Abstract
The μ opioid receptor (OR), a member of the class A subfamily of G-protein coupled receptors (GPCRs), is a major target for the treatment of pain. G-protein biased μ-OR agonists promise to be developed as analgesics. Thus, TRV130, the first representative μ-OR ligand with G-protein bias, has entered into phase III clinical trials. To identify the detailed G-protein-biased activation and inactivation mechanisms of the μ-OR, we constructed five μ-OR systems that were in complexes with the G-protein-biased agonists TRV130 and BU72, the antagonists β-FNA and naltrexone, as well as the free receptor. We performed a series of conventional molecular dynamics simulations and analyses of G-protein-biased activation and inactivation mechanisms of μ-OR. Our results, together with previously reported mutation results, revealed the operating mode of the activation switch composed of residues W6.48 and Y7.43 (Ballesteros/Weinstein numbering), the activity of which was responsible for down- and up-regulation, respectively, of the β-arrestin signaling, which in turn affected G-protein-biased activation of μ-OR. TRV130 was found to stabilize W6.48 by interacting with Y7.43. In addition, we obtained useful information regarding μ-OR-biased activation, such as strong stabilization of W7.35 through a hydrophobic ring interaction in the TRV130 system. These findings may facilitate understanding of μ-OR biased activation and the design of new biased ligands for GPCRs.
Keywords: G-protein-coupled receptor, μ opioid receptor, molecular dynamics, activation switch, G-protein bias, β-arrestin signaling, TRV130, BU72, β-FNA, naltrexone
Introduction
G protein-coupled receptors (GPCRs) are the largest superfamily of drug targets1 and have gained attention in recent years. In contrast to the G-protein pathway, which elicits therapeutic effects, the β-arrestin pathway is the main cause of side effects caused by ligands targeting GPCRs2. Therefore, ligands that down-regulate or inhibit β-arrestin binding are of great interest3. Although many advances have been made in describing the mechanism of β-arrestin signaling, numerous problems remain regarding GPCR functional selectivity, particularly the dynamic propagation of biased signaling4,5. To design new ligands with a suitable signaling bias, additional studies are necessary.
Opioid receptors (ORs) are members of the class A (rhodopsin-like) subfamily of GPCRs, which includes three subtypes, μ, and κ, and are important targets for analgesia6. Many studies have shown that the analgesic effect of μ-OR occurs through activation of the Gi signal, whereas the downstream β-arrestin pathway of the receptor can lead to constipation, respiratory depression, addiction and other adverse effects7,8. Therefore, a G-protein biased μ-OR ligand is a major focus in designing new analgesics9,10.
TRV13011, the first representative μ-OR ligand with G-protein bias (Figure 1), has entered into phase III clinical trials12. The ligand was obtained through a series of methods involving biological screening, analysis of structure-activity relationships and structural modification. Compared with morphine and other traditional opioids, this ligand can induce multiple binding conformations in its mode of action13. Recently, Schneider et al14 have studied the binding mode of TRV130 through residue mutation experiments and long-term molecular dynamics (MD) simulations, but little information on the dynamic mechanism exists to explain the functional selectivity of TRV130.
Figure 1.
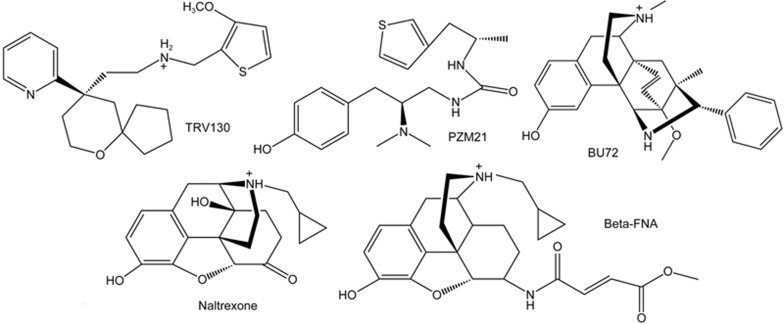
Two-dimensional structures of μ-OR G-protein-biased agonists TRV130 and PZM21, μ-OR agonist BU72, and μ-OR antagonists naltrexone (NTX) and β-FNA.
Recent studies of the mechanism of opioids15,16 suggest that the mechanism of functional selectivity of ORs is highly complex, involving a wide range of interactions. Hothersall et al17 have found that the W7.35A mutation (Ballesteros/Weinstein numbering18) down-regulates β-arrestin biased signaling of the ligand DAMGO, whereas the Y7.43F mutant exhibits abrogated β-arrestin signaling. Using residue mutation experiments, Sun et al19 have confirmed that W6.48 significantly affects the β-arrestin pathway but not G-protein signaling. Fenalti et al20 have performed a series of mutations and have found that residues D2.50, N3.35, F3.41, N7.45 and N7.49 are closely associated with β-arrestin signaling.
In our previous study21, we have found that residues E6.58 and I6.55 function as an activation switch for κ-OR that determines receptor activation. The activation switch is first triggered by large fluctuation of I6.55 and then is immediately induced by the motion of E6.58, which finally leads to rearrangement of each transmembrane helix. However, because only one agonist was introduced in the simulation, it was not possible for us to further investigate the detailed mechanism of the G-protein biased effects of ORs in that study.
Here, on the basis of agonist BU72-bound22,23 and antagonist β-FNA-bound24μ-OR crystal structures, we constructed five μ-OR systems: the G-protein biased agonist TRV130, two crystal complexes, the antagonist naltrexone (NTX) and the free receptor (Table 1), to further study the G-protein biased mechanism of μ-OR at the atomic level. We performed sub-microsecond unbiased molecular dynamics (MD) simulations on these μ-OR systems, focusing on the dynamic motions of the activation switch comprising residues W6.48 and Y7.43. Furthermore, we performed a detailed comparison of various interaction modes of the activation switch between TRV130 and another G-protein biased agonist, PZM21 (Figure 1). In addition, we identified several residues that may also be crucial for TRV130 G-protein bias. Therefore, our simulations provide a better understanding of the activation and G-protein biased activation of μ-OR, and may be used for new G-protein biased ligand design.
