Abstract
Polysome-profiling is commonly used to study translatomes and applies laborious extraction of efficiently translated mRNA (associated with >3 ribosomes) from a large volume across many fractions. This property makes polysome-profiling inconvenient for larger experimental designs or samples with low RNA amounts. To address this, we optimized a non-linear sucrose gradient which reproducibly enriches for efficiently translated mRNA in only one or two fractions, thereby reducing sample handling 5–10-fold. The technique generates polysome-associated RNA with a quality reflecting the starting material and, when coupled with smart-seq2 single-cell RNA sequencing, translatomes in small tissues from biobanks can be obtained. Translatomes acquired using optimized non-linear gradients resemble those obtained with the standard approach employing linear gradients. Polysome-profiling using optimized non-linear gradients in serum starved HCT-116 cells with or without p53 showed that p53 status associates with changes in mRNA abundance and translational efficiency leading to changes in protein levels. Moreover, p53 status also induced translational buffering whereby changes in mRNA levels are buffered at the level of mRNA translation. Thus, here we present a polysome-profiling technique applicable to large study designs, primary cells and frozen tissue samples such as those collected in biobanks.
INTRODUCTION
Protein levels are modulated via a series of mechanisms including transcription, mRNA-splicing (1), -transport (2), -localization (3), -stability (4), –translation (2) and protein-stability (5). Notably, mRNA translation is the most energy consuming process in the cell (6) and its tight control is therefore essential (7). Consistently, mRNA translation was suggested as the predominant post-transcriptional mechanism impacting protein levels (8,9) although the relative contribution of different mechanisms affecting protein levels is context dependent (10,11). Moreover dysregulation of translation is associated with pathologies as diverse as fibrosis (12), cancer (13) and neurodegenerative disease (14–17). Thus, there is a need to study translatomes (i.e. the transcriptome-wide pool of efficiently translated mRNA) to obtain a more complete understanding of how gene expression is modulated in both health and disease.
Regulation of mRNA translation can be global, by affecting mRNAs transcribed from essentially all genes; selective, by targeting mRNAs from a gene subset; or specific, by affecting mRNA copies from a single gene (14,18). Studies of translatomes can be used to explore the latter two contexts as global changes in translation cannot be assessed using relative quantification methods such as RNA sequencing (RNAseq) or DNA-microarrays (19). Translation can be divided into four phases: initiation, elongation, termination and recycling (20). Although the elongation phase can be regulated by e.g. cellular stress (21), most described modulation of translation occurs at the initiation step, where mRNAs are recruited to ribosomes (20,22). When translation is regulated via changes in initiation, a change in the proportion of all mRNA copies from a single gene that are efficiently translated is observed (19). Such changes appear to be mediated via two modes of regulation: a large change in translational efficiency from almost complete association to almost complete dissociation with polysomes (on-off regulation); or a less dramatic modulation of translational efficiency largely contained within polysomes (19). For example, following inhibition of the mammalian/mechanistic target of rapamycin (mTOR), mRNAs harbouring a 5′ Terminal Oligopyrimidine Tracts element (TOP-mRNAs) in their 5′ un-translated regions (5′UTRs) show on-off regulation while mRNAs, e.g. encoding mitochondria-related proteins show a shift in translational efficiency while still largely being associated with polysomes (19). Importantly, both modes of regulation lead to a change in the proportion of mRNA associated with >3 ribosomes (19). This property underlies selection of mRNAs associated with >3 ribosomes to represent the pool of efficiency translated mRNA during polysome-profiling.
Polysome- and ribosome-profiling are commonly used to study translatomes (18). Polysome-profiling involves isolation of cytosolic extracts followed by sedimentation in a linear sucrose gradient (commonly 5–50% sucrose). During centrifugation, mRNAs sediment according to how many ribosomes they associate with and, following fractionation, efficiently translated mRNAs (i.e. those fractions containing mRNA associated with >3 ribosomes) can be identified and pooled. The mRNA-pool is then quantified using either DNA-microarrays or RNAseq to derive data on translatomes. The >3 ribosome cutoff for isolation of efficiently translated mRNA could potentially result in that mRNAs whose change in translational efficiency does not involve a transition across this threshold cannot be identified. Detailed studies of mTOR sensitive translation indicate that many mRNAs will shift across the >3 ribosome cutoff (19). Moreover, as ribosome association is normally distributed (19), even a shift in mean ribosome association from 1 to 3 ribosomes will involve a change in the proportion associated with >3 ribosomes (i.e. as a result of the shift of the tails of the distributions). For the same reason, shifts from e.g. 5 to 10 ribosomes also involves a small shift in the amount of mRNA associated with >3 ribosomes. Thus selection of >3 ribosomes to represent efficiently translated mRNA has an underpinning for studies of mammalian cells but it cannot be excluded that some shifts cannot be observed (those largely occurring within very high ribosome association). During ribosome-profiling, ribosome-protected fragments (RPFs) are generated by applying a mild RNase treatment and isolated using gel purification (23). RPFs are then identified and quantified using RNAseq to reveal nucleotide resolution ribosome location. Such data is most commonly used to decipher patterns of ribosome positioning (24,25), but can also be used to assess changes in translational efficiency.
Thus, in contrast to polysome-profiling where efficiently translated mRNAs associated with >3 ribosomes are quantified (i.e. an mRNA perspective), ribosome-profiling quantifies ribosome-association (i.e. a ribosome-perspective). We recently showed that this leads to a bias during ribosome-profiling when mRNAs showing large changes in translational efficiency and those showing smaller shifts contained within polysomes are regulated under the same condition. Under these settings ribosome-profiling will, to a larger extent as compared to polysome-profiling, bias towards identification of abundant mRNAs showing larger changes as differentially translated (26). This difference in bias between polysome- and ribosome-profiling originates from that, during ribosome-profiling, translational efficiency is indirectly inferred by the number of RPFs, and thus the magnitude of the shift in ribosome association is directly proportional to the fold-changes estimated. In contrast, during polysome profiling, translational efficiency is directly estimated from amounts of efficiently translated mRNAs associated with >3 ribosomes which results in a less pronounced bias (26). One limitation of polysome-profiling is that information regarding which part of the mRNA is translated is lacking. Indeed, some mRNAs such as SLC45A4 can shift between translating the main open reading frame (ORF) to an upstream ORF (uORF) (27). Such changes in ribosome location can only be resolved using ribosome profiling. Ribosome-profiling can in turn not be used to link changes in translational efficiency to transcription start site usage as untranslated regions (UTRs) are degraded during isolation of RPFs (19). In contrast polysome-profiling allows for exploration of the impact of 5′UTRs on translational efficiency (19). Thus, polysome- and ribosome-profiling are complementary methods necessary to enhance our understanding of how translatomes are modulated where polysome-profiling is preferred for unbiased studies of changes in translational efficiency.
A technical challenge during polysome-profiling, however, is that the pool of efficiently translated mRNA is collected in a large volume (often >3 ml) spread across 5–10 fractions. Such RNA is commonly isolated from each fraction separately and pooled during re-suspension of purified RNA pellets. For small samples, however, such extensive dilution is problematic as it may cause sample loss. Moreover, isolation of RNA from many fractions is labor intensive and, in larger experimental setups (e.g. large in vitro experiments or studies of clinical cohorts involving hundreds of samples resulting in thousands of fractions that need to be pooled), may introduce a risk of mistakes such as erroneous pooling of fractions and sample mislabeling. Therefore, approaches simplifying collection of efficiently translated mRNAs (i.e. associated with >3 ribosomes) are warranted. Herein, we describe an optimized non-linear sucrose gradient which collects efficiently translated mRNA (associated with >3 ribosomes) in only one or two fractions to reduce sample handling 5–10-fold and time needed for RNA extraction by 10–20-fold. By coupling isolation of such mRNA with RNAseq methods developed for single-cells, we show that it is possible to derive data on translatomes from small tissue samples such as those collected in biobanks. Importantly, this approach produces very similar data on translatomes as compared to the standard linear gradient approach (28). Thus, polysome-profiling can now be applied to small samples from tissues or primary cells where RNA amount is limited.
