Abstract
In the brome mosaic virus (BMV) virion, the coat protein (CP) selectively contacts the RNA motifs that regulate translation and RNA replication (Hoover et al., 2016. J. Virol. 90, 7748). We hypothesize that the unstructured N-terminal arm (NTA) of the BMV CP can specifically recognize RNA motifs. Using ion mobility spectrometry-mass spectrometry, we demonstrate that peptides containing the NTA of the CP were found to preferentially bind to an RNA hairpin motif that directs the initiation of BMV RNA synthesis. RNA binding causes the peptide to change from heterogeneous structures to a single family of structures. Fluorescence anisotropy, fluorescence quenching and size exclusion chromatography experiments all confirm that the NTA can specific recognize the RNA motif. The peptide introduced into plants along with BMV virion increased accumulation of the BMV CP and accelerated the rate of minus-strand RNA synthesis. The intrinsically disordered BMV NTA could thus specifically recognize BMV RNAs to affect viral infection.
INTRODUCTION
Brome mosaic virus (BMV) is a multipartite RNA virus that has served as a model system for RNA virus infection and virion assembly (1). Motifs within the BMV genomic RNAs that direct RNA replication and translation have been mapped and characterized (2). A tRNA-like structure present at the 3′ terminal region of all of the BMV genomic RNAs directs the synthesis of the minus-strand RNA (3,4). Within this structure, Stem-loop C (SLC) binds the BMV replication proteins and enables the initiation of RNA-dependent RNA synthesis at the 3′ terminus of the BMV RNA (5).
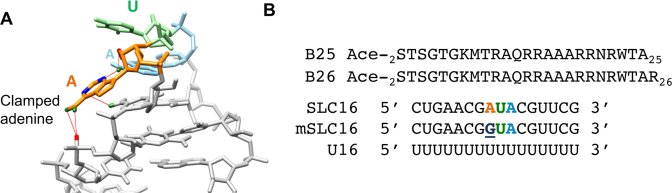
SLC forms a hairpin with the loop nucleotides of 5′AUA3′. Nuclear magnetic resonance (NMR) spectroscopy revealed that the 5′-most adenylate projects out of the loop and is stabilized by a network of interactions to form a clamped adenine motif (CAM) (Figure 1A) (6). Substitution of the clamped adenine with a guanine severely reduced RNA-dependent RNA synthesis by the BMV replicase in vitro and debilitated BMV RNA replication in plant cells (6,7).
Figure 1.
The clamp adenine motif structure and the sequences of the peptides and RNA used in the study. (A) The structure of the CAM. The solution structure is derived from PDB ID: 1ESH. (B) The sequences of the peptides and RNAs used in this study. ‘Ace’ denotes an acetyl group.
Viruses typically have small genomes, requiring viral proteins to serve multiple functions (8). The BMV coat protein (CP) that encapsidates the viral genome also regulates translation from BMV RNAs and RNA replication (9). Within the BMV genomic RNA, several regulatory motifs, including SLC, are contacted by the CP in virions (10). A screen of proteins that can bind to SLC also revealed that the BMV CP was capable of recognizing the CAM in SLC (11).
The BMV CP has a globular domain flanked by an extended N-terminal arm (NTA) and a shorter C-terminal tail (12). The globular domain from CP subunits interact to form the protein shell of the virion. The C-terminal tail interacts with other CP subunits while the NTA contains a high proportion of positively charged amino acid residues and contacts the encapsidated RNA (10,12,13). The NTA is predicted to lack a stable structure. We hypothesize that the disordered NTA can specifically recognize RNA motifs in BMV RNAs to regulate the viral infection process.
Ion mobility spectrometry-mass spectrometry (IMS-MS) has become a valuable tool to study structures and protein interactions with ligands (14,15). By employing electrospray ionization to gently introduce molecules into the gas phase with minimal perturbations of structure, IMS-MS enables populations of a molecule in solution state to be sampled (16–20). IMS-MS has been used to document changes in peptide and protein conformations upon binding to metals and drugs (21–25). Protein–DNA complex interactions have also been studied by IMS-MS techniques (26–28). Peptides complexed to nucleic acids have been more challenging to analyze using IMS-MS due to the charge differences between peptides and nucleic acids. Nucleic acids are usually electrosprayed in negative ion mode, which is less efficient when compared to the positive mode (29–32). Loo and coworkers have shown that the buffer ammonium acetate can neutralize the negative charge and allow for analysis of oligonucleotides in positive ion mode (32).
We used IMS-MS to demonstrate that the BMV CP NTA can preferentially recognize SLC that contains the CAM in the gas phase. Comparable interactions were observed in the solution state using fluorescence anisotropy and quenching, and size exclusion chromatography (SEC). The N-terminal eight residues of the NTA are required to recognize the RNA. The peptide also increased the translation of the BMV CP and the rate of BMV RNA accumulation during infection.
MATERIALS AND METHODS
Peptide and RNAs
Peptides were chemically synthesized by Elim Biopharmaceuticals, Inc. (Hayward, CA, USA) or by Ontores Biotechnologies (Zhejiang, China). The masses of the peptides were confirmed by mass spectrometry and the purity by high pressure (or high performance) liquid chromatography analysis. RNAs SLC16, mSLC16 and U16 were chemically synthesized and purified by Integrated DNA Technologies, Inc. (Coralville, IA, USA). The masses were independently confirmed by mass spectrometry. All RNA molecules were annealed at 90°C for 3 min and cooled on ice in order to encourage intra-molecular folding of the RNAs prior to analysis.
Ion mobility spectrometry-mass spectrometry
The peptides and RNAs were stored in 100 mM ammonium acetate. Peptides B25, B26, R14/15K or Δ8 were mixed with RNAs SLC16 or mSLC16 in the appropriate ratios and incubated at room temperature for 10 min before analysis. IMS-MS analyses were performed on a Synapt G2-S system (Waters Corp., Manchester, UK) using conditions described in Ruotolo et al. and Jacobs et al. (22,33). Briefly, ions were introduced into the instrument through infusion by a Syringe pump (KD Scientific, Inc. Holliston, MA, USA) at a rate of 30 μl/min. Source and desolvation temperatures were set to 25°C. Samples were ionized at 2.0 kV through electrospray ionization and guided into the instrument through the source with the cone voltage set to 30 V and the extraction voltage set to 50 V. The trap traveling wave height was set to 8.0 V at 150 m/s, the IMS cell was set to a height of 25 V at 250 m/s and the transfer cell traveling wave was set to 8.0 V at 150 m/s. Collision cross sections were determined as previously described, using ubiquitin, cytochrome C and myoglobin as calibrants (33,34).
All mass spectra and ion mobility distributions were analyzed with Mass Lynx v4.1 (Waters Corporation, Manchester, UK) and OriginPro 2016 (OriginLab Corporation, New Hampton, MA, USA). Deconvolution of m/z spectra were performed using MaxEnt, an algorithm in Mass Lynx 4.1 software, which uses a maximum entropy approach, allowing for determination of the approximate mass of the ions (35).
Molecular modeling
Molecular Dynamic simulations were performed using the Insight II software suite (Accelrys, Inc., San Diego, CA, USA). Simulation in vacuo annealing of B25 was performed using the AMBER force field (19,36). Charge was assigned to two of the following for each simulation: R11, R14, R15, R19, R20 and R22. Structures were equilibrated at 300 K for 5 ns. Simulated unfolding was induced by equilibrating the native structure to 1000 K for 2.5 ns. A total of 150 trial geometries were collected, and the cross sections were calculated using the trajectory method implemented within the Mobcal suite of programs (37).
