Abstract
Deciphering the functions of long non-coding RNAs (lncRNAs) is facilitated by visualization of their subcellular localization using in situ hybridization (ISH) techniques. We evaluated four different ISH methods for detection of MALAT1 and CYTOR in cultured cells: a multiple probe detection approach with or without enzymatic signal amplification, a branched-DNA (bDNA) probe and an LNA-modified probe with enzymatic signal amplification. All four methods adequately stained MALAT1 in the nucleus in all of three cell lines investigated, HeLa, NHDF and T47D, and three of the methods detected the less expressed CYTOR. The sensitivity of the four ISH methods was evaluated by image analysis. In all three cell lines, the two methods involving enzymatic amplification gave the most intense MALAT1 signal, but the signal-to-background ratios were not different. CYTOR was best detected using the bDNA method. All four ISH methods showed significantly reduced MALAT1 signal in knock-out cells, and siRNA-induced knock-down of CYTOR resulted in significantly reduced CYTOR ISH signal, indicating good specificity of the probe designs and detection systems. Our data suggest that the ISH methods allow detection of both abundant and less abundantly expressed lncRNAs, although the latter required the use of the most specific and sensitive probe detection system.
INTRODUCTION
In the last decades, non-coding RNAs (ncRNAs) including small ncRNAs like microRNA (miRNA), long ncRNAs (lncRNA) (1), and more recently circular RNAs (circRNA) (2) have attracted much attention and added yet a layer of complexity to the regulatory machinery involved in gene expression. The number of lncRNAs (3) is much higher than both the number of protein coding mRNAs and miRNAs (4–8). The expression of lncRNA is tightly regulated by key developmental, metabolic and/or external stimuli, suggesting important functional roles (1,9). However, despite their poor conservation among species compared to mRNAs (10–12) and their disputed coding potential (13,14), roles of lncRNAs in biological and pathological processes are continuously being identified, revealing their involvement in transcription, signaling and intracellular trafficking (15–18).
Information regarding the expression dynamics of lncRNAs and their subcellular localization are important to help identifying their biological functions. The expression of lncRNAs is often tissue-specific and even cell-type specific (19–21), and therefore the localization of lncRNAs within a complex tissue can provide important insight on their role in physiological and pathological situations. On average, lncRNAs are shorter and contain more repeats than mRNA molecules (21,22), and lncRNAs are generally expressed at lower levels than mRNAs (23), making the lncRNAs challenging to detect by in situ hybridization (ISH) methods.
In recent years, major advances in probe technologies and detection methods have been made to improve ISH methods for RNA visualization. These technologies include fluorophore-labeled multiple oligo probe sets (24,25), LNA probes (26) and branched-DNA (bDNA) probes (27–29).
A multiple probe set contain up to 48 antisense fluorophore-labeled DNA oligonucleotides (oligos in the following) that are designed and selected for unique sequences in the target RNA molecule and individually labeled with fluorophores (24,25). The oligos in a multiple DNA probe set hybridize along the entire RNA molecule, which provides sufficient label density to allow visualization of the RNA molecules (30–36).
The incorporation of modified oligonucleotides, such as locked nucleic acid (LNA) (26) or 2′-O-methyl (2OMe) (37,38), into DNA oligos significantly increase the specificity and binding affinity of oligonucleotide probes to RNA targets. The ISH methods, based on hapten-labeled LNA oligos have been found to be highly advantageous in the detection of miRNA in experimental and clinical tissue samples (39,40), whereas only a few attempts have been reported for detection of mRNAs (41) and lncRNAs (42,43). The DNA-LNA chimeric probes comprise typically 18–22 nt, and can be single or double labeled with haptens, like digoxigenin or carboxyfluorescein (FAM). Subsequent visualization of the probe is performed with enzyme-conjugated antibodies and chromogenic or fluorogenic substrates. The use of a single oligo probe, optimally designed and with minimum cross-binding to other RNAs, reduces the risk of off-target probe hybridization and the use of LNA probes, instead of pure DNA probes, increases the specificity of the hybridization (26,44–47).
A third recently established ISH method is based on bDNA technology. Here, two antisense DNA oligonucleotides, comprising linker sequences and called double-Z probes, are designed to bind adjacent sequences as pairs on the target sequence. Dependent on the length of the RNA target, up to 20 probe pairs may be designed into a single bDNA probe set (27–29). The linker sequences of the primary paired probes form a template for a second DNA oligo that can bind only if the two paired probes have hybridized in tandem on the same RNA molecule. This probe design provides a high level of specificity. The second detecting oligo forms yet another template for additional detection oligos, which together forms branches of DNA. The last step in the procedure is the addition of DNA oligos that can be either fluorophore-labeled or enzyme-conjugated, eventually resulting in 8000- to 96 000-fold signal amplification (27–29,48).
In this study, we have evaluated the performance of different fluorescence based ISH methods for the detection of two different lncRNAs; the highly abundant metastasis associated in lung adenocarcinoma transcript 1 (MALAT1) target and the less abundant CYTOR (previously called long intergenic non-coding RNA 152 or linc00152). For the comparison of fluorescence, we have used fluorophores with similar spectral characteristics including probes labeled directly with Quasar-570 and Cy3 and the use of tyramide signal amplification (TSA) systems with Cy3-labeled TSA substrates for peroxidase. The TSA technology has in itself strongly contributed to the increase of sensitivity of ISH methods (49,50).
In this study we have compared four different ISH methods: the multiple DNA probe detection approach using fluorophore labeled probe sets, a modified version of the multiple DNA probe detection approach, to the best of our knowledge for the first time demonstrating the ability to combine this with enzymatic amplification using the TSA system, a bDNA probe detection format and a single oligo probe detection system using LNA-modified oligos with enzymatic amplification.
MATERIALS AND METHODS
Cell culture and treatment
HeLa, NHDF dermal fibroblast and MCF7 breast adenocarcinoma cell lines were purchased from ATCC. Cells were grown in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% fetal bovine serum (FBS), 0.2 mM Ultraglutamine and 1% Penicillin/Streptomycin. T47D breast cancer cells were a kind gift from Dr Agla Fridriksdóttir, Institute for Cellular and Molecular Medicine, University of Copenhagen, and were grown in RPMI-1640 supplemented with 10% FBS, 0.25% Glucose, 2 mM of Ultraglutamine, 10 μg/ml of insulin and 0.2% Penicillin/Streptomycin. A549 lung adenocarcinoma cells and the different MALAT1 clones (51,52) were grown in DMEM containing 10% FBS, 0.2 mM Ultraglutamine and 1% Penicillin/Streptomycin. All cell lines were maintained at 37°C with 5% CO2. For ISH experiments, cells were plated on Nunc™ Lab-Tek™ II Chamber Slide system. Cells were fixed overnight in 10% neutral-buffered formalin and subsequently washed and stored in PBS. Before in situ hybridization analyses, cells were permeabilized by incubation with 70% ethanol for 1h at 4°C.
In situ hybridization
Custom Stellaris® ISH probes were designed against human MALAT1, human CYTOR using the Stellaris RNA ISH Probe Designer (Biosearch Technologies, Inc., Petaluma, CA, USA) at http://www.biosearchtech.com/stellarisdesigner (Stellaris Probe Designer version 4.2). A probe set against mouse TNFα was used as negative control. This probe was chosen due to its low homology (74%) with the human TNFα, and expected low expression of TNFα mRNA in the cultured human cells. The sequences of probes are provided as supplementary information. Cells were hybridized with Stellaris RNA ISH probes against MALAT1, CYTOR or mTNFα labeled either with either Quasar 570 or 6-FAM, following the manufacturer's instructions available online at http://www.biosearchtech.com/ with minor modifications. In brief, cells were incubated with Quasar 570-labeled probes at 25 nM or FAM-labeled probes at 2.5 nM in hybridization buffer (10% formamide, 2× SSC) and hybridized overnight at 42°C. Cells were washed in hybridization buffer for 2 × 30 min at 42°C and briefly in 0.1× SSC. Cells hybridized with fluorescein-labeled probes were first incubated in 3% hydrogen peroxide to block potential endogenous peroxidase, and then the probes were detected with peroxidase-conjugated anti-fluorescein-Ab (Roche applied Sciences, Mannheim, Germany) diluted 1:400 followed by addition of Cy3-labeled TSA substrate for 10 minutes (Perkin Elmer, Waltham, MA, USA). All cells were mounted with ProLong® Gold Antifade Mountant containing DAPI nuclear stain (ThermoFisher Scientific).