Table 1. Systems prepared for the MD simulations.
| System ID | μ-OR state | Ligand | POPC | Na+ | Cl− | Water |
|---|---|---|---|---|---|---|
| A | Active | BU72 | 106 | 51 | 65 | 11033 |
| B | Active | TRV130 | 106 | 51 | 65 | 11032 |
| C | Active | NTX | 106 | 51 | 65 | 11030 |
| D | Active | - | 106 | 51 | 64 | 11039 |
| E | Inactive | β-FNA | 104 | 52 | 66 | 11001 |
Materials and methods
Protein preparation
There are two crystal structures of μ-OR available in the Protein Data Bank (PDB), both from mice. Interestingly, one receptor structure is bound to the agonist BU72 and is in the active state (PDB entry code: 5C1M)22, whereas the other structure is complexed with the antagonist β-FNA and is in the inactive form (PDB entry code: 4DKL)24.
In 5C1M, the receptor was engineered with a G-protein mimetic camelid antibody fragment22. To perform MD simulations on the wild-type receptor, the G-protein part was removed from the crystal structure and the engineered residue YCM was mutated back to the original Cys in the N-terminus. In 4DKL, the receptor was engineered with part of the intracellular loop between transmembrane helices TM5 and TM6 replaced by T4 lysozyme (T4L)24. Similarly, T4L and other unnecessary parts were removed from 4DKL, and the IL3 loop was reconstructed by adding the missing residues with the loop refinement protocol in Discovery Studio 3.5 (Aecelrys Inc, San Diego, CA, USA). The 10 loop models generated were clustered as a dendrogram on the basis of the pairwise main-chain root-mean-square deviation (RMSD) of the loop region, and the model with the lowest RMSD value was selected for loop reconstruction25. A Na+ ion located at the allosteric pocket position20 was exactly positioned according to the crystal structure of the inactive NTI-complexed δ-OR (PDB entry code: 4N6H).
The Protein Preparation Tool (ProPrep) in the Schrödinger 2012 software package was then used to prepare the above integrated three-dimensional structures of the μ-OR. Residues Asn, Gln, and His were automatically checked for their protonated states in ProPrep. The PROPKA tool in Maestro was utilized to add hydrogen atoms into the structures at a physiological pH environment with an optimized hydrogen bond network.
Molecular docking
For the conformations of β-FNA and BU72 in complex with μ-OR, the agonist TRV130 and antagonist NTX were sketched in Maestro and subjected to a Monte Carlo Multiple Minimum conformational search under the OPLS_2005 force field. Implicit solvent was used for water molecules (Surface Generalized Born model). The output conformations were utilized as the starting point for molecular docking.
The active state of μ-OR in 5C1M was selected for docking ligands TRV130 and NTX, and it was also used as the free receptor by removal of the ligand BU72. Molecular docking was performed using the XP mode of Glide 5.826,27. Residue D3.32 was defined as the grid core to generate the pocket Grid file of μ-OR by 20 Å with the Receptor Grid Generation module. The Van der Waals (vdW) scaling was set to 0.8 for nonpolar atoms of the receptor and ligand in the initial Glide docking. The number of docking output per docking run was set as 10 000 poses during docking. The most reasonable conformation of the ligand was selected on the basis of two criteria. First, the protonated nitrogen should form an electrostatic interaction with D3.32, which is a well-known essential action for opioid ligands. Second, the docking score was used to rank the remaining poses.
MD preparations
Five systems (Table 1), TRV130-μ-OR, BU72-μ-OR, NTX-μ-OR, and β-FNA-μ-OR complexes and an apo-μ-OR system, were constructed for the simulations. A POPC (1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine) bilayer with a surface area of 75×75 Å2 on the X-Y plane was generated under the CHARMM36 force field in the VMD program (Version 1.9.1). For each system, our in-house program was first used to embed the receptor into the POPC bilayer pre-aligned in the OPM (Orientations of Proteins in Membranes) database28,29,30,31. Thereafter, pre-equilibrated POPC lipids coupled with TIP3P water molecules in a box ∼75×75×100 Å3 were used to solvate the protein (Table 1). Water molecules in the bilayer and lipid molecules within 0.85 Å of the heavy atoms on the protein structure were removed. Neutral systems with 0.15 mol/L NaCl were produced through Na+ and Cl− ions in the water phase.
Paramchem webserver32,33,34 (https://cgenff.paramchem.org/), a program coupled with the CHARMM force field, was used to generate ligand force field parameters. We input small molecules with a correct configuration into the Paramchem webserver and then obtained the force field parameters of the compounds through a script. Because β-FNA is complexed with μ-OR through a covalent bond, we then defined ligand β-FNA and residue K5.39 as one residue in the force field.
MD simulations
All simulations were performed on the high-performance computing cluster of the State Key Laboratory of Bioreactor Engineering by using Gromacs V.4.6.535 and the CHARMM36 parameters for all compositions. In the first step, a 10 000-step energy minimization was implemented with 1000.0 kJ/mol/nm as the force threshold for each system. Then, 100 ps initial equilibration was produced to gradually heat the systems from 0 K to 310 K at a constant volume and temperature at 310 K. The systems were subjected to an additional 1 ns equilibration at constant pressure and temperature (NPT ensemble; 310 K, 1 atm) with three thermostats (stabilizing temperature independently for protein-ligand system, the lipid POPCs and water-ions system) at 2 ps time steps. The cut off value for vdW and short-range electrostatic interactions was set at 12 Å. The Particle Mesh Ewald (PME) summation scheme was used to compute long-range electrostatic interactions. Finally, the production MD simulations of five systems were performed for 300 ns under NPT conditions with a Nose-Hoover thermostat for temperature coupling and Parrinello-Rahman pressure coupler methods. The integrator leap-frog algorithm was used, and the time step was set as 2 fs.
Analysis of MD simulations
RMSD and RMSF (root-mean-square fluctuation) calculations, hydrogen bond analysis, torsion angle and distance evolutions were produced with the program Gromacs. The interval time of trajectory calculations, including RMSD, torsion angle, distance and hydrogen bond analysis, was 200 ps in these 300 ns simulations. RMSD values were calculated by comparing the initial simulation conformations. All smooth curves in Results were fitting groups, which were identical to the corresponding angle/RMSD calculations in color.
Torsion angle analysis was performed with the g_angle tool under dihedral angle style in Gromacs. The average distance between residues W6.48 and Y7.43 was measured with the g_dist tool. Water tracking was performed by using the minimum distance method, which was assessed with the g_mindist tool.
Results
Initial docking modes of μ-OR with agonist TRV130 and antagonist NTX
Agonist BU72-bound and antagonist β-FNA-bound μ-OR complexes were directly used for unbiased MD simulations, and the receptor bound with BU72 was used for ligand docking.