Fluorescence anisotropy and quenching
Measurements of fluorescence anisotropy and quenching were conducted using a FluoroMax-4 Spectrofluorometer (HORIBA Scientific, Edison, NJ, USA) with an excitation wavelength of 289 nm and emission wavelength of 361 nm and a slit width of 10 nm. All fluorescence data presented were the averaged values of at least three independently measured datasets and their standard deviations were all within 10% or less. Anisotropy values were determined 20 times to generate each data point, with standard errors of <15%. All experiments were done at 22°C in a pH 6.5 buffer containing 10 mM sodium phosphate and 100 mM ammonium acetate.
Size exclusion chromatography (SEC)
SEC experiments were conducted at room temperature (∼22°C) in a sterilized 10 mM sodium phosphate buffer at pH 6.5 with 100 mM NaCl, using the FPLC AKTA Pure system, with Unicorn software version 6.4 and a Superdex 3/70 size exclusion column (GE Healthcare, Wauwatosa, WI, USA). The Superdex column is designed to resolve proteins between 3 and 70 kD. The capillary loops used had capacity from 140 to 500 μl. The FPLC instrument was equilibrated with two column volumes (ca. 47 ml) of degassed deionized MilliQ water, followed by two column volumes of the elution buffer, eluted at a flow rate of 0.5 ml/min.
NMR spectroscopy
In order to determine the initial conformation of peptide and RNA, a 1D imino proton experiment was performed for B25 and U16 RNA at 10°C. A 2D 15N-1H HSQC experiment was done on B25 where the glycine and alanine residues were labeled with 15N. All experiments were done at 10°C in 10 mM sodium phosphate buffer at pH 6.5 and 100 mM NaCl, using a Bruker 800 MHz NMR spectrometer.
Plant growth
Wheat (Triticum aestivum Spring var.) seeds were sown into autoclaved soil in 4′ diameter pots. The pots were incubated in a constant temperature incubator at 25°C with fluorescent lights at 3000 lux with a 16 h light and 8 h dark cycle. Seedlings 7 days after sowing were used for BMV infections.
Analysis of BMV CP accumulation
Infected wheat seedlings were harvested over a time course. The samples were cut into ca. 1 mm pieces using a clean razor blade and 150 mg of leaf tissue was homogenized with pestles that fit into 1.5 ml micro-centrifuge tubes. Insoluble material was removed by centrifugation at 10 000 x g for 10 min. The supernatant was mixed with an equal volume of 2× Laemmli sample buffer, heated to 70°C for 5 min and separated by sodium dodecylsulphate-polyacrylamide gel electrophoresis. CP was detected by western blotting as described in Hoover et al. (10).
Real-time RT-PCR analysis
TRIzol reagent (Life Technologies Inc., Gaithersburg, MD, USA) was used to extract total RNA from 150 mg of wheat leaves. The solubilized material was extracted with a 50% volume of chloroform followed by adjustment of the mixture with 0.3 M ammonium acetate and precipitation of the RNA with isopropanol. RNA concentration was determined by ultraviolet spectroscopy and confirmed by denaturing agarose gel electrophoresis.
The levels of replicated BMV RNAs were quantified using reverse transcription and real-time polymerase chain reaction (PCR) as described in Hoover et al. (10). Briefly, 1 μg of RNA was used for reverse transcription using SuperScript™ II Reverse Transcriptase as defined by the manufacturer (Invitrogen, Waltham, MA, USA). PCR was performed for 32 cycles with forward and reverse primers that will amplify nucleotides (nt) 100–232 of RNA1 and nt 371–499 of RNA2.
RESULTS
The BMV NTA can specifically recognize the CAM on SLC
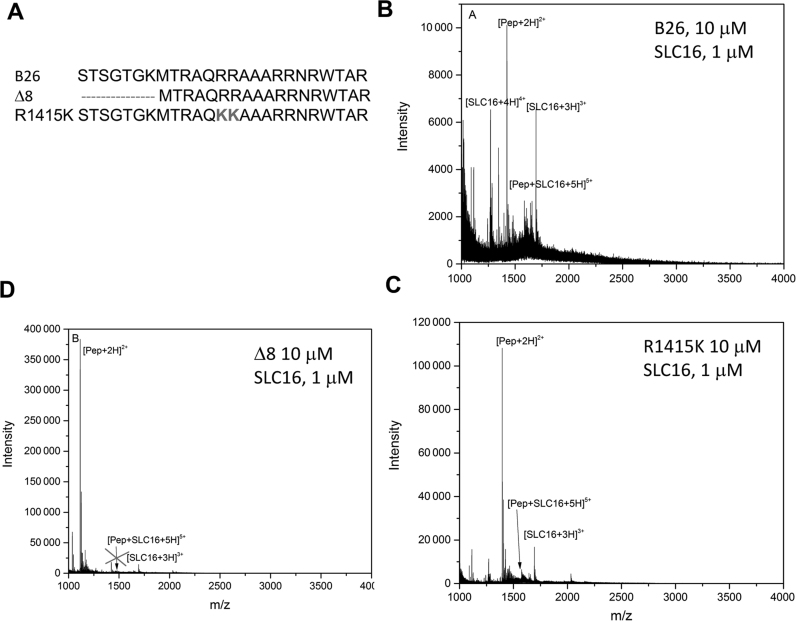
To examine the interaction between the NTA of the BMV CP and regulatory motifs in the BMV RNA, we needed to identify the sequence that likely contains this activity. In the cryo-EM structure of the BMV virion, the CP exists as an asymmetric trimer. The N-terminal 25 residues of subunit B and the N-terminal 26 residues of subunit C of the trimer had electron densities that were indistinguishable from those of the encapsidated RNAs (38). The BMV CP has a propensity to be cleaved, likely by cellular proteases. Mass spectrometric analysis of BMV virions that suffered proteolysis revealed that a prominent cleavage site occurs C-terminal to residue 26 (Supplementary Figure S1). These results suggest that the first 25 or 26 residues of the BMV CP contact the RNA. Because the N-terminal methionine of the BMV CP is removed in the mature CP (39), we synthesized peptides that contained residues 2–25 and 2–26 of the BMV CP, respectively naming them B25 and B26 (Figure 1B).