In situ hybridization using double-FAM labeled LNA probes (Exiqon, Vedbaek, Denmark) was performed as described previously (53). The fixed and permeabilized cells were pre-hybridized in hybridization buffer (53) and then hybridized at 55°C for 1 h with LNA probes for MALAT1: 5′-ACATTGCCTACCACTCTAAGA-3′ (Predicted RNA Tm = 84°C) and CYTOR: 5′-ATTCGATCAAGTGTGTCATAGA-3′ (Predicted RNA Tm = 82°C) and 5′-ATGTACACACGACTAAGAAGGA-3′ (Predicted RNA Tm = 80°C), and with a scramble probe 5′-TGTAACACGTCTATACGCCCA-3′ (Predicted RNA Tm = 87°C) as negative control, all at 25 nM. The FAM-labeled probes were detected as described above for the FAM-labeled Stellaris probes.
Branched-DNA probe in situ hybridization was performed using the ViewRNA® ISH Cell Assay Kit (Affymetrix, Santa Clara, CA, USA), essentially according to the manufacturer’s instructions. After pre-treatment, the Type 1 bDNA probes against MALAT1 and CYTOR and the negative control probe against bacterial dihydrodipicolinate reductase (dapB) mRNA (all at 10 nM) were hybridized for 3 h at 40°C. The bDNA probes were detected using Alexa® Fluor 546 dyes, according to the manufacturer’s instructions. Cells were mounted with ProLong Gold Antifade Mountant with DAPI (ThermoFisher Scientific).
Detailed information regarding the splice variant of each target and the location and number of probes used in each assay is provided in Table 1. Additionally, a file containing all the experimental information, according to the MISFISHIE guidelines (54) is provided as Supplementary Data.
Table 1. Detailed information about the lncRNA-variants studied and the types and number of probes used to detect both MALAT1 and CYTOR.
| Stellaris | ViewRNA | LNA + TSA | Stellaris + TSA | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Size of transcript (nt) | Region covered (nt) | Number of probes | Region covered (nt) | Number of probes | Region covered (nt) | Number of probes | Region covered (nt) | Number of probes | |
| MALAT1 (NR_002819) | 8687 | (Throughout the lncRNA) | 48 | 5983–7050 | 20 pairs | 6260–6281 | 1 (See sequence) | (Throughout the lncRNA) | 48 |
| CYTOR (NR_024206) | 518 | (Throughout the lncRNA) | 16 | 23–487 | 10 pairs | 166–188 | 1 (See Sequence) | (Throughout the lncRNA) | 16 |
Imaging and image analysis
Images were acquired using a Zeiss Axio Imager Z1 epi-fluorescence microscope equipped with an AxioCam MRm CCD camera and a Plan-APOCHROMAT 63x/1.4 objective (Zeiss, Oberkochen, Germany). For imaging of the CYTOR ISH signals using the ViewRNA probe technology, an optical sectioning system (ApoTome, Zeiss) was used to acquire 11 z-stack images at 0.24 μm intervals. The 11 z-stack images were subsequently displayed using the maximum intensity projection mode of the AxioVision40 version 4.8.2.0 image processing software (Zeiss) to create a single image representing signals from all 11 z-stacks. Within the same experiment, images were acquired at the same exposure conditions.
The ImageJ software (55) was used to both obtain mean pixel intensities for MALAT1 ISH signals and to count numbers of the CYTOR ISH focal spots as follows. For MALAT1 quantification, regions of interest were defined by encircling the nuclei and the cytoplasm of individual cells followed by a measurement of the fluorescence mean pixel intensity of both nuclei and cytoplasm corrected for the slide noise (signal outside the cells). Similarly, the fluorescence signal of the negative control probes was measured in the nuclei and cytoplasm of individual cells. For quantification of the CYTOR ISH signal in nuclear and cytoplasmic regions, all images were identically processed by thresholding, and the number of spots and foci was counted. For both MALAT1 and CYTOR, quantification results were obtained from at least six different images from a minimum of three independent experiments, with more than 70 cells measured per experimental condition.
siRNA transfection
For silencing of CYTOR the following siRNA oligonucleotide was synthesized: 5′- GGAAUGCAGCUGAAAGAUU-3′. MISSION® siRNA Universal Negative Control (Sigma-Aldrich) was used as a negative control. The siRNA’s were transfected at a final concentration of 30 nM by reverse transfection using RNAiMAX (Invitrogen) according to the manufacturer’s instructions.
Total RNA isolation and RT-PCR
Total RNA from all the different cell lines, except for MCF-7, was extracted using the RNeasy mini kit (Qiagen, Hilden, Germany) according to manufacturer’s instructions. From MCF-7 cells, total RNA was isolated using Trizol reagent (Invitrogen) and treated with TURBO DNase (Ambion, Lifetechnologies). RNA reverse transcription was performed using the reverse transcriptase from the TaqMan miRNA reverse transcription kit (Applied Biosystem, Vilnius, Lithuania) with random primers (Thermo Fisher Scientific, Waltham, MA, USA). For MALAT-1, quantitative polymerase chain reactions (qPCRs) were performed using Ampliqon RealQ Plus 2× Master Mix Green (Ampliqon A/S, Odense, Denmark), according to manufacturer’s instructions using a StepOnePlus™ Real-Time PCR System instrument (Applied Biosystems). Previously published primers for MALAT-1 (51) were used, and the housekeeping gene GAPDH was used for normalization.
For CYTOR, qPCRs were performed using SYBR Green PCR Fast PCR Master Mix 2× (Applied Biosystems) using a StepOnePlus Real-Time PCR System instrument (Applied Biosystems) using the following two primers: CYTOR-F: 5′-TCTTCACAGCACAGTTCCTGG-3′ and CYTOR-R: 5′-AGGTAGAGGTGCTGGAGGG-3′. The housekeeping genes GAPDH and HPRT1 were used for normalization of qRT-PCR data. All the qPCR experiments were conducted according to the MIQE guidelines (56).
RESULTS
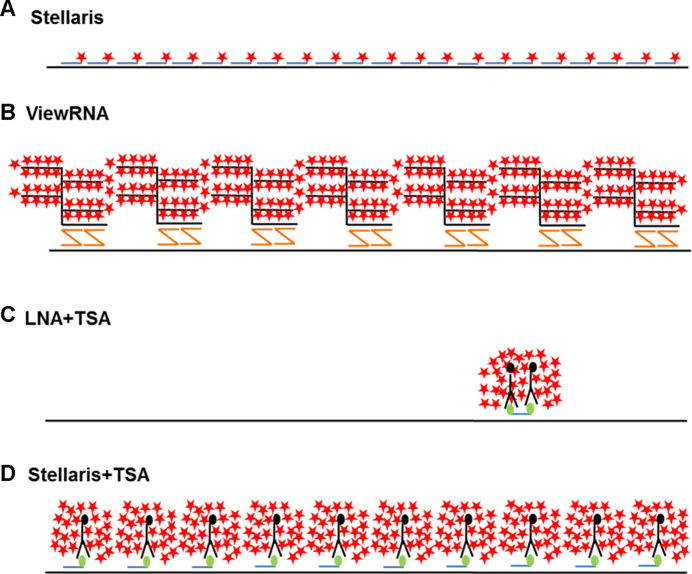
For detection of MALAT1 and CYTOR, we obtained commercially available multiple singly-labeled probes (Stellaris®), which were either Quasar 570-labeled for direct detection or FAM-labeled for subsequent enzyme-enhanced TSA detection. To test the bDNA probe technology we used commercially available custom-made ViewRNA® probe, and for single probe detection, we designed double-FAM-labeled LNA probes. The coverage of the three probe systems of the MALAT1 sequence, as well as the principle of probe detection is illustrated in Figure 1.
Figure 1.
Schematic representation of ISH technologies used in this study. (A) Multiple DNA probe (Stellaris) technology without enzymatic signal amplification (24,25), (B) bDNA (ViewRNA) technology (27), (C) LNA probe technology with enzymatic amplification (26) and (D) multiple DNA probe (Stellaris) technology with enzymatic signal amplification.
Comparison of ISH methods for detection of MALAT1
MALAT1 is expressed in a vast range of normal tissues (57) and increased expression levels are found in most human cancers (reviewed in 58). MALAT1 is a highly abundant transcript derived from multiple promoters and three different variants have been described (51). MALAT1 specifically localizes to nuclear speckles, where it is believed to influence transcription and RNA splicing (59,60). The abundant expression and particular nuclear localization make MALAT1 an ideal lncRNA for development and evaluation of ISH methods. Prior to comparing the performance of the individual ISH-methods cell culture conditions for HeLa, NHDF and T47D cell lines were established and a common fixation procedure, and individual optimization of the four ISH methods concerning probe concentrations and hybridization conditions was performed.