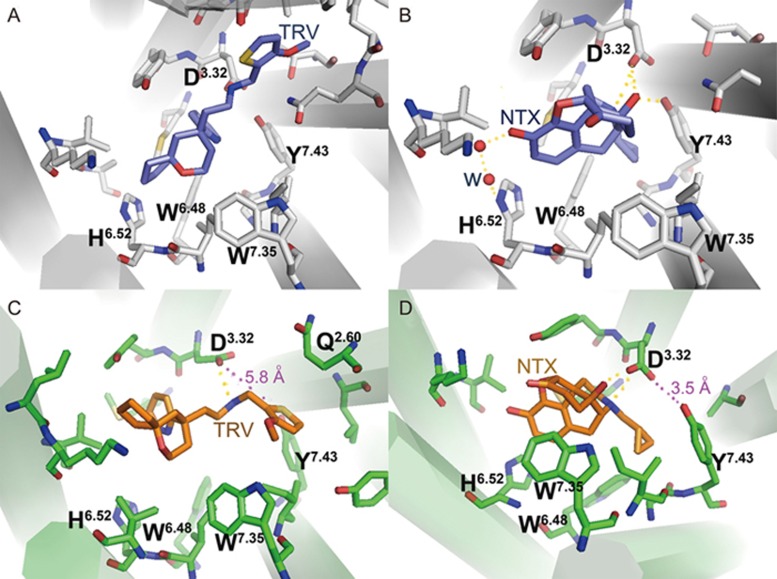
As observed from the docking modes (Figure 2A, 2B), the protonated nitrogen ions of both TRV130 and NTX formed electrostatic interactions with residue D3.32. The cyclopropyl group of antagonist NTX, similarly to that of β-FNA in 4DKL, inserted into the space between residues W6.48 and Y7.43 and simultaneously interacted with the two residues. The phenolic hydroxyl group of NTX interacted with H6.52 through two water molecules. In the TRV130 system, owing to a ring structure replacing the polar group at this position, TRV130 formed interactions with the key residue H6.52 through the ring structure. On the basis of the docking modes, we constructed TRV130-μ-OR and NTX-μ-OR systems for unbiased MD simulations.
Figure 2.
Presentation of the initial docking poses (A, B), key residues and final 300 ns frames (C, D) in the TRV130-μ-OR and NTX-μ-OR complexes. The backbones of the proteins are colored green (300 ns) and gray (initial), whereas those of the ligands are colored orange (300 ns) and slate-blue (initial). The hydrogen bonding interactions are shown as yellow dotted lines, whereas the measured distances are shown as purple dotted lines.
Overview of the five unbiased MD simulations
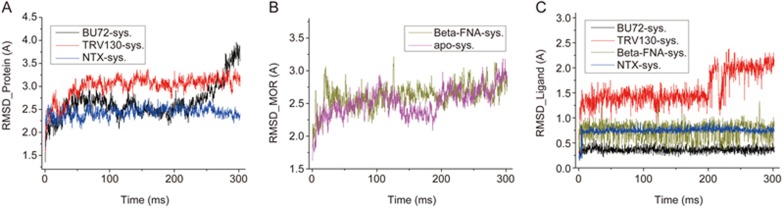
As shown in Figure 3A and 3B, the RMSD values of the five MD simulations reached the equilibrium stage in approximately 50 ns. In the TRV130 system, the RMSD value of the receptor fluctuated from 2.7–3.5 Å as compared with its initial structure. From 50–200 ns, the RMSD fluctuations of TRV130 were from 1 to 1.7 Å (Figure 3C) and were mostly stable from 1.7–2.7 Å from 260–300 ns, thus indicating a large conformational change of TRV130 from 200–260 ns, as also reflected through the conformational superimposition results (Figure 2C). Moreover, the local protein structure was altered with these large conformational changes of ligands, particularly at the region composed of residues Q2.60, D3.32 and Y7.43, where a tri-joint structure was formed by hydrogen bonds with one another in either the active or inactive state of the receptor. However, this tri-joint structure was lost in the final 300 ns TRV130-μ-OR complex, and the distance between residues Y7.43 and D3.32 reached 5.8 Å, which was too far to form a hydrogen bond (HB) again.
Figure 3.
RMSD evolutions of proteins (A, B) and ligands (C) in the five systems during the simulations.
As compared with the initial structure, the RMSD values of the receptor in other systems under the equilibrium stage fluctuated from 2.3–3.2 Å. However, the RMSD value of the BU72 system began to increase at 250 ns. Through the superposition, we found that the RMSD increase was closely related to conformational changes of the extracellular N-terminal loop region (Supplementary Figure S1A). Ligands BU72 and NTX exhibited high stability, and their RMSD values were stabilized at 0.4 and 0.7 Å, respectively, whereas those of β-FNA fluctuated in the range of 0.4 and 1.2 Å, probably because of the swing of the covalently bound residue K5.39.
The dynamics of “message” residues D3.32 and H6.52 in different systems
The crystal structure of β-FNA-bound μ-OR24 demonstrates that residues D3.32 and H6.52 form direct interactions with the ligand, thereby determining the “message”36,37 roles of the two residues. In the simulations, we first investigated the performance of D3.32 in each system. This residue in both agonist systems stably formed one HB with the ligands (Table 2). However, for the antagonists, 1–2 stable HBs were formed in the NTX system, and almost no interaction was found in the β-FNA system, possibly because of the covalent binding of the ligand. The interactions between D3.32 and water molecules in the antagonist models were generally more numerous than those in the agonist models. For example, 6–7 HBs were stably formed in β-FNA system, whereas 3 steady HBs were found in both agonist models.
Table 2. Occupancies of the key hydrogen bonds in the five μ-OR systems. All data were produced with the program Gromacs.
