Abstract
Aim:
Present hospital based study was carried out at our tertiary care centre with an aim to study the distribution of Cryptosporidium species subtypes in patients with complaints of diarrhea.
Background:
Cryptosporidium species are one of the important causative agents of parasitic diarrhea, amongst which Cryptosporidium hominis (C.hominis) and Cryptosporidium parvum (C.parvum) are the two major species that are associated with human cryptosporidiosis.
Methods:
Four hundred and fifty (n=450) diarrheic patients complaining of different types of diarrhea were enrolled in the present study. Both microscopic and molecular diagnostic methods were used for the detection as well as for identification of Cryptosporidium species and its speciation and subtyping.
Results:
Forty one (n=41) and forty three (n=43) patients were positive for Cryptosporidium species by microscopy and Polymerase chain reaction (PCR) assay respectively. Of these 43 cases, 70% (30/43) were identified as C. hominis and 21% (9/43) was as C. parvum, 7% (3/43) was as Cryptosporidium felis (C.felis) and 2% (1/43) as Cryptopsoridium viatorum (C. viatorum) respectively . Upon subtyping of C. hominis and C. parvum, 16 subtypes belonging to 8 different subtype families could be identified. The frequency of different families were Ia (13%, 5/39), Ib (15%, 6/39), Id (18%, 7/39), Ie (30%, 12/39) and IIa (5%, 2/39), IIc (8%, 3/39), IId (8%, 3/39) and IIe (3%, 1/39).
Conclusion:
Our study results strongly suggest and reinforces the fact that most of the human cryptosporidiosis is anthroponotic and we expect that present molecular epidemiological data will provide more insight to unravel the changing clinical paradigm of human cryptosporidiosis at large.
Key Words: Cryptosporidium, Genetic variations, Glycoprotein gp60, phylogeny
Introduction
In recent times, Cryptosporidium species are one of the most important causative agents of diarrhea in both immunocompetent and immunocompromised individuals. Humans can acquire infections either by direct contact with infected persons (anthroponotic transmission) or from animals (zoonotic transmission) or by ingestion of contaminated food or water (1,2). Human cryptosporidiosis is primarily caused by Cryptosporidium hominis (C. hominis) and Cryptosporidium parvum (C. parvum); however other species such as C. meleagridis, C. felis and C. canis have also been reported to cause human cryptosporidiosis but with much lower frequency (3).
It has been observed that the distribution of Cryptosporidium spp. in humans varies to some extent with geographic locations and socioeconomic conditions of a particular country. C. hominis is the most common species that infect humans in most of the developing and developed nations (3-6) except Middle Eastern countries, where C. parvum is the most common infecting species (7). Apart from above variables infecting Cryptosporidium spp. does vary with age of the infected individuals (8, 9). C. hominis has been commonly reported in children compared to C. parvum albeit some studies that defy such association (10, 11).
Human cryptosporidiosis in immunocompetent individuals may present with three major clinical presentations such as i) asymptomatic carriage, ii) acute diarrhea and with iii) persistent or chronic diarrhea. The incubation period may vary from three to fourteen days and the illness is usually self-limiting (12). However, Cryptosporidiosis tends to be much more severe in immune-compromised individuals with impaired or defective cell-mediated or humoral immunity or both (13, 14).
In recent years various studies have delineated the causal association of different clinical manifestations of cryptosporidiosis with that of infecting Cryptosporidium species, subtype family or subtypes (11,12, 15-17). The present hospital based cross sectional study was conducted to detect and subtype Cryptosporidium species from different group of patients attending our tertiary care centre with complaints of diarrhea.
Methods
Ethical approval
Ethical approval for the present study was obtained by institutional ethical committee of All India Institute of Medical Sciences, New Delhi, India. All the participants were apprised about the study protocol. Informed consent was obtained from all adult patients as well as and from guardians/ Parents of Children enrolled in the study. During the meetings, enrolled individuals were informed that their participation was voluntarily and they have all the rights to withdraw from the study at any point of time without giving any reason, however withdrawal from the study did not affect the treatment and medical care provided to these patients. Patients’ personal details were kept confidential.
Study groups/Patients
The present study was conducted in the Department of Microbiology. Study population were enrolled from both in-patient and out-patient departments of Gastroenterology and Human Nutrition, Paediatrics, Internal Medicine and Dr. B. R. Ambedkar Institute of Rotary Cancer Hospital (IRCH) of our tertiary care referral health centre. The patients who had received any luminal anti- parasitic treatment in last three weeks were excluded from the study.