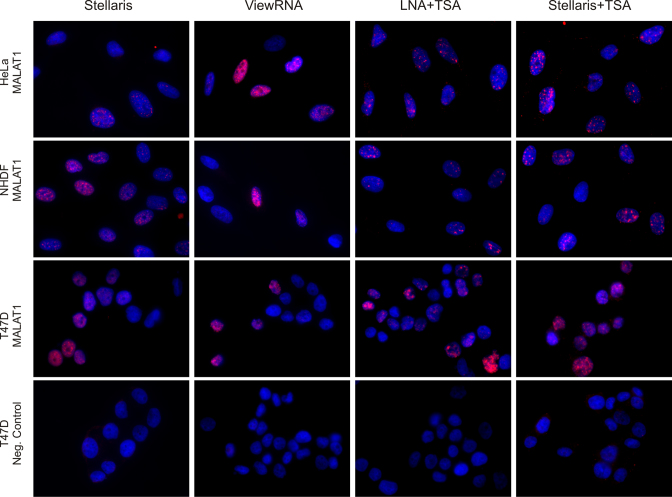
Figure 2 shows representative images of the MALAT1 ISH results in the different cell lines using optimized hybridization conditions. We generally observed a highly specific nucleus-associated signal in all cell lines; however, with a high level of heterogeneity between individual cells. The multiple probe direct detection method (Stellaris) gave a distinct nuclear signal in NHDF and T47D cells and a weaker signal in HeLa cells. The bDNA (ViewRNA) probe resulted in nuclear staining in HeLa and T47D cells, and a weak signal in the NHDF cells. The LNA probe gave an intense nuclear signal in all three cell lines, and similarly, the enzyme-enhanced Stellaris probe also detected MALAT1 in the nuclear structures in all three cell lines. This signal was consistent with its known localization in the nuclear speckles (59,60). None of the negative control probes used in the four respective methods resulted in a nuclear staining pattern similar to that of the MALAT1 probes (Figure 2).
Figure 2.
Detection of MALAT1 in HeLa, NHDF and T47D cells. Detection of MALAT1 (red stain) in cells hybridized with a Quasar 570-labeled Stellaris probe (Stellaris), a bDNA probe (ViewRNA®), a FAM-labeled LNA oligo-probe and a FAM-labeled Stellaris probe with subsequent peroxidase mediated detection using Cy3 TSA (LNA+TSA and Stellaris+TSA, respectively). Cells were counter-stained with DAPI (blue stain). Images were acquired at the same exposure conditions. Scale bar: 10 μm.
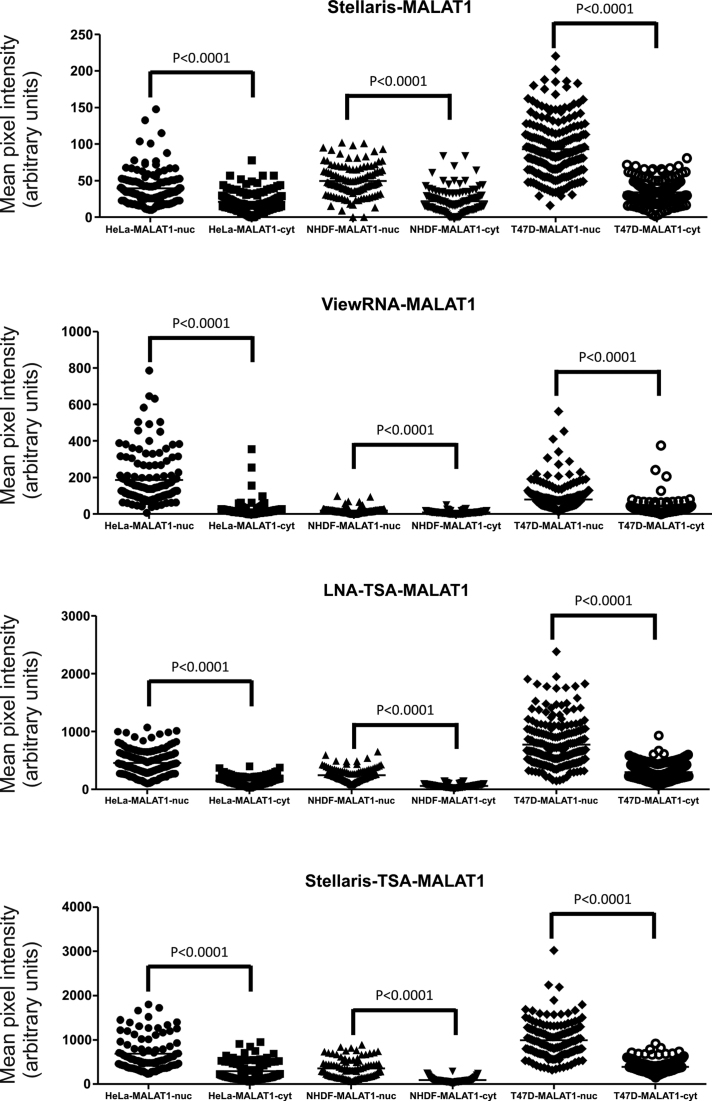
Using digital images acquired at identical and non-saturating exposure times, we quantitatively evaluated and compared the performance of each of the ISH methods by image analysis. Figure 3 shows the distribution of the mean pixel intensities in the nucleus (nuc) and the cytoplasm (cyt), respectively, of individual cells recorded from images across the different cell lines and the different ISH methods, and the associated quantitative parameters for each method and cell line is summarized in Table 2. In all cases, a significant increase was measured in nucleus-associated MALAT1 signal compared to signal in the cytoplasm. For comparison, a similar analysis was applied to mean pixel intensities obtained for the negative control probes for each of the ISH-methods (Supplementary Figure S1 and Table S1), and no difference in the signal intensities between nucleus and cytoplasmic signal was found. The T47D cell line in three of the four ISH methods expressed the highest average signal intensity in the nucleus (Table 2); however, in addition, the T47D cells also showed the highest cytoplasmic signal corresponding to high background using all methods.
Figure 3.
Comparative quantification of MALAT1 in HeLa, NHDF and T47D cells using four different ISH methods. Quantification of signal intensity in individual cells is depicted as a vertical scatter plot. Each individual dot represents the signal of the mean pixel intensities in the nucleus (nuc) and the cytoplasm (cyt), respectively, of individual cells recorded from images acquired at identical exposure times across the different cell lines and the different ISH methods. The statistical differences of the MALAT1 signal in the nuclei and the cytoplasm were determined using a paired t-test.
Table 2. Quantification of the Malat-1 ISH-signal in three different cell lines using four different probe technologies.
| HeLa | NHDF | T47D | ||
|---|---|---|---|---|
| Stellaris | MALAT1 nuc | 39 ± 2 N = 129 | 50 ± 2 N = 114 | 93 ± 23 N = 211 |
| MALAT1 cyt | 20 ± 1 N = 129 | 22 ± 2 N = 114 | 34 ± 1 N = 211 | |
| nuc-cyt | 19 | 28 | 59 | |
| nuc/cyt ratio | 1.9 | 2.3 | 2.7 | |
| ViewRNA | MALAT1 nuc | 186 ± 12 N = 132 | 18 ± 2 N = 117 | 78 ± 5 N = 227 |
| MALAT1 cyt | 24 ± 4 N = 132 | 8 ± 1 N = 117 | 28 ± 2 N = 227 | |
| nuc-cyt | 162 | 10 | 50 | |
| nuc/cyt ratio | 7.6 | 2.3 | 2.8 | |
| LNA+TSA | MALAT1 nuc | 456 ± 19 N = 139 | 246 ± 10 N = 123 | 774 ± 27 N = 215 |
| MALAT1 cyt | 150 ± 7 N = 139 | 63 ± 3 N = 123 | 299 ± 9 N = 215 | |
| nuc-cyt | 307 | 183 | 475 | |
| nuc/cyt ratio | 3.1 | 3.9 | 2.6 | |
| Stellaris+TSA | MALAT1 nuc | 693 ± 40 N = 91 | 357 ± 23 N = 88 | 991 ± 34 N = 149 |
| MALAT1 cyt | 292 ± 22 N = 91 | 91 ± 6 N = 88 | 393 ± 12 N = 149 | |
| nuc-cyt | 401 | 266 | 598 | |
| nuc/cyt ratio | 2.4 | 3.9 | 2.5 |
The mean signal intensity ± the standard error of mean is given based on the quantification of individual cells (N). In each cell, the mean pixel intensity has been determined in the nucleus (nuc) and the cytoplasm (cyt), respectively. The nuc-cyt value has been calculated by subtracting the signal of the cytoplasm from the signal of nucleus. In addition, the ratio of the signal in the nucleus and the cytoplasm (nuc/cyt ratio) has been calculated.
The two probe technologies without enzymatic amplification, Stellaris and ViewRNA, resulted in 4- to 20-fold lower ISH signal intensities than the probe technologies relying on enzymatic amplification. However, since the background signal, defined as the cytoplasmic signal was equally reduced, the signal-to-background ratio (i.e. nuc/cyt ratio) was only marginally affected for the two methods. The HeLa cells showed the highest variation in the nuc/cyt ratio among the different methods, with the ViewRNA resulting in a ratio of 7.6, and Stellaris in a ratio of 1.89. For comparison, the NHDF cells showed nuc/cyt ratios between 2.3 and 3.9. For the T47D cells, the nuc/cyt ratios across the methods were almost identical (2.5–2.7).