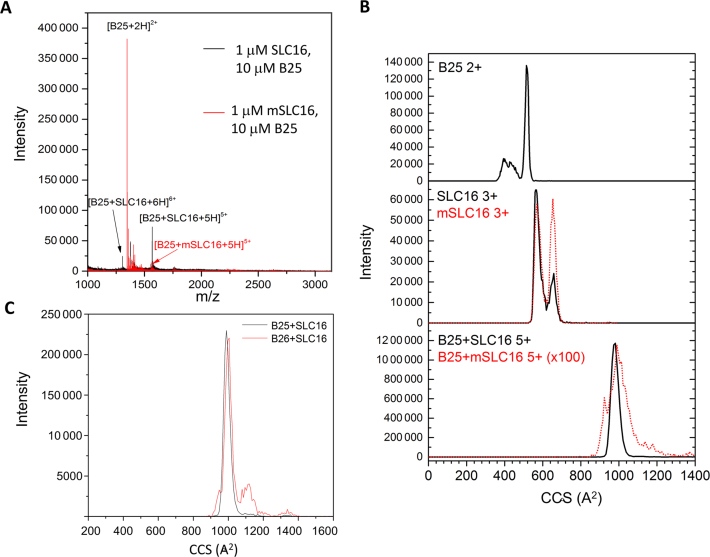
The binding of peptides B25 and B26 to an RNA oligonucleotide that contains the CAM named SLC16, was analyzed using IMS-MS. For specificity, we also analyzed their binding to mSLC16, where the only change from SLC16 was the substitution of the clamped adenine with a guanine. For B25, the B25/SLC16 complex 5+ charge state was in ∼50% abundance relative to the most intense peak, while the B25/mSLC16 complex 5+ charge state was <5% abundance (Figure 2A). The recognition of the RNA was dependent upon RNA concentration. At a molar ratio of 10 peptides to 1 RNA and a final RNA concentration of 1 μM RNA, no free RNA was detected. Increasing the RNA concentration to 2.5 μM allowed the free RNA to be seen in the mass spectra. At a molar ratio of 1 peptide to 1 RNA (10 μM relative to 10 μM B25), the B25/RNA mass spectra had similar peak distributions. These results could be explained by higher RNA concentrations that allow more non-specific, possibly electrostatic interactions with B25.
Figure 2.
IMS-MS analysis of the NTA binding to RNA. (A) Mass spectra for B25 complexed to either SLC16 or mSLC16. (B) Cross section distributions of B25, RNAs and the B25/RNA complex. The mSLC16 data are superimposed as a dotted line. Notably, the B25/mSLC16 cross section distribution is enhanced 100-fold in order for it to be appreciably visible with the B25/SLC16 cross section distribution. (C) A comparison of the B25–SLC16 complex and the B26–SLC16 complex.
The collision cross sections of B25, SLC16 and the B25/SLC16 complex were examined (Figure 2B and Supplementary Figure S2). In the absence of RNA, B25 exists in two distinct conformations, a broad distribution of compact conformations, 354 to 478 Å2. In 2D NMR that examine for backbone N–H bonds that form with protein secondary structure, few crosspeaks were observed, indicating that B25 was in a highly disordered state (Supplementary Figure S3). The sharp peak observed in IMS-MS suggests an elongated conformation at 509 Å2. The elongated conformation is likely a product of ion activation in the ion-transfer region of the Synapt G2-S, which has been reported previously (40–42). Increasing the transmission energy can cause activation of proteins in the gas phase as the protein moves from one cell of the instrument to the next. Such activation often leads to extended conformations in the gas phase and their cross sections may not reflect structures that are present in solution. The region of conformational heterogeneity suggests that the experimental conditions of low transmission voltage along with lower traveling wave magnitudes and velocities allow us to observe conformations that are similar to those that are present in solution (33).
The distributions of collision cross sections for SLC16 shows three families of structures, a compact family centered of 564 Å2, an extended conformation at 657 Å2 and a small, unresolved one at 608 Å2 (Figure 2C). While the distribution of the cross sections for mSLC16 is similar to that of SLC16, it shows much higher intensity for the extended conformation at 657 Å2. The interaction of B25 and SLC16 results in a single, sharp feature at 980 Å2. B26 complexed with SLC16 shows nearly the same profile as did B25 complexed to SLC16 (Figure 2C).
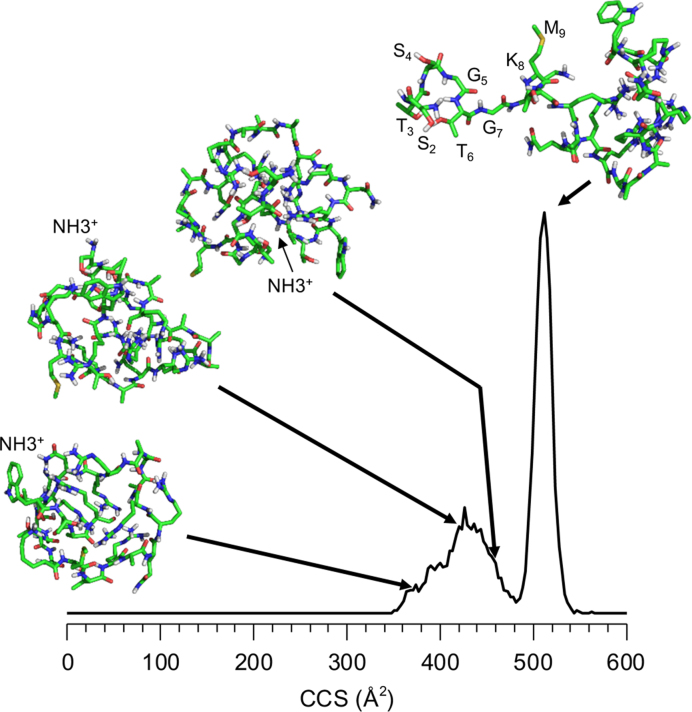
Molecular modeling was performed to examine the structure(s) of B25 emerging from solution. In total, 125 candidate structures fell within the range of the region of conformational heterogeneity. Of these, three were superimposed on the mobility distribution of doubly protonated B25 (Figure 3). Changing the location of the charge affected the structure of B25 due to a change in Coulombic interactions that will cause rearrangements of the structure. Notably, while the structures are quite compact and are stabilized only by hydrogen bonds between the side chains and the peptide backbone and by van der Walls forces, none show stable secondary structure characteristics. An additional 25 structures were produced to determine the structure of the elongated conformation. A representative structure having a calculated cross section in good agreement with the experimental measurement is also shown in Figure 3. The N-terminus ca. eight residues of B25 in this structure is exposed to solution, in contrast to the structures that are more globular (Figure 3). The elongated structure was produced only when simulation temperature was increased to 1000 K. The large amount of energy needed to generate structure suggests that this structure is partially unfolded, likely from the heating of the ions as they are transferred into the IMS cell of the Synapt (40–42).
Figure 3.
Molecular dynamic simulations of the NTA peptide. Selected conformations are shown, drawn to their calculated cross section in the distribution. The N-terminus of each structures is labeled. The sharp peak at 509 Å2 shows that the N-terminal eight residues of B25 is exposed to the solution. This feature was induced by simulating heating of the peptide to cause the unfolding of the peptide.
Fluorescence anisotropy and quenching studies
Fluorescence anisotropy and quenching measurements were used to independently examine whether the NTA of the BMV CP can recognize SLC16 in solution (for review, see 43,44). B26 contains a tryptophan residue at amino acid 23, allowing its intrinsic fluorescence to be monitored. Binding of a RNA molecule to the peptide should increase its anisotropy due to the increase in its size (45). Given that B26 is highly basic in charge and that it exhibits non-specific binding to RNA in IMS-MS when the peptide is at high concentration relative to the RNA, non-specific electrostatic interactions with RNA molecules will contribute to the binding affinity along with structure-specific attractive interactions. To assess non-specific interactions with B25, we examined the binding of an oligoribonucleotide named U16 in which all 16 nt are uridylates. The 1D imino proton NMR spectroscopy of U16 did not show noticeable imino proton peaks that forms with base pairing, indicating that U16 lacks stable secondary structures (Supplementary Figure S3) and thus serves as a useful control to assess the contribution that general electrostatic interactions have on NTA binding to RNA.
