Abstract
Here we report an efficient method to generate multiple co-structures of the A2A G protein-coupled receptor (GPCR) with small-molecules from a single preparation of a thermostabilised receptor crystallised in Lipidic Cubic Phase (LCP). Receptor crystallisation is achieved following purification using a low affinity “carrier” ligand (theophylline) and crystals are then soaked in solutions containing the desired (higher affinity) compounds. Complete datasets to high resolution can then be collected from single crystals and seven structures are reported here of which three are novel. The method significantly improves structural throughput for ligand screening using stabilised GPCRs, thereby actively driving Structure-Based Drug Discovery (SBDD).
Introduction
Many of the world’s top selling drugs target G protein-coupled receptors (GPCRs)1 for indications including inflammaory, neurological, gastrointestinal, cardiovascular and respiratory diseases2. Structural data on this clinically relevant membrane protein superfamily has increased dramatically over the last decade, resulting from pioneering research from a number of groups3,4. High resolution crystal structures are now available for almost all major GPCR classes and are transformative from a pharmaceutical perspective, with several drug candidates generated by structure-based drug design (SBDD) techniques5,6. Nevertheless, GPCR crystallography throughput lags behind that of soluble targets (e.g. kinases)7, in part due to the inherent conformational flexibility and instability of GPCRs when removed from the native cell membrane environment. To overcome this, receptors have been thermostabilised by introducing a small number of targeted point mutations using the StaR® 8, SABRE9 or CHESS10 technologies, or other mutagenesis approaches11–13. These mutations enhance apparent thermostability and stabilise receptors in a specific pre-defined conformation, and detergent-resistant form14. Such approaches were instrumental in solving structures of members of class B and C GPCRs2,15,16. Receptors stabilised using the StaR® technology rely less on stability conferred by high affinity ligands to increase the chance of crystallogenesis. Co-crystal structures are thus obtainable even with low affinity compounds and fragments17 identified in early stages of discovery projects. This provides a unique opportunity to apply soaking techniques, successfully utilised for soluble targets (e.g. kinases), to GPCR crystals grown in meso by lipidic cubic phase crystallisation (LCP). The reliable production of multiple co-structures on a regular basis, in step with the medicinal chemistry cycle time, is fully enabling for SBDD.
Here, we report an in meso crystal soaking method developed to improve the crystallographic throughput for our work with the adenosine A2A receptor (A2AR), including drug discovery activities. Previously, each ligand complex structure required a separate, bespoke A2AR-ligand protein preparation. Now a single protein preparation can yield high resolution structural data for A2AR in complex with up to a dozen different ligands. This also significantly minimises ligand amounts required to generate co-structures compared to using bespoke A2AR-ligand protein preparations.
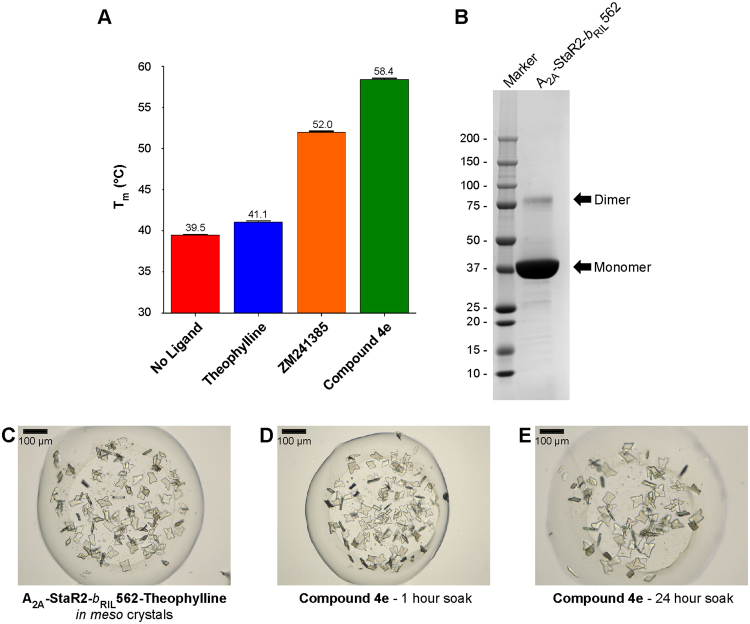
Theophylline binds to the thermostabilised receptor used for crystallisation (A2A-StaR2-b RIL562), with relatively low affinity (pK D = 5.71), and with fast kinetics18, whereas potent A2AR-selective antagonists such as 1,2,4-triazine derivatives19, typically bind with higher affinity (pK D > 8) and exhibit slow off-rates. Despite its low affinity for A2A-StaR2-b RIL562, theophylline provides some thermostabilisation to the receptor in comparison to apo protein (Fig. 1A). This in meso soaking method uses theophylline as a low affinity carrier ligand, present throughout purification, to provide crystallisation-grade A2A-StaR2-b RIL562 (Fig. 1B). The A2A-StaR2-b RIL562-Theophylline complex readily crystallises in meso yielding thick ~60 µm long plates (Fig. 1C), typically diffracting to 2.0 Å and containing a theophylline molecule in the A2AR orthosteric binding site20 (Fig. 2A). Crystals with theophylline have also been used previously to generate a structure with another xanthine, PSB3620.