The ‘nuc-cyt’ values provided information about the dynamic intensity range of the ISH signal for comparison of the different methods (Table 2), and were obtained by subtracting the average signal intensity in the cytoplasm from the average signal intensity of the nuclei. The Stellaris+TSA method showed the highest dynamic range in all cell lines. In the NHDF and T47D cells, the ViewRNA method showed the lowest dynamic range of all methods. By adding the enzymatic TSA amplification to the Stellaris probe technology, the dynamic range was increased 10- to 20-fold.
Quantitative real-time RT-qPCR analyses showed that the T47D cells expressed 20-fold more MALAT1 than the HeLa and NHDF cells, in which similar levels were measured (Supplementary Figure S2). A simple correlation analysis of the qPCR and the mean intensities of the ISH signal showed that the LNA-based ISH method had the best correlation to the qPCR-based expression levels (data not shown).
Specificity analyses of the MALAT1 ISH methods using MALAT1 KO cells
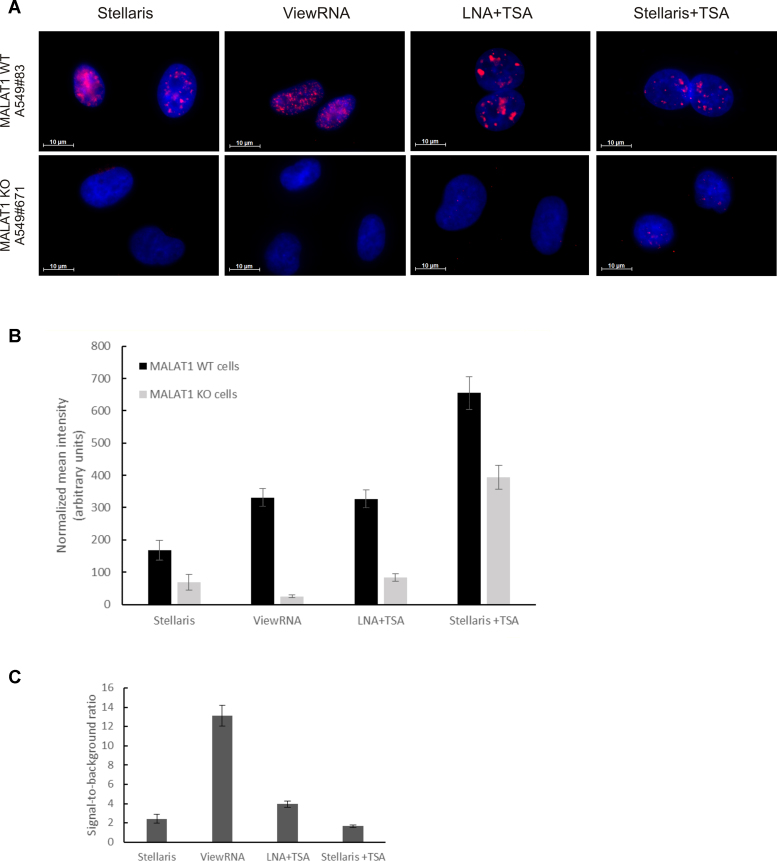
To assess the specificity of the MALAT1 ISH methods, we used a loss-of-function cellular model for MALAT1, in which MALAT1 had been stably silenced using Zinc Finger Nucleases (51,52). ISH analysis was performed using the four different detection methods in three different MALAT1 wild-type clones (MALAT1 WT) and in three knock-out clones (MALAT1 KO). All four ISH methods stained MALAT1 in the nucleus consistent with a localization in nuclear speckles and showed reduced signal in the MALAT1 KO cell lines (Figure 4A and B; Supplementary Figure S3). Calculating the ratio between the nuclear signals obtained in MALAT1 WT cells and in MALAT1 KO cells revealed that bDNA probe-based ISH method provided the best specificity (Figure 4C).
Figure 4.
Analysis of MALAT1 ISH methods in MALAT1 knock-out cell lines. (A) Images of the ISH signal (red stain) for MALAT1 in the A549 WT and in the MALAT1 knock-out cell line A549#671 using Quasar 570-labeled and FAM-labeled Stellaris probes, bDNA (ViewRNA), and LNA probes as indicated. Cells were counter-stained with DAPI (blue stain). Images were acquired at the same exposure conditions. Scale bar: 10 μm. (B) Depiction of the average nuclear ISH signal obtained with the different ISH methods in three A549 wild-type clones (WT, #10, #83) and three A549 MALAT1 knock-out cell lines (#671, #752, #772). The diagram shows the mean intensity of the nuclear MALAT1 signal measured against the mean intensity of the cytoplasmic ISH signal in each individual cell. Data represents average ± SEM of two experiments (n ≥ 30 cells for each condition/experiment). (C) Depiction of the ratio between the average nuclear signals obtained in A459 MALAT1 WT cells and in the MALAT1 knock-out A549 cells using the four different ISH-methods.
Comparison of ISH methods for detection of CYTOR
CYTOR/linc00152 has been described in several cancers including gastric cancer (61) and hepatocellular carcinoma (62) and functional studies showed the involvement of CYTOR in cell cycle regulation, apoptosis, epithelial-to-mesenchymal transition and cell migration and invasion (63). There are five different annotated transcript variants of CYTOR, and in this study we have focused on variant three which is the most abundant in MCF7 cells (unpublished results). Initially, we assessed the expression level of CYTOR and compared it with the expression level of the highly abundant MALAT1 in the three cell lines by real-time RT-PCR (Supplementary Figure S2). CYTOR is expressed between 100- and 350-fold lower than MALAT1 in HeLa and NHDF cells and more than 2000 fold lower than MALAT1 in T47D cells. The NHDF cells expressed the highest levels, and the HeLa cells the lowest.
The four different ISH methods were then employed for CYTOR detection. The Stellaris direct probe approach did not give any detectable signal above background (Supplementary Figure S4); while the other three methods to a varying degree gave signal in all three cell lines (Figure 5). No significant signal was observed in cells hybridized with the negative control probes.
Figure 5.
Detection of CYTOR in HeLa, NHDF and T47D cells. Detection of CYTOR (red stain) in cells hybridized a bDNA probe (ViewRNA), a FAM-labeled LNA oligo-probe and a FAM-labeled Stellaris probe with subsequent peroxidase-mediated detection using Cy3 TSA (LNA + TSA and Stellaris + TSA, respectively). Cells were counter-stained with DAPI (blue stain). Images were acquired at the same exposure conditions. Scale bar: 10 μm.
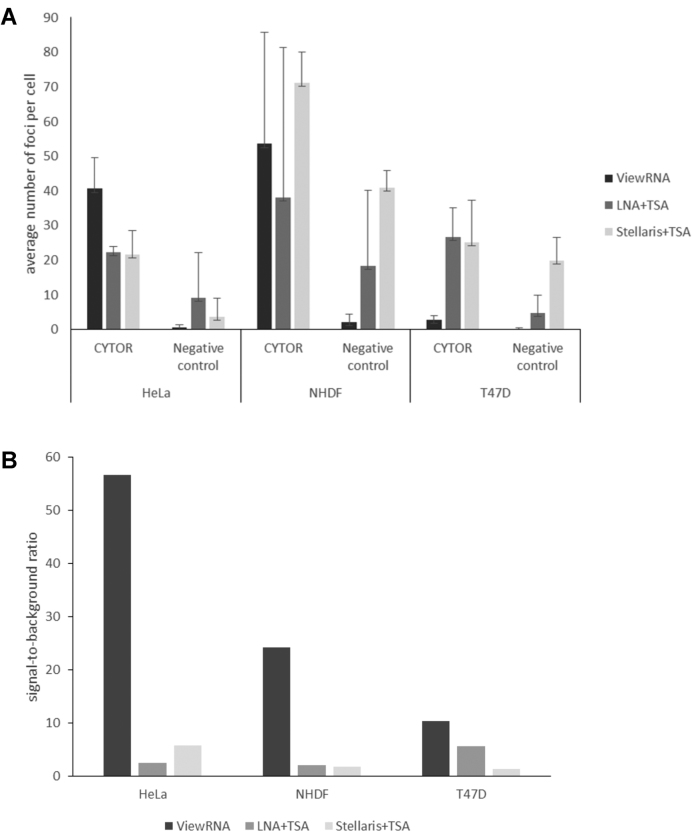
The ISH signals observed with probes against CYTOR, different from the nuclei-associated MALAT1 signals, appeared as small single spots or foci both in the nuclei and the cytoplasm, and we found that the image analysis approach used for assessment of MALAT1 was not useful for a quantitative assessment of CYTOR ISH performance. Instead, the numbers of CYTOR foci present in the nucleus and in the cytoplasm in individual cells were counted using image analysis. The results of these analyses are shown in Figure 6A, where the average number of foci per cell in all three cell lines are depicted using ViewRNA, the LNA+TSA and the Stellaris+TSA probes for CYTOR and the respective negative control probes. With the ViewRNA probes, NHDF cells presented an average of 50 foci per cell. The HeLa cells in average had 40 foci and the T47D cells <5 foci per cell (Figure 6A). In the NHDF cells, most of the positive foci were located in the cytoplasm, whereas in the HeLa cells, CYTOR ISH signals were equally distributed in the nucleus and the cytoplasm (Figure 5). Interestingly, the bDNA probe technology (ViewRNA) presented the best signal-to-background ratios in all three cell lines (Figure 6B), primarily because the background signal was almost absent. With the TSA-enhanced ISH methods, a high level of background was observed, which significantly reduced the signal-to-background ratio for these methods.