MATERIALS AND METHODS
Preparation of cytosolic lysates from cell lines
HCT-116 p53+/+, HCT-116 p53−/− (kindly provided by Galina Selivanova, Karolinska Institutet) and MCF7 (ATCC HTB-22™) cell lines were cultured in Dulbecco's Modified Eagle Medium (DMEM) supplemented with 10% fetal bovine serum, 1% penicillin/streptomycin and 1% l-glutamine (Gibco, Life Technologies). Briefly, cells (1 × 106) were seeded in 15-cm cell culture dishes (Corning), harvested at 80% confluency (HCT-116 p53+/+ and p53−/− cells were also serum starved [0.1% fetal bovine serum for 16 h prior to harvest]), lysed in hypotonic lysis buffer (5 mM Tris–HCl, pH 7.5, 2.5 mM MgCl2, 1.5 mM KCl, 100 μg/ml cycloheximide, 2 mM DTT, 0.5% Triton, 0.5% sodium deoxycholate; all from Sigma Aldrich) and the cytosolic extract was loaded onto the sucrose gradient. A detailed protocol described previously (28) was applied with the modification that instead of adding cycloheximide to the media, the cell media was discarded, plates were placed on ice and immediately washed in an ice-cold solution of 1× phosphate-buffered saline (PBS) and cycloheximide (100 μg/ml; Sigma Aldrich).
Preparation of cytosolic lysates from tissues
All breast cancer samples were collected in the A.C. Camargo Cancer Center biobank (São Paulo, Brazil) under informed consent (ethical permission 1844/13). To prevent tissue thawing and subsequent RNA degradation, all materials including tools and plastics were kept in liquid nitrogen during sample processing. Tissues were pulverized using a BioPulverizer (United Laboratory Plastics) followed by grinding in a liquid nitrogen-proof container until a fine powder was obtained. The powder was collected and kept on dry ice or stored at −80°C. A modified hypotonic lysis buffer with a 10-fold higher concentration of cycloheximide (as compared to above) was used. Also the RNase inhibitor (RNaseOUT, Invitrogen) was replaced by Recombinant RNasin® Ribonuclease Inhibitor (Promega). To prevent clogging, a modified 1000 μl tip (cut to get a wider entry channel) was used to add 500–1000 μl (depending on the size of the tissue sample) ice-cold lysis buffer to the previously collected powder and mixed until homogenization. The sample was further homogenized using an ice-cold Dounce homogenizer (60 strokes using both the loose and tight pestle) in the presence of 0.5% Triton and 0.5% sodium deoxycholate. The homogenate was centrifuged at maximum speed (21 500 RCF) for 2 min at 4°C in a table top centrifuge and 50 μl supernatant was collected and diluted in 450 μl of nuclease-free water (referred to as cytosolic RNA). TRI-reagent® (Sigma Aldrich) was added and cytosolic RNA samples were stored at −80°C. The remaining cytosolic lysate was immediately loaded on sucrose gradients.
Preparation of optimized non-linear sucrose gradients
A 5X gradient buffer (100 mM HEPES pH 7.6, 500 mM KCl, 25 mM MgCl2; all from Sigma Aldrich) was used to prepare sucrose solutions needed for the gradient: 5% (w/v), 34% (w/v) and 55% (w/v) (1× final concentration of gradient buffer [v/v] adjusted with water). A gradient cylinder (BioComp) was used to draw a line on each centrifuge tube at the highest level of the cylinder (corresponds to about 5.5 ml when using the Open-Top Polyclear Centrifuge Tubes [14 × 89 mm, SETON Scientific, Part No. 7030]). A 1000 μl pipette was then used to add 2 ml of the 5% sucrose solution to the tube. Then, a syringe with a layering needle (BioComp) was used to add the 34% sucrose solution at the bottom of the tube (i.e. below the 5% sucrose solution) until the surface of the 5% solution reached the drawn line. Finally, a 55% sucrose solution was added from the bottom of the tube until the interface between the 34% and 55% solutions reached the drawn line. The tubes were then capped with rate zonal caps (BioComp) and stored at 4°C for 2 h before use (to standardize the time between gradient preparation and loading of samples). All reagents were nuclease-free.
Preparation of linear sucrose gradients
The linear gradients were prepared as described (28).
Sample loading onto sucrose gradients and fractionation
First, 500 μl of sucrose solution was removed from the top of the gradient without disturbing gradient composition. The cytosolic lysate (∼500 μl) was then layered on the surface of the gradient. To precisely balance the samples for ultracentrifugation we used 1× hypotonic lysis buffer when needed. The samples were centrifuged at 209 815 RCF for 2 h at 4°C in a SW 41 Ti rotor and a Beckman Coulter Ultracentrifuge Optima L-90K. Samples were eluted using either the gradient station (BioComp) or the Biologic LP pump (Bio-Rad) coupled to a Model EM-1 Econo UV detector (BioRad). Fractions (∼500 μl) were collected with either a Piston Gradient Fractionator (BioComp) coupled with a fraction collector (Gilson) or a model 2110 fraction collector (Bio-Rad). The precise location of the fractions along the UV-tracing was monitored using either the gradient profiler v1.25 (BioComp) software or the LP Data View v1.03 (Bio-Rad). TRI-reagent® was immediately added to each fraction and fractions were kept on ice prior to storage at −80°C.
RNA extraction
For the optimized non-linear gradient, fractions containing the center of the peak and the fraction towards the bottom of the centrifugation tube (referred to as fractions 0 and +1 in the results section) were pooled and RNA was extracted using Tri-reagent®. For cell lines profiled using linear gradients, fractions corresponding to mRNA associated with >3 ribosomes (fractions 17–25 for the current setup, resulting in 9 tubes with a volume of ∼500 ul each) where extracted separately using Tri-reagent® (according to the protocol of the manufacturer) and then pooled during resuspension of RNA pellets. RNA extraction was performed differently depending on the source of the RNA (cell line or tissue sample). RNA from cell lines was extracted using Tri-reagent® and further purified with RNAeasy MinElute Cleanup Kit (Qiagen) according to manufacturer's recommendations. For tissue samples, the extraction protocol was modified to maximize the amount of RNA recovered: After standard phase separation with chloroform, the upper aqueous phase was transferred to a mixture of 2 volumes of ethanol 99.5% and linear acrylamide (15 μg/ml; Life Technologies). The mixture was centrifuged at 4°C for 40 min in a table top centrifuge at maximum speed (21 500 RCF) and the pellet was dissolved in 100 μl of RNase free water. The RNA was further purified using the RNAeasy MinElute Cleanup Kit (Qiagen) according to manufacturer's protocol. RNA quantity was measured by target-specific fluorescence (Qubit, Life Technologies) and its quality assessed with on-chip electrophoresis using an Agilent Bioanalyzer 2100. The latter applies an algorithm to assign an RNA integrity number (RIN) to each sample. An RIN of 10 indicates perfectly intact RNA.