Figure 1.
A2A-StaR2-b RIL562 Crystal Soaking. (A) Bar chart showing the melting temperature of A2A-StaR2-b RIL562 in its apo form or in the presence of theophylline, ZM241385 or Compound 4e, reflecting the relative stability of each protein preparation. (B) SDS-PAGE of concentrated A2A-StaR2-b RIL562 protein prior to crystallisation. (C) Crystals of the A2A-StaR2-b RIL562-Theophylline complex. (D) A2A-StaR2-b RIL562-Theophylline crystals following a 1 hour soak in 1 mM Compound 4e. (E) A2A-StaR2-b RIL562-Theophylline crystals following a 24 hour soak in 1 mM Compound 4e.
Figure 2.
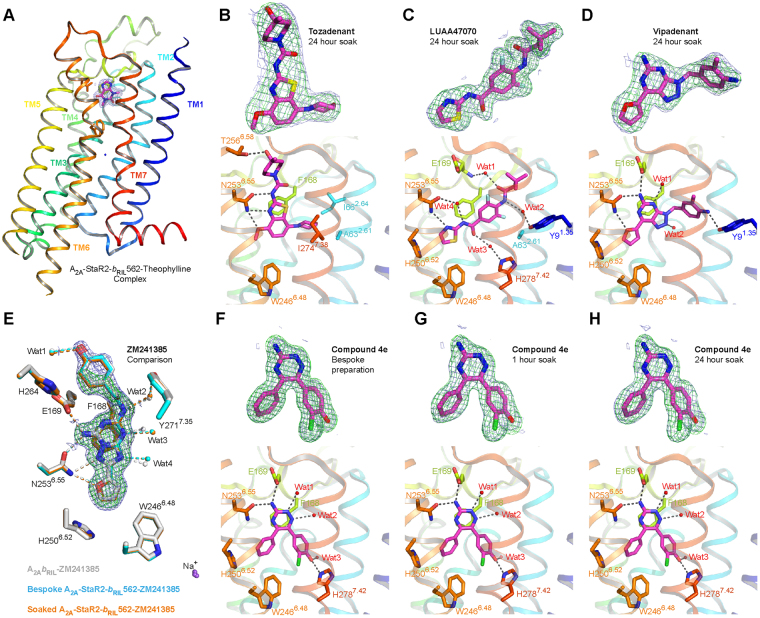
Structure of A2A-StaR2-b RIL562-ligand complexes. (A) Structure of the A2A-StaR2-b RIL562-Theophylline complex (PDB: 5MZJ) shown in cartoon, with helices coloured differently from blue (helix 1) to red (helix 8). Theophylline is shown as sticks within the 1.0 σ contoured 2mFo-dFc electron density maps (blue mesh) carved around the ligand. Interesting orthosteric binding site residues are shown as sticks. 1.0 σ contoured 2mFo-dFc and 3.5 σ contoured mFo-dFc ligand omit electron density maps (blue and green meshes respectively) reflecting the quality of ligand (purple sticks) fitting are shown in the top panel, whereas the lower panel provides interaction details between A2A-StaR2-b RIL562 binding site residues (sticks) with Tozadenant (B), LUAA47070 (C) or Vipadenant (D). In these figures, water molecules are represented as red spheres whereas hydrogen bonding is highlighted by dotted lines. An overlay of structures of ZM241385 in complex with A2A-StaR2-b RIL562 from either a bespoke preparation (PDB: 5UI4) (cyan) or from a soaking experiment (orange), and with A2A b RIL562 (PDB: 4EIY) (white) depicts the high degree of conservation in positioning of orthosteric binding site residues (E). Residues and water molecules involved in ligand binding within a 5 Å radius are represented as sticks and as spheres respectively. Hydrogen bonds are shown as dotted lines and the 1.0 σ contoured 2mFo-dFc and 3.5 σ contoured mFo-dFc ligand omit electron density maps corresponding to ZM241385 from the soaking experiment are represented as blue and green meshes respectively, carved around the ligand. Similarly electron density maps and interactions are shown for A2A-StaR2-b RIL562-Compound 4e generated from bespoke crystallisation (F) or from either 1 hour (G) or 24 hour (H) soaks of A2A-StaR2-b RIL562-Theophylline crystals with Compound 4e.