Clinical specimens
Three (n=3) consecutive stool samples on three consecutive days were obtained from four hundred and fifty (n=450) patients with diarrhea (cases) and two hundred (n=200) individuals without diarrhea (controls). Diarrhea was defined as the passage of liquid or unformed stool at an increased frequency and was further categorized as ‘acute’ if the diarrhea was for a period of less than two weeks and ‘persistent’ if it was present for two to four weeks. ‘Chronic’ diarrhea was defined when the complaints were for more than four weeks’ duration (http://www.who.int/topics/diarrhoea/en/) (WHO, 2009). All (n=450) cases were comprised of both clinically apparent immunocompetent and immunocompromised patients that included HIV sero-positives, transplant recipients, patients with malignant diseases or other immunodeficiency disorders. Controls enrolled in the study included healthy individuals without having any gastrointestinal complaints. Three consecutive samples obtained from each patient during the study were considered as ‘one’ for the convenience of subsequent analyses.
Clinical manifestations
Information regarding clinical manifestations was recorded by interviewing the patients as well as from caregivers in case of paediatric population. Data included relevant gastrointestinal symptoms such as abdominal pain, fever, general malaise, nausea, vomiting, frequency and duration of diarrhea and presence of blood or mucus in the stools. Duration of diarrhea, vomiting, fever, abdominal pain and weight loss were included in the study for subsequent analysis.
Statistical analysis
Statistical analysis was performed using STATA 12.2 software and wherever applicable p-values were calculated. The discriminatory power of gp60 gene was calculated online using http://insilico.ehu.es/mini_tools/discriminatory_power/.
Microscopic examination
Processing of fecal samples was carried out depending upon the consistency. In case of watery, samples direct wet-mounts (both in normal saline and 3% Lugol’s iodine) and fecal smears were prepared, whereas for the formed stools, additional standard formol-ether concentration technique was used. Wet mounts as well as fecal smears were prepared from the sediments for examination (18). Fecal smears were subjected to Modified Ziehl-Neelsen (mZN) staining (19).
Cryptosporidium spp. detection, identification and subtyping by molecular methods
Extraction of genomic DNA
Immediately after collection, all the clinical samples (n=650; ie. 450 cases and 200 controls) were subjected to DNA extraction using commercially available QIaAmp stool minikit (Qiagen, Germany) as per the manufacturer’s instructions, with some modifications. The modifications included (i) mechanical disruption of oocysts using glass beads, (ii) incubation of clinical sample at 95ºC for 60 minutes after addition of lysis buffer, (iii) incubation at 70ºC for 30 minutes after addition of Proteinase K and (iv) DNA elution was done in 50µl of elution buffer instead of 200µl recommended in the protocol. The extracted DNA was stored at -80°C till further analysis.
SSU-rRNA PCR-RFLP assay
PCR assay targeting small subunit ribosomal RNA (SSU rRNA) gene of Cryptosporidium spp. was performed a using nested protocol. The external round of PCR assay was performed using SSU-F1 (5’- TCTAGAGCTAATACATGCG-3’) as the forward primer and SSU-R1 (5’-CCCATTTCCTTCGAAACAGGA-3’) as the reverse primer, whereas, SSU-F2 (5’-GGAAGGGTTGTATTTATTAGATAAAG-3’) and SSU-R2 (5’-CTCATAAGG TGCTGAAGGAGTA-3’) were used as the forward and reverse primer for the nested round respectively (20). The reaction mixtures for both external and internal rounds of PCR assay contained 2.5µl of 10X buffer, 3µl of 25mM MgCl2, 2µl of 10mM dNTPs, 0.5µl of 20pm of each primers and 0.8µl of 3 units of TaqDNA polymerase.PCR assays were performed by an initial denaturation at 95ºC for 10 minutes followed by 35 cycles of denaturation at 94ºC for 10 seconds. Annealing was carried out at 61ºC (57ºC for internal round) for 10 seconds, followed by extension at 72ºC for 15 sec and final extension at 72ºC for 10 minutes. The assays were completed with a final hold step at 4ºC in C1000TM Bio-Rad thermocycler (Bio-Rad, USA). The intensity and size of all amplicons were assessed by electrophoresis in 1.5% ethidium bromide stained agarose gels using Tris Borate EDTA (65 mMTris-HCl, 27 mM boric acid, 1 mM Ethylene diamide tetra acetate, pH 9; Bio-Rad, USA) as the buffer and 100bp molecular marker (Fermentas).
PCR-RFLP assay was then performed using SspI and AseI (New England Biolabs, USA) restriction enzymes for genotyping of Cryptosporidium spp. Six microlitre (6µl) of PCR product was digested with 6 units of each enzyme in separate reactions at 37ºC for a period of 4 hours.
Subtyping using gp60 gene
DNA samples from all PCR-confirmed C. hominis and C. parvum cases were subjected to further amplification of a 850-bp fragment of the Cryptosporidium specific gp60 gene using F1 5’-ATAGTCTCCGCTGTATTC-3’and R1 5’-GGAAGGAACGATGTATCT-3’ as external primers and F2 5’-TCCGCTGTATTCTCAGCC-3’ and R2 5’ GCAGAGGAACCAGCATC-3’ as internal primers (21). Annealing was carried out at 50ºC for 60 seconds.