Figure 6.
Comparative quantification of CYTOR detection in HeLa, NHDF and T47D cells using three different ISH methods. (A) The average number of ISH signal foci per cell were obtained with the CYTOR probes (ViewRNA, LNA + TSA and Stellaris + TSA) and the respective negative control probes (bacterial dapB ViewRNA probe, scramble LNA probe and murine TNFα Stellaris probe). (B) The average signal-to-noise ratio of the three different ISH methods in the three cell lines are depicted by dividing the CYTOR signal with the signal of the respective negative control probes.
Specificity analyses of the CYTOR ISH method
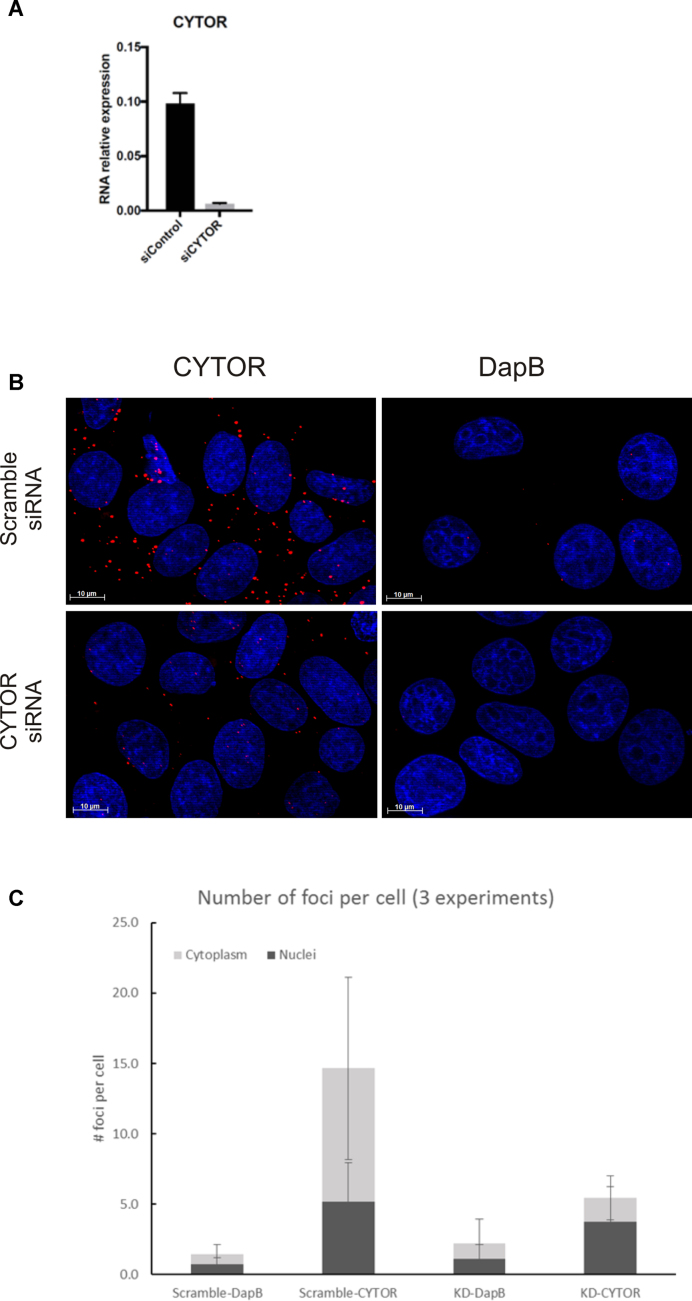
Focusing on using the ViewRNA probe technology, we assessed the specificity of the CYTOR ISH signal in MCF7 cells exposed to siRNA silencing CYTOR. In the MCF7 cells, a 15-fold reduction of CYTOR was determined by RT-qPCR after siRNA silencing (Figure 7A). In the MCF7 cells, we found similar ISH signal foci representing CYTOR as in the reference cell lines (Figure 7B), and therefore quantified the ISH signals using the same image analysis procedure as above. The quantification showed that CYTOR was almost 2-fold more expressed in the cytoplasm than in the nucleus (Figure 7C), and that a significant reduction of the total number of CYTOR ISH foci was measured in cells transfected with CYTOR siRNAs compared to cells transfected with a scramble siRNA (Figure 7C). Counting the number of foci in the nuclei and in the cytoplasm separately, we found a significant reduction of ISH signal foci in the cytoplasm after siRNA treatment, whereas the number in the nuclei largely remained unchanged, suggesting that the siRNA treatment was most efficient in the cytoplasm (Figure 7C).
Figure 7.
Evaluation of CYTOR ISH in CYTOR knock-down cell lines. (A) RT-qPCR measurements of CYTOR expression in MCF7 cells transfected with a siRNA with a scramble sequence or against CYTOR. GAPDH mRNA levels were used to normalize gene expression. (B) CYTOR ISH signal (red stain) and dapB ISH signal (negative control) in MCF7 cells transfected with scramble- or CYTOR-specific siRNA. Cells were counter-stained with DAPI (blue stain), scale bar: 10 μm. (C) Quantification of the CYTOR ISH signal (number of positive spots or foci per cell) in the nucleus and in the cytoplasm of the siRNA treated cells. Data represents average ± SEM of at least three experiments (n ≥ 30 cells for each condition/experiment).
DISCUSSION
In this study, we evaluated four ISH probe technologies, a Quasar-570-conjugated multiple probe set (Stellaris), a bDNA (ViewRNA) probe set and two modified ISH probes, a hapten-labeled Stellaris probe set and an LNA-modified probe both detected with TSA, to visualize and quantify two lncRNAs, MALAT1 and CYTOR in cultured cells. All four ISH methods were capable of detecting the characteristic nuclear signal of MALAT1, and three of the methods detected the weaker expressed CYTOR transcript. The two methods using TSA resulted in a much higher signal intensity compared to both the bDNA probe technology and the Quasar-570-conjugated probe set. Thus, the enzymatic signal enhancement through TSA-enabled visualization of MALAT1 using a single LNA probe and using the hapten-labeled Stellaris probe set. The lower level of CYTOR was best detected with the bDNA probe, suggesting that probe strategy and signal amplification are key factors in ISH analyses of lncRNAs.
We used MALAT1 as a model for evaluation of the different ISH methods because MALAT1’s characteristic localization in nuclear speckles provided an immediate assessment of the probes’ specificity. All four established methods resulted in the typical MALAT1 ISH signal. In MALAT1 knock-out cells, generated by MALAT1 silencing (51), the ISH signal was significantly reduced for all four ISH methods. The strongest effect of the MALAT1 knock-out was observed with the bDNA and LNA probes, whereas the Stellaris Quasar-570-conjugated probe set and the Stellaris probe set with TSA showed the smallest reduction in signal. These observations can be explained by the inherent specificity of the probes and the risk of exhibiting off-target binding. The bDNA method has an in-built restriction in which signal can only be generated if two detecting oligos bind to adjacent sequences on the RNA target, while the LNA probe is designed to have minimum off-targets using Exiqon’s probe design tool. Also the Stellaris probe sets are designed to exhibit minimum off-target effects, but the risk that one or more out of up to 48 probes show off-target binding is higher. Cabili et al. reported issues with off-target binding in a subset of the Stellaris probes analyzed (64). The requirement for probe specificity is even higher for RNAs expressed at lower level as the CYTOR. We found that CYTOR was best detected using the ViewRNA probe set and showed localization in both cytoplasm and nuclei. The ISH signal obtained in the cytoplasmic compartment was significantly reduced in siRNA-treated cells, whereas the nuclear signal appeared unaffected. Since the nuclear CYTOR ISH signal was well over background, our observation may be related to the efficiency of siRNA knock-down being less efficient in the nuclear compartment. Indeed, it has been reported that in general siRNA-mediated silencing is more efficient for lncRNAs residing in the cytoplasm (65). By RT-qPCR the CYTOR expression level was reduced 15-fold after the siRNA treatment and the siRNA treatment is therefore not a full depletion of the transcript. The observations taken together suggest that specificity analyses of ISH methods applied to cultured cells should include both RT-qPCR to establish the expression level and knock-out or know-down cells to evaluate off-target binding. It should be added that Cabili et al. (64) used a two-probe system with two differently labeled non-overlapping Stellaris probes, which represented an elegant approach to confirm the specificity of Stellaris probes.