The utility of the in meso soaking system for diverse ligands from chemical series other than xanthines was then investigated. A2A-StaR2-b RIL562-Theophylline crystals were soaked in mother liquor supplemented with A2A antagonists Tozadenant 21 (pK D = 8.4), LUAA47070 22 (pK D = 6.5) or Vipadenant 23 (pK D = 9.0), and their diffraction characterised. Crystals from these experiments diffracted in spacegroup C222 1 to 2.0–3.1 Å resolution (Table 1). Tozadenant, LUAA47070 and Vipadenant are all well defined in the electron density maps (Fig. 2B–D). For these ligands the basal region of the orthosteric site is delimited by Trp2466.48, which engages in Van der Waals contacts to the Tozadenant benzothiazole ring, the LUAA47070 thiazole ring or the Vipadenant furan ring (Fig. 2B–D). These ligands explore different regions at the apical end of the orthosteric site. The 4-hydroxy,4-methylpiperidine moiety of Tozadenant sits upright on the benzothiazole ring, and hydrogen bonds to Thr2566.58. The N,2,2-trimethylpropanamide group of LUAA47070 extends obliquely towards transmembrane helix 1 (TM1), and engages in water-mediated contacts with ECL2 Glu169 and Tyr91.35. Further this structure shows how the experimentally defined water mediated interactions of the amide group of LUAA47070 to both Asn2536.55 and His2787.42 contribute to this ligand binding pose. Finally, the Vipadenant 2-methylaniline moiety points laterally towards TM1, and is hydrogen-bonded to Tyr91.35. We find that, despite adopting a range of orientations in the orthosteric binding site, ligands from different chemical series can be effectively soaked into A2A-StaR2-b RIL562-Theophylline crystals, and used in crystallographic structural studies to identify their binding modes. Contrary to poorly diffracting, bespoke A2A-StaR2-b RIL562-Tozadenant crystals, likely resulting from the disruption of the salt bridge between extracellular loop 2 (ECL2) Glu169 and ECL3 His264, interfering with crystal packing, co-crystals from soaking experiments yielded good quality structural data, highlighting the versatility of the in meso soaking system.
Table 1.
Data collection and refinement statistics.
| Compound 4e (1 hour soak) 5OM1 | Compound 4e (24 hour soak) 5OM4 | Compound 4e (Bespoke) 5OLZ | Tozadenant (24 hour soak) 5OLO | Vipadenant (24 hour soak) 5OLH | LUAA47070 (24 hour soak) 5OLV | ZM241385 (24 hour soak) 5OLG | |
|---|---|---|---|---|---|---|---|
| Data collection | |||||||
| Space group | C2221 | C2221 | C2221 | C2221 | C2221 | C2221 | C2221 |
| Cell dimensions | |||||||
| a, b, c (Å) | 39.54, 179.85, 140.32 | 39.47, 179.11, 140.03 | 39.37, 179.25, 140.07 | 39.38, 181.10, 141.77 | 39.40, 179.33, 141.14 | 39.43, 180.77, 140.90 | 39.45, 179.39, 139.60 |
| α, β, γ (°) | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 |
| Resolution (Å) | 33.83–2.10 (2.16–2.10)a | 32.92–2.00 (2.05–2.00)a | 33.71–1.90 (1.94–1.90)a | 38.48–3.10 (3.21–3.10)a | 29.82–2.60 (2.72–2.60)a | 76.08–2.00 (2.05–2.00)a | 46.53–1.85 (1.89–1.85)a |
| R pim | 0.061 (0.564)a | 0.059 (0.635)a | 0.040 (0.583)a | 0.078 (0.559)a | 0.097 (0.547)a | 0.059 (0.624)a | 0.068 (0.935)a |
| I/σ(I) | 10.0 (1.5)a | 10.3 (1.3)a | 11.3 (1.4)a | 8.3 (1.5)a | 8.3 (1.8)a | 8.0 (1.3)a | 7.7 (1.0)a |
| CC 1/2 | 0.997 (0.524)a | 0.998 (0.439)a | 0.999 (0.421)a | 0.997 (0.480)a | 0.988 (0.519)a | 0.997 (0.420)a | 0.994 (0.372)a |
| Completeness (%) | 100.0 (100.0)a | 98.4 (98.8)a | 99.3 (99.6)a | 99.9 (100.0)a | 99.5 (98.7)a | 98.5 (94.1)a | 99.7 (100)a |
| Redundancy | 6.6 (6.7)a | 6.3 (6.5)a | 4.7 (4.8)a | 5.7 (5.9)a | 6.9 (5.1)a | 3.6 (3.7)a | 6.2 (6.4)a |
| Refinement | |||||||
| Resolution (Å) | 33.83–2.10 | 32.92–2.00 | 33.71–1.90 | 38.48–3.10 | 29.82–2.60 | 41.68–2.00 | 41.31–1.86 |
| No. reflections | 56388 | 63810 | 74939 | 9645 | 15827 | 34169 | 41376 |
| R work/R free | 0.1882/0.2097 | 0.1831/0.2049 | 0.1727/0.1963 | 0.1987/0.2448 | 0.2000/0.2487 | 0.1801/0.2081 | 0.1921/0.2332 |
| No. atoms | |||||||
| Protein | 3082 | 3083 | 3104 | 2983 | 3047 | 3083 | 3097 |
| Ligand | 21 | 21 | 21 | 28 | 24 | 24 | 25 |
| Solvent | 687 | 727 | 684 | 397 | 488 | 607 | 597 |
| B factors | |||||||
| Protein | 40.55 | 39.22 | 45.33 | 80.98 | 43.21 | 40.19 | 36.81 |
| Ligand | 19.40 | 17.37 | 20.13 | 49.00 | 21.26 | 18.28 | 27.17 |
| Solvent | 55.49 | 56.88 | 59.69 | 87.00 | 51.60 | 51.83 | 49.20 |
| R.m.s. deviations | |||||||
| Bond lengths (Å) | 0.002 | 0.003 | 0.004 | 0.002 | 0.003 | 0.009 | 0.014 |
| Bond angles (°) | 0.92 | 0.93 | 0.99 | 0.74 | 0.75 | 1.19 | 1.54 |
aValues in parentheses are for highest-resolution shell. All data presented above were collected from single crystals except for the A2A-StaR2-b RIL562-Vipadenant complex, where data was merged from three different crystals.