| HB No | BU72 system (%) |
TRV130 system (%) |
NTX system (%) |
β-FNA system (%) |
Apo system (%) |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| D2.50-water | D3.32-ligand | D3.32-water | D2.50-water | D3.32-ligand | D3.32-water | D2.50-Water | D3.32-ligand | D3.32-water | D2.50-water | D3.32-ligand | D3.32-water | D2.50-water | D3.32-Water | |
| 0 | 0.1 | 12.4 | --a | -- | 1.3 | -- | -- | 7.3 | -- | -- | 97.8 | -- | -- | -- |
| 1 | 11.4 | 86.4 | 0.6 | 0.4 | 92.5 | -- | 3.0 | 42.6 | 0.2 | 3.8 | 2.2 | -- | 0.7 | -- |
| 2 | 42.8 | 1.2 | 10.9 | 2.4 | 6.2 | 1.3 | 15.2 | 38.3 | 6.0 | 46.7 | -- | 0.2 | 6.7 | -- |
| 3 | 43.2 | -- | 50.3 | 27.4 | -- | 41.8 | 32.9 | 11.8 | 37.6 | 48.0 | -- | 0.8 | 21.8 | -- |
| 4 | 2.4 | -- | 32.8 | 57.9 | -- | 26.0 | 39.8 | -- | 41.8 | 1.4 | -- | 2.2 | 55.4 | 1.1 |
| 5 | -- | -- | 5.2 | 11.6 | -- | 18.5 | 8.7 | -- | 10.9 | -- | -- | 13.0 | 14.8 | 8.5 |
| 6 | -- | -- | 0.3 | 0.2 | -- | 9.8 | 0.3 | -- | 1.9 | -- | -- | 34.7 | 0.6 | 26.4 |
| 7 | -- | -- | -- | -- | -- | 1.5 | -- | -- | 1.5 | -- | -- | 37.4 | -- | 36.8 |
| 8-9 | -- | -- | -- | 0.3 | 11.6 | -- | 27.2 | |||||||
aThe symbol “--” represents no related hydrogen bond interaction.
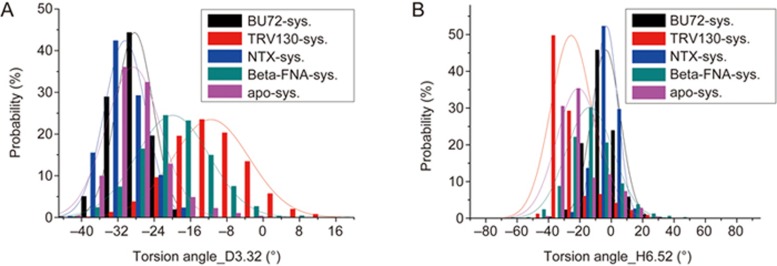
At the end of the simulation, the side chain of D3.32 significantly changed in conformation (Figure 2), as was also revealed by the RMSF values (Supplementary Table S1). Therefore, we performed a detailed analysis on the torsion angle of the D3.32 side chain (Figure 4). Although D3.32 formed one HB with each of BU72 and TRV130, the residue underwent a large conformational twist in the TRV130 system. Viewed from the distribution of the torsion angle, the dominant torsion angle was approximately -12° and occupied approximately 23% of the simulation time in the TRV130 system, whereas the dominant torsion angle was approximately -28° and occupied approximately 45% of the simulation time in the BU72 system, thus indicating that the stability of D3.32 in the TRV130 system was lower than in the BU72 system, and this residue was substantially twisted.
Figure 4.
The distributions of the torsion angles of the “message” residues D3.32(A) and H6.52 (B) in the five systems.
The performance of another “message” residue H6.52 was also investigated in detail. Because there is no polar group at this position of TRV130, H6.52 interacted through its ring structure, and thus the most stable torsion angle of this residue was approximately -38° in TRV130 system, whereas it was approximately 10° in other systems.
The operation modes of activation switch residues W6.48 and Y7.43
From the existing μ-OR ligands38, a ligand with a cyclopropyl group on the protonated nitrogen atom, such as NTX, performs as an antagonist, whereas no such a group at this position makes oxymorphine exhibit agonistic activity. Given that the interaction site20,24 of the cyclopropyl group is located in the space between W6.48 and Y7.43, we propose that residues W6.48 and Y7.43 may act as a paired activation switch in μ-OR, which is called the activation switch residue here. A recent study has illustrated that inhibition of W6.48 up-regulates G-protein activation selectivity19, because the 6.48 position is strongly correlated with β-arrestin biased activation. Moreover, the 7.43 position is also related to G-protein biased activation of μ-OR17. Thus, we focused on the performance of these two residues in each system (Figure 5).
Figure 5.
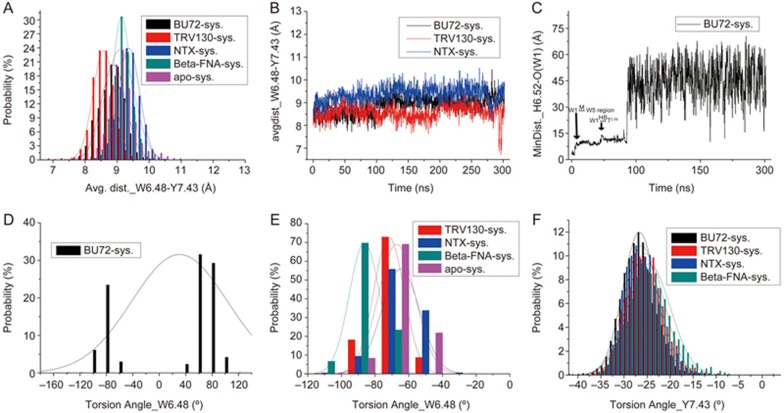
The average distance distributions between the activation switch residues W6.48 and Y7.43 in the five systems (A) and evolutions of these average distances in the BU72, TRV130 and NTX systems (B). (C) Detailed evolutions of the minimum distances between residue H6.52 and water W1 in the BU72 system. (D, E) The distributions of the side chain torsion of “switching” residue W6.48 in the five systems. (F) The distributions of the side chain torsion of important residue Y7.43 in the four ligand-bound systems.
The average distance (AD) between W6.48 and Y7.43 was approximately 8.5 Å in the agonist TRV130 system, whereas it was 9.2 Å in the β-FNA system, reaching up to 9.4 Å in NTX system. As observed from the evolution of AD with simulation time, the shortest distance between Y7.43 and W6.48 reached 6.8 Å in the TRV130 system (Figure 5B), whereas it was only 7.4 Å in the apo system (Supplementary Figure S2A).
In the BU72 system, the AD value between W6.48 and Y7.43 increased from 8.5 Å to approximately 9 Å at 100 ns (Figure 5B). Further investigation showed that the side chain of W6.48 (Figure 5D) reversed, thus potentially increasing the distance between them. Furthermore, we found that water molecule W1 (Supplementary Figure S3) moved to the W5 position (Figure 5C), and the movement was across the position of W6.48, thus potentially causing its side chain to reverse. The side chain reversal of W6.48 has also been observed in the simulations performed by Huang et al22.
The performance of W6.48 was different among the five systems (Figure 5E). In the TRV130 system, W6.48 was more stable than in the other systems, and the torsion angle of its side chain was mostly stabilized at -70°. In antagonist models, the stability of W6.48 was higher in the β-FNA system than in the NTX system.
Interestingly, the dynamics of Y7.43 was completely opposite from that of W6.48 in each system (Figure 5F). Y7.43 was most stable in the BU72 system, whereas large volatility was observed in TRV130/β-FNA systems. As observed from the TRV130 system at 300 ns (Figure 2C), the ring structure of TRV130 formed direct interactions with Y7.43, a result consistent with findings from residue mutation experiments by Schneider et al14. These interactions led to a large swing of the residue; similarly, the cyclopropyl group of β-FNA interacted with Y7.43, thereby increasing the activity of Y7.43. Moreover, Y7.43 in the apo system presented the largest swing. The aromatic ring even reversed at approximately 80 ns (Supplementary Figure S2), and the torsion angle of Y7.43 also fluctuated within a large range.
The different performances of “address” residue W7.35 and key residue Y7.53
The 7.35 position in δ-OR is a non-polar aliphatic residue Leu, but the residue at that site is replaced by an aromatic residue, Trp/Tyr, in μ/κ-OR, respectively. The diversity of this position determines its relationship with opioid selectivity, thus making it an “address” site39. A recent study has shown that this position is also highly correlated with the functional selectivity of μ-OR17. In the W7.35A mutant, DAMGO increased the relative β-arrestin activity, which directly reflected the significance of W7.35 in the functional selectivity of μ-OR. Moreover, as observed from the docking mode of DAMGO17, the ligand did not form an efficient interaction with W7.35, thus suggesting that good stability of the residue might favor G-protein signaling.
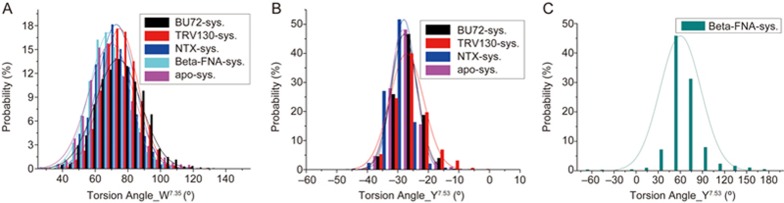
Although the side chain torsion angle of W7.35 in each of the five systems was stabilized at approximately 70° on average (Figure 6), the percentage of the main conformation in the TRV130 system was approximately 1/4 higher than that of the BU72 system, thus indicating that the residue had better stability in the agonist TRV130 system. As observed from the superposition of TRV130-μ-OR complexes at 300 ns with the initial conformation, the ring structure of TRV130 formed hydrophobic interactions with W7.35, thereby increasing the stability of W7.35. Furthermore, the residue showed good stability in the antagonist systems.
Figure 6.
The distributions of torsion angles of the “address” residue W7.35(A) and key residue Y7.53(B, C) in the five systems.
In the antagonistic β-FNA system, the torsion angle of Y7.53 was completely different from that in the other systems. This torsion angle should be directly associated with the active and inactive crystal structures (Supplementary Figure S4). Furthermore, the fluctuation range of the Y7.53 torsion angle in the β-FNA model was particularly large, from -60 to 180°, thereby suggesting that Y7.53 is highly active. The performance of Y7.53 was diverse, even among the four activated state models, and it was more volatile in the TRV130 system (Figure 6B). Because Y7.53 is located in the G-protein binding region, such variations also indicated that the G-protein binding pattern induced by TRV130 may be different from that of BU72.
The dynamic changes in key structural water molecules in each system
The 'NPxxY' motif, a key motif in the activation40,41,42 and G-protein binding43 of family A GPCRs, is closely associated with water clusters44. According to the active crystal structure of μ-OR 5C1M (Supplementary Figure S4), the NPxxY region is directly connected with the internal water molecular environment, and there are a few water molecules around. Furthermore, N7.49 is involved in the formation of an antagonistic sodium-mediated allosteric pocket containing multiple water molecules. Therefore, we selected two important water molecules, W3 and W4 (Figure 7), located at the center position of μ-OR (Supplementary Figure S3), for detailed analysis and comparison of the dynamic changes between the agonist BU72 system and fully antagonistic β-FNA system.
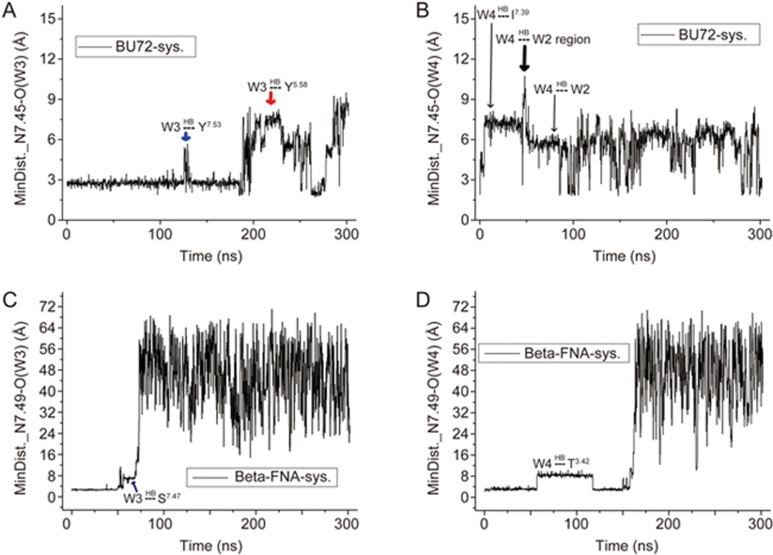
Figure 7.
Detailed evolutions of the minimum distances between residue N7.45 and water W3 (A) and W4 (B) in the BU72 system. Detailed evolutions of the minimum distances between residue N7.49 and water W3 (C) and W4 (D) in the β-FNA-μ-OR system. 'HB' means 'to form a hydrogen bonding (HB) interaction'.
As observed from the trajectory, W3 in the BU72 system belonged to the intracellular environment (Figure 7A), whereas the molecule in the β-FNA system was affiliated with the extracellular section (Figure 7C). The W4 molecule in both systems belonged to the extracellular environment. Given that the NPxxY region in the intracellular side of the antagonistic state was surrounded by a large number of hydrophobic residues (Supplementary Figure S4), such as I2.43, L2.46, M6.36 and L7.56, we propose that the NPxxY region may be a separation area in an activated state between external and interior water molecules in the cell, yet this separation area in the completely antagonistic state was much greater than that in the active state.
In addition, the motion of W4 suggested that water molecules in the binding pocket of the BU72 system may be much more active than in the β-FNA system. The W4 molecule in the BU72 system almost always changed in position; however, the molecule's position changed only twice in the first 150 ns of the β-FNA system, and each interval was approximately 50 ns. The water molecule was located in the sodium binding allosteric site, thus also indicating that the water molecules inside the pocket had better stability.
The antagonistic sodium-mediated allosteric pocket is mainly composed of a few polar residues: D2.50, N3.35, S3.39, N7.45, S7.46 and N7.4920. Therefore, we performed a detailed analysis of the number of water molecules in the pocket of each system (Tables 2 and 3). On the one hand, we directly computed the amounts of water molecules by sampling every 100 ns. The statistical results showed that 4 water molecules were contained in the antagonistic β-FNA system, but the largest number was 3 in other systems, thus indicating that the allosteric pocket was larger in fully antagonistic μ-OR than in the others. On the other hand, because residue D2.50 is located at the core section of the allosteric site, we investigated the number of water molecules in the pocket through HB calculations between water molecules and D2.50. The statistical results showed that D2.50 formed 4 stable HBs with water molecules in TRV130, NTX and apo systems, particularly in the TRV130 system, and the proportion of 4 HB formed reached 57.9%, whereas the most stable number was 3 in the BU72 system. Because D2.50 may be affected by the coupling of a sodium ion, only the 3 most stable HBs were observed in β-FNA system.
Table 3. The number statistics of the structural water molecules located at the sodium allosteric position with a100 ns interval time in the five μ-OR systems.
| No | BU72 system | TRV130 system | NTX system | β-FNA system | Apo system |
|---|---|---|---|---|---|
| 0 ns | 1 | 1 | 1 | 4 | 3 |
| 100 ns | 2 | 3 | 1 | 4 | 3 |
| 200 ns | 3 | 3 | 3 | 4 | 3 |
| 300 ns | 3 | 3 | 2 | 4 | 3 |
Discussion
In this study, we performed a series of MD simulations and comparative analyses of the atomic-level conformational changes of μ-OR in G-protein biased activation and inactivation, including the dynamics of crucial “message-address" residues, crucial residues for selective μ-OR activation and inactivation and important structural water molecules. This information may be helpful for further understanding the biased interaction mechanism of μ-OR.
TRV130, a G-protein biased agonist, had diverse effects in many aspects, as compared with the agonist BU72. First, the triplet structure composed of residues Q2.60, D3.32 and Y7.43 was abrogated by TRV130 binding. Second, the formation of the direct interaction with Y7.43 induced W6.48 and Y7.43 to maintain proximity to each other, thus further enhancing the stability of W6.48. Third, the ring structure of TRV130 also stabilized W7.35. Moreover, the conformation of Y7.53 was greatly influenced by the binding of TRV130. These effects may be closely related to ligand G-protein bias. Moreover, according to the experimental results17,19, the above interactions down-regulated β-arrestin signaling, thereby increasing the G-protein bias of TRV130.
In this paper, owing to time limitations, we simulated only one G-protein biased agonist. A new G-protein biased ligand, PZM2145, was discovered in 2016 and has exhibited good activity in many types of μ-OR activation effects. As observed from the docking mode45, the phenolic hydroxyl group of PZM21 is located in the W6.48 side, close to the activation switch W6.48 and Y7.43. Therefore, we suggest that PZM21 may stabilize W6.48 through a direct interaction and then significantly down-regulate the β-arrestin signal and ultimately improve the G-protein bias. However, our simulation results and residue mutation experiments14 showed that the crucial sites of TRV130, in contrast to those of PZM21, are located between residues Y7.43 and Y2.64 . To better understand the functionally selective mechanism of μ-OR, it is necessary to simulate the dynamic mechanism of PZM21 and to perform a comprehensive analysis to reveal more information for designing the ligand with G-protein bias, which will be performed in the future.
Through the above analyses, we observed that the operation of the activation switch determined the receptor activation. Additionally, the operation mode of the activation switch probably plays a key role in the binding of β-arrestin, thereby up-regulating or down-regulating G-protein signal transduction. These findings may be applied to new drug design. For example, we the activation switch of a target can be identified, then a reasonable mode of action can be selected, and finally a variety of design experiments can be performed.
Okude et al13, by using NMR and Met labeling methods, have shown that the TRV130 G-protein bias occurs through multiple conformational changes. They have also determined that both the N3.35A and F3.41A mutations lead to a significant increase of β-arrestin signal for TRV130 binding. These two residues are located in TM3, and we observed the differences in the TRV130 system compared with the BU72 and antagonist systems (Figure 2 and 4). Because the two residues are far from the ligand binding pocket, we attributed the differences to the mode of action of TRV130.
The μ-OR antagonists blocked the interaction between the activation switch residues W6.48 and Y7.43, and we observed a notable diversity between the agonist and antagonist action. In addition, the effect of D3.32 occurred at a different point. The residue in the agonist system mainly interacted with the ligand through one hydrogen bond, but in the antagonist system, either no interaction occurred, such as for β-FNA, or more hydrogen bonds were found, such as in the NTX system. Therefore, we propose that these two aspects are a critical distinction between μ-OR agonists and antagonists.
Although the interaction between W6.48 and Y7.43 was separated by the cyclopropyl group of β-FNA and NTX, the load-bearing residues were opposite from each other in the two antagonist systems, as observed from the different effects induced by the ligands. There were also diverse effects of these two ligands in other aspects, such as the influence on message residue D3.32. In addition, W7.35 was well stabilized by the two antagonists, as well as TRV130, thus ensuring their subtype selectivity on μ-OR to a certain extent.
Huang22, Shim46 and Schneider14 have all performed MD simulations on the activation mechanism of μ-OR, but their analyses have been performed only for a single residue, such as W6.48 or D3.32. From existing interaction mechanisms of opioids21, particularly agonists, the key roles were often realized through more than one residue, together with dynamic associations. According to our experimental results, residues W6.48 and Y7.43 act as a paired activation switch.
The key structural water molecules are an indispensable part of the activation and antagonistic process of ORs, and their dynamic motions also reflect some structural information. We observed that the NPxxY area was a separation area between the intracellular and extracellular environment in the activated μ-OR, and yet the separation area was much greater in the completely antagonistic state than that in the active state of the receptor. Furthermore, more water molecules existed in the sodium-controlled allosteric pocket of the antagonistic β-FNA system, and they were much more stable than those in the activated BU72 system.
In addition, the derivatives of Salvinorin47,48 do not contain a protonated nitrogen ion. Thus the activation and antagonistic mechanism must be further reconsidered for this special type of opioid.
Conclusions
In this study, we performed a series of MD simulations and analyses of the G-protein biased activation and inactivation mechanisms of μ-OR. Five μ-OR systems were constructed: a G-protein biased agonists TRV130 and BU72, antagonists β-FNA and NTX, and the free receptor. We propose that residues W6.48 and Y7.43 function as a paired activation switch, in agreement with related mutation experiments. Operation of this activation switch is a critical factor for μ-OR activation, whereas up-regulation or down-regulation of the β-arrestin signal is probably induced by the interaction mode between the ligand and activation switch. Furthermore, we predicted the appropriate activation mode of another G-protein biased ligand, PZM21, on the basis of the above switch. Together with the docking mode, we hypothesized that PZM21 stabilized W6.48 through a direct interaction, then significantly down-regulated β-arrestin signal, and finally improved the G protein bias. In addition, we obtained diverse information about μ-OR activation and inactivation, including the conformational varieties of the triplet structure, residues W7.35 and Y7.53, and structural water molecules. Thus, our simulations should provide helpful information for new biased ligand design, such as identification of an activation switch on a target, and subsequent selection of a more reasonable mode of action, and performance of a variety of experiments.
Author contribution
Jian-xin CHENG performed the molecular dynamics simulations; Jian-xin CHENG and Yun TANG designed the study and analyzed the data; Yun TANG was responsible for the project; Jian-xin CHENG, Tao CHENG, Wei-hua LI, Gui-xia LIU, Wei-liang ZHU and Yun TANG contributed to writing and commenting on the manuscript.
Acknowledgments
All simulations were performed on the high-performance computing cluster kindly provided by Prof Bang-ce YE of the State Key Laboratory of Bioreactor Engineering, East China University of Science and Technology. This work was supported by the National Natural Science Foundation of China (Grants 81673356 and U1603122) and the 111 Project (Grant B07023).
Footnotes
Supplementary files are available at the website of Acta Pharmacologica Sinica.
Supplementary Information
Supplementary Table and Figures
References
- Lagerstrom MC, Schioth HB. Structural diversity of G protein-coupled receptors and significance for drug discovery. Nat Rev Drug Discov 2008; 7: 339–57. [DOI] [PubMed] [Google Scholar]
- McDonald PH, Chow CW, Miller WE, Laporte SA, Field ME, Lin FT, et al. β-Arrestin 2: a receptor-regulated MAPK scaffold for the activation of JNK3. Science 2000; 290: 1574–7. [DOI] [PubMed] [Google Scholar]
- Whalen EJ, Rajagopal S, Lefkowitz RJ. Therapeutic potential of β-arrestin and G protein-biased agonists. Trends Mol Med 2011; 17: 126–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurevich VV, Gurevich EV. The molecular acrobatics of arrestin activation. Trends Pharmacol Sci 2004; 25: 105–11. [DOI] [PubMed] [Google Scholar]
- Thomsen AR, Plouffe B, Cahill TJ, Shukla AK, Tarrasch JT, Dosey AM, et al. GPCR-G protein-β-arrestin super-complex mediates sustained G protein signaling. Cell 2016; 166: 907–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruchas MR, Roth BL. New technologies for elucidating opioid receptor function. Trends Pharmacol Sci 2016; 37: 279–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohn LM, Gainetdinov RR, Lin FT, Lefkowitz RJ, Caron MG. Mu-opioid receptor desensitization by beta-arrestin-2 determines morphine tolerance but not dependence. Nature 2000; 408: 720–3. [DOI] [PubMed] [Google Scholar]
- Raehal KM, Walker JK, Bohn LM. Morphine side effects in beta-arrestin 2 knockout mice. J Pharmacol Exp Ther 2005; 314: 1195–201. [DOI] [PubMed] [Google Scholar]
- Thompson GL, Kelly E, Christopoulos A, Canals M. Novel GPCR paradigms at the mu-opioid receptor. Br J Pharmacol 2015; 172: 287–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burford NT, Traynor JR, Alt A. Positive allosteric modulators of the mu-opioid receptor: a novel approach for future pain medications. Br J Pharmacol 2015; 172: 277–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen XT, Pitis P, Liu G, Yuan C, Gotchev D, Cowan CL, et al. Structure-activity relationships and discovery of a G protein biased mu opioid receptor ligand, [(3-methoxythiophen-2-yl)methyl]({2-[(9R)-9-(pyridin-2-yl)-6-oxaspiro-[4.5]decan- 9-yl]ethyl})amine (TRV130), for the treatment of acute severe pain. J Med Chem 2013; 56: 8019–31. [DOI] [PubMed] [Google Scholar]
- Viscusi ER, Webster L, Kuss M, Daniels S, Bolognese JA, Zuckerman S, et al. A randomized, phase 2 study investigating TRV130, a biased ligand of the mu-opioid receptor, for the intravenous treatment of acute pain. Pain 2016; 157: 264–72. [DOI] [PubMed] [Google Scholar]
- Okude J, Ueda T, Kofuku Y, Sato M, Nobuyama N, Kondo K, et al. Identification of a conformational equilibrium that determines the efficacy and functional selectivity of the mu-opioid receptor. Angew Chem Int Ed Engl 2015; 54: 15771–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider S, Provasi D, Filizola M. How oliceridine (TRV-130) binds and stabilizes a mu-opioid receptor conformational state that selectively triggers G protein signaling pathways. Biochemistry 2016; 55: 6456–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeWire SM, Yamashita DS, Rominger DH, Liu G, Cowan CL, Graczyk TM, et al. A G protein-biased ligand at the mu-opioid receptor is potently analgesic with reduced gastrointestinal and respiratory dysfunction compared with morphine. J Pharmacol Exp Ther 2013; 344: 708–17. [DOI] [PubMed] [Google Scholar]
- Soergel DG, Subach RA, Burnham N, Lark MW, James IE, Sadler BM, et al. Biased agonism of the mu-opioid receptor by TRV130 increases analgesia and reduces on-target adverse effects versus morphine: A randomized, double-blind, placebo-controlled, crossover study in healthy volunteers. Pain 2014; 155: 1829–35. [DOI] [PubMed] [Google Scholar]
- Hothersall JD, Torella R, Humphreys S, Hooley M, Brown A, McMurray G, et al. Residues W320 and Y328 within the binding site of the mu-opioid receptor influence opiate ligand bias. Neuropharmacology 2017; 118: 46–58. [DOI] [PubMed] [Google Scholar]
- Ballesteros JA, Weinstein H. Integrated methods for the construction of three-dimensional models and computational probing of structure-function relations in G protein-coupled receptors. Methods Neurosci 1995; 25: 366–428. [Google Scholar]
- Sun X, Laroche G, Wang X, Agren H, Bowman GR, Giguere PM, et al. Propagation of the allosteric modulation induced by sodium in the delta-opioid receptor. Chemistry 2017; 23: 4615–24. [DOI] [PubMed] [Google Scholar]
- Fenalti G, Giguere PM, Katritch V, Huang XP, Thompson AA, Cherezov V, et al. Molecular control of delta-opioid receptor signalling. Nature 2014; 506: 191–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J, Sun X, Li W, Liu G, Tu Y, Tang Y. Molecular switches of the kappa opioid receptor triggered by 6'-GNTI and 5'-GNTI. Sci Rep 2016; 6: 18913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, Manglik A, Venkatakrishnan AJ, Laeremans T, Feinberg EN, Sanborn AL, et al. Structural insights into μ-opioid receptor activation. Nature 2015; 524: 315–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neilan CL, Husbands SM, Breeden S, Ko MC, Aceto MD, Lewis JW, et al. Characterization of the complex morphinan derivative BU72 as a high efficacy, long-lasting mu-opioid receptor agonist. Eur J Pharmacol 2004; 499: 107–16. [DOI] [PubMed] [Google Scholar]
- Manglik A, Kruse AC, Kobilka TS, Thian FS, Mathiesen JM, Sunahara RK, et al. Crystal structure of the μ-opioid receptor bound to a morphinan antagonist. Nature 2012; 485: 321–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spassov VZ, Flook PK, Yan L. LOOPER: a molecular mechanics-based algorithm for protein loop prediction. Protein Eng Des Sel 2008; 21: 91–100. [DOI] [PubMed] [Google Scholar]
- Friesner RA, Banks JL, Murphy RB, Halgren TA, Klicic JJ, Mainz DT, et al. Glide: a new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J Med Chem 2004; 47: 1739–49. [DOI] [PubMed] [Google Scholar]
- Halgren TA, Murphy RB, Friesner RA, Beard HS, Frye LL, Pollard WT, et al. Glide: a new approach for rapid, accurate docking and scoring. 2. Enrichment factors in database screening. J Med Chem 2004; 47: 1750–9. [DOI] [PubMed] [Google Scholar]
- Lomize AL, Pogozheva ID, Mosberg HI. Anisotropic solvent model of the lipid bilayer. 2. Energetics of insertion of small molecules, peptides, and proteins in membranes. J Chem Inf Model 2011; 51: 930–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomize MA, Pogozheva ID, Joo H, Mosberg HI, Lomize AL. OPM database and PPM web server: resources for positioning of proteins in membranes. Nucleic Acids Res 2012; 40: D370–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klauda JB, Venable RM, Freites JA, O'Connor JW, Tobias DJ, Mondragon-Ramirez C, et al. Update of the CHARMM all-atom additive force field for lipids: validation on six lipid types. J Phys Chem B 2010; 114: 7830–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J Mol Graph 1996; 14: 33–8. [DOI] [PubMed] [Google Scholar]
- Vanommeslaeghe K, Hatcher E, Acharya C, Kundu S, Zhong S, Shim J, et al. CHARMM general force field: A force field for drug-like molecules compatible with the CHARMM all-atom additive biological force fields. J Comput Chem 2010; 31: 671–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanommeslaeghe K. MacKerell ADJr. Automation of the CHARMM General Force Field (CGenFF) I: bond perception and atom typing. J Chem Inf Model 2012; 52: 3144–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanommeslaeghe K, Raman EP. MacKerell ADJr. Automation of the CHARMM General Force Field (CGenFF) II: assignment of bonded parameters and partial atomic charges. J Chem Inf Model 2012; 52: 3155–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess B, Kutzner C, van der Spoel D, Lindahl E. GROMACS 4: algorithms for highly efficient, load-balanced, and scalable molecular simulation. J Chem Theory Comput 2008; 4: 435–47. [DOI] [PubMed] [Google Scholar]
- Chavkin C, Goldstein A. Specific receptor for the opioid peptide dynorphin: structure--activity relationships. Proc Natl Acad Sci U S A 1981; 78: 6543–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filizola M, Devi LA. Grand opening of structure-guided design for novel opioids. Trends Pharmacol Sci 2013; 34: 6–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasternak GW, Pan YX. Mu opioids and their receptors: evolution of a concept. Pharmacol Rev 2013; 65: 1257–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granier S, Manglik A, Kruse AC, Kobilka TS, Thian FS, Weis WI, et al. Structure of the delta-opioid receptor bound to naltrindole. Nature 2012; 485: 400–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritze O, Filipek S, Kuksa V, Palczewski K, Hofmann KP, Ernst OP. Role of the conserved NPxxY(x)5,6F motif in the rhodopsin ground state and during activation. Proc Natl Acad Sci U S A 2003; 100: 2290–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trzaskowski B, Latek D, Yuan S, Ghoshdastider U, Debinski A, Filipek S. Action of molecular switches in GPCRs--theoretical and experimental studies. Curr Med Chem 2012; 19: 1090–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenkamp RE, Teller DC, Palczewski K. Rhodopsin: a structural primer for G-protein coupled receptors. Arch Pharm (Weinheim) 2005; 338: 209–16. [DOI] [PubMed] [Google Scholar]
- Liu R, Nahon D, le Roy B, Lenselink EB, AP IJ. Scanning mutagenesis in a yeast system delineates the role of the NPxxY(x)(5,6)F motif and helix 8 of the adenosine A(2B) receptor in G protein coupling. Biochem Pharmacol 2015; 95: 290–300. [DOI] [PubMed] [Google Scholar]
- Sun X, Agren H, Tu Y. Functional water molecules in rhodopsin activation. J Phys Chem B 2014; 118: 10863–73. [DOI] [PubMed] [Google Scholar]
- Manglik A, Lin H, Aryal DK, McCorvy JD, Dengler D, Corder G, et al. Structure-based discovery of opioid analgesics with reduced side effects. Nature 2016; 537: 185–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim J, Coop A. MacKerell ADJr. Molecular details of the activation of the mu opioid receptor. J Phys Chem B 2013; 117: 7907–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filizola M, Devi LA. Structural biology: how opioid drugs bind to receptors. Nature 2012; 485: 314–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Wacker D, Mileni M, Katritch V, Han GW, Vardy E, et al. Structure of the human kappa-opioid receptor in complex with JDTic. Nature 2012; 485: 327–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table and Figures