Preparation of smart-seq2 RNAseq libraries and sequencing
Smart-seq2 was performed as previously described (29) using 10 ng (when possible, otherwise less) of RNA as starting material from five tissue samples (cytosolic RNA and polysome-associated RNA isolated using the optimized non-linear gradient) and four biological replicates of HCT-116 p53+/+ and HCT-116 p53−/− cells serum starved (0.1% fetal bovine serum) for 16 h (cytosolic RNA, polysome-associated RNA isolated using the optimized non-linear gradient and polysome-associated RNA isolated using the linear gradient). Quality of RNAseq libraries was evaluated by on-chip electrophoresis using an Agilent Bioanalyzer 2100. Sequencing was performed on RNAseq libraries from cell lines and tissues separately. Prior to sequencing, all RNAseq libraries were adjusted to a concentration of 10 nM and then pooled, clustered using cBot clustering and sequenced on the HiSeq2500 platform (HiSeq Control Software 2.2.58/RTA 1.18.64, Illumina) with a 1 × 50 bases setup using ‘HiSeq SBS Kit v4′ chemistry. After each cycle, image analysis and base calling was performed using the CASAVA software suit. Technical quality of the RNA sequencing was assessed using MultiQC (30).
Analysis of RNAseq data
RNAseq reads from the breast cancer tissues and 4 biological replicates of serum starved HCT-116 p53+/+ and HCT-116 p53−/− cells were mapped to the human reference genome GRCh38 using Bowtie (31) (settings: –a –m 1 –best –strata –n 2 –l 28). The rpkmforgenes script (32) was used to quantify gene expression (with options -readCount, -fulltranscript and -onlycoding) based on RefSeq annotation. The reads per kilobase per million mapped read (RPKM) output from the rpkmforgenes script was used to assess quality of breast cancer RNAseq libraries. For analysis of HCT-116 cells, genes with zero count(s) or overlapping gene variants were excluded from the analysis leading to a total of 8675 quantified genes. Raw counts were scaled using TMM normalized library sizes (33) and log2 counts per million were computed using the voom function of the limma R package (34). To explore whether the two gradient methods led to similar gene expression patterns, principal component analysis was performed after centering per gene. Each sucrose gradient method was then considered separately to assess differential expression of polysome-associated mRNA between HCT-116 p53+/+ and HCT-116 p53−/− cell lines. Differential expression analysis was performed using t-tests applying RVM (Random Variance Model) (35), including the replicate number as factor in the models as implemented in the anota2seq R package (available at Bioconductor). P-values were adjusted using the Benjamini–Hochberg (BH) method (36) and a false discovery rate (FDR) < 0.1 was considered significant. The correlation of log2 fold changes between HCT-116 p53+/+ and HCT-116 p53−/− obtained using linear and optimized sucrose gradients was assessed using the Spearman rank correlation coefficient. The different modes of regulation of gene expression associated with p53 status were then identified using the optimized gradient data. To this end, polysome-associated mRNA from optimized non-linear sucrose gradients and cytosolic mRNA data were re-normalized as described above. A previously described gene signature of p53 transcriptional targets (37) was used to characterize expected differences between HCT-116 p53+/+ versus HCT-116 p53−/− cells. Anota2seq allows for identification of differences in translational efficiency affecting protein levels and buffering. The replicate number was included as a covariate in the linear models. Unrealistic models of differential translation and buffering were excluded (the filtering criteria in the anota2seqSelSigGenes function were as follows: maxSlope = 1.5, minSlope = -0.5, deltaPT = log2(1.2), deltaP = log2(1.2) for translation and maxSlope = 0.5, minSlope = -1.5, deltaT = log2(1.2), deltaTP = log2(1.2) for buffering. Genes with an FDR < 0.25 (a relaxed FDR threshold was used to obtain a sufficiently large number of identifiers in each regulatory pattern to allow for sensitive Gene Ontology [GO] enrichment analysis) were selected for GO enrichment analysis. Only GO terms (from the Biological Processes ontology) with 5 to 500 genes were considered for hypergeometric tests using GOstats (35) where ‘conditional’ was set to FALSE (the structure of the GO graph is not considered in the tests). Only processes annotated to at least five genes in a regulatory pattern were considered. The significant GO terms (FDR < 0.05) of each set (translation up, translation down, buffering up, buffering down, abundance up [i.e. congruent up-regulation of cytosolic and polysome-associated mRNA levels] and abundance down) were visualized in a heatmap (row dendrogram shows unsupervised clustering using default method of the gplots::heatmap.2 function) (36). All analyses were done using R version 3.3.1.
RESULTS
Design of an optimized non-linear sucrose gradient for isolation of efficiently translated mRNA
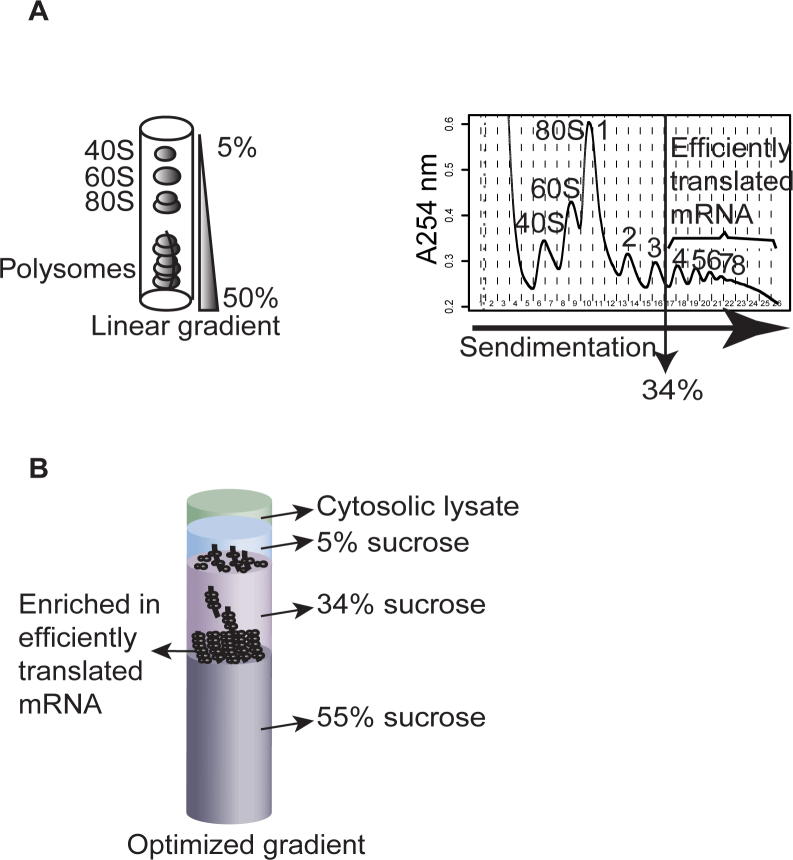
Because polysome-profiling with linear gradients requires extraction of RNA from many fractions per sample to isolate efficiently translated mRNA (Figure 1A), we considered alternative approaches. As many mRNAs show continuous shifts in translational efficiency within polysomes, pelleting ribosomes would not allow for estimates of their changes in translational efficiencies as such mRNA would be pelleted when associated with at least 1 ribosome. Instead we reasoned that a high percentage sucrose solution below an intermediate concentration sucrose solution could potentially enrich for efficiently translated mRNAs (associated with >3 ribosomes) at the surface of the high percentage sucrose – if the optimal sucrose concentrations were identified. This would allow for elution of efficiently translated mRNA in a smaller volume as compared to the standard linear gradient. While 55% sucrose essentially halts polysome sedimentation, we searched to identify a second sucrose concentration that would allow for such enrichment at the 55% sucrose surface. We therefore examined the linear relationship between the log2 number of associated ribosomes and sedimentation distance (19). This allowed us to calculate the sucrose concentration that separates mRNAs associated with three ribosomes from those associated with four ribosomes in a linear gradient (Figure 1B; equals to 34% sucrose). To facilitate entry into the gradient, we added a third sucrose layer of 5% sucrose on the top of the 34% sucrose. We next attempted to determine the appropriate volumes of each of the sucrose layers. The objective was to position the surface of the 55% sucrose close to the top of the tube (to reduce time needed for elution) while still allowing a sufficient volume of 34% sucrose for good separation between efficiently (>3 ribosomes) and less efficiently translated mRNA. A highly reproducible approach for generating layers of sucrose is to start by adding the lowest concentration of sucrose solution to the tube and then add increasingly higher sucrose concentrations at the bottom of the tube; thereby displacing the lower concentration sucrose solution(s) towards the top of the centrifuge tube (28). While the first layer of 5% sucrose can be added by volume directly to the tube, additional volumes for the remaining layers are best determined by monitoring the interface between layers and let these reach a certain position in the tube. It is essential that this position can be reproducibly indicated on the tube and we therefore used the same approach as when making a linear gradient using the BioComp gradient maker, whereby a cylinder is used to indicate the desired level on the tube (28). Our initial test of the optimized non-linear gradient indicated separation of 40S, 60S ribosomal subunits and the 80S monosome followed by a peak at the interface between the 34% and 55% sucrose solutions (Figure 2A).