The validity of structural results obtained by the in meso soaking method was checked using ZM241385 24 (pK D = 8.6), a well-characterised A2AR antagonist that increases A2A-StaR2-b RIL562 stability by ~12 °C (Fig. 1A). The crystal structure of the receptor in complex with ZM241385 resulting from in meso soaking, was compared with similar complexes obtained from bespoke crystallisation setups using either A2A-StaR2-b RIL56225 or A2A-b RIL56226 (Fig. 2E). Overlaying these structures shows a remarkably similar structural conformation of residues in the orthosteric located within 5 Å of the ligand with an all atom r.m.s.d. of only 0.074 Å (soaked v/s bespoke A2A-StaR2-b RIL562 (PDB: 5IU4)) or 0.118 Å (soaked v/s bespoke A2A-b RIL562 (PDB: 4EIY)) (Fig. 2E). Such a high degree of structural conservation across different crystallisation methods (and A2AR constructs) benchmarks and underlines the robustness of the in meso soaking system described here.
To determine the feasibility of using the in meso soaking method system to support optimisation of novel A2AR antagonists for drug discovery, Compound 4e, a 1,2,4-triazine derivative19, was investigated. Compound 4e is a low nanomolar affinity ligand (pK D = 9.6) for A2AR and increases A2A-StaR2-b RIL562 stability by ~19 °C when compared to apo protein (Fig. 1A) and co-crystals were generated using either a bespoke protein preparation or by soaking A2A-StaR2-b RIL562-Theophylline crystals in mother liquor supplemented with Compound 4e for 1 or 24 hours (Fig. 1D,E). Crystal morphology remained unchanged regardless of soaking times (Fig. 1D,E) and crystals from these three experiments diffracted to 1.9–2.1 Å in spacegroup C222 1. Structures generated from bespoke crystallisation or from the soaking experiments were essentially equivalent (r.m.s.d ~0.1 Å over 297 residues). Compound 4e was well defined in electron density maps from the resultant three co-structures and binds in the same orientation in the orthosteric site (Fig. 2F–H), displaying similar B factors (17.8–19.8 Å2) (Table 1). Compound 4e sits lower in the orthosteric site than theophylline, with the triazine ring π stacking against Phe168 from ECL2, while also engaging in polar contacts with an extensive water network. The amine moiety on the triazine ring is further hydrogen-bonded to ECL2 Glu169 and Asn2536.55, whereas the hydroxyl group on the chlorophenol ring makes a hydrogen bond with His2787.43. In the basal region of the orthosteric site, the ligand benzyl ring makes Van der Waals interactions with Trp2466.48.
A pairwise comparison of residues located within 5 Å of all the different liganded structures presented here demonstrates all atom r.m.s.d. values ranging from 0.48 Å (between the A2A-StaR2-b RIL562-Compound 4e and -LUAA47070 structures) to 1.05 Å (between the A2A-StaR2-b RIL562-ZM241385 and -Tozadenant structures). Altogether, most of the mobility stems from Tyr2717.35, involved in water-mediated interactions with ZM241385, and from Glu169 in ECL2 and His264 which adopt different rotamer orientations in the A2A-StaR2-b RIL562-Tozadenant structure compared to the other ligand complexes.
In drug development, high-throughput X-ray crystallography expedites the elaboration of novel hits into lead compounds and drug candidates by providing multiple high resolution views of ligand-receptor complexes, which are key for understanding critical intermolecular interactions alongside interpretation of ligand-induced receptor conformational changes27. The accelerated availability of multiple receptor-ligand complexes provides a data-rich starting point for SBDD and medicinal chemistry28 which, when correlated with in vitro biological activity, allows rapid incorporation of molecular modifications towards increasing ligand affinity for the binding site or improvement of their absorption, distribution, metabolism, excretion and toxicity (ADMET) properties.
We have demonstrated that an in meso ligand soaking methology can rapidly and efficiently yield multiple high-resolution co-crystal structures from a diverse set of ligands in complex with a given GPCR. Such soaking techniques have also been employed in-house for other discovery projects. The method described here has general applicability to further discovery campaigns with stabilised membrane proteins using LCP crystallisation setups, provided high quality crystals exist for the target in complex with low affinity stabilising carrier ligands with fast off-rates.
Methods
StaR generation
The thermostabilisation of the human A2A receptor (resulting in A2A-StaR2) using a mutagenesis approach8, has been previously described29.