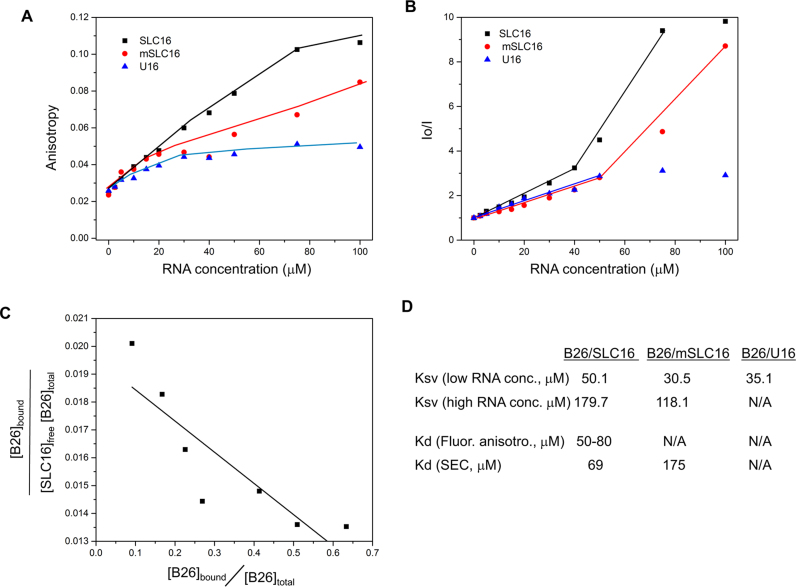
The anisotropy of B26 did not change significantly at low RNA concentrations for SLC16, mSLC16 or U16 (Figure 4A). At a 1:3 to 1:10 ratio of B26 to SLC16, the anisotropy of B26 exhibited the highest increase, reaching its saturation value (about 0.11) at 1:15. At comparable concentrations of mSLC16, the anisotropy of B26 was at a reduced level when compared to SLC16. Saturation was not reach even at a 1:20 ratio of B26 and mSLC16 (the highest anisotropy was 0.085). The anisotropy of B26 was minimal with U16, reaching a plateau after the 1:6 ratio (0.045). These results are consistent with the results from IMS-MS in that both B25 and B26 have structure-driven recognition of SLC16, confirming that RNA-peptide binding maintains its native characteristics even when transitioned to gas-phase in IMS-MS.
Figure 4.
B25 preferentially binds SLC16 in solution. (A) Fluorescence anisotropy of B26 with RNAs. The complex of B26 with SLC16 (black squares), mSLC16 (red circles) and U16 RNA (blue triangles) were measured with increasing concentrations of the RNA and a fixed concentration of B26 (5.0 μM). All measurements were obtained at ca. 22°C. (B) Quenching of the fluorescence signal of B26 by RNA. Both SLC16 and mSLC16 showed two distinct phases of quenching, which is not observed with U16 RNA. The slope of each linear plot corresponds to the Stern–Volmer constant (Ksv) for the B26–RNA interactions. (C) Scatchard plot based on B26–SLC16 anisotropy measurements. The data from 5 to 50 μM that show appreciable change in anisotropy was used. The fraction of the bound B26 was calculated based on the assumption that the binding being one to one. Saturation value of anisotropy of bound B26 is 0.11 and for unbound B26 is 0.024. The slope of the equation corresponds to −1/Kd and its Y-axis corresponds to 1/Kd. The Kd values were calculated from both parameters. (D) Dissociation constants (Kd) were calculated based on fluorescence anisotropy as well as SEC and Ksv (10−3 μM−1) for B26-RNA interactions. Low-RNA concentration ranges from 2.5 to 40 μM, while high RNA concentration ranges from 40 to 100 μM. Kd is not available for B26-mSLC16 and B26–U16 from the anisotropy measurement since their saturated values were not available. The Kd from SEC was not determined for B26–U16 mixture since the peak area of the B26–U16 complex was negligible.
RNA binding is also expected to quench the fluorescence signal of the peptide by increasing its static quenching effect (46). Fluorescence quenching due to B26 interaction with the RNA was examined. Both SLC16 and mSLC16 show two distinct phases of quenching as a function of RNA concentration (Figure 4B). A phase of linear but slowly increasing quenching was observed at low RNA concentrations and switched to a phase of fast linear increase at higher RNA concentrations. Notably, the slopes for both phases were significantly higher for SLC16 than for mSLC16. U16 exhibits only a slow linear increase at low RNA concentrations, reaching the saturation stage at higher RNA concentrations. Given that U16 shows little binding to B26 in the anisotropy measurements, the rapid quenching at a higher RNA concentration is likely to be the result of structure-specific binding of B26 to a cognate RNA.
The Kd of SLC16 for B26 was determined using a Scatchard plot based on the anisotropy data (47). One to one binding between B26 and RNA was assumed, given their comparable molecular sizes. As shown in Figure 4C, the plot fits reasonably well to a line, from which the Kd was calculated to be between 50 and 80 μM. In addition, the Ksv for each B25/RNA interaction was calculated using the fluorescence quenching data (Figure 4D). Ksv represents accessibility of the RNA to the B26 as a quencher, and the presence of the second-phase quenching in both SLC16 and mSLC16 suggests that the quenching caused by binding to B26 becomes prominent at higher RNA concentration. SLC16 shows significantly higher quenching of B26 when compared to mSLC16 (Figure 4D), which is consistent with higher affinity binding to B26, evidenced by the anisotropy measurements.
Size exclusion chromatography
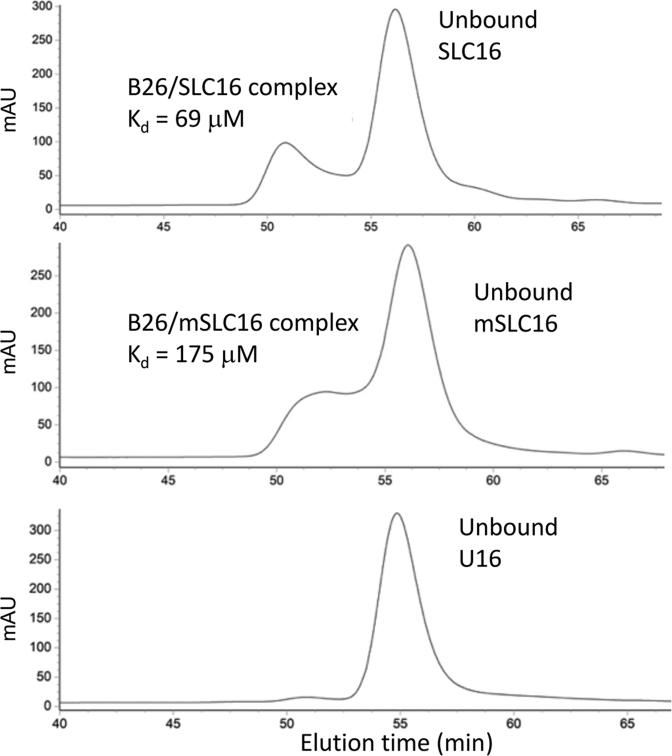
We used SEC to confirm NTA binding to SLC16. RNA bound by a peptide should elute more quickly than unbound RNA. B26 mixed with SLC16 at a molar ratio of 1:1 (5 μM of each molecule) resulted in a peak that eluted with an earlier retention time than did the peak of the unbound RNA (Figure 5). This peak was confirmed by gel electrophoresis and by fluorescence spectroscopy to contain both RNA and B26 peptide (data not shown). B26 mixed with mSLC16 resulted in an early eluting peak that was broader and weaker than the peak from the B25/SLC16 complex, suggesting reduced interactions. No additional peak was detected in a mixture of B26 and U16 (Figure 5). The Kd was calculated from the area of these two peaks (Supplementary Table S1 and Figure S4D). The B26/SLC16 had a Kd of 69 μM while the B26/mSLC16 complex had a Kd of 175 μM. The calculated Kd of B26/SLC16 by SEC is comparable to the one calculated from the fluorescence anisotropy data (Kd = 50–80 μM).