Figure 1.
The linear and optimized non-linear gradients. (A) Polysome profiling using a linear 5% to 50% sucrose gradient. Cytoplasmic RNA is extracted and loaded on the linear gradient. Following ultracentrifugation, 40S and 60S ribosome subunits, the 80S monosomes, and polysomes are separated (schematics show UV tracing at 254 nm across the sucrose gradient). Efficiently translated mRNA is isolated from polysome-fractions containing mRNA associated with more than 3 ribosomes. This corresponds to a sucrose concentration of 34%. (B) The optimized non-linear sucrose gradient is made of layers of 5%, 34% and 55% sucrose. The purpose is to collect mRNA associated with more than three ribosomes at the surface of the 55% sucrose solution.
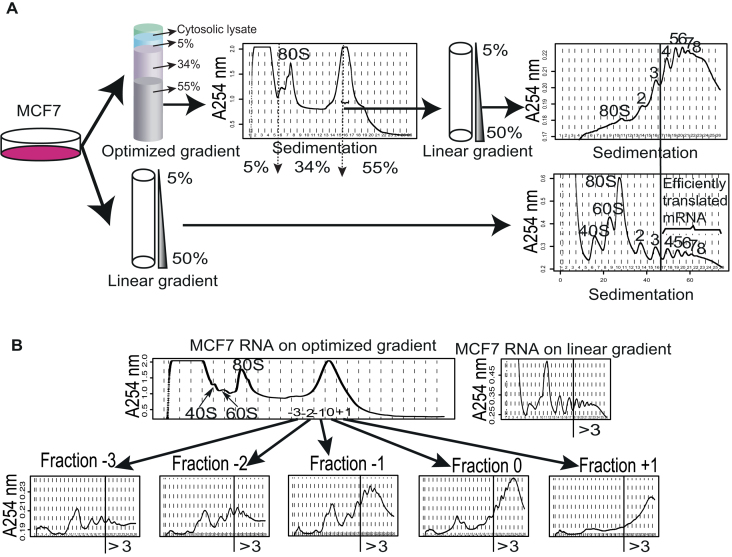
Figure 2.
Identification of fractions from the optimized non-linear sucrose gradient that reproducibly contain mRNA associated with >3 ribosomes. (A) Cytosolic lysates from MCF7 cells were sedimented on an optimized non-linear sucrose gradient or a linear sucrose gradient in parallel. Subsequently, the nature of the large high peak from the optimized non-linear sucrose gradient was explored using a linear sucrose gradient which indicated a strong enrichment of mRNA associated with >3 ribosomes (compare upper right tracing to lower right tracing). The positions of the sucrose layers for the optimized non-linear gradient are indicated by arrows in the tracing. (B) Multiple fractions from the optimized non-linear gradient were evaluated on linear gradients similar to (A). The fraction at the center of the peak was designated 0 and fractions towards the right or left of fraction 0 were assigned positive or negative numbers, respectively. Shown is also the profile from a linear gradient processed in parallel. The vertical lines indicate fractions containing >3 ribosomes (designated ‘>3’ in figure).
Reproducible isolation of efficiently translated mRNA using the optimized gradient
We noticed that the width of the peak at the interface between the 34% and 55% sucrose layers increased with the time between preparation of the gradient and centrifugation of the sample, consistent with that a local gradient is formed at the interface between the 34% and 55% sucrose solutions. We therefore standardized the time between preparation of the gradient and layering of the sample to 2 h (which is sufficient to prepare lysates to load onto the gradient). We next sought to explore the nature of the fractions surrounding the large peak at the 34% and 55% sucrose interface to assess whether this peak indeed is enriched for mRNA associated with >3 ribosomes. To this end, we used a large batch of cells (12 plates [15 cm] of MCF7 cells) and sedimented their lysate on the optimized non-linear gradient. The major area of the peak between the 34% and the 55% sucrose was spread across 3 fractions (Figure 2A). We designated the fraction containing the center of the peak as 0 and those toward the 34% sucrose as −1 (etc.) while those toward the 55% sucrose as +1 (etc.; Figure 2B). The vast amount of RNA used as input allowed us to collect five fractions surrounding the peak between the 34% and the 55% sucrose solutions and dilute these 7-fold to enable a run of each fraction separately on the standard 5–50% linear sucrose gradient (diluted to prevent samples from sinking into the linear gradient; Figure 2A). In addition, we generated a new cytosolic lysate (from 2 additional plates of MCF7 cells cultured in parallel with those used for the non-linear gradient) and loaded this on a linear gradient as control. As shown in Figure 2B, fraction 0 and +1 are strongly enriched for mRNA with >3 ribosomes while fraction −1, although still enriched for efficiently translated mRNA, show relatively more mRNA associated with <3 ribosomes. This pattern was observed in two additional independent experiments (Supplementary Figure S1). We therefore concluded that collection of fractions containing the center of the peak (i.e. 0) and the one further towards the bottom of the tube (i.e. +1) allows for isolation of a pool of mRNA that is strongly enriched for those associated with >3 ribosomes. This allows collection of efficiently translated RNA in just two fractions. After pooling these fractions (i.e. 0 and +1) and splitting them into one sample for processing and one sample as backup, the procedure allows for downstream processing of a single tube containing efficiently translated mRNA.
The optimized non-linear gradient allows for consistent isolation of high quality RNA
A concern is that application of the optimized non-linear gradient leads to a reduction in the amount of isolated efficiently translated mRNA. To assess this, and to validate that high quality RNA can be reproducibly obtained, we used two cell lines that differ in their p53 status (HCT-116 p53+/+ and HCT-116 p53−/−) (Supplementary Figure S2A). We serum-starved the cells (16 h) as translation is commonly modulated during cellular stress and this setup would allow us to assess whether p53 status affected such responses. Serum starvation had a comparable effect on global translation for the two cell lines as judged by a similar reduction in polysome-associated RNA coupled with an increase in 80S and free ribosome subunits (Supplementary Figure S2B). To allow for a rigid comparison between the standard linear gradient and the optimized non-linear gradient, we prepared cytosolic lysates from six plates (15 cm) from each cell type and divided the lysates equally between the optimized non-linear gradient and the linear gradient. We then collected fractions corresponding to mRNA associated with >3 ribosomes from the linear gradient and peak 0 and +1 from the optimized non-linear gradient. We repeated the experiment four times and measured the RNA quantity. The optimized non-linear gradient and linear gradient allowed for isolation of similar amounts of efficiently translated mRNA (Figure 3A). Assessment of the quality of extracted mRNA by RNA Integrity Number (RIN; Agilent Bioanalyzer) showed that both gradients allow for consistent isolation of essentially perfectly intact RNA (RIN > 9.5; Figure 3B). Thus, the optimized non-linear gradient and linear gradient show similar performance in terms of quality and quantity of the isolated efficiently translated RNA.