Expression, membrane preparation and protein purification
The A2A-StaR2-b RIL562 construct has been described previously25 and harbours eight thermostabilising mutations (A54L2.52, T88A3.36, R107A3.55, K122A4.43, L202A5.63, L235A6.37, V239A6.41 and S277A7.42) as well as a mutation to remove a glycosylation site (N154A). The construct further comprises an Apocytochrome b RIL562 fusion between transmembrane helices 5 and 6 and a C-terminal decahistidine tag. The receptor was expressed using the Bac to Bac Expression System (Invitrogen) in Trichoplusa ni Tni PRO cells using ESF 921 medium (Expression Systems) supplemented with 5% (v/v) fetal bovine serum (Sigma-Aldrich) and 1% (v/v) Penicillin/Streptomycin (PAA Laboratories). Cells were infected at a density of 2.6 × 106 cells/ml with virus at an approximate multiplicity of infection of 1. Cultures were grown at 27 °C with constant shaking and harvested by centrifugation 48 hours post infection. All subsequent protein protein purification steps were carried out at 4 °C unless otherwise stated.
For each protein preparation, cells from 2 L cultures were resuspended in 40 mM TRIS buffer at pH 7.6 supplemented by 1 mM EDTA and Complete EDTA-free protease inhibitor cocktail tablets (Roche). Cells were disrupted at ~15 000 psi using a microfluidizer (Processor M-110L Pneumatic, Microfluidics). Membranes pelleted by ultra-centrifugation at 200 000 g for 50 minutes, were subjected to a high salt wash in a buffer containing 40 mM Tris pH 7.6, 1 M NaCl and Complete EDTA-free protease inhibitor cocktail tablets, before they were centrifuged at 200,000 g for 50 minutes. Washed membranes were resuspended in 50 mL 40 mM Tris pH 7.6 supplemented with Complete EDTA-free protease inhibitor cocktail tablets and stored at −80 °C until further use.
Protein preparations intended for soaking experiments were carried out in the presence of theophylline whereas the bespoke preparation of A2A-StaR2-b RIL562 in complex with Compound 4e was done in the presence of 5 µM ligand.
Membranes were thawed, resuspended in a total volume of 150 ml with 40 mM Tris–HCl pH 7.6, Complete EDTA-free protease inhibitor cocktail tablets (Roche), 3 mM theophylline (Sigma Aldrich) (or 5 µM Compound 4e), and incubated for 2 hours at room temperature. Membranes were then solubilized by addition of 1.5% n-Decyl-β-D-maltopyranoside (DM, Anatrace), and incubation for 2 hours at 4 °C, followed by centrifugation at 145 000 g for 60 min to harvest solubilised material.
The solubilised material was applied to a 5 ml Ni-NTA (nickel-nitrilotriacetic acid) Superflow cartridge (Qiagen) pre-equilibrated in 40 mM Tris pH 7.4, 200 mM NaCl, 0.15% DM, 1 mM theophylline (or 5 µM Compound 4e). The column was washed with 25 column volumes of buffer 40 mM Tris pH 7.4, 200 mM NaCl, 0.15% DM, 70 mM imidazole, 1 mM theophylline (or 5 µM Compound 4e) and then the protein was eluted with 40 mM Tris pH 7.4, 200 mM NaCl, 0.15% DM, 280 mM imidazole, 1 mM theophylline (or 5 µM Compound 4e).
Collected fractions were analyzed by SDS PAGE and fractions containing A2a-StaR2-b RIL562 were pooled and concentrated using an Amicon Ultra Ultracell 50 K ultrafiltration membrane to a final volume of ~800 µl. The protein sample was ultra-centrifuged at 436 000 g for 10 minutes before being applied to a Superdex200 size exclusion column (GE Healthcare) pre-equilibrated with 40 mM Tris pH 7.4, 200 mM NaCl, 0.15% DM, 1 mM theophylline (or 5 µM Compound 4e). Eluted fractions containing the protein were analyzed by SDS PAGE, pooled and concentrated to ~35 mg/ml using an Amicon Ultra Ultracell 50 K ultrafiltration membrane and subjected to an ultra-centrifugation at 436 000 g prior to crystallisation. Protein concentrations were measured using the DC assay (Bio-Rad), and confirmed using quantitative amino acid analysis.
Thermal unfolding experiments
A2A-StaR2-b RIL562 purified in DM in the presence of 500 µM theophylline was used for thermal unfolding experiments. The protein was diluted in 40 mM Tris pH 7.4, 200 mM NaCl, 0.15% DM to a final concentration of 0.2 mg/ml. Following heavy dilution (~70-fold) of the protein in a buffer without ligand, the sample was considered to be in an apo-like state. Samples were supplemented with the respective ligands to a final concentration of 50 µM, with a final DMSO concentration of 5% (v/v). The control sample was supplemented with DMSO to a final concentration of 5% (v/v). Samples were incubated 30 minutes on ice before being loaded into UV capillaries (NanoTemper Technologies) and experiments were carried out using the Prometheus NT.48. The temperature gradient was set to an +1 °C/min from 20 °C to 90 °C. Protein unfolding was measured by detecting the temperature-dependent change in tryptophan fluorescence at emission wavelengths of 330 and 350 nm. The experiment was repeated four times and data analysed with the one-way analysis of variance (ANOVA) with Dunnett’s post-test. Tm values obtained for the three ligands are statistically different from the control sample with p < 0.001.