In order to compare the sensitivity of the methods we used image analysis, where the total nuclear MALAT1 ISH signal was measured relative to that in the cytoplasm (nuc/cyt ratios). The image analysis approach identified variation in the mean pixel intensity of the MALAT1 ISH signal, which we considered as representative expression estimates of the different cell lines and allowed a promiscuous comparison of the ISH methods. The image analysis for all four ISH methods measured highest expression levels in the T47D cells, which was in agreement with our RT-qPCR findings. We observed a discrete MALAT1 ISH signal in HeLa cells using the Quasar-570 conjugated oligo probes using a 63× objective. This finding appears surprising since Cabili et al. found abundant MALAT1 signal in their HeLa cells (64), but may be explained by the use of Alexa-594 labeled Stellaris® probes and/or their use of a 100× objective for evaluation of the expression. Attempts to increase the sensitivity of the multiple probe set by adding antibody-based probe detection followed by enzymatic signal amplification (TSA) allowed better visualization of the ISH signal, but also increased the background level, which disturbed the overall sensitivity of the assay. Thus, the background levels were also elevated and only a marginal improvement of the signal-to-background ratio was seen. However, enzymatic signal amplification was very efficient in the detection of the LNA probe for MALAT1, for which the image analysis-based expression estimates showed the best association with the RT-qPCR data.
To obtain quantitative expression estimates for CYTOR in the three cell lines, we applied a spot-based ISH signal detection and counted the number of signal foci in both nuclei and cytoplasm. The detection of spots is limited by the diffraction limit of the microscope, which in this case is ∼0.5 μm using a 63× objective with a numerical aperture of 1.4 in the Cy3/Quasar570/Alexa Fluor 546-fluorescence channel. This approach is similar to the one employed by Cabili et al. (64), who counted the number of ISH signal spots for a variety of lncRNAs after hybridization with Stellaris probes on cultured cells. In this study, we counted ISH signal foci for CYTOR after hybridization with the bDNA probe both within the nuclei and in the cytoplasm, and found ISH signal in both regions. The foci-based CYTOR expression estimates identified highest total levels in the NHDF cells among the three cell lines studied, which was in agreement with our RT-qPCR findings. The low level of ISH signal obtained by the other ISH methods is likely associated with the low expression level of CYTOR. The RT-qPCR data indicated a more than 100-fold lower expression level than MALAT1. Factors such as the secondary and tertiary structure of the RNA and the interaction with RNA-binding proteins can render the target sites inaccessible to the probes (66), which may help to explain the poor signal since the enzyme-based signal amplification methods also resulted in weak CYTOR ISH signal.
The Quasar-570-conjugated Stellaris probe set and the ViewRNA probe set have been reported to allow single molecule detection (24–25,27–29), meaning that each spot of a certain size represent a single RNA molecule. A conglomerate of spots may fuse into single foci. In this study, all nuclear MALAT1-positive spots are likely to represent more than single molecules, whereas the single spots identified in the CYTOR ISH are more likely to represent single molecules. The TSA-based ISH methods for detection of MALAT1 resulted in larger spots and are considered unlikely to represent distinct single molecules. The use of TSA-based systems to detect long transcripts is not widely used in cultured cells, despite some early references (67,68), but it has been used to detect miRNAs in several studies (69–71). TSA is a peroxidase-based amplification system that in immunohistochemistry has been reported to amplify signal 100-fold. The amplification method may not provide staining at single molecule resolution, because the reactive product from the enzymatic reaction diffuses and bind tyrosine residues of neighbouring proteins. Obviously, unspecific bound probe will also cause amplification and increased background signals. The control of hybridization specificity is therefore important for this staining method.
The subcellular localization of lncRNAs has been found to be dramatically different from that of mRNAs (30). Whereas most mRNAs are transferred to the cytoplasm, most lncRNAs are located in the nuclei. In the nuclei, the lncRNAs are associated with chromatin, where the transcripts bind chromatin-modifying protein complexes and act in regulating gene expression (30). A vast majority of the lncRNAs studied by Cabili et al. (64) localized to the nuclei, including MALAT1, and around 40% the studied lncRNAs were also seen in the cytoplasm. For most of the lncRNAs, the localization of lncRNAs was noted to be dependent on the cell cycle, thus, Cabili et al. reported that mitotic cells lost the nuclear signal (64). As mentioned above, we found CYTOR ISH signal both in the cytoplasm and in the nuclei in all the four untreated cell lines. Compared to MALAT1, CYTOR is a relatively poorly described lncRNA. In the few studies published, the localization of the CYTOR transcript has been reported in the cytoplasm of gastric cancer cell lines (72), and in the nuclei of a hepatocellular carcinoma (73) and lung adenocarcinoma cell line (74), suggesting that CYTOR may have different functions in different cell types.
Compared to each other the four ISH methods presented both benefits and limitations, which can be evaluated in relation to ease of handling, time, signal properties, sensitivity and specificity of the assays. At one end is the multiple probe technology (Stellaris) involving a simple and fast ‘one-step detection method’, which, however, also shows limitations in specificity, with risk of off-target binding, and sensitivity since it does not include signal amplification except for the multiplication of fluorophore labeled probes (up to 48 per target). Hence, the Stellaris method would be preferred for long and abundant targets, where highly specific probe sets can be designed. In the other end, the bDNA ISH method uses a more laborious five-step detection method, which, however, can be automated. The bDNA method, has a high level of specificity because of the double-Z probe design, and has in this study shown a high level of sensitivity inherent to the five-step signal amplification procedure that in the end in theory can give a 8000 times higher number of labels per target compared to a single fluorophore-labeled probe. This very localized signal amplification is in fact claimed to allow for detection of single molecules, and the spotty signal can be counted as such. Supported by the results presented in this study, the bDNA method is recommended especially for low abundant lncRNA targets. The two remaining methods that were tested both rely on enzymatic amplification using TSA-system. The LNA+TSA-based method is, with its three-step procedure less complex than the bDNA-ISH method, and the use of LNA-containing probes has proven to increase specificity compared to using DNA-probes. The use of the TSA-system provides higher signal intensities compared to both Stellaris and bDNA-ISH methods; however, with an associated higher background signal as well. The LNA+TSA method may be chosen for both low and high abundant lncRNAs, but a panel of probe designs is required to find the best probe. The Stellaris+TSA method is an elaboration of the conventional Stellaris-method introducing an enzymatic amplification step using the TSA-system similar to the LNA+TSA, and, hence, has more or less the same complexity in handling. Similar to the direct Stellaris approach, this procedure also presents potential limitations regarding specificity due to off-target binding. The use of the TSA-system in association with the Stellaris-probes gives much higher signal intensities compared to the direct approach, but like with the LNA+TSA method also with an accompanied increase in background signal. In this study, we have shown that the modification of the Stellaris direct ISH-method actually improved the performance of the Stellaris probes to detect the less expressed CYTOR target, which could not be detected by the direct method. We recommend this method to be used in cases where gene-targeted knock-out cells are available to confirm reliability and specificity of the detection system.
In conclusion, we have reported a comparison of ISH methods to detect lncRNAs in cultured cells, as exemplified by MALAT1 and CYTOR. We evaluated assay specificity by referring to control probes, knock-down cells and RT-qPCR data, and evaluated assay sensitivity using different image analysis approaches. For the two targets, MALAT1 and CYTOR, representing high and weakly expressed transcripts, respectively, we found that all four ISH methods allowed detection of MALAT1 and, except from the direct Stellaris method also CYTOR. Our findings suggest that knowledge about a transcript, in terms of expression level and subcellular localization, is advantageous for selecting the optimal ISH method for a given target.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank Trine Møller for her excellent technical assistance.
Footnotes
Present address: Tony Gutschner, Faculty of Medicine, Martin-Luther-University Halle-Wittenberg, Halle (Saale), Germany.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
RNATRAIN Marie Curie Initial Training Network [project #607720]. Funding for open access charge: Bioneer A/S.
Conflict of interest statement. The authors declared no potential conflicts of interest with respect to the authorship and/or publication of this article. S.D. is co-owner of siTOOLs Biotech GmbH, Martinsried, Germany.