Figure 3.
Assessing the quality and quantity of RNA isolated with linear or optimized non-linear gradients. (A) Mean (error bars correspond to standard deviations) amount of RNA (log2 ng) from quadruplicate experiments using linear or optimized non-linear gradients for both HCT-116 p53+/+ and HCT-116 p53−/− cells (the cytosolic lysate obtained from three 15 cm plates of 80% confluent cells was applied to each gradient). (B) Obtained RNA integrity numbers (RIN) for samples processed using linear and optimized non-linear gradients (same experiment as A).
The optimized non-linear gradient and the linear gradient generate similar data on translatomes
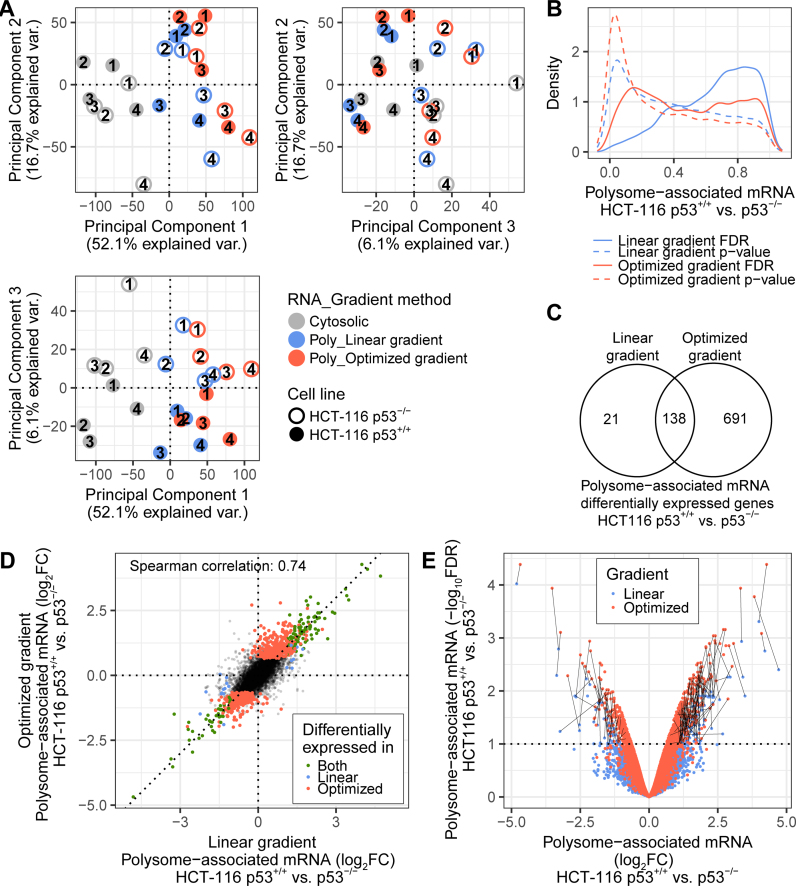
Ideally, data on translatomes from the optimized non-linear gradient should be similar to those obtained using the standard approach employing linear gradients. To evaluate this, we determined translatomes from serum starved (16 h) HCT-116 cells with and without p53 (as described above). To mimic a situation observed in tissue samples or primary cells, where obtained RNA amounts are often limited, we employed smart-seq2 developed for single-cell RNAseq (29) and used 10 ng of RNA as input. Smart-seq2 libraries were prepared using efficiently translated RNA obtained using linear gradients and optimized non-linear gradients; and cytosolic RNA as input. Following sequencing and data processing, we used principal component analysis to explore the major sources of variation in the data set. Absence of a highly ranked PCA component (by percentage of explained variance) showing differences between techniques used to prepare efficiently translated RNA would indicate that methods are comparable. As expected during analysis of polysome-profiling data, the first component capturing the main source of variance (52.1%) relates to RNA source such that cytosolic mRNA samples separate from polysome-associated mRNA samples. The second (16.7% of the variance) and third (6.1% of the variance) principal components separate samples according to replicate (notably the need to adjust for run-bias during analysis was recently identified (12)) and p53 status, respectively (Figure 4A). Thus, the absence of a highly ranked component showing differences between the optimized non-linear gradient and the linear gradient is consistent with that gradients produce similar data on translatomes. To further substantiate this observation, we compared gene expression between HCT-116 p53+/+ and p53−/− cells using data from polysome-associated mRNA isolated from the optimized non-linear sucrose gradient or the linear gradient separately. At an FDR threshold of 0.1, the optimized gradient approach was associated with more differential expression as compared to the linear gradient method (Figure 4B). Moreover, nearly all mRNAs identified by the linear gradient were also identified by the optimized non-linear gradient (Figure 4C). Furthermore, the fold-changes obtained between HCT-116 p53+/+ and p53−/− cells when applying the two approaches showed high correlation (Spearman coefficient: 0.74, Figure 4D). Consistently, the difference between the two techniques was the obtained FDRs rather than the fold changes such that lower FDRs were obtained when using the optimized non-linear gradient (Figure 4E). This is consistent with that the optimized non-linear gradient generated data of lower variance as compared to the linear gradient. We therefore conclude that the optimized non-linear gradient will provide similar data on translatomes as compared to the standard linear gradient.
Figure 4.
Linear and optimized non-linear gradients produce similar data on translatomes. (A) A projection of all samples in the three first components of the principal component analysis. Replicate numbers are indicated within circles. (B) For both gradient methods, an analysis of differential polysome-associated mRNA levels between HCT-116 p53+/+ and HCT-116 p53−/− cell lines was performed. Shown are density plots of p-values (dashed lines) and false discovery rates (plain lines) for both gradient methods. (C) Venn diagram showing the overlap of genes identified using both gradient methods (mRNAs with an FDR<0.1 were considered differentially associated with polysomes). (D) Scatter plot showing log2 fold changes using data from the optimized non-linear gradient vs. the linear gradient. Colors correspond to genes differentially polysome-associated and identified by both gradient methods (green); the linear gradient only (blue); the optimized non-linear gradient only (red); or none of the methods (black). (E) Volcano plot for each gradient method (linear gradient in blue, optimized non-linear gradient in red). For genes considered significant by both methods, black lines join the data per mRNA. Poly = Polysome-associated mRNA; FDR = False Discovery Rate; FC = Fold Change.