Figure 5.
Size exclusion chromatograms of mixtures of B25 and RNAs. A mixture of SLC16-B26 (Top), mSLC16-B26 (Middle) or U16-B26 (Bottom) was separated using a Superdex 3/70 size exclusion column. The peaks for unbound RNAs were identified by comparison to the standard chromatograms of each of free RNA samples. For the SLC16-B26 and mSLC16-B26, one additional peak showed up earlier than the peak for unbound RNA and they were confirmed to be peaks for a RNA–peptide complex. U16 and B26 did not form a peak of appreciable intensity.
The first eight residues of the NTA are required for its binding to SLC
Residues in the NTA of the BMV CP have previously been shown to affect RNA binding. Arginines at residues 14 and 15 substituted with lysines reduced the encapsidation of the BMV RNA4 into virions (12,48). Removal of the N-terminal 8-residues of the NTA significantly affected the accumulation of BMV RNA replication products and the encapsidation of the BMV RNAs in virions (10,48). Peptide R1415K has arginines at positions 14 and 15 replaced with lysines had comparable recognition of SLC16 in IMS-MS as did B25 (Figure 6C). The mobility distribution of the bound species has a single large peak with a cross section of 1005 Å2 and a small shoulder at 1119 Å2. Peptide Δ8 that removed residues 2–8 failed to form a complex with SLC16. The recognition of SLC16 thus requires the first eight residues of the BMV CP NTA.
Figure 6.
Comparison of binding to SLC16 by B26 and peptides with changes in the NTA. (A) B26 exhibits sharp peak (at 1005 Å2) binding to SLC16, similar to that seen in Figure 3. (B) Δ8 does not bind to SLC16 when mixed in the same concentrations as the WT NTA, indicated by the lack of a peak of the peptide–RNA species. (C) Peptide R1415 retains comparable binding to SLC16 as B26.
B25 peptide and BMV infection
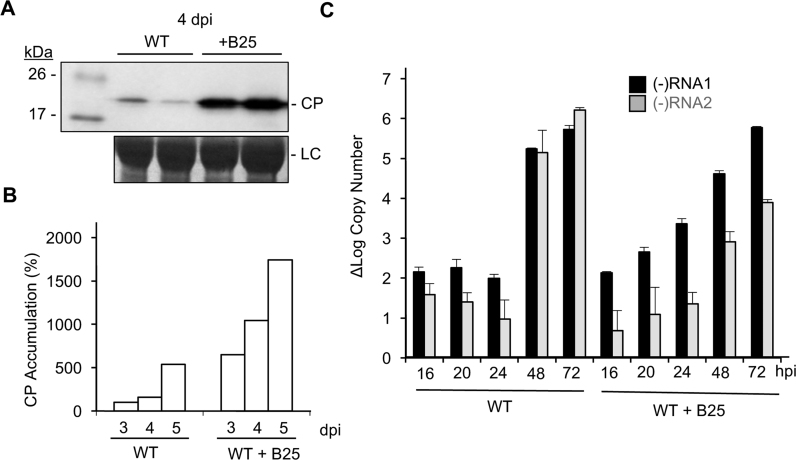
The ability of B25 to recognize a specific RNA motif led us to examine whether the peptide could affect BMV infection. Barley protoplasts transfected with a mixture of 100:1 of a peptide containing the NTA and BMV genomic RNA produced an abundance of BMV CP by 7 h post-transfection while protoplasts transfected with a polar peptide did not (Supplementary Figure S5). To examine the effects of the NTA further, wheat seedlings were inoculated with either BMV virions or BMV virions with 100 molar excess of B25. Plants were harvested over a time course and their lysates were examined by western blot for the accumulation of the BMV CP. Plants inoculated with both virion and B25 consistently had higher accumulation of the BMV CP in vivo at the time points examined (Figure 7A and B). The results were reproducible in two independent experiments.
Figure 7.
Effects of B25 on infection. (A) Accumulation of the BMV CP in wheat seedlings inoculated with WT virions or a mixture of B26 and WT virions for 120 h. Each sample was taken from a separately inoculated pot of wheat seedlings. BMV CP was analyzed by western blots from the lysate of the total wheat seedlings. BMV CP accumulation was undetectable at 24 and 48 h post-inoculation (hpi), indicating the signal is from CP accumulation during infection. LC denotes the loading control. (B) Bar graph representing the amount of BMV CP accumulation in plant leaves inoculated with BMV virions and either with or without B25. The amount of the CP present in plants inoculated with only the WT BMV was normalized as 100%. (C) (−)-strand RNA1 (black) and (−)-strand RNA2 (gray) accumulation at 16, 20, 24, 48 and 72 hpi in plants infected with WT virion or a mixture of B25 and WT virion. The gray bars are from seedlings infected with WT virions and the black bars show results from mixture of B25 and WT virions. The range for one standard deviation and P-value are shown at each time point.
We examined whether B25 could affect the accumulation of the replicated BMV RNAs in plants. Real-time RT-PCR was used to quantify minus-strand RNAs that are indicative of BMV replication. Wheat inoculated with only the WT virion had the expected kinetics of minus-strand RNA accumulation wherein (−)-strand RNA1 was synthesized at a higher rate than (−)-strand RNA2 at the early time points (Figure 7C) (10). Furthermore, the amount of (−)-strand RNA accumulated at a higher rate between 24 and 48 h. This result was reproducible in two independent experiments. Plants inoculated with both the virion and a 100 molar excess of B25 exhibited a more rapid accumulation of (−)-strand RNA1 and RNA2. Together, these results show that B25 can increase the rate of BMV gene expression and replication.
DISCUSSION
The recognition of viral RNAs by viral proteins is critically important for the proper timing of gene expression and viral RNA encapsidation. In this work, we found that the NTA of the BMV CP can preferentially recognize an RNA element that directs BMV RNA replication. An intrinsically disordered sequence in the BMV CP being the element that specifically recognizes the promoter for RNA synthesis was unexpected, as the CP is thought to bind RNA non-specifically. However, mass spectral analysis shows that the CAM in SLC that directs the initiation of minus-strand RNA synthesis is required for efficient recognition by the CP NTA (Figure 1). The recognition is confirmed using fluorescence anisotropy and quenching (Figure 4), and by SEC (Figure 5). Furthermore, the first eight residues of the CP NTA are needed for the recognition.
Binding of the NTA to SLC16 affects the conformations of both the peptide and the RNA. In the apo state, both the peptide and SLC16 have some degree of conformational heterogeneity, with the peptide forming a large family of conformationally dynamic structures as would be expected of an intrinsically disordered molecule (Figure 3). Two-dimensional 15N-1H HSQC NMR spectra of NTA confirm that B25 has flexible conformation (Supplementary Figure S3). From the molecular dynamic simulations, every one of the 125 candidate structures fell within this region in the mobility distribution. This is expected, as the differences between some of these structures entailed slight positional changes in the side chains of the amino acids. In addition, the location of the charges did not seem to matter since these structures still showed similar cross sections to each other. All simulated structures for the NTA did not form stable secondary structures, even when bound to RNA.