Crystallisation
The A2A-StaR2-b RIL562 in complex with either theophylline or Compound 4e was crystallized in lipidic cubic phase at 20 °C. Concentrated protein was mixed with monoolein (Nu-Chek) supplemented with 10% (w/w) cholesterol (Sigma Aldrich) and 10 µM theophylline (or 5 µM Compound 4e) using the twin-syringe method30. The final protein:lipid ratio was 40:60 (w/w). 40 nl boli were dispensed onto 96-well Laminex Glass Bases (Molecular Dimensions ltd) using a Mosquito LCP crystallization robot (TTP Labtech) and overlaid with 800 nL precipitant solution. Glass bases were sealed using Laminex Film covers (Molecular Dimensions Ltd). 60–80 µm long plate-shaped crystals grew within 2 weeks in 0.l M tri-sodium citrate pH 5.3–5.4, 0.05 M sodium thiocyanate, 29–32% PEG400, 2% (v/v) 2,5-hexanediol and 0.5 mM theophylline (or 5 µM Compound 4e).
In meso soaking and crystal harvesting
For soaking experiments, incisions were made into the Laminex cover over base wells containing crystals identified for harvesting and these wells were flooded with 10 µL motherliquor supplemented by 1 mM ligand. The crystals are soaked in motherliquor with a final ligand concentration of 925 µM, and a final theophylline concentration of 74 µM. Flooded wells were then re-sealed using Crystal Clear Sealing Tape (Hampton Research), and plates were incubated for 1 hour or 24 hours at 20 °C. Single crystals were mounted in LithoLoops (Molecular Dimensions Ltd) and flash-frozen in liquid nitrogen without the addition of further cryoprotectant.
Diffraction data collection and processing
X-ray diffraction data were measured on a Pilatus 6 M detector at beamline I24 (Diamond Light Source) using a 6 × 9 μm beam size of for crystals of A2A-StaR2-b RIL562 in complex with Compound 4e, Tozadenant or LUAA47070. Complete datasets were acquired from a single crystal for each of these complexes at wavelengths 0.96857 Å (Compound 4e and LUAA47070) or 0.96862 Å (Tozadenant), using an unattenuated beam and 0.2° oscillation per frame, with an exposure of 0.1 second per degree of oscillation. Diffraction data for the A2A-StaR2-b RIL562-Vipadenant complex were acquired from 3 different crystals on an Eiger 16 M detector at beamline X06SA (Swiss Light Source) at a wavelength of 1 Å, using 10% beam transmission and 0.1° oscillation per frame, with an exposure of 1 second per degree of oscillation. The A2A-StaR2-b RIL562-ZM241385 data was collected from a single crystal on an Eiger 16 M detector at beamline × 06SA at a wavelength of 1 Å, using 20% beam transmission and 0.25° oscillation per frame, with an exposure of 0.24 second per degree of oscillation. Data from individual crystals were integrated using XDS 31, merged and scaled using AIMLESS 32 from the CCP4 suite33. Data collection statistics are reported in Table 1.
Structure solution and refinement
The structures of the different A2A-StaR2-b RIL562-ligand complexes were solved by molecular replacement (MR) with Phaser 34 using the A2A-StaR2-b RIL562-theophylline complex structure20 as the search model (PDB code: 5MZJ). Iterative rounds of model refinement performed using phenix.refine 35, were interspersed with manual model building in COOT 36. Both xray and B-factor restraint weights were optimised in phenix.refine, and 2 TLS groups corresponding to the receptor and to the b RIL562 respectively were defined during refinement. Refinement was with positional and individual isotropic B-factor refinement. The final models were validated using MolProbity37. The final refinement statistics are presented in Table 1. Structure figures were generated using PyMOL38. The three structures of the A2A-StaR2-b RIL562-Compound 4e reported here generated from crystals grown using LCP, are comparable with the previously reported A2A-StaR2-Compound 4e structure (PDB: 3UZC)19 solved from crystals grown using the vapour diffusion technique, with an all atom r.m.s.d. of 0.91 Å over 276 residues.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request. Co-ordinates and structure factors have been deposited in the Protein Data Bank under the accession codes 5OM1, 5OM4, 5OLZ, 5OLV, 5OLO, 5OLH and 5OLG.
Acknowledgements
The research leading to these results has received support from the Innovative Medicines Initiative Joint Undertaking under K4DD (www.k4dd.eu), grant agreement no. 115366, resources of which are composed of financial contribution from the European Union’s Seventh Framework Programme (FP7/2007–2013) and EFPIA companies’ in kind contribution. More info: www.imi.europa.eu. We thank D. Axford, R. Owen and D. Sherrell at I24, Diamond Light Source, Oxford, UK and M. Wang at beamline X06SA, Swiss Light Source, Villigen, Switzerland for technical support. We thank colleagues at Heptares Therapeutics Ltd. for suggestions and comments.