P53 status affects gene expression via multiple mechanisms including translational buffering
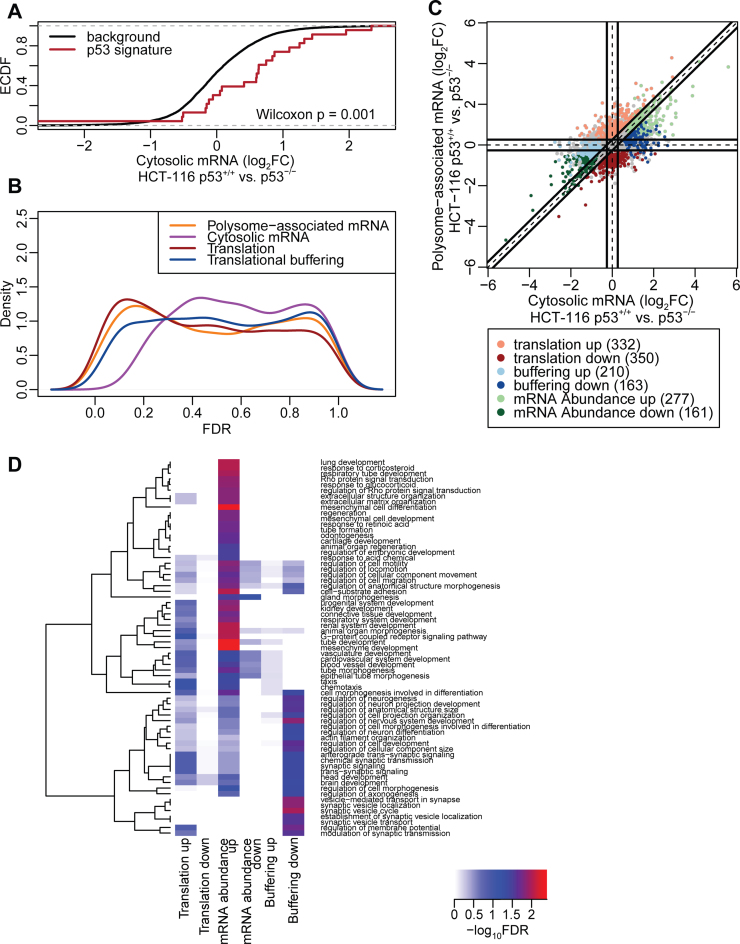
Next we sought to determine how p53 status affects gene expression under serum starvation at multiple levels including mRNA abundance (i.e. congruent changes in cytosolic and polysome-associated mRNA levels—which is consistent with altered transcription or mRNA stability); and translational efficiency affecting protein levels (i.e. a larger change in polysome-associated mRNA as compared to cytosolic mRNA) or buffering (wherein polysome-associated mRNA levels remain largely unchanged despite altered cytosolic mRNA levels). Translational buffering is distinct in the sense that it acts to maintain protein levels constant while the former two modes of regulation will influence protein levels. The recently developed anota2seq algorithm efficiently identifies and separates these three regulatory modes and was employed for analysis. As expected, genes whose transcription was previously identified as sensitive to p53 activity (37) showed increased cytosolic mRNA levels in p53+/+ as compared to p53−/− cells (Figure 5A). Interestingly, changes in polysome-associated mRNA quantified from the optimized non-linear sucrose gradient were more abundant, as judged by the number of mRNAs showing low FDRs for p53-status dependent expression, compared to changes in cytosolic mRNA levels (Figure 5B). Consistently, more mRNAs showed modulation in translational efficiency affecting protein levels (682 mRNAs) as compared to changes in mRNA abundance (438 mRNAs; Figure 5C; Supplementary Table S1). Intriguingly many changes in cytosolic mRNA levels were buffered at the level of mRNA translation (373 mRNAs; Figure 5C; Supplementary Table S1). Thus, the optimized non-linear gradient in combination with Smart-seq2 and anota2seq analysis efficiently interrogates the impact on p53 status on multiple gene expression programs and highlights a key role of mRNA translation in regulating gene expression under cellular stress induced by serum starvation. To gain insights into whether different modes of regulation target distinct cellular functions (i.e. a division of labor pattern) or whether there is a large overlap with regards to which functions are affected, we searched for an over-representation of genes with shared functions as defined by the gene ontology consortium (38). Indeed, mRNAs that showed congruently increased cytosolic and polysome-associated mRNA levels in p53+/+ cells were enriched for functions including those related to extracellular matrix, development and migration. Moreover, genes whose cytosolic mRNA levels were buffered at the level of translation were enriched for neural related functions (Figure 5D; Supplementary Table S2). Thus, there appears to be selectivity with regards to which cellular functions are targeted by which gene expression mechanism (Figure 5D).
Figure 5.
Analysis of the p53 translatome of HCT-116 cells using an optimized non-linear gradient. (A) Cumulative distributions of fold-changes for cytosolic mRNA between HCT-116 p53+/+ and HCT-116 p53−/− cells for all genes (background) and a set of genes described as transcriptionally induced by p53. (B) Densities of FDRs for analysis of polysome-associated mRNA, cytosolic mRNA and changes in translational efficiency affecting protein levels or buffering (HCT-116 p53+/+ versus HCT-116 p53−/− cells). (C) Scatter plot of polysome-associated mRNA log2 fold changes versus cytosolic mRNA log2 fold changes. Regulated are indicated. (D) Heatmap showing GO term enrichment among genes regulated via abundance (i.e. congruent changes in cytosolic and polysome-associated mRNA) or genes whose translational efficiency affects protein levels or buffering. –log10 FDR of the hypergeometric tests are colour-coded in the heatmap and an unsupervised clustering was applied to GO terms. FDR = False Discovery Rate; FC = Fold Change.
Polysome-profiling of bio-banked breast cancer tissues using optimized non-linear gradients and smartSeq2
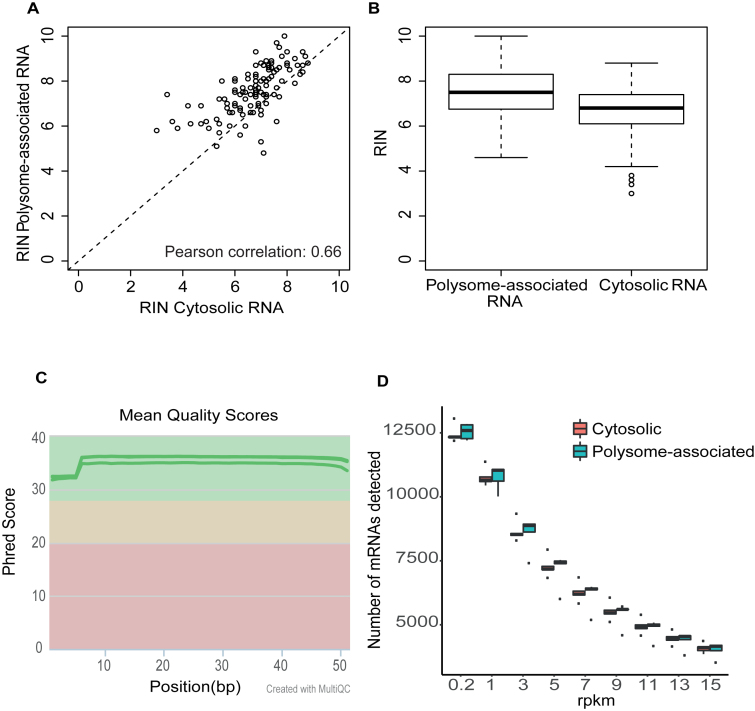
To comprehensively evaluate the non-linear sucrose gradient for isolation of efficiently translated mRNA from biobanked tissue samples, we identified a cohort of 161 breast cancer tissues and applied the optimized non-linear gradient to isolate their efficiently translated mRNA. The sizes of the tissue samples were not recorded in the biobank but estimated to vary between <30 to ∼100 mg. We could consistently identify the 0 and +1 fractions in the profiles obtained from the non-linear gradient (Supplementary Figure S3). These fractions were pooled and split into one assay- and one backup-sample. The assay-sample was then subjected to RNA extraction and the quality of the isolated cytosolic and polysome-associated RNA was assessed. There was a correlation between the RINs of the two RNA pools (Pearson correlation = 0.66, Figure 6A). RINs for efficiently translated mRNA were higher (i.e. indicating more intact RNA) as compared to those observed in cytosolic input samples (Figure 6B). This shows that a low RIN for the pool of efficiently translated RNA is not caused by the isolation technique, but rather by lower initial RNA quality in those tissue samples. Many of these samples generated very low RNA amounts (<1 ng/μl and <10 ng in total) and hence application of protocols adopted for single cell sequencing would be essential to generate translatomes. Indeed, application of Smart-seq2 to a subset of these samples generated RNAseq libraries amendable for RNAseq (Supplementary Figure S3). To ensure that these libraries indeed are of sufficient quality to comprehensively quantify translatomes we performed RNA sequencing of efficiently translated and cytosolic RNA from the set of five breast cancer tissues from Supplementary Figure S3 (Supplementary Figure S4). This revealed high technical quality of the resulting sequencing data (Figure 6C). Moreover, we obtained high coverage of the breast cancer translatomes as judged by that mRNAs from >12 000 genes showed an RPKM >0.2 (which represent a lower limit of detection (32)) and >10 000 genes had an RPKM >1 (Figure 6D). Thus, combining the optimized non-linear gradient with single cell RNAseq protocols allows for comprehensive exploration of translatomes in small tissue samples from biobanks.