NTA binding to SLC16 results in a more homogeneous family of conformations, suggesting that B25 and SLC16 lock each other into a set conformation. Evidence of this is seen in both cross section distributions for the peptide and RNA (Supplementary Figure S3). Similar results were observed in the elution of the NTA peptide complexed with RNA (Figure 5). At this point, we cannot assign a specific structure for the NTA for this region. Several structures were generated from molecular dynamics simulations and sufficient differences between the candidate structures make it difficult to assess the accurate structure. Nonetheless, it is possible that the structures formed could serve to nucleate binding of other viral proteins, possibly to establish functionally important complexes for RNA replication. The CP for alfalfa mosaic virus was shown through northwestern blotting to contact both the viral RNA and the polymerase that could impact minus-strand RNA synthesis (49).
Mutant Δ8 removes the first eight amino acids from the peptide sequence. These eight amino acids have been implicated in regulating CP binding to RNA in the BMV virion. The removal of the eight residues perturbed virion formation (10,48). In addition, a serine at residue 6 of the BMV CP could be phosphorylated to affect both the release of the BMV RNA from the virion and BMV RNA synthesis (10). The recognition of RNA by this region of the CP NTA could then be regulated by additional cellular kinases and/or phosphatases that regulate the post-translational modification of the BMV CP.
This study shows that structure-specific attractive interaction serves as a main molecular driving force for RNA–peptide binding. RNA is negatively charged due to the phosphate groups in its backbone. B26 has seven arginine residues that would attract any RNA molecules through general electrostatic interactions. Electrostatic interactions, however, are not the primary driving force for specific recognition of SLC. U16, which lacks stable structure, resulted in much weaker binding to B26, compared to both SLC16 and mSLC16. Furthermore, peptide Δ8 contains seven arginine residues and did not bind to the RNAs while B25, which also contains seven positively charged residues, exhibit structure-specific recognition of SLC16. Electrostatic properties of the BMV CP were previously found to be insufficient to explain the encapsidation of BMV RNAs (12). And they cannot account for the recognition of the SLC by the NTA of the BMV CP.
The difference in NTA interaction among the three RNAs tested becomes more prominent as RNA concentration increases, demonstrating that the specific RNA structure plays an important role in its binding to NTA. Moreover, IMS-MS, Fluorescence spectroscopy, and SEC studies all indicate that SLC16 shows stronger binding to the peptides than mSLC16. Given that both SLC16 and mSLC16 form distinct tri-loop conformations (50), it is clear that the CAM presented by SLC16 serves as a key recognition motif for SLC16’s binding to the NTA peptides and a deviation from the original CAM structure in mSLC16 negatively affects this binding to the peptides. In IMS-MS, non-specific binding interactions between the NTA and mSLC16 likely result in complexes that tend to dissociate or partially unfold upon transition into the gas phase, suggesting that their binding is sub-optimal compared to the binding of SLC16. In the SEC study, the broader elution peak for NTA–mSLC16 mixture is also consistent with their binding being less stable (Figure 5). With the more specific interactions such as those with the NTA and SLC16, the RNA–peptide complex remains unbroken through additional binding interactions when transitioned into the gas phase in IMS-MS.
The Kd of the NTA for the CAM motif in SLC16 from both fluorescence anisotropy and SEC was in a range of 50–80 μM, suggesting relatively weak binding. Binding specificity of the CAM motif of SLC16 becomes more prominent at higher concentrations of either RNA or the NTA. It should be noted that the interactions between BMV molecules necessary for replication take place in a highly localized, membrane-encased, structure that has tight packing of the viral protein and RNAs (51). The BMV CP is also produced in large amounts during infection and can form higher-order structures when the proteins contain their globular domain. In the BMV virions, the CP preferentially contacted SLC (10). However, whether the seemingly weak binding may be sufficient for biologically meaningful specificity to the BMV RNA in infected cells remain to be determined.
Intrinsically disordered regions of proteins tend to have high specificity, but low affinity for interaction with ligands (52). Importantly, intrinsically disordered regions of proteins often have multiple binding partners and act as hubs to mediate a network of interactions (53–55). It is possible that the NTA of the BMV CP could recognize RNA motifs in addition to SLC to contribute to the regulation of viral gene expression. The BMV CP also preferentially contacts specific RNA sequences, including the motifs that regulate translation (9,10). We note that a significant portion of the CPs of both non-enveloped and enveloped spherical positive-strand RNA viruses are structurally flexible (56). Indeed, the MS2 capsid protein has been shown to selectively bind RNA motifs to result in the packaging of the viral RNA (57). These features may have evolved to convey specific recognition of motifs in the cognate RNAs for a subset of RNA viruses.
The addition of B25 to BMV virions was found to affect the accumulation of BMV protein and also accelerate the rate of BMV minus-strand RNA replication. It is possible that the peptide binds RNA to form a platform for the recognition of the BMV proteins to affect the infection. We cannot rule out that at least some of the effects B25 had in enhancing BMV replication could be through action on cellular molecules. The NTA of the capsid protein from Turnip crinkle virus (TCV) interacted with resistance protein HRT in Arabidopsis to alter the infection process (58). The TCV CP can also bind defense-related RNAs to improve infection (59,60).
Finally, IMS-MS was used to monitor peptide binding to RNA and the specific recognition of the RNA by a peptide. The results of these interactions in the gas phase are consistent with the results from fluorescence and SEC that investigates binding in solution phase. IMS-MS could be used to gain an understanding on how RNA structure is affected by the binding of the peptide as well as how the conformational flexibility of both the peptide and the RNA play roles in their biological functions. IMS was able to capture a large conformational change in the peptides upon binding to RNA. We note that this conformational change does not average the changes in a population of molecules, as is the case with NMR. Furthermore, IMS-MS works on the millisecond timescale, it is able to monitor the interactions that are not always measurable with other solution-based methods. This work demonstrates that IMS-MS can be used to investigate the properties of molecular interactions, especially that of conformationally flexible molecules.
Supplementary Material
ACKNOWLEDGEMENTS
The work is dedicated to the memory of Dr. Ignacio (Nacho) Tinoco, an amazing scientist and an even better mentor. We thank Laura Kao for editing this manuscript.
Footnotes
Present addresses:
Alexander Jacobs, Department of Chemistry, University of Florida, Gainesville, FL 32611, USA.
Haley Hoover, Baxter Healthcare Corp., 1 Baxter Pkwy, Deerfield, IL 60015, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Robert and Majorie Mann Fellowship (to A.J.); Indiana University Biology Department Floyd Summer Fellowship (to H.H.). Funding for open access charge: Kao laboratory of Indiana University.
Conflict of interest statement. The authors have no financial interests or connections that might raise question of bias in the work reported or the conclusions of the work.