Author Contributions
R.K.Y.C. devised initial soaking experiments, performed LCP crystallization, designed crystal optimization, performed in meso soaking experiments, collected and processed X-ray diffraction data, solved and refined the structures. E.S. established the protein expression and purification protocols and performed LCP crystallization. T.G. performed and optimized protein purification. P.R. and T.G. optimized and performed in meso soaking experiments, collected and processed X-ray diffraction data and solved and refined structures. P.R. refined the final structures. Project management was carried out by A.S.D., J.C.E., G.A.B., R.M.C., and F.H.M. The manuscript was prepared by P.R., A.S.D. and F.H.M.
Competing Interests
The authors are shareholders of Sosei Group Corporation and declare competing financial interests.
Footnotes
Prakash Rucktooa, Robert K. Y. Cheng and Elena Segala contributed equally to this work.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Cooke RM, Brown AJ, Marshall FH, Mason JS. Structures of G protein-coupled receptors reveal new opportunities for drug discovery. Drug discovery today. 2015;20:1355–1364. doi: 10.1016/j.drudis.2015.08.003. [DOI] [PubMed] [Google Scholar]
- 2.Jazayeri A, et al. Extra-helical binding site of a glucagon receptor antagonist. Nature. 2016;533:274–277. doi: 10.1038/nature17414. [DOI] [PubMed] [Google Scholar]
- 3.Stevens RC, et al. The GPCR Network: a large-scale collaboration to determine human GPCR structure and function. Nature reviews. Drug discovery. 2013;12:25–34. doi: 10.1038/nrd3859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xiang J, et al. Successful Strategies to Determine High-Resolution Structures of GPCRs. Trends in pharmacological sciences. 2016;37:1055–1069. doi: 10.1016/j.tips.2016.09.009. [DOI] [PubMed] [Google Scholar]
- 5.Congreve M, Dias JM, Marshall FH. Structure-based drug design for G protein-coupled receptors. Progress in medicinal chemistry. 2014;53:1–63. doi: 10.1016/B978-0-444-63380-4.00001-9. [DOI] [PubMed] [Google Scholar]
- 6.Shoichet BK, Kobilka BK. Structure-based drug screening for G-protein-coupled receptors. Trends in pharmacological sciences. 2012;33:268–272. doi: 10.1016/j.tips.2012.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van Montfort RL, Workman P. Structure-based design of molecular cancer therapeutics. Trends in biotechnology. 2009;27:315–328. doi: 10.1016/j.tibtech.2009.02.003. [DOI] [PubMed] [Google Scholar]
- 8.Robertson N, et al. The properties of thermostabilised G protein-coupled receptors (StaRs) and their use in drug discovery. Neuropharmacology. 2011;60:36–44. doi: 10.1016/j.neuropharm.2010.07.001. [DOI] [PubMed] [Google Scholar]
- 9.Schutz M, et al. Directed evolution of G protein-coupled receptors in yeast for higher functional production in eukaryotic expression hosts. Scientific reports. 2016;6:21508. doi: 10.1038/srep21508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Scott DJ, Kummer L, Egloff P, Bathgate RA, Pluckthun A. Improving the apo-state detergent stability of NTS1 with CHESS for pharmacological and structural studies. Biochimica et biophysica acta. 2014;1838:2817–2824. doi: 10.1016/j.bbamem.2014.07.015. [DOI] [PubMed] [Google Scholar]
- 11.Miller JL, Tate CG. Engineering an ultra-thermostable beta(1)-adrenoceptor. Journal of molecular biology. 2011;413:628–638. doi: 10.1016/j.jmb.2011.08.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alexandrov AI, Mileni M, Chien EY, Hanson MA, Stevens RC. Microscale fluorescent thermal stability assay for membrane proteins. Structure. 2008;16:351–359. doi: 10.1016/j.str.2008.02.004. [DOI] [PubMed] [Google Scholar]
- 13.Yasuda, S. et al. Hot-Spot Residues to be Mutated Common in G Protein-Coupled Receptors of Class A: Identification of Thermostabilizing Mutations Followed by Determination of Three-Dimensional Structures for Two Example Receptors. The journal of physical chemistry. B, 10.1021/acs.jpcb.7b02997 (2017). [DOI] [PubMed]
- 14.Serrano-Vega MJ, Magnani F, Shibata Y, Tate CG. Conformational thermostabilization of the beta1-adrenergic receptor in a detergent-resistant form. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:877–882. doi: 10.1073/pnas.0711253105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hollenstein K, et al. Structure of class B GPCR corticotropin-releasing factor receptor 1. Nature. 2013;499:438–443. doi: 10.1038/nature12357. [DOI] [PubMed] [Google Scholar]
- 16.Dore AS, et al. Structure of class C GPCR metabotropic glutamate receptor 5 transmembrane domain. Nature. 2014;511:557–562. doi: 10.1038/nature13396. [DOI] [PubMed] [Google Scholar]
- 17.Christopher JA, et al. Biophysical fragment screening of the beta1-adrenergic receptor: identification of high affinity arylpiperazine leads using structure-based drug design. Journal of medicinal chemistry. 2013;56:3446–3455. doi: 10.1021/jm400140q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Segala E, Errey JC, Fiez-Vandal C, Zhukov A, Cooke RM. Biosensor-based affinities and binding kinetics of small molecule antagonists to the adenosine A(2A) receptor reconstituted in HDL like particles. FEBS letters. 2015;589:1399–1405. doi: 10.1016/j.febslet.2015.04.030. [DOI] [PubMed] [Google Scholar]
- 19.Congreve M, et al. Discovery of 1,2,4-triazine derivatives as adenosine A(2A) antagonists using structure based drug design. Journal of medicinal chemistry. 2012;55:1898–1903. doi: 10.1021/jm201376w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cheng RKY, et al. Structures of Human A1 and A2A Adenosine Receptors with Xanthines Reveal Determinants of Selectivity. Structure. 2017 doi: 10.1016/j.str.2017.06.012. [DOI] [PubMed] [Google Scholar]
- 21.Flohr, A., Moreau, J.-L., Poli, S. M., Riemer, C. & Steward, L. 4-hydroxy-4-methyl-piperidine-1-carboxylic acid (4-methoxy-7-morpholin-4-yl-benzothiazol-2-yl)-amide. US 20050261289 A1 (2008).