Figure 6.
The optimized non-linear gradients isolates efficiently translated mRNA from biobank tissues with an RNA integrity reflecting the starting material sufficient for generation of high coverage translatomes. (A) Scatter plot of RIN obtained from polysome-associated mRNA (using the optimized non-linear gradient) and the corresponding cytosolic mRNA. (B) Boxplot of RIN for polysome-associated and cytosolic mRNA (same samples as in (A). (C) Phred scores across the length of the RNAseq read for sequencing libraries obtained using efficiently translated and cytosolic mRNA from 5 breast cancer tissues (i.e. same as Supplementary Figure S3) as input material for smartSeq2. (D) Boxplots of numbers of genes whose transcribed mRNAs are detected at different RPKM thresholds among cytosolic mRNA (N = 5) and efficiently translated mRNA (N = 5) isolated from breast cancer samples (i.e. same as Supplementary Figure S3).
DISCUSSION
Many studies show that translational control can have dramatic effects on the proteome (22). This mode of regulation is prevalent following e.g. various stresses and modulation of key cellular pathways such as the mTOR pathway (18). Given the pivotal role of cell signaling and stress in human diseases, the translatome is expected to be modulated under a range of pathological conditions. Yet, translatomes are vastly understudied as compared to transcriptomes. This includes human tumors where such data are, to our knowledge, lacking (18). This likely reflects that while genome wide approaches for measurements of transcriptomes are relatively easy to apply, methods for studying translatomes have not scaled well to larger sample collections. Although polysome-profiling is the preferred method for studies of changes in translational efficiency as compared to ribosome-profiling when the precise location of ribosomes is of lesser importance (19), both these techniques are highly laborious and therefore challenging to apply in large studies. Here, we focused on one aspect of polysome-profiling which makes it inconvenient, namely that the pool of efficiently translated RNA is obtained in a large volume distributed across multiple fractions that need to be collected and pooled (28). When studying translatomes in small tissue samples or primary cells (39,40), this poses a major limitation by leading to extensive dilution of the efficiently translated mRNA, which may cause sample loss and technical variability that may undermine reproducible quantification of the translatome. We addressed these issues by introducing an optimized non-linear sucrose gradient, which allows for consistent isolation of efficiently translated mRNA associated with >3 ribosomes in only one or two fractions. The ability to isolate efficiently translated mRNA enriched for mRNA associated with >3 ribosomes is important to capture shifts in translational efficiency that are largely contained within polysomes. Changes in translation of such genes will be more difficult to detect using e.g. pull-down or pelleting approaches, as association with one ribosome is sufficient for isolation of such RNA. As a result, those approaches will likely be biased towards identification of mRNAs showing on-off regulation. By applying these approaches we assessed the effect of p53 status under cellular stress induced by serum starvation. It is important to consider that although the cells we used differ in p53 status they likely exhibit additional genetic and epigenetic differences accumulated during their in vitro culturing. As a result, the changes in gene expression that we observe may not be directly linked to p53. Nevertheless, consistent with previous literature, we identify a prominent role for changes in translational efficiency affecting protein levels depending on p53 status/activity (41–43). In addition, we highlight translational buffering as a prevalent mechanism that will further decouple mRNA levels from protein levels. Although translational buffering was initially identified in yeast (44–46), it has been observed in a limited set of mammalian models (10,47). This raises the possibility that translational buffering is an overlooked mode of regulation that the cell can utilize to maintain protein levels constant despite fluctuations in mRNA levels. Such a role of post-transcriptional control of gene expression was previously suggested from studies showing that protein levels are more consistent across species as compared to mRNA levels (48,49). Thus, translational buffering may be one underlying factor for this observation, yet the mechanisms and scope of translational buffering remain obscure.
We also introduced single-cell sequencing protocols for quantification of efficiently translated mRNA, as the amounts of purified RNA can be scarce. Similar to when obtaining translatomes from limited numbers of primary cells (40), this is essential to allow for studies of translatomes using biobank tissue samples as the amount of RNA isolated from such samples appears to be insufficient for standard RNAseq protocols. Although the presented approach alleviates some of the issues with polysome-profiling it does not circumvent the need of sucrose gradients which is still a limiting factor when planning large studies of translatomes. Nevertheless, this approach made it possible for us to process >150 tissue samples. Thus, polysome-profiling can now be applied to collections of small tissue samples or primary cells.
AVAILABILITY
The dataset has been deposited at the gene expression omnibus (GSE99909).
Supplementary Material
ACKNOWLEDGEMENTS
We acknowledge support from Science for Life Laboratory, the Knut and Alice Wallenberg Foundation, the National Genomics Infrastructure funded by the Swedish Research Council, and Uppsala Multidisciplinary Center for Advanced Computational Science for assistance with massively parallel sequencing and access to the UPPMAX computational infrastructure. We acknowledge the A.C. Camargo Cancer Center Biobank for the support in the storage and evaluation of human samples.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR online.
FUNDING
Swedish Research Council; Swedish Childhood Cancer Foundation; Swedish Cancer Society; Cancer Society in Stockholm; Wallenberg Academy Fellows Program; STRATCAN (to O.L.); Swedish Childhood Cancer Foundation (to L.M.); STINT to initiate international collaboration (to O.L. and G.H.); FAPESP [2014/15550-9 to GNMH, 2013/03315-2 to FCSL, 2014/04513-5 to HMB, 2015/15451-3 to MR]. Funding for open access charge: Swedish Research Council.
Conflict of interest statement. None declared.