REFERENCES
- 1. Kao C.C., Sivakumaran K.. Brome mosaic virus, good for an RNA virologist's basic needs. Mol. Plant Pathol. 2000; 1:91–97. [DOI] [PubMed] [Google Scholar]
- 2. Choi S.K., Hema M., Gopinath K., Santos J., Kao C.C.. Replicase binding sites on plus- and minus-strand BMV RNAs and identification of their roles in RNA replication in plant cells. J. Virol. 2004; 78:13420–13429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Dreher T.W., Hall T.C.. Mutational analysis of the tRNA mimicry of brome mosaic virus RNA. J. Mol. Biol. 1998; 201:41–53. [DOI] [PubMed] [Google Scholar]
- 4. Felden B., Florentz C., Giege R., Westof E.. Solution structure of the 3′-end of brome mosaic virus genomic RNAs. Conformational mimicry with canonical tRNAs. J. Mol. Biol. 1994; 234:508–518. [DOI] [PubMed] [Google Scholar]
- 5. Chapman M.R., Kao C.C.. A minimal RNA promoter for minus-strand RNA synthesis by the brome mosaic virus polymerase complex. J. Mol. Biol. 1999; 286:709–720. [DOI] [PubMed] [Google Scholar]
- 6. Kim C.-H., Kao C.C., Tinoco I. Jr. RNA motifs that determine specificity between a viral replicase and its promoter. Nat. Struct. Mol. Biol. 2000; 7:415–423. [DOI] [PubMed] [Google Scholar]
- 7. Sivakumaran K., Bao Y., Roossinck M.J., Kao C.. Recognition of the core RNA promoter for minus-strand RNA synthesis by the replicases of brome mosaic virus and cucumber mosaic virus. J. Virol. 2000; 74:10323–10331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ni P., Kao C.C.. Non-encapsidation activities of the capsid proteins of positive-strand RNA viruses. Virology. 2013; 446:123–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yi G., Letteney E., Kim C.-H., Kao C.C.. Brome mosaic virus capsid protein regulates accumulation of viral replication proteins by binding to the replicase assembly RNA element. RNA. 2009; 15:615–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hoover H., Vaughan R., Wang J., Middleton S., Ni P., Kao C.C.. Phosphorylation regulates brome mosaic virus RNA encapsidation and infection. J. Virol. 2016; 90:7748–7760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhu J., Gopinath K., Murali A., Yi G.H., Hayward S.D., Zhu H., Kao C.C.. RNA-binding proteins that inhibit RNA virus infection. Proc. Natl. Acad. Sci. U.S.A. 2007; 104:3129–3134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ni P., Wang Z., Ma X., Das N.C., Sokol P., Chiu W., Dragnea B., Hagan M., Kao C.C.. An examination of the electrostatic interactions between the N-terminal tail of the Brome mosaic virus coat protein and encapsidated RNAs. J. Mol. Biol. 2012; 419:284–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Rao A.L., Grantham G.L.. Molecular studies on bromovirus capsid protein. II. Functional analysis of the amino-terminal arginine-rich motif and its role in encapsidation, movement, and pathology. Virology. 1996; 226:294–305. [DOI] [PubMed] [Google Scholar]
- 14. Bohrer B.C., Merenbloom S.I., Koeniger S.L., Hilderbrand A.E., Clemmer D.E.. Biomolecule Analysis by Ion Mobility Spectrometry. Annu. Rev. Anal. Chem. 2008; 1:293–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Niu S., Rabuck J.N., Ruotolo B.T.. Ion mobility-mass spectrometry of intact protein–ligand complexes for pharmaceutical drug discovery and development. Curr. Opin. Chem. Biol. 2013; 17:809–817. [DOI] [PubMed] [Google Scholar]
- 16. Fenn J.B., Mann M., Meng C.K., Wong S.F., Whitehouse C.M.. Electrospray ionization for mass spectrometry of large biomolecules. Science. 1989; 246:64–71. [DOI] [PubMed] [Google Scholar]
- 17. Pierson N.A., Chen L., Valentine S.J., Russell D.H., Clemmer D.E.. Number of solution states of bradykinin from ion mobility and mass spectrometry measurements. J. Am. Chem. Soc. 2011; 133:13810–13813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ruotolo B.T., Robinson C.V.. Aspects of native proteins are retained in vacuum. Curr. Opin. Chem. Biol. 2006; 10:402–408. [DOI] [PubMed] [Google Scholar]
- 19. Shi H., Pierson N.A., Valentine S.J., Clemmer D.E.. Conformation types of ubiquitin [M+ 8H] 8+ ions from water: methanol solutions: evidence for the N and A states in aqueous solution. J. Phys. Chem. B. 2012; 116:3344–3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Shi L., Holliday A.E., Shi H., Zhu F., Ewing M.A., Russell D.H., Clemmer D.E.. Characterizing intermediates along the transition from polyproline I to polyproline II using ion mobility spectrometry-mass spectrometry. J. Am. Chem. Soc. 2014; 136:12702–12711. [DOI] [PubMed] [Google Scholar]
- 21. Glover M.S., Dilger J.M., Zhu F., Clemmer D.E.. The binding of Ca2+, Co2+, Ni2+, Cu2+, and Zn2+ cations to angiotensin I determined by mass spectrometry based techniques. Int. J. Mass Spectrom. 2013; 354:318–325. [Google Scholar]
- 22. Jacobs A.D., Chang F.M., Morrison L., Dilger J.M., Wysocki V.H., Clemmer D.E., Giedroc D.P.. Resolution of stepwise cooperativities of copper binding by the homotetrameric copper-sensitive operon repressor (CsoR): Impact on structure and stability. Angew. Chem. Int. Ed. Engl. 2015; 54:12795–12799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mehmood S., Marcoux J., Gault J., Quigley A., Michaelis S., Young S.G., Carpenter E.P., Robinson C.V.. Mass spectrometry captures off-target drug binding and provides mechanistic insights into the human metalloprotease ZMPSTE24. Nat. Chem. 2012; 8:1152–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Pacholarz K.J., Garlish R.A., Taylor R.J., Barran P.E.. Mass spectrometry based tools to investigate protein-ligand interactions for drug discovery. Chem. Soc. Rev. 2012; 41:4335–4355. [DOI] [PubMed] [Google Scholar]
- 25. Schmidt C., Robinson C.V.. Dynamic protein ligand interactions – insights from MS. FEBS J. 2014; 281:1950–1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Atmanene C., Chaix D., Bessin Y., Declerck N., Van Dorsselaer A., Sanglier-Cianferani S.. Combination of noncovalent mass spectrometry and traveling wave ion mobility spectrometry reveals sugar-induced conformational changes of central glycolytic gene repressor/DNA complex. Anal. Chem. 2010; 82:3597–3605. [DOI] [PubMed] [Google Scholar]
- 27. Jurneczko E., Cruickshank F., Porrini M., Clarke D.J., Campuzano I.D.G., Morris M., Nikolova P.V., Barran P.E.. Probing the conformational diversity of cancer-associated mutations in p53 with ion-mobility mass spectrometry. Angew. Chem. Int. Ed. 2013; 52:4370–4374. [DOI] [PubMed] [Google Scholar]
- 28. Ma X., Shah S., Zhou M., Park C.K., Wysocki V.H., Horton N.C.. Structural analysis of activated SgrAI–DNA oligomers using ion mobility mass spectrometry. Biochemistry. 2013; 52:4373–4381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Greig M., Griffey R.H.. Utility of organic bases for improved electrospray mass spectrometry of oligonucleotides. Rapid Commun. Mass Spectrom. 1995; 9:97–102. [DOI] [PubMed] [Google Scholar]