Sequencing
The positive amplified product obtained using both SSU-rRNA and gp60 gene PCR assay was excised from agarose gel and purified using QIAquick gel extraction kit (Qiagen, Valencia, USA) according to the manufacturer’s instructions. The PCR fragments after purification were further subjected to sequencing using an automated DNA sequencer (ABI Prism 310) using BigDye Terminator Chemistry. DNA chromatograms were examined using BioEdit software versions 7.1.3. Both forward and reverse sequences were pair-wise aligned along with the reference sequences using Clustal W software and were manually refined to obtain a better consensus sequence.
Phylogenetic analysis
To further support the results of subtyping, two neighbour-joining trees were built using Kimura two-parameter model in MEGA 6, each for C. hominis and C. parvum. The reliability of all phylogenetic groupings was determined through a bootstrap resampling analysis (1,000 replicates). Reference sequences of both C. hominis and C. parvum subtypes identified from Indian studies form Kolkata (22) and Chandigarh (23) were included from the NCBI GenBank database as controls for the alignment.
Nucleotide sequence Accession numbers
Representative sequences of the Cryptosporidium spp. at SSU-rRNA and gp60 genes in this survey were submitted to the GenBank under accession numbers KX056082-KX056097, KX174306-KX174309 and KU169226 -KU169235.
Results
Study group
Of the total four hundred and fifty (n=450) cases enrolled, three hundred and seven (n=307) were adults and one hundred and forty three (n=143) were children with a mean age of 35.75±13.4 years and 5.28±3.44 years, respectively. A total of two hundred age-matched healthy individuals comprising of 100 adults (males 55, females 45) and 100 children (males 57, females 43) without any gastro-intestinal disorders were enrolled as controls. The mean age of the adults and children was 29.7±12.5 and 5.7±3.8 years, respectively. Distribution of these patients along with their underlying condition has been shown in Table 1.
Table 1.
Distribution of Cryptosporidium spp. in adults and children according to underlying disease conditions
| Underlying conditions | Total Number (n=307) Adults |
Cryptosporidium Positive (n=26) (%) | Total number (n=143)Children |
Cryptosporidium Positive (n=17) (%) |
|---|---|---|---|---|
| HIV seropositives | 53 | 9 (17%) | 15 | 6 (40%) |
| Malignant Diseases | 9 | 1 (11%) | 32 | 2 (6%) |
| Transplant Recipients | 57 | 16 (28%) | 5 | 2 (40%) |
| Primary immunodeficiency diseases | 10 | - | 9 | 2 (22%) |
| Secondary immunodeficiency diseases | 10 | - | 8 | 1 (13%) |
| Chronic disease | 14 | - | 2 | 1 (50%) |
| No apparent immunocompromised condition | 154 | - | 72 | 3 (42%) |
Cryptosporidium spp. detection, identification and subtyping
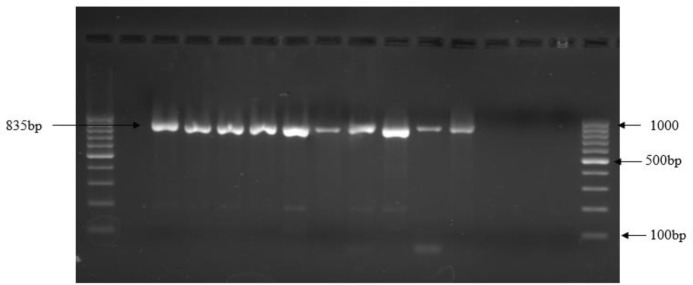
Oocysts of Cryptosporidium spp. measuring 4-6 µm by micrometry were detected upon microscopic examinations of modified acid-fast stained smears of stool samples. Only 41 cases were positive for oocysts of Cryptosporidium spp. by microscopy. Using SSU-rRNA PCR assay, two additional cases of cryptosporidiosis were detected making it to a total 43 cases of human cryptosporidiosis (Fig. 1; Table 2).
Figure 1.
PCR assay for small subunit ribosomal (SSU rRNA) RNA gene of Cryptosporidium species
Table 2.