REFERENCES
- 1. Quinodoz S., Guttman M.. Long noncoding RNAs: an emerging link between gene regulation and nuclear organization. Trends Cell. Biol. 2014; 11:651–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Barrett S.P., Salzman J.. Circular RNAs: analysis, expression and potential functions. Development. 2016; 143:1838–1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lyer M.K., Niknafs Y.S., Malik R., Singhal U., Sahu A., Hosono Y., Barrette T.R., Prensner J.R., Evans J.R., Zhao S. et al. . The landscape of long noncoding RNAs in the human transcriptome. Nat. Genet. 2015; 47:199–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Clamp M., Fry B., Kamal M., Xie X., Cuff J., Lin M.F., Kellis M., Lindblad-Toh K., Lander E.S.. Distinguishing protein-coding and noncoding genes in the human genome. Proc. Natl. Acad. Sci. U.S.A. 2007; 104:19428–19433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Pheasant M., Mattick J.S.. Raising the estimate of functional human sequences. Genome Res. 2007; 17:1245–1253. [DOI] [PubMed] [Google Scholar]
- 6. Taft R.J., Pheasant M., Mattick J.S.. The relationship between non-protein-coding DNA and eukaryotic complexity. Bioessays. 2007; 29:288–299. [DOI] [PubMed] [Google Scholar]
- 7. Mercer T.R., Dinger M.E., Mattick J.S.. Long non-coding RNAs: insights into functions. Nat. Rev. Genet. 2009; 10:155–159. [DOI] [PubMed] [Google Scholar]
- 8. Ponting C.P., Oliver P.L., Reik W.. Evolution and functions of long noncoding RNAs. Cell. 2009; 136:629–641. [DOI] [PubMed] [Google Scholar]
- 9. Vance K.W., Ponting C.P.. Transcriptional regulatory functions of nuclear long noncoding RNAs. Trends Genet. 2014; 30:348–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Pang K.C., Frith M.C., Mattick J.S.. Rapid evolution of noncoding RNAs: lack of conservation does not mean lack of function. Trends Genet. 2006; 22:1–5. [DOI] [PubMed] [Google Scholar]
- 11. Pollard K.S., Salama S.R., King B., Kern A.D., Dreszer T., Katzman S., Siepel A., Pedersen J.S., Bejerano G., Baertsch R. et al. . Forces shaping the fastest evolving regions in the human genome. PLoS Genet. 2006; 2:e168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Prabhakar S., Noonan J.P., Pääbo S., Rubin E.M.. Accelerated evolution of conserved noncoding sequences in humans. Science. 2006; 314:786. [DOI] [PubMed] [Google Scholar]
- 13. Derrien T., Johnson R., Bussotti G., Tanzer A., Djebali S., Tilgner H., Guernec G., Martin D., Merkel A., Knowles G. et al. . The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012; 22:1775–1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Niazi F., Valadkhan S.. Computational analysis of functional long noncoding RNAs reveals lack of peptide-coding capacity and parallels with 3′ UTRs. RNA. 2012; 18:825–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Khanduja J.S., Calvo I.A., Hill I.T., Motamedi M.. Nuclear noncoding RNAs and genome stability. Mol. Cell. 2016; 63:7–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Roundtree I.A., He C.. RNA epigenetics–chemical messages for posttranscriptional gene regulation. Curr. Opin. Chem. Biol. 2016; 30:46–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zhao X.Z., Lin J.D.. Long noncoding RNAs: a new regulatory code in metabolic control. Trends Biochem. Sci. 2015; 40:586–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Angrand P.O., Vennin C., Le Bourhis X., Adriaenssens E.. The role of long non-coding RNAs in genome formatting and expression. Front. Genet. 2015; 6:165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Djebali S., Davis C.A., Merkel A., Dobin A., Lassmann T., Mortazavi A., Tanzer A., Lagarde J., Lin W., Schlesinger F. et al. . Landscape of transcription in human cells. Nature. 2012; 489:101–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hangauer M.J., Vaughn I.W., McManus M.T.. Pervasive transcription of the human genome produces thousands of previously unidentified long intergenic noncoding RNAs. PLoS Genet. 2013; 9:e1003569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cabili M.N., Trapnell C., Goff L., Koziol M., Tazon-Vega B., Regev A., Rinn J.L.. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011; 25:1915–1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kelley D., Rinn J.. Transposable elements reveal a stem cell-specific class of long noncoding RNAs. Genome Biol. 2012; 13:R107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mercer T.R., Gerhardt D.J., Dinger M.E., Crawford J., Trapnell C., Jeddeloh J.A, Mattick J.S., Rinn J.L.. Targeted RNA sequencing reveals the deep complexity of the human transcriptome. Nat. Biotechnol. 2012; 30:99–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Femino A.M., Fay F.S., Fogarty K., Singer R.H.. Visualization of single RNA transcripts in situ. Science. 1998; 280:585–590. [DOI] [PubMed] [Google Scholar]
- 25. Raj A., van den Bogaard P., Rifkin S.A., van Oudenaarden A., Tyagi S.. Imaging individual mRNA molecules using multiple singly labeled probes. Nat. Methods. 2008; 5:877–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Thomsen R., Nielsen P.S., Jensen T.H.. Dramatically improved RNA in situ hybridization signals using LNA-modified probes. RNA. 2005; 11:1745–1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Player A.N., Shen L.P., Kenny D., Antao V.P., Kolberg J.A.. Single-copy gene detection using branched DNA (bDNA) in situ hybridization. J. Histochem. Cytochem. 2001; 49:603–612. [DOI] [PubMed] [Google Scholar]
- 28. Battich N., Stoeger T., Pelkmans L.. Image-based transcriptomics in thousands of single human cells at single-molecule resolution. Nat. Methods. 2013; 10:1127–1133. [DOI] [PubMed] [Google Scholar]
- 29. Wang H., Su N., Wang L.C., Wu X., Bui S., Nielsen A., Vo H.T., Luo Y., Ma X.J.. Quantitative ultrasensitive bright-field RNA in situ hybridization with RNAscope. Methods Mol. Biol. 2014; 1211:201–212. [DOI] [PubMed] [Google Scholar]
- 30. Khalil A.M., Guttman M., Huarte M., Garber M., Raj A., Rivea Morales D., Thomas K., Presser A., Bernstein B.E., van Oudenaarden A. et al. . Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc. Natl. Acad. Sci. U.S.A. 2009; 106:11667–11672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wang K.C., Yang Y.W., Liu B., Sanyal A., Corces-Zimmerman R., Chen Y., Lajoie B.R., Protacio A., Flynn R.A., Gupta R.A. et al. . A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011; 472:120–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bumgarner S.L., Neuert G., Voight B.F., Symbor-Nagrabska A., Grisafi P., van Oudenaarden A., Fink G.R.. Single-cell analysis reveals that noncoding RNAs contribute to clonal heterogeneity by modulating transcription factor recruitment. Mol. Cell. 2012; 45:470–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mohammad F., Pandey R.R., Nagano T., Chakalova L., Mondal T., Fraser P., Kanduri C.. Kcnq1ot1/Lit1 noncoding RNA mediates transcriptional silencing by targeting to the perinucleolar region. Mol. Cell. Biol. 2008; 28:3713–3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Carpenter S., Aiello D., Atianand M.K., Ricci E.P., Gandhi P., Hall L.L., Byron M., Monks B., Henry-Bezy M., Lawrence J.B. et al. . long noncoding RNA mediates both activation and repression of immune response genes. Science. 2013; 341:789–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kretz M., Siprashvili Z., Chu C., Webster D.E., Zehnder A., Qu K., Lee C.S., Flockhart R.J., Groff A.F., Chow J., Johnston D. et al. . Control of somatic tissue differentiation by the long non-coding RNA TINCR. Nature. 2013; 493:231–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hacisuleyman E., Goff L.A., Trapnell C., Williams A., Henao-Mejia J., Sun L., McClanahan P., Hendrickson D.G., Sauvageau M., Kelley D.R. et al. . Topological organization of multichromosomal regions by the long intergenic noncoding RNA Firre. Nat. Struct. Mol. Biol. 2014; 21:198–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Molenaar C., Marras S.A., Slats J.C.M., Truffert J.C., Lemaitre M., Raap A.K., Dirks R.W., Tanke H.J.. Linear 2′ O-methyl RNA probes for the visualization of RNA in living cells. Nucleic Acids Res. 2001; 29:E89–E90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Majlessi M., Nelson N.C., Becker M.M.. Advantages of 2′-O-methyl oligoribonucleotide probes for detecting RNA targets. Nucleic Acids Res. 1998; 26:2224–2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wienholds E., Kloosterman W.P., Miska E., Alvarez-Saavedra E., Berezikov E., de Bruijn E., Horvitz H.R., Kauppinen S., Plasterk R.H.. MicroRNA expression in zebrafish embryonic development. Science. 2005; 309:310–311. [DOI] [PubMed] [Google Scholar]