- 30. Stults J.T., Marsters J.C., Carr S.A.. Improved electrospray ionization of synthetic oligodeoxynucleotides. Rapid Commun. Mass Spectrom. 1991; 5:359–363. [Google Scholar]
- 31. McLuckey S.A., Habibi-Goudarzi S.. Decompositions of multiply charged oligonucleotide anions. J. Am. Chem. Soc. 1993; 115:12085–12095. [Google Scholar]
- 32. Sannes-Lowery K.A., Mack D.P., Hu P., Mei H.-Y., Loo J.A.. Positive ion electrospray ionization mass spectrometry of oligonucleotides. J. Am. Soc. Mass Spectrom. 1997; 8:90–95. [Google Scholar]
- 33. Ruotolo B.T., Benesch J.L., Sandercock A.M., Hyung S.-J., Robinson C.V.. Ion mobility–mass spectrometry analysis of large protein complexes. Nat. Protocols. 2008; 3:1139–1152. [DOI] [PubMed] [Google Scholar]
- 34. Salbo R., Bush M.F., Naver H., Campuzano I., Robinson C.V., Pettersson I., Jørgensen T.J., Haselmann K.F.. Traveling‐wave ion mobility mass spectrometry of protein complexes: accurate calibrated collision cross‐sections of human insulin oligomers. Rapid Commun. Mass Spectrom. 2012; 26:1181–1193. [DOI] [PubMed] [Google Scholar]
- 35. Ferroge A.G., Seddon M.J., Skilling J., Ordsmith N.. The application of ‘MaxEnt’ to high resolution mass spectrometry. Rapid Commun. Mass Spectrom. 1992; 6:765–770. [Google Scholar]
- 36. Counterman A.E., Clemmer D.E.. Cis-Trans signatures of proline-containing tryptic peptides in the gas phase. Anal. Chem. 2002; 74:1946–1951. [DOI] [PubMed] [Google Scholar]
- 37. Mesleh M.F., Hunter J.M., Shvartsburg A.A., Schatz G.C., Jarrold M.F.. Structural information from ion mobility measurements: effects of the long-Range Potential. J. Phys. Chem. 1996; 100:16082–16086. [Google Scholar]
- 38. Wang Z., Hryc C., Bammes B., Afonine P.V., Jakana J., Chen D.H., Liu X., Baker M.L., Kao C., Ludtke S.J. et al. . An atomic model of brome mosaic virus using direct electron detection and real-space optimization. Nat. Commun. 2014; 5:4808–4819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Running W.E., Ni P., Kao C.C., Reilly J.P.. Chemical reactivity of brome mosaic virus capsid protein. J. Mol. Biol. 2012; 423:79–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Merenbloom S.I., Flick T.G., Williams E.R.. How hot are your ions in TWAVE Ion mobility spectrometry?. J. Am. Soc. Mass Spectrom. 2012; 23:553–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Shvartsburg A.A., Smith R.D.. Fundamentals of traveling wave ion mobility spectrometry. Anal. Chem. 2008; 80:9689–9699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Morsa D., Gabelica V., De Pauw E.. Effective temperature of ions in traveling wave ion mobility spectrometry. Anal. Chem. 2011; 83:5775–5782. [DOI] [PubMed] [Google Scholar]
- 43. Jing M., Bowser M.T.. Methods for measuring aptamer-protein equilibria: a review. Anal. Chem. Acta. 2011; 686:9–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Blakeley B., DePorter S., Mohan U., Burai R., Tolbert B., McNaughton B.. Methods for identifying and characterizing interactions involving RNA. Tetrahedron. 2012; 68:8837–8855. [Google Scholar]
- 45. LeTilly V., Royer C.A.. Fluorescence anisotropy assays implicate protein-protein interactions in regulating trp repressor DNA binding. Biochemistry. 1993; 32:7753–7758. [DOI] [PubMed] [Google Scholar]
- 46. Yuan T., Weiljie A.M., Vogel H.J.. Tryptophan fluorescence quenching by methionine and selenomethionine residues of calmodulin: orientation of peptide and protein binding. Biochemistry. 1998; 37:3187–3195. [DOI] [PubMed] [Google Scholar]
- 47. Moller M., Denicola A.. Study of protein-ligand binding by fluorescence. Biochem. Mol. Biol. Educ. 2002; 30:309–312. [Google Scholar]
- 48. Ni P., Vaughan R.C., Tragesser B., Hoover H., Kao C.. The plant host can affect the encapsidation of brome mosaic virus (BMV) RNA: BMV virions are surprisingly heterogeneous. J. Mol. Biol. 2014; 425:1061–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Reichert V.L., Choi M., Petrillo J.E., Gehrke L.. Alfalfa mosaic virus coat protein bridges RNA and RNA-dependent RNA polymerase in vitro. Virology. 2007; 364:224–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kim C.H., Tinoco I. Jr. Structural and thermodynamic studies on mutant RNA motifs that impair the specificity between a viral replicase and its promoter. J. Mol. Biol. 2001; 307:827–839. [DOI] [PubMed] [Google Scholar]
- 51. Kopek B.G., Perkins G., Miller D.J., Ellisman M.H., Ahlquist P.. Three-dimensional analysis of a viral RNA replication comple reveals a virus-induced mini-organelle. PLoS Biol. 2007; 5:e220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Dunker K.A., Brown C.J., Lawson J.D., Iakoucheva L.M., Obradovic Z.. Intrinsic disorder and protein function. Biochemistry. 2002; 21:6573–6582. [DOI] [PubMed] [Google Scholar]
- 53. Hsu W.L., Oldfield C., Meng J., Huang F., Xue B., Uversky V.N., Romero P., Dunker A.K.. Exploring the binding diversity of intrinsically disordered proteins involved in one-to-many binding. Pac. Symp. Biocomput. 2013; 2012:116–127. [Google Scholar]
- 54. Dunker A.K., Cortese M.S., Romero P., Lakoucheva L.M., Uversky V.N.. Flexible nets: the roles of intrinsic disorder in protein interaction networks. FEBS J. 2005; 272:5129–5148. [DOI] [PubMed] [Google Scholar]
- 55. Oldfield C.J., Meng J., Yang J.V., Yang M.Q., Uversky V.N., Dunker A.K.. Flexible nets: disorder and induced fit in the association of p53 and 14-3-3 with their partners. BMC Genomics. 2008; 9:S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Ivanyi-Nagy R., Lavergne J.P., Gabus C., Ficheus D., Darlix J.L.. RNA chapteroning and intrinsic disorder in the core proteins of Flaviviridae. Nucleic Acids Res. 2006; 36:712–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Rolfsson O., Middleton S., Manfield I.W., White S.J., Fan B., Vaughan R., Ranson N.A., Dykeman E., Twarock R., Ford J. et al. . Direct evidence for packaging signal-mediated assembly of bacteriophage MS2. J. Mol. Biol. 2016; 428:431–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Kang S.H., Qu F., Morris T.J.. A spectrum of HRT-dependent hypersensitive responses elicited by the 52-amino acid N-terminus of turnip crinkle virus capsid protein and its mutants. Virus Res. 2015; 200:30–34. [DOI] [PubMed] [Google Scholar]
- 59. Qu F., Ren T., Morris T.J.. The coat protein of Turnip crinkle virus suppresses post-transcriptional gene silencing at an early step. J. Virol. 2003; 77:511–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Thomas C.L., Leh V., Lederer C., Maule A.J.. Turnip crinkle virus coat protein mediates suppression of RNA silencing in Nicotiana benthamiana. Virology. 2003; 306:33–41. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.