Distribution of subtypes within the subtype families
| Subtype family | No. of Subtypes | Distribution of Subtypes |
|---|---|---|
| Ia | 5 | IaA17R2 (1) IaA20R2 (2) IaA23G1R1 (1) IaA29G2R2 (1) |
| Ib | 6 | IbA9G3 (5) IbA10G2 (1) |
| Id | 7 | IdA15G1 (5) IdA14 (2) |
| Ie | 12 | IeA11G3T3 (5) IeA11G3T1 (2) IeA13G3T3 (2) IeA13G3T1 (3) |
| IIa | 2 | IIaA15G3 (2) |
| IIc | 3 | IIc5G3 (3) |
| IId | 3 | IIdA15G1 (3) |
| IIe | 1 | IIeA7G1 (1) |
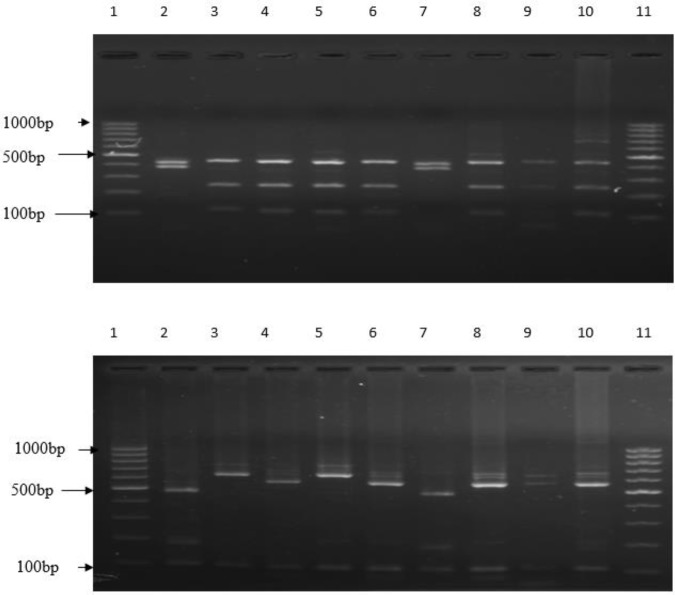
Of these 43 positive samples, 70% (n=30; 30/43) were C. hominis and 21% (n=9; 9/43) were C. parvum. Only 7% (n=3; 3/43) were C. felis and 2% (n=1; 1/43) was C. viatorum (Fig. 2a and 2b). None of the stool samples from healthy controls were positive for Cryptosporidium spp. either by microscopy and/or PCR assay.
Figure 2(a and b).
PCR-RFLP analysis using SspI and AseI restriction enzyme for differentiating Cryptosporidium species
Sequence analysis of gp60 gene could be done for 30 C. hominis and 9 C. parvum and that could detect eight subtype families. These subtype families were Ia (13%, 5/39), Ib (15%, 6/39), Id (18%, 7/39), Ie (30%, 12/39), IIa (5%, 2/39), IIc (8%, 3/39), IId (8%, 3/39) and IIe (3%, 1/39). Amongst various subtypes, the frequency of occurrence of subtypes IbA9G3, IdA15G1 and IeA11G3T3 was relatively higher compared to others (Table-2).
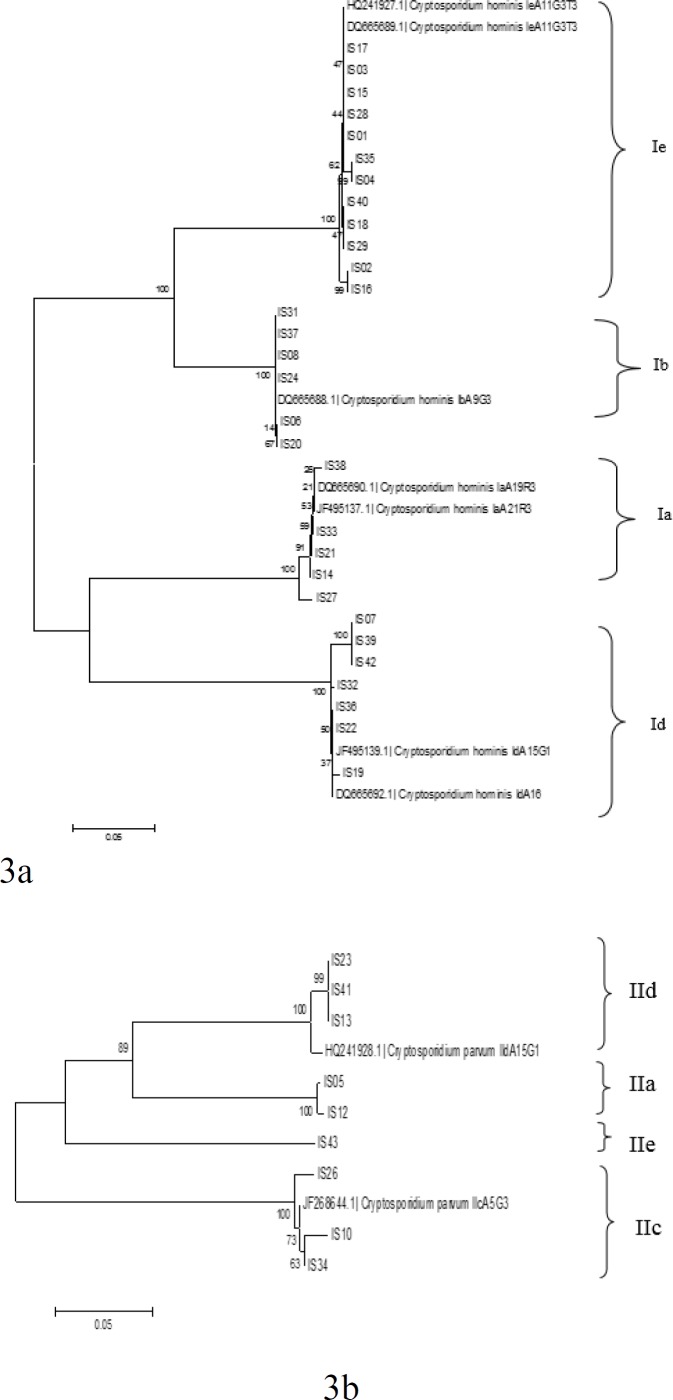
C. hominis subtypes identified in the present study formed two different clades upon phylogenetic analysis (Fig. 3). C. hominis subtype family Ie and Ib were grouped together in one clade whereas subtype family Ia and Id were in another. Separation of these two-subtype family (Ie and Ib and Ia and Id) in two different clades was statistically reliable (bootstrap values 100%). Similarly, C. parvum also formed two clades with IIc subtypes in one and IIa, IId and IIe clustered in another. The discriminatory power (D) for gp60 subtyping was 0.83 in the present study.
Figure 3(a and b).
Phylogenetic relationship of C. hominis and C. parvum identified in the present study. Phylogenetic analysis inferred by neighbor-joining analysis of the gp60gene sequence based on evolutionary distances calculated using the Kimura two-parameter model. Bootstrap values were obtained using 1,000 pseudo replicates
Clinical manifestations
When the clinical manifestations of infected individuals were correlated with different infecting subtypes, it was observed that all patients infected with subtype family Ia, Id and IIc had chronic diarrhea, whereas 83% (5/6) patients infected with Ib subtype family subtype family complained of vomiting and appetite loss. Patients infected with Ie subtype family showed higher frequency of passage of stools in a day. Specific clinical manifestation could not be ascertained in patients infected with other subtypes Clinical manifestations between the subtypes and their statistical association has been shown in Table 3.
Table 3.
Clinical presentation in patients infected with different subtypes of Cryptosporidium hominis and Cryptosporidium parvum
| Clinical manifestations | Ia (n=5) | Ib (n=6) | Id (n=7) | Ie (n=12) | IIa (n=2) | IIc (n=3) | IId (n=3) | IIe (n=1) | p-values |
|---|---|---|---|---|---|---|---|---|---|
| Chronic diarrhea | 5 (100)* | 3 (50) | 7 (100) | 7 (58) | 1 (50) | 3 (100) | - | - | 0.09 |
| Frequency more than 5 | 2 (40) | 2 (33) | 2 (29) | 8 (66) | 1 (50) | 2 (67) | - | 1 (100) | 0.5 |
| Blood in stools | 1 (20) | 1 (17) | - | - | - | - | - | - | 0.88 |
| Mucus in stools | 3 (60) | 1 (17) | 2 (29) | 3 (25) | - | - | 2 (67) | - | 0.36 |
| Abdominal pain | 2 (40) | 1 (17) | 3 (43) | 6 (50) | - | 2 (67) | - | 1 (100) | 0.56 |
| Bloating | 2 (40) | 2 (33) | 4 (57) | 4 (33) | - | 2 (67) | - | 1 (100) | 0.66 |
| Fever | 2 (40) | 3 (50) | 4 (57) | 5 (42) | - | 2 (67) | 1 (33) | 1 (100) | 0.88 |
| Vomiting | 1 (20) | 5 (83) | 2 (29) | 8 (66) | - | 1 (33) | 1 (33) | 1 (100) | 0.14 |
| Appetite loss | 2 (40) | 5 (83) | 2 (29) | 2 (17) | - | 2 (67) | 1 (33) | 1 (100) | 0.11 |
| Weight loss | 4 (80) | 1 (17) | 3 (43) | 5 (42) | - | 1 (33) | 1 (100) | 0.31 | |
| Ulcers | - | 1 (17) | 2 (29) | 1 (8.3) | - | 2 (67) | - | 1 (100) | 0.09 |
No. (%)
Discussion
The findings obtained in the present study showed that patients attending our tertiary care centre were infected with diverse population of Cryptosporidium species including C. felis and C. viatorum. C. hominis was the most common species (70%) responsible for cryptosporidiosis in our study followed by C. parvum. Similar reports of higher prevalence of C. hominis compared to other species have been reported in other developing countries including Brazil (24), Peru (10, 11), Kenya (25), Vietnam (26), Iran (27) and Haiti (28) and India (22, 29,30).