- 22.Sams AG, et al. Discovery of phosphoric acid mono-{2-[(E/Z)-4-(3,3-dimethyl-butyrylamino)-3,5-difluoro-benzoylimino]-thiazol-3 -ylmethyl} ester (Lu AA47070): a phosphonooxymethylene prodrug of a potent and selective hA(2A) receptor antagonist. Journal of medicinal chemistry. 2011;54:751–764. doi: 10.1021/jm1008659. [DOI] [PubMed] [Google Scholar]
- 23.Gillespie RJ, et al. Antagonists of the human A(2A) adenosine receptor. 4. Design, synthesis, and preclinical evaluation of 7-aryltriazolo[4,5-d]pyrimidines. Journal of medicinal chemistry. 2009;52:33–47. doi: 10.1021/jm800961g. [DOI] [PubMed] [Google Scholar]
- 24.Poucher SM, et al. The in vitro pharmacology of ZM 241385, a potent, non-xanthine A2a selective adenosine receptor antagonist. British journal of pharmacology. 1995;115:1096–1102. doi: 10.1111/j.1476-5381.1995.tb15923.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Segala E, et al. Controlling the Dissociation of Ligands from the Adenosine A2A Receptor through Modulation of Salt Bridge Strength. Journal of medicinal chemistry. 2016;59:6470–6479. doi: 10.1021/acs.jmedchem.6b00653. [DOI] [PubMed] [Google Scholar]
- 26.Liu W, et al. Structural basis for allosteric regulation of GPCRs by sodium ions. Science. 2012;337:232–236. doi: 10.1126/science.1219218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Blundell TL, Jhoti H, Abell C. High-throughput crystallography for lead discovery in drug design. Nature reviews. Drug discovery. 2002;1:45–54. doi: 10.1038/nrd706. [DOI] [PubMed] [Google Scholar]
- 28.Ferreira LG, Dos Santos RN, Oliva G, Andricopulo AD. Molecular docking and structure-based drug design strategies. Molecules. 2015;20:13384–13421. doi: 10.3390/molecules200713384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dore AS, et al. Structure of the adenosine A(2A) receptor in complex with ZM241385 and the xanthines XAC and caffeine. Structure. 2011;19:1283–1293. doi: 10.1016/j.str.2011.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Caffrey M, Cherezov V. Crystallizing membrane proteins using lipidic mesophases. Nature protocols. 2009;4:706–731. doi: 10.1038/nprot.2009.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kabsch W. Xds. Acta crystallographica. Section D, Biological crystallography. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Evans P. Scaling and assessment of data quality. Acta crystallographica. Section D, Biological crystallography. 2006;62:72–82. doi: 10.1107/S0907444905036693. [DOI] [PubMed] [Google Scholar]
- 33.Winn MD, et al. Overview of the CCP4 suite and current developments. Acta crystallographica. Section D, Biological crystallography. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McCoy AJ, et al. Phaser crystallographic software. Journal of applied crystallography. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Afonine PV, et al. Towards automated crystallographic structure refinement with phenix.refine. Acta crystallographica. Section D, Biological crystallography. 2012;68:352–367. doi: 10.1107/S0907444912001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta crystallographica. Section D, Biological crystallography. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen VB, et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta crystallographica. Section D, Biological crystallography. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schrodinger, L. L. C. The PyMOL Molecular Graphics System, Version 1.8 (2015).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request. Co-ordinates and structure factors have been deposited in the Protein Data Bank under the accession codes 5OM1, 5OM4, 5OLZ, 5OLV, 5OLO, 5OLH and 5OLG.