- 40. Jørgensen S., Baker A., Møller S., Nielsen B.S.. Robust one-day in situ hybridization protocol for detection of microRNAs in paraffin samples using LNA probes. Methods. 2010; 52:375–381. [DOI] [PubMed] [Google Scholar]
- 41. Darnell D.K., Antin P.B.. LNA-based in situ hybridization detection of mRNAs in embryos. Methods Mol. Biol. 2014; 1211:69–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zhang Z., Zhu Z., Watabe K., Zhang X., Bai C., Xu M., Wu F., Mo Y.Y.. Negative regulation of lncRNA GAS5 by miR-21. Cell Death Differ. 2013; 20:1558–1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zhang A., Zhao J.C., Kim J., Fong K.W., Yang Y.A., Chakravarti D., Mo Y.Y., Yu J.. LncRNA HOTAIR enhances the androgen-receptor-mediated transcriptional program and drives castration-resistant prostate Cancer. Cell Rep. 2015; 13:209–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Silahtaroglu A.N., Tommerup N., Vissing H.. FISHing with locked nucleic acids (LNA): evaluation of different LNA/DNA mixmers. Mol. Cell. Probes. 2003; 17:165–169. [DOI] [PubMed] [Google Scholar]
- 45. Avril-Sassen S., Goldstein L.D., Stingl J., Blenkiron C., Le Quesne J., Spiteri I., Karagavriilidou K., Watson C.J., Tavaré S., Miska E.A. et al. . Characterisation of microRNA expression in post-natal mouse mammary gland development. BMC Genomics. 2009; 10:548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Thomsen R., Nielsen P.S., Jensen T.H.. Dramatically improved RNA in situ hybridization signals using LNA-modified probes. RNA. 2005; 11:1745–1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kloosterman W.P., Wienholds E., de Bruijn E., Kauppinen S., Plasterk R.H.. In situ detection of miRNAs in animal embryos using LNA-modified oligonucleotide probes. Nat. Methods. 2006; 3:27–29. [DOI] [PubMed] [Google Scholar]
- 48. Player A.N., Shen L.P., Kenny D., Antao V.P., Kolberg J.A.. Single copy gene detection using branched DNA (bDNA) in situ hybridization. J. Histochem. Cytochem. 2001; 49:603–612. [DOI] [PubMed] [Google Scholar]
- 49. Bobrow M.N., Harris T.D., Shaughnessy K.J., Litt G.J.. Catalyzed reporter deposition, a novel method of signal amplification. Application to immunoassays. J. Immunol. Methods. 1989; 125:279–285. [DOI] [PubMed] [Google Scholar]
- 50. Kwon S. Single-molecule fluorescence in situ hybridization: quantitative imaging of single RNA molecules. BMB Rep. 2013; 46:65–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Gutschner T., Baas M., Diederichs S.. Noncoding RNA gene silencing through genomic integration of RNA destabilizing elements using zinc finger nucleases. Genome Res. 2011; 21:1944–1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Gutschner T., Hämmerle M., Eissmann M., Hsu J., Kim Y., Hung G., Revenko A., Arun G., Stentrup M., Gross M. et al. . The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Cancer Res. 2013; 73:1180–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Søe M.J., Møller T., Dufva M., Holmstrøm K.. A Sensitive Alternative for MicroRNA In Situ Hybridizations Using Probes of 2′-O-Methyl RNA + LNA. J. Histochem. Cytochem. 2011; 59:661–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Deutsch E.W., Ball C.A., Berman J.J., Bova G.S., Brazma A., Bumgarner R.E., Campbell D., Causton H.C., Christiansen J.H., Daian F. et al. . Minimum information specification for in situ hybridization and immunohistochemistry experiments (MISFISHIE). Nat. Biotechnol. 2008; 26:305–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Abramoff M.D., Magalhaes P.J., Ram S.J.. Image processing with ImageJ. Biophotonics Int. 2004; 11:36–42. [Google Scholar]
- 56. Bustin S.A., Benes V., Garson J.A., Hellemans J., Huggett J., Kubista M., Mueller R., Nolan T., Pfaffl M.W., Shipley G.L. et al. . The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009; 55:611–622. [DOI] [PubMed] [Google Scholar]
- 57. Ji P., Diederichs S., Wang W., Böing S., Metzger R., Schneider P.M., Tidow N., Brandt B., Buerger H., Bulk E. et al. . MALAT-1, a novel noncoding RNA, and thymosin4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene. 2003; 22:8031–8041. [DOI] [PubMed] [Google Scholar]
- 58. Wei Y., Niu B.. Role of MALAT1 as a prognostic factor for survival in various cancers: a systematic review of the literature with meta-analysis. Dis. Markers. 2015; 2015:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Hutchinson J.N., Ensminger A.W., Clemson C.M., Lynch C.R., Lawrence J.B., Chess A.. A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains. BMC Genomics. 2007; 8:39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Nakagawa S., Ip J.Y., Shioi G., Tripathi V., Zong X., Hirose T., Prasanth K.V.. Malat1 is not an essential component of nuclear speckles in mice. RNA. 2012; 18:1487–1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Cao W.J., Wu H.L., He B.S., Zhang Y.S., Zhang Z.Y.. Analysis of long non-coding RNA expression profiles in gastric cancer. World J. Gastroenterol. 2013; 19:3658–3664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Li J., Wang X., Tang J., Jiang R., Zhang W., Ji J., Sun B.. HULC and linc00152 act as novel biomarkers in predicting diagnosis of hepatocellular carcinoma. Cell. Physiol. Biochem. 2015; 37:687–696. [DOI] [PubMed] [Google Scholar]
- 63. Zhao J., Liu Y., Zhang W., Zhou Z., Wu J., Cui P., Zhang Y., Huang G.. Long non-coding RNA Linc00152 is involved in cell cycle arrest, apoptosis, epithelial to mesenchymal transition, cell migration and invasion in gastric cancer. Cell Cycle. 2015; 14:3112–3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Cabili M.N., Dunagin M.C., McClanahan P.D., Biaesch A., Padovan-Merhar O., Regev A., Rinn J.L., Raj A.. Localization and abundance analysis of human lncRNAs at single-cell and single-molecule resolution. Genome Biol. 2015; 16:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Lennox K.A., Behlke M.A.. Cellular localization of long non-coding RNAs affects silencing by RNAi more than by antisense oligonucleotides. Nucleic Acids Res. 2016; 44:863–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Kubota K., Ohashi A., Imachi H., Harada H.. Improved in situ hybridization efficiency with locked-nucleic-acid-incorporated DNA probes. Appl. Environ. Microbiol. 2006; 72:5311–5317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Punnonen E.L., Fages C., Wartiovaara J., Rauvala H.. Ultrastructural localization of beta-actin and amphoterin mRNA in cultured cells: application of tyramide signal amplification and comparison of detection methods. J. Histochem. Cytochem. 1999; 47:99–112. [DOI] [PubMed] [Google Scholar]
- 68. Bobrow M.N., Moen P.T. Jr. Tyramide signal amplification (TSA) systems for the enhancement of ISH signals in cytogenetics. Curr. Protoc. Cytom. 2001; doi:10.1002/0471142956.cy0809s11. [DOI] [PubMed] [Google Scholar]
- 69. Lu J., Tsourkas A.. Quantification of miRNA abundance in single cells using locked nucleic acid-FISH and enzyme-labeled fluorescence. Methods Mol. Biol. 2011; 680:77–88. [DOI] [PubMed] [Google Scholar]
- 70. Nielsen B.S., Møller T., Holmstrøm K.. Chromogen detection of microRNA in frozen clinical tissue samples using LNA™ probe technology. Methods Mol. Biol. 2014; 1211:77–84. [DOI] [PubMed] [Google Scholar]
- 71. Sempere L.F. Fully automated fluorescence-based four-color multiplex assay for co-detection of microRNA and protein biomarkers in clinical tissue specimens. Methods Mol. Biol. 2014; 1211:151–170. [DOI] [PubMed] [Google Scholar]
- 72. Zhou J., Zhi X., Wang L., Wang W., Li Z., Tang J., Wang J., Zhang Q., Xu Z.. Linc00152 promotes proliferation in gastric cancer through the EGFR-dependent pathway. J. Exp. Clin. Cancer Res. 2015; 34:135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Ji J., Tang J., Deng L., Xie Y., Jiang R., Li G., Sun B.. LINC00152 promotes proliferation in hepatocellular carcinoma by targeting EpCAM via the mTOR signaling pathway. Oncotarget. 2015; 6:42813–42824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Chen Q.N., Chen X., Chen Z.Y., Nie F.Q., Wei C.C., Ma H.W., Wan L., Yan S., Ren S.N., Wang Z.X.. Long intergenic non-coding RNA 00152 promotes lung adenocarcinoma proliferation via interacting with EZH2 and repressing IL24 expression. Mol. Cancer. 2017; 16:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.