Differences in geographical distribution of C. hominis and C. parvum are generally considered as a reflection of differences in both sources of infection and routes of transmission (5). Transmission of C. hominis is essentially anthroponotic and that of C. parvum excluding the subtype families IIc and IIe are primarily zoonotic. The predominance of C. hominis in most of the developing countries suggests that anthroponotic transmission is more common and important than zoonotic transmission in the epidemiology of human cryptosporidiosis (5). Certain subtype such as “If” could not be identified in the present study although such subtype has been reported from one of the study from India (29). The distribution of C. parvum subtypes in this study suggests the likely occurrence of zoonotic transmission at a much lower frequency. C. parvum subtype family IId was identified in only three (n=3) cases. Subtype family IId of C. parvum is more commonly found in sheep and goats (31), but has also been reported from amongst calves in China, Egypt, and Sweden (32-34).
The existence of many subtypes within C. hominis and C. parvum subgenotype families in the present study reinforces the complexity of Cryptosporidium spp. transmission, and its association with different clinical presentation (5). Many subtype alleles were observed in the phylogenetic analysis for C. hominis subtype families. The high heterogeneity of C. hominis shown in the phylogenetic tree is thought to express intensive and stable anthroponotic transmission of cryptosporidiosis in the present study. Similarly, recent study from Nigeria showed high heterogeneity of C. hominis compared to C. parvum (35). The discriminatory power for gp60 subtyping is high in the study setting (D = 0.83) making it a suitable tool for the detection of outbreaks in human-to-human transmission settings (8). Amongst C. parvum species, only one subtype allele was observed from all four subtypes identified in the present study. From earlier studies conducted in India one subtype allele of IIc (IIcA5G3) and IIe(IIeA7G1) as well as two alleles of IId have been reported (16,23).
Variations in the clinical presentations were observed among C. hominis and C. parvum subtype families (Table -3). Various studies have shown the association of subtype family with diarrhea (10,11), abdominal pain (34), fever and dehydration (36). In the present study, we could observe association of Ia and Id subtype with chronic diarrhea. Cama et al., had reported the association of Ia with diarrhea and Ib with vomiting, however the results were not statistically significant. Infection with Ib subtype family has been reported to be much more virulent than other subtype families (10,11). Recent study from Ethiopia showed significant association of C. parvum subtype family IIa with diarrhea (37)
Present study reported four Cryptosporidium species and 16 subtypes belonging to 8 subtype families of C. hominis and C. parvum in Indian patients. The study results provided updated molecular evidence on the diversity and frequency of the C. hominis and C. parvum subtypes currently circulating in symptomatic individuals seeking medical care at our tertiary care centre. The results may have further implications in better understanding of the changing epidemiology and transmission dynamics of cryptosporidiosis.
Acknowledgment
Authors would like to thank Indian Council of Medical Research (ICMR), Government of India, for providing financial support to carry out this work. First author is thankful to Council of Scientific and Industrial Research (CSIR), Government of India for providing the fellowship grant during the study period.
Conflict of interests
The authors declare that they have no conflict of interest.
References
- 1.Fayer R. Cryptosporidium: a water-borne zoonotic parasite. Vet Parasitol. 2004;126:37–56. doi: 10.1016/j.vetpar.2004.09.004. [DOI] [PubMed] [Google Scholar]
- 2.Xiao L, Ryan UM. Cryptosporidiosis: an update in molecular epidemiology. Curr Opin Infect Dis. 2004;17:483–90. doi: 10.1097/00001432-200410000-00014. [DOI] [PubMed] [Google Scholar]
- 3.Xiao L, Feng Y. Zoonotic cryptosporidiosis. FEMS Immunol Med Microbiol. 2008;52:309–23. doi: 10.1111/j.1574-695X.2008.00377.x. [DOI] [PubMed] [Google Scholar]
- 4.Xiao L. Molecular epidemiology of cryptosporidiosis: an update. Exp Parasitol. 2010;124:80–9. doi: 10.1016/j.exppara.2009.03.018. [DOI] [PubMed] [Google Scholar]
- 5.Nazemalhosseini-Mojarad E, Feng Y, Xiao L. The importance of subtype analysis of Cryptosporidium spp in epidemiological investigations of human cryptosporidiosis in Iran and other Mideast countries. Gastroenterol Hepatol Bed Bench. 2012;5:67–70. [PMC free article] [PubMed] [Google Scholar]
- 6.Sulaiman IM, Hira PR, Zhou L, Al-Ali FM, Al-Shelahi FA, Shweiki HM, et al. Unique endemicity of cryptosporidiosis in children in Kuwait. J Clin Microbiol. 2005;43:2805–09. doi: 10.1128/JCM.43.6.2805-2809.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ryan U, Fayer R, Xiao L. Cryptosporidium species in humans and animals: current understanding and research needs. Parasitol. 2014;141:1667–85. doi: 10.1017/S0031182014001085. [DOI] [PubMed] [Google Scholar]
- 8.Wielinga PR, De Vries A, Van der Goot TH, Mank T, Mars MH, Kortbeek LM, et al. Molecular epidemiology of Cryptosporidium in humans and cattle in The Netherlands. Int J Parasitol. 2008;38:809–17. doi: 10.1016/j.ijpara.2007.10.014. [DOI] [PubMed] [Google Scholar]
- 9.Chalmers RM, Hadfield SJ, Jackson CJ, Elwin K, Xiao L, Hunter P. Geographic linkage and variation in Cryptosporidium hominis. Emerg Infect Dis. 2008;14:496–98. doi: 10.3201/eid1403.071320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cama VA, Ross JM, Crawford S, Kawai V, Chavez-Valdez R, Vargas D, et al. Differences in clinical manifestations among Cryptosporidium species and subtypes in HIV-infected persons. J Infect Dis. 2007;196:684–91. doi: 10.1086/519842. [DOI] [PubMed] [Google Scholar]
- 11.Cama VA, Bern C, Roberts J, Cabrera L, Sterling CR, Ortega Y, et al. Cryptosporidium species and subtypes and clinical manifestations in children, Peru. Emerg Infect Dis. 2008;14:1567–74. doi: 10.3201/eid1410.071273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cama VA, Ross JM, Crawford S, Kawai V, Chavez-Valdez R, Vargas D, et al. Differences in clinical manifestations among Cryptosporidium species and subtypes in HIV-infected persons. J Infect Dis. 2007;196:684–91. doi: 10.1086/519842. [DOI] [PubMed] [Google Scholar]
- 13.Navin TR, Juranek DD. Cryptosporidiosis: clinical, epidemiologic, and parasitologic review. Rev Infect Dis. 1984;6:313–27. doi: 10.1093/clinids/6.3.313. [DOI] [PubMed] [Google Scholar]
- 14.Fayer R, Ungar BL. Cryptosporidium spp. and cryptosporidiosis. Microbiol Rev . 1986;50:458–83. doi: 10.1128/mr.50.4.458-483.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Matos O, Alves M, Xiao L, Cama V, Antunes F. Cryptosporidium felis and C. meleagridis in persons with HIV, Portugal. Emerg Infect Dis. 2004;10:2256–57. doi: 10.3201/eid1012.031068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ajjampur SS, Liakath FB, Kannan A, Rajendran P, Sarkar R, Moses PD, et al. Multisite study of cryptosporidiosis in children with diarrhea in India. J Clin Microbiol. 2010;48:2075–81. doi: 10.1128/JCM.02509-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chalmers RM, Elwin K, Thomas AL, Joynson DH. Infection with unusual types of Cryptosporidium is not restricted to immunocompromised patients. J Infect Dis. 2002;185:270–71. doi: 10.1086/338196. [DOI] [PubMed] [Google Scholar]
- 18.Casemore DP, Armstrong M, Sands RL. Laboratory diagnosis of cryptosporidiosis. J Clin Pathol. 1985;38:1337–41. doi: 10.1136/jcp.38.12.1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garcia LS, Shum AC, Bruckner DA. Evaluation of a new monoclonal antibody combination reagent for direct fluorescence detection of Giardia cysts and Cryptosporidium oocysts in human fecal specimens. J Clin Microbiol. 1992;30:3255–57. doi: 10.1128/jcm.30.12.3255-3257.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xiao L, Escalante L, Yang C, Sulaiman I, Escalante AA, Montali RJ, et al. Phylogenetic analysis of Cryptosporidium parasites based on the small-subunit rRNA gene locus. Appl Environ Microbiol. 1999;65:1578–83. doi: 10.1128/aem.65.4.1578-1583.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alves M, Xiao L, Sulaiman I, Lal AA, Matos O, Antunes F. Subgenotype analysis of Cryptosporidium isolates from humans, cattle, and zoo ruminants in Portugal. J Clin Microbiol. 2003;41:2744–47. doi: 10.1128/JCM.41.6.2744-2747.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gatei W, Das P, Dutta P, Sen A, Cama V, Lal AA, et al. Multilocus sequence typing and genetic structure of Cryptosporidium hominis from children in Kolkata, India. Infect Genet Evol. 2007;7:197–05. doi: 10.1016/j.meegid.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 23.Sharma P, Sharma A, Sehgal R, Malla N, Khurana S. Genetic diversity of Cryptosporidium isolates from patients in North India. Int J Infect Dis. 2013;17:e601–05. doi: 10.1016/j.ijid.2012.12.003. [DOI] [PubMed] [Google Scholar]
- 24.Bushen OY, Kohli A, Pinkerton RC, Dupnik K, Newman RD, Sears CL, et al. Heavy cryptosporidial infections in children in northeast Brazil: comparison of Cryptosporidium hominis and Cryptosporidium parvum. Trans R Soc Trop Med Hyg. 2007;101:378–84. doi: 10.1016/j.trstmh.2006.06.005. [DOI] [PubMed] [Google Scholar]
- 25.Nyamwange CI, Mkoji G, Mpoke S, Nyandieka HS. Cryptosporidiosis and its genotypes among children attending Moi teaching and referral hospital in Eldoret, Kenya. East Afr Med J. 2012;89:11–9. [PubMed] [Google Scholar]
- 26.Gatei W, Greensill J, Ashford RW, Cuevas LE, Parry CM, Cunliffe NA, et al. Molecular analysis of the 18S rRNA gene of Cryptosporidium parasites from patients with or without human immunodeficiency virus infections living in Kenya, Malawi, Brazil, the United Kingdom, and Vietnam. J Clin Microbiol. 2003;41:1458–62. doi: 10.1128/JCM.41.4.1458-1462.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zavvar M, Sadraei J, Emadi H, Pirestani M. The use of a nested PCR-RFLP technique, based on the parasite's 18S ribosomal RNA, to characterize Cryptosporidium isolates from HIV/AIDS patients. Ann Trop Med Parasitol. 2008;102:597–01. doi: 10.1179/136485908X311876. [DOI] [PubMed] [Google Scholar]
- 28.Raccurt CP, Brasseur P, Verdier RI, Li X, Eyma E, Stockman CP, et al. Human cryptosporidiosis and Cryptosporidium spp in Haiti. Trop Med Int Health. 2006;11:929–34. doi: 10.1111/j.1365-3156.2006.01631.x. [DOI] [PubMed] [Google Scholar]
- 29.Muthusamy D, Rao SS, Ramani S, Monica B, Banerjee I, Abraham OC, et al. Multilocus genotyping of Cryptosporidium sp isolates from human immunodeficiency virus-infected individuals in South India. J Clin Microbiol. 2006;44:632–34. doi: 10.1128/JCM.44.2.632-634.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rao Ajjampur SS, Asirvatham JR, Muthusamy D, Gladstone BP, Abraham OC, Mathai D, et al. Clinical features & risk factors associated with cryptosporidiosis in HIV infected adults in India. Indian J Med Res. 2007;126:553–57. [PMC free article] [PubMed] [Google Scholar]
- 31.Quílez J, Vergara-Castiblanco C, Monteagudo L, del Cacho E, Sánchez-Acedo C. Host association of Cryptosporidium parvum populations infecting domestic ruminants in Spain. Appl Environ Microbiol. 2013;79:5363–71. doi: 10.1128/AEM.01168-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Amer S, Zidan S, Feng Y, Adamu H, Li N, Xiao L. Identity and public health potential of Cryptosporidium spp in water buffalo calves in Egypt. Vet Parasitol. 2013;191:123–27. doi: 10.1016/j.vetpar.2012.08.015. [DOI] [PubMed] [Google Scholar]
- 33.Silverlås C, Bosaeus-Reineck H, Näslund K, Björkman C. Is there a need for improved Cryptosporidium diagnostics in Swedish calves? Int J Parasitol. 2013;43:155–61. doi: 10.1016/j.ijpara.2012.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ng JS, Pingault N, Gibbs R, Koehler A, Ryan U. Molecular characterisation of Cryptosporidium outbreaks in Western and South Australia. Exp Parasitol. 2010;125:325–28. doi: 10.1016/j.exppara.2010.02.012. [DOI] [PubMed] [Google Scholar]
- 35.Ukwah BN, Ezeonu IM, Ezeonu CT, Roellig D, Xiao L. Cryptosporidium species and subtypes in diarrheal children and HIV-infected persons in Ebonyi and Nsukka, Nigeria. J Infect Dev Ctries. 2017;11:173–79. doi: 10.3855/jidc.8034. [DOI] [PubMed] [Google Scholar]
- 36.Iqbal J, Khalid N, Hira PR. Cryptosporidiosis in Kuwaiti children: association of clinical characteristics with Cryptosporidium species and subtypes. J Med Microbiol. 2011;60:647–52. doi: 10.1099/jmm.0.028001-0. [DOI] [PubMed] [Google Scholar]
- 37.Adamu H, Petros B, Zhang G, Kassa H, Amer S, Ye J, et al. Distribution and clinical manifestations of Cryptosporidium species and subtypes in HIV/AIDS patients in Ethiopia. PLoS Negl Trop Dis. 2014;8:e2831. doi: 10.1371/journal.pntd.0002831. [DOI] [PMC free article] [PubMed] [Google Scholar]