REFERENCES
- 1. Bava F.A., Eliscovich C., Ferreira P.G., Minana B., Ben-Dov C., Guigo R., Valcarcel J., Mendez R.. CPEB1 coordinates alternative 3′-UTR formation with translational regulation. Nature. 2013; 495:121–125. [DOI] [PubMed] [Google Scholar]
- 2. Rousseau D., Kaspar R., Rosenwald I., Gehrke L., Sonenberg N.. Translation initiation of ornithine decarboxylase and nucleocytoplasmic transport of cyclin D1 mRNA are increased in cells overexpressing eukaryotic initiation factor 4E. Proc. Natl. Acad. Sci. U.S.A. 1996; 93:1065–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Holt C.E., Schuman E.M.. The central dogma decentralized: new perspectives on RNA function and local translation in neurons. Neuron. 2013; 80:648–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lindstein T., June C.H., Ledbetter J.A., Stella G., Thompson C.B.. Regulation of lymphokine messenger RNA stability by a surface-mediated T cell activation pathway. Science. 1989; 244:339–343. [DOI] [PubMed] [Google Scholar]
- 5. Deschênes-Simard X., Lessard F., Gaumont-Leclerc M.F., Bardeesy N., Ferbeyre G.. Cellular senescence and protein degradation: Breaking down cancer. Cell Cycle. 2014; 13:1840–1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Roux P.P., Topisirovic I.. Regulation of mRNA translation by signaling pathways. Cold Spring Harb. Perspect. Biol. 2012; 4:a012252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Morita M., Gravel S.P., Hulea L., Larsson O., Pollak M., St-Pierre J., Topisirovic I.. mTOR coordinates protein synthesis, mitochondrial activity and proliferation. Cell cycle. 2015; 14:473–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kristensen A.R., Gsponer J., Foster L.J.. Protein synthesis rate is the predominant regulator of protein expression during differentiation. Mol. Syst. Biol. 2013; 9:689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Schwanhausser B., Busse D., Li N., Dittmar G., Schuchhardt J., Wolf J., Chen W., Selbach M.. Global quantification of mammalian gene expression control. Nature. 2011; 473:337–342. [DOI] [PubMed] [Google Scholar]
- 10. Jovanovic M., Rooney M.S., Mertins P., Przybylski D., Chevrier N., Satija R., Rodriguez E.H., Fields A.P., Schwartz S., Raychowdhury R. et al. . Immunogenetics. Dynamic profiling of the protein life cycle in response to pathogens. Science. 2015; 347:1259038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Liu Y., Beyer A., Aebersold R.. On the dependency of cellular protein levels on mRNA abundance. Cell. 2016; 165:535–550. [DOI] [PubMed] [Google Scholar]
- 12. Parker M.W., Rossi D., Peterson M., Smith K., Sikstrom K., White E.S., Connett J.E., Henke C.A., Larsson O., Bitterman P.B.. Fibrotic extracellular matrix activates a profibrotic positive feedback loop. J. Clin. Invest. 2014; 124:1622–1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bhat M., Robichaud N., Hulea L., Sonenberg N., Pelletier J., Topisirovic I.. Targeting the translation machinery in cancer. Nat. Rev. Drug Discov. 2015; 14:261–278. [DOI] [PubMed] [Google Scholar]
- 14. Roffe M., Beraldo F.H., Bester R., Nunziante M., Bach C., Mancini G., Gilch S., Vorberg I., Castilho B.A., Martins V.R. et al. . Prion protein interaction with stress-inducible protein 1 enhances neuronal protein synthesis via mTOR. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:13147–13152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ma T., Trinh M.A., Wexler A.J., Bourbon C., Gatti E., Pierre P., Cavener D.R., Klann E.. Suppression of eIF2alpha kinases alleviates Alzheimer's disease-related plasticity and memory deficits. Nat. Neurosci. 2013; 16:1299–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Moreno J.A., Radford H., Peretti D., Steinert J.R., Verity N., Martin M.G., Halliday M., Morgan J., Dinsdale D., Ortori C.A. et al. . Sustained translational repression by eIF2alpha-P mediates prion neurodegeneration. Nature. 2012; 485:507–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bellato H.M., Hajj G.N.. Translational control by eIF2alpha in neurons: Beyond the stress response. Cytoskeleton (Hoboken, N.J.). 2016; 73:551–565. [DOI] [PubMed] [Google Scholar]
- 18. Piccirillo C.A., Bjur E., Topisirovic I., Sonenberg N., Larsson O.. Translational control of immune responses: from transcripts to translatomes. Nat. Immunol. 2014; 15:503–511. [DOI] [PubMed] [Google Scholar]
- 19. Gandin V., Masvidal L., Hulea L., Gravel S.P., Cargnello M., McLaughlan S., Cai Y., Balanathan P., Morita M., Rajakumar A. et al. . nanoCAGE reveals 5′ UTR features that define specific modes of translation of functionally related MTOR-sensitive mRNAs. Genome Res. 2016; 26:636–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hershey J.W., Sonenberg N., Mathews M.B.. Principles of translational control: an overview. Cold Spring Harb. Perspect. Biol. 2012; 4:a011528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Leprivier G., Remke M., Rotblat B., Dubuc A., Mateo A.R., Kool M., Agnihotri S., El-Naggar A., Yu B., Somasekharan S.P. et al. . The eEF2 kinase confers resistance to nutrient deprivation by blocking translation elongation. Cell. 2013; 153:1064–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ruggero D. Translational control in cancer etiology. Cold Spring Harb. Perspect. Biol. 2013; 5:a012336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ingolia N.T., Ghaemmaghami S., Newman J.R., Weissman J.S.. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009; 324:218–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Andreev D.E., O’Connor P.B., Loughran G., Dmitriev S.E., Baranov P.V., Shatsky I.N.. Insights into the mechanisms of eukaryotic translation gained with ribosome profiling. Nucleic Acids Res. 2017; 45:513–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ingolia N.T. Ribosome footprint profiling of translation throughout the genome. Cell. 2016; 165:22–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Masvidal L., Hulea L., Furic L., Topisirovic I., Larsson O.. mTOR-sensitive translation: cleared fog reveals more trees. RNA Biol. 2017; 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Andreev D.E., O’Connor P.B., Fahey C., Kenny E.M., Terenin I.M., Dmitriev S.E., Cormican P., Morris D.W., Shatsky I.N., Baranov P.V.. Translation of 5′ leaders is pervasive in genes resistant to eIF2 repression. eLife. 2015; 4:e03971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gandin V., Sikstrom K., Alain T., Morita M., McLaughlan S., Larsson O., Topisirovic I.. Polysome fractionation and analysis of mammalian translatomes on a genome-wide scale. J. Visual. Exp.: JoVE. 2014; doi:10.3791/51455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Picelli S., Faridani O.R., Bjorklund A.K., Winberg G., Sagasser S., Sandberg R.. Full-length RNA-seq from single cells using Smart-seq2. Nat. Protoc. 2014; 9:171–181. [DOI] [PubMed] [Google Scholar]
- 30. Ewels P., Magnusson M., Lundin S., Kaller M.. MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics. 2016; 32:3047–3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Langmead B., Trapnell C., Pop M., Salzberg S.L.. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009; 10:R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ramskold D., Wang E.T., Burge C.B., Sandberg R.. An abundance of ubiquitously expressed genes revealed by tissue transcriptome sequence data. PLoS Comput. Biol. 2009; 5:e1000598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Robinson M.D., Oshlack A.. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010; 11:R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., Smyth G.K.. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015; 43:e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wright G.W., Simon R.M.. A random variance model for detection of differential gene expression in small microarray experiments. Bioinformatics. 2003; 19:2448–2455. [DOI] [PubMed] [Google Scholar]
- 36. Benjamini Y., Hochberg Y.. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. 1995; 57:289–300. [Google Scholar]
- 37. Mills J.R., Malina A., Lee T., Di Paola D., Larsson O., Miething C., Grosse F., Tang H., Zannis-Hadjopoulos M., Lowe S.W. et al. . RNAi screening uncovers Dhx9 as a modifier of ABT-737 resistance in an Emu-myc/Bcl-2 mouse model. Blood. 2013; 121:3402–3412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Gene Ontology Consortium Gene ontology consortium: going forward. Nucleic Acids Res. 2015; 43:D1049–D1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bjur E., Larsson O., Yurchenko E., Zheng L., Gandin V., Topisirovic I., Li S., Wagner C.R., Sonenberg N., Piccirillo C.A.. Distinct translational control in CD4+ T cell subsets. PLoS Genet. 2013; 9:e1003494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Mao Y., van Hoef V., Zhang X., Wennerberg E., Lorent J., Witt K., Masvidal L., Liang S., Murray S., Larsson O. et al. . IL-15 activates mTOR and primes stress-activated gene expression leading to prolonged antitumor capacity of NK cells. Blood. 2016; 128:1475–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Loayza-Puch F., Drost J., Rooijers K., Lopes R., Elkon R., Agami R.. p53 induces transcriptional and translational programs to suppress cell proliferation and growth. Genome Biol. 2013; 14:R32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zaccara S., Tebaldi T., Pederiva C., Ciribilli Y., Bisio A., Inga A.. p53-directed translational control can shape and expand the universe of p53 target genes. Cell Death Differ. 2014; 21:1522–1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Marcel V., Catez F., Diaz J.J.. p53, a translational regulator: contribution to its tumour-suppressor activity. Oncogene. 2015; 34:5513–5523. [DOI] [PubMed] [Google Scholar]
- 44. McManus C.J., May G.E., Spealman P., Shteyman A.. Ribosome profiling reveals post-transcriptional buffering of divergent gene expression in yeast. