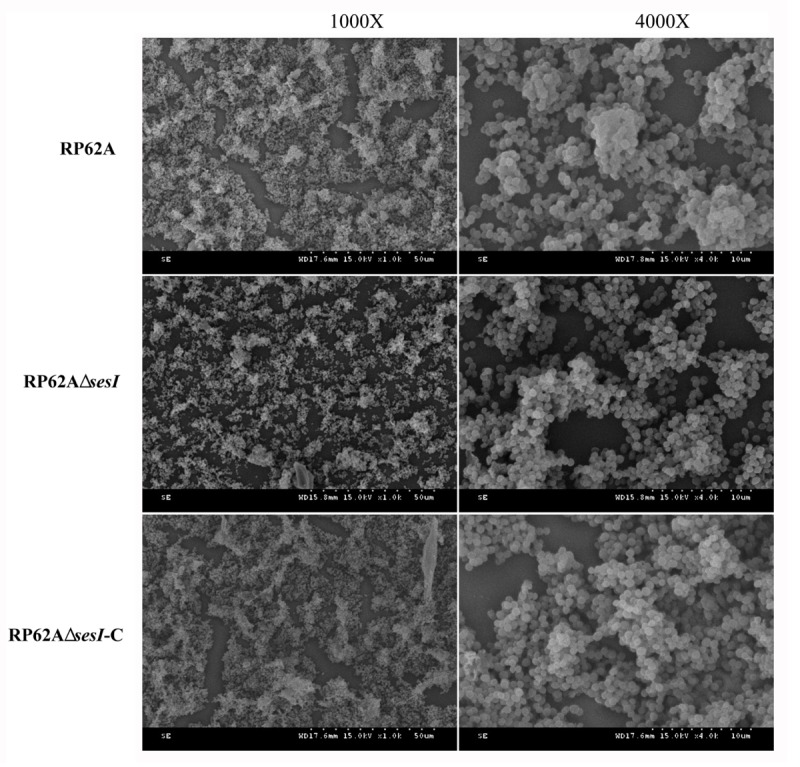
Scanning electron microscope study. Transmission electron micrograph of biofilm formation in mature stage of wild type (RP62A), isogenic sesI mutant (RP62AΔsesI), and sesI-complemented (RP62AΔsesI-C) strains.
Impact of sesI on S. epidermidis Murein Hydrolase Activity
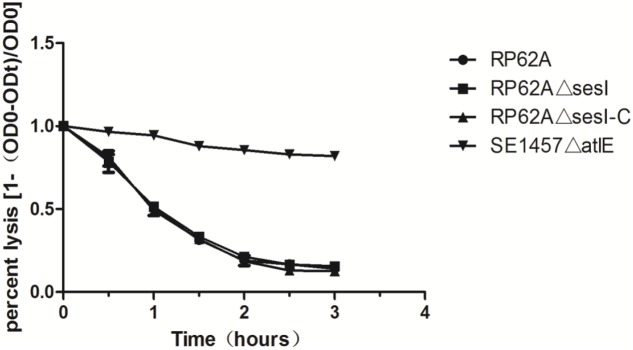
Triton X-100 induced autolysis of bacterial cells was carried out to investigate the effect of sesI on autolysis in S. epidermidis. No difference was found between S. epidermidis RP62AΔsesI and its parent strain in the Triton X-100 induced autolysis, while the negative control of the atlE knockout mutant was resistant to autolysis (Figure 4).
FIGURE 4.
Comparison of autolytic ability of wild type (RP62A), isogenic sesI mutant (RP62AΔsesI), and sesI-complemented (RP62AΔsesI-C) strains.
Discussion
The pathogenicity of S. epidermidis is mostly due to its ability to colonize implanted biomaterials and form biofilm on polymeric surfaces. The biofilm formation of S. epidermidis is a multifactorial process. The cell wall anchored protein (CWA) produced by Gram-positive bacteria can interact with the host extracellular matrix proteins, which can promote the adhesion function of bacteria. The cell wall associated surface protein containing the LPXTG motif can induce the surface protein to anchor to the peptidoglycan layer of the cell wall (Becker et al., 2014). Of the 11 known cell wall anchoring proteins containing the LPXTG motif, only 5 are known for their functions (Bowden et al., 2005). In the process of biofilm formation, Bhp can promote the initial adhesion and aggregation of bacteria. Aap protein is necessary for the aggregation of bacteria on the surface of polyethylene, thereby promoting the formation of a visible biofilm. SdrF and SdrG are members of the Sdr family of proteins, which are essential for the adhesion of SdrG to the human Fg matrix (Buttner et al., 2015). SesC proteins play an important role in the early stages of biofilm formation, thereby contributing to the chronic and persistent infection of the material (Khodaparast et al., 2016).
In the present study, we found that the prevalence of sesI among S. epidermidis invasive isolates was significantly higher than that among colonizing isolates. Previous study has reported that sesI was not found among the normal S. epidermidis flora of healthy individuals, but was detected in approximately 50% of invasive isolates associated with invasive infections (Soderquist et al., 2009), indicating that sesI may be a marker of the invasive capacity of S. epidermidis. For the sesI-positive S. epidermidis invasive isolates, having a higher resistance to clinically antimicrobial agents enables them to highly adapt to the nosocomial environment and to become important pathogen in healthcare setting.
Surface-active antiseptics and mupirocin are frequently used as part of comprehensive strategies designed to reduce Staphylococci colonization and infection in healthcare settings, and emergence of resistance to these antimicrobial compounds threatens hospital infection control efforts (Lepainteur et al., 2013). The mupA was detected by polymerase chain reaction in 63% of coagulase-negative Staphylococci isolates isolated from catheter-associated bloodstream infections in very preterm neonates (Lepainteur et al., 2013). In the present study, a relatively high prevalence of mupA found among sesI-positive S. epidermidis invasive isolates raises a challenge for the efforts to control hospital-acquired infections by decolonization of S. epidermidis isolates.
It has been reported that the majority of invasive S. epidermidis strains contain the ica operon in their genome. It has been verified that icaA is associated with biofilm formation of Staphylococci (Becker et al., 2014). Du et al. (2013) reported that 41.3% (43/104) of the clinical MRSE isolates isolated from blood, catheter and other sterile specimens of patients were found to carry icaA. In this present study, the prevalence of icaA among sesI-positive S. epidermidis isolates was higher relative to sesI-negative isolates. The accumulation-associated protein (Aap) encoded by aap confers intercellular adhesion and plays a role in skin colonization by its N-terminal domain A mediating adherence to human corneocytes (Macintosh et al., 2009; Salgueiro et al., 2017). The positive rate of aap among sesI-positive isolates was significantly higher than that among sesI-negative isolates (56.6%) (P < 0.05) in the present study. Previous study reported that atlE, encoding a surface-associated autolysin/adhesin which mediates initial adherence of bacterial cells to the polymer surfaces, was found in a total of 96.9% of invasive isolates (Heilmann et al., 1997). In the present study, more than 95% of sesI-positive and sesI-negative S. epidermidis isolates were positive for atlE. Interestingly, all atlE-positive isolates were also positive for another multifunctional autolysin/adhesingene aae encoding Aae with bacteriolytic activity that binds to fibrinogen, vitronectin, and fibronectin (Heilmann et al., 2003). Arginine catabolic mobile element (ACME), a genomic island which contributes to the enhanced capacity of S. epidermidis to colonize human skin and mucosal surfaces has been found in both commensal and invasive S. epidermidis isolates (Cherifi et al., 2013). Previous reports showed that >70% of S. epidermidis invasive isolates were positive for ACME-arcA (Cherifi et al., 2013; Du et al., 2013). In the present study, ACME-arcA was also found in both sesI-positive and sesI-negative S. epidermidis invasive isolates. The prevalence of ACME-arcA among sesI-positive isolates in the present study was similar to the result reported by Miragaia et al. (2009), in which 51% of S. epidermidis isolates from a widespread geographical origin were positive for ACME. IS256 is involved in biofilm formation, the transcriptional regulation of resistance genes and the inactivation of global gene regulators (Maki and Murakami, 1997; Ziebuhr et al., 1999; Conlon et al., 2004). The hospital-associated S. epidermidis invasive isolates belonging to ST2 were all positive for icaA and IS256 (Li et al., 2009). In the present study, the prevalence of IS256 among sesI-positive isolates was significantly higher than that among sesI-negative isolates. In contrast, bhp, a gene homologous to bap and assumed to promote biofilm formation (Becker et al., 2014), was found more frequent among sesI-negative isolates than sesI-positive isolates. Taken together, our data showed that sesI-positive S. epidermidis invasive isolates harbored more virulence-associated genes relative to sesI-negative isolates, which is similar to the previous study (Salgueiro et al., 2017).
In the present study, majority of sesI-positive S. epidermidis invasive isolates belonged to ST2, while no sesI-negative invasive isolates belonged to this clone, indicating that there was strong association between carriage of sesI and ST2 clone. ST2 was by far the most prevalent clonal type for hospital-associated invasive S. epidermidis isolates (Miragaia et al., 2007; Li et al., 2009; Iorio et al., 2012; Du et al., 2013). Previous study showed the hospital-associated invasive S. epidermidis isolates belonging to ST2 were all positive for IS256 and icaA (Li et al., 2009). Du et al. (2013) reported that IS256 and icaA were detected in the majority of the predominant clinical ST2 MRSE isolates. Our study found that high prevalence of virulence-associated genes and resistance to non-lactam antimicrobials, along with a high prevalence of resistance to surface-active antiseptics used for decolonization, contribute to the spread of prevalent ST2 sesI-positive S. epidermidis clone in hospital environments.
We speculate that icaA and IS256, as well as sesI, are important factors which help ST2 to spread and become successfully a predominant clone in the healthcare setting.
The knockout and recovery of sesI gene did not affect the growth and resistance of bacteria. In order to explore the role of sesI in the biofilm aggregation stage, we constructed an in vitro aggregation experiment based on the construction of sesI mutant and compared the roles of the mutant and wild strains in the aggregation stage. Performing Gram staining experiment, the results showed that the distribution of mutant strains was relatively sparse, scattered and did not aggregate with each other. However, the Staphylococcus epidermidis RP62AΔsesI-C accumulated and overlapped each other, and its ability of accumulation also returned to the wild type strain level. Therefore, it has proved sesI played a major role in the gathering phase. In addition, Staphylococcus epidermidis RP62A, RP62AΔsesI and RP62AΔsesI-C have been tested in semi-quantitative in vitro biofilms, we found that there was no significant difference in biofilm formation stage between the mutant and the wild type strain (P > 0.05). Biofilm formation ability of each strain was enhanced after the environmental factors changed (added 4% NaCl, 4% ethanol and 0.5% Glu), but there was no noticeable change among the three strains. By SEM, the formation of a dense and overlapping biofilm mass, indicating that during the mature stage, the formation of biofilm did not change between the mutant strain and the wild type. Our data showed that the sesI gene promoted the initiation adhesion and aggregation phases of biofilm formation, but not the maturation stage of biofilm formation.
Taken together, these results demonstrate that sesI, along with antimicrobial and other biofilm-associated genes enables S. epidermidis easier for colonization and adhesion and contributes to the spread of S. epidermidis, especially ST2 clone.
Author Contributions
XQ, YJ, JD, ZH, SW, YG, and JL performed the laboratory measurements. FY and LW made substantial contributions to conception and design. LH, LW, and FY revised the manuscript critically for important intellectual content. LH and LW participated in experimental design and data analysis. FY drafted the manuscript. All authors read and approved the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank José R. Mediavilla from International Center for Public Health, Rutgers, The State University of New Jersey, United States, for revising the manuscript.
Abbreviations
- CLSI
Clinical and Laboratory Standards Institute
- MLST
multi-locus sequence typing
- MRSE
methicillin-resistant Staphylococcus epidermidis
- MSSE
methicillin-susceptible Staphylococcus epidermidis
- ST
sequence type
Footnotes
Funding. This study was supported by grants from the Natural Science Fund of China (81472011).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2017.02574/full#supplementary-material
References
- Bae T., Schneewind O. (2006). Allelic replacement in Staphylococcus aureus with inducible counter-selection. Plasmid 55 58–63. 10.1016/j.plasmid.2005.05.005 [DOI] [PubMed] [Google Scholar]
- Barbier F., Lebeaux D., Hernandez D., Delannoy A. S., Caro V., Francois P., et al. (2011). High prevalence of the arginine catabolic mobile element in carriage isolates of methicillin-resistant Staphylococcus epidermidis. J. Antimicrob. Chemother. 66 29–36. 10.1093/jac/dkq410 [DOI] [PubMed] [Google Scholar]
- Becker K., Heilmann C., Peters G. (2014). Coagulase-negative staphylococci. Clin. Microbiol. Rev. 27 870–926. 10.1128/CMR.00109-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowden M. G., Chen W., Singvall J., Xu Y., Peacock S. J., Valtulina V., et al. (2005). Identification and preliminary characterization of cell-wall-anchored proteins of Staphylococcus epidermidis. Microbiology 151 1453–1464. 10.1099/mic.0.27534-0 [DOI] [PubMed] [Google Scholar]
- Bowden M. G., Visai L., Longshaw C. M., Holland K. T., Speziale P., Hook M. (2002). Is the GehD lipase from Staphylococcus epidermidis a collagen binding adhesin? J. Biol. Chem. 277 43017–43023. 10.1074/jbc.M207921200 [DOI] [PubMed] [Google Scholar]
- Buttner H., Mack D., Rohde H. (2015). Structural basis of Staphylococcus epidermidis biofilm formation: mechanisms and molecular interactions. Front. Cell. Infect. Microbiol. 5:14. 10.3389/fcimb.2015.00014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cala C., Amodio E., Di Carlo E., Virruso R., Fasciana T., Giammanco A. (2015). Biofilm production in Staphylococcus epidermidis strains, isolated from the skin of hospitalized patients: genetic and phenotypic characteristics. New Microbiol. 38 521–529. [PubMed] [Google Scholar]
- Catalanotti P., Lanza M., Del Prete A., Lucido M., Catania M. R., Galle F., et al. (2005). Slime-producing Staphylococcus epidermidis and S. aureus in acute bacterial conjunctivitis in soft contact lens wearers. New Microbiol. 28 345–354. [PubMed] [Google Scholar]
- Cherifi S., Byl B., Deplano A., Nonhoff C., Denis O., Hallin M. (2013). Comparative epidemiology of Staphylococcus epidermidis isolates from patients with catheter-related bacteremia and from healthy volunteers. J. Clin. Microbiol. 51 1541–1547. 10.1128/JCM.03378-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- CLSI (2014). Performance Standards for Antimicrobial susceptibility Testing, 24th Informational Supplement (M100-S24). Wayne, PA: Clinical and Laboratory Standards Institute. [Google Scholar]
- Conlon K. M., Humphreys H., O’gara J. P. (2004). Inactivations of rsbU and sarA by IS256 represent novel mechanisms of biofilm phenotypic variation in Staphylococcus epidermidis. J. Bacteriol. 186 6208–6219. 10.1128/JB.186.18.6208-6219.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du X., Zhu Y., Song Y., Li T., Luo T., Sun G., et al. (2013). Molecular analysis of Staphylococcus epidermidis strains isolated from community and hospital environments in China. PLOS ONE 8:e62742. 10.1371/journal.pone.0062742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heilmann C., Hussain M., Peters G., Gotz F. (1997). Evidence for autolysin-mediated primary attachment of Staphylococcus epidermidis to a polystyrene surface. Mol. Microbiol. 24 1013–1024. 10.1046/j.1365-2958.1997.4101774.x [DOI] [PubMed] [Google Scholar]
- Heilmann C., Thumm G., Chhatwal G. S., Hartleib J., Uekotter A., Peters G. (2003). Identification and characterization of a novel autolysin (Aae) with adhesive properties from Staphylococcus epidermidis. Microbiology 149 2769–2778. 10.1099/mic.0.26527-0 [DOI] [PubMed] [Google Scholar]
- Iorio N. L., Caboclo R. F., Azevedo M. B., Barcellos A. G., Neves F. P., Domingues R. M., et al. (2012). Characteristics related to antimicrobial resistance and biofilm formation of widespread methicillin-resistant Staphylococcus epidermidis ST2 and ST23 lineages in Rio de Janeiro hospitals, Brazil. Diagn. Microbiol. Infect. Dis. 72 32–40. 10.1016/j.diagmicrobio.2011.09.017 [DOI] [PubMed] [Google Scholar]
- Khodaparast L., Khodaparast L., Shahrooei M., Stijlemans B., Merckx R., Baatsen P., et al. (2016). The Possible Role of Staphylococcus epidermidis LPxTG surface protein SesC in biofilm formation. PLOS ONE 11:e0146704. 10.1371/journal.pone.0146704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepainteur M., Royer G., Bourrel A. S., Romain O., Duport C., Doucet-Populaire F., et al. (2013). Prevalence of resistance to antiseptics and mupirocin among invasive coagulase-negative staphylococci from very preterm neonates in NICU: the creeping threat? J. Hosp. Infect. 83 333–336. 10.1016/j.jhin.2012.11.025 [DOI] [PubMed] [Google Scholar]
- Li M., Wang X., Gao Q., Lu Y. (2009). Molecular characterization of Staphylococcus epidermidis strains isolated from a teaching hospital in Shanghai, China. J. Med. Microbiol. 58 456–461. 10.1099/jmm.0.007567-0 [DOI] [PubMed] [Google Scholar]
- Li T., Song Y., Zhu Y., Du X., Li M. (2013). Current status of Staphylococcus aureus infection in a central teaching hospital in Shanghai, China. BMC Microbiol. 13:153. 10.1186/1471-2180-13-153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liduma I., Tracevska T., Bers U., Zilevica A. (2012). Phenotypic and genetic analysis of biofilm formation by Staphylococcus epidermidis. Medicina 48 305–309. [PubMed] [Google Scholar]
- Macintosh R. L., Brittan J. L., Bhattacharya R., Jenkinson H. F., Derrick J., Upton M., et al. (2009). The terminal A domain of the fibrillar accumulation-associated protein (Aap) of Staphylococcus epidermidis mediates adhesion to human corneocytes. J. Bacteriol. 191 7007–7016. 10.1128/JB.00764-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maki H., Murakami K. (1997). Formation of potent hybrid promoters of the mutant llm gene by IS256 transposition in methicillin-resistant Staphylococcus aureus. J. Bacteriol. 179 6944–6948. 10.1128/jb.179.22.6944-6948.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miragaia M., De Lencastre H., Perdreau-Remington F., Chambers H. F., Higashi J., Sullam P. M., et al. (2009). Genetic diversity of arginine catabolic mobile element in Staphylococcus epidermidis. PLOS ONE 4:e7722. 10.1371/journal.pone.0007722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miragaia M., Thomas J. C., Couto I., Enright M. C., De Lencastre H. (2007). Inferring a population structure for Staphylococcus epidermidis from multilocus sequence typing data. J. Bacteriol. 189 2540–2552. 10.1128/JB.01484-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery C. P., Boyle-Vavra S., Roux A., Ebine K., Sonenshein A. L., Daum R. S. (2012). CodY deletion enhances in vivo virulence of community-associated methicillin-resistant Staphylococcus aureus clone USA300. Infect. Immun. 80 2382–2389. 10.1128/IAI.06172-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami K., Minamide W., Wada K., Nakamura E., Teraoka H., Watanabe S. (1991). Identification of methicillin-resistant strains of staphylococci by polymerase chain reaction. J. Clin. Microbiol. 29 2240–2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onishi M., Urushibara N., Kawaguchiya M., Ghosh S., Shinagawa M., Watanabe N., et al. (2013). Prevalence and genetic diversity of arginine catabolic mobile element (ACME) in clinical isolates of coagulase-negative staphylococci: identification of ACME type I variants in Staphylococcus epidermidis. Infect. Genet. Evol. 20 381–388. 10.1016/j.meegid.2013.09.018 [DOI] [PubMed] [Google Scholar]
- Otto M. (2004). Virulence factors of the coagulase-negative staphylococci. Front. Biosci. 9:841–863. 10.2741/1295 [DOI] [PubMed] [Google Scholar]
- Otto M. (2012). Molecular basis of Staphylococcus epidermidis infections. Semin. Immunopathol. 34 201–214. 10.1007/s00281-011-0296-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pintens V., Massonet C., Merckx R., Vandecasteele S., Peetermans W. E., Knobloch J. K., et al. (2008). The role of sigmaB in persistence of Staphylococcus epidermidis foreign body infection. Microbiology 154 2827–2836. 10.1099/mic.0.2007/015768-0 [DOI] [PubMed] [Google Scholar]
- Salgueiro V. C., Iorio N. L., Ferreira M. C., Chamon R. C., Dos Santos K. R. (2017). Methicillin resistance and virulence genes in invasive and nasal Staphylococcus epidermidis isolates from neonates. BMC Microbiol. 17:15. 10.1186/s12866-017-0930-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahrooei M., Hira V., Khodaparast L., Khodaparast L., Stijlemans B., Kucharikova S., et al. (2012). Vaccination with SesC decreases Staphylococcus epidermidis biofilm formation. Infect. Immun. 80 3660–3668. 10.1128/IAI.00104-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahrooei M., Hira V., Stijlemans B., Merckx R., Hermans P. W., Van Eldere J. (2009). Inhibition of Staphylococcus epidermidis biofilm formation by rabbit polyclonal antibodies against the SesC protein. Infect. Immun. 77 3670–3678. 10.1128/IAI.01464-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang S., Chen G., Wu Y., Du L., Zhao Z. (2005). Rapid diagnosis of bacterial sepsis with PCR amplification and microarray hybridization in 16S rRNA gene. Pediatr. Res. 58 143–148. 10.1203/01.PDR.0000169580.64191.8B [DOI] [PubMed] [Google Scholar]
- Soderquist B., Andersson M., Nilsson M., Nilsdotter-Augustinsson A., Persson L., Friberg O., et al. (2009). Staphylococcus epidermidis surface protein I (SesI): a marker of the invasive capacity of S. epidermidis? J. Med. Microbiol. 58 1395–1397. 10.1099/jmm.0.008771-0 [DOI] [PubMed] [Google Scholar]
- Thomas J. C., Vargas M. R., Miragaia M., Peacock S. J., Archer G. L., Enright M. C. (2007). Improved multilocus sequence typing scheme for Staphylococcus epidermidis. J. Clin. Microbiol. 45 616–619. 10.1128/JCM.01934-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziebuhr W., Krimmer V., Rachid S., Lossner I., Gotz F., Hacker J. (1999). A novel mechanism of phase variation of virulence in Staphylococcus epidermidis: evidence for control of the polysaccharide intercellular adhesin synthesis by alternating insertion and excision of the insertion sequence element IS256. Mol. Microbiol. 32 345–356. 10.1046/j.1365-2958.1999.01353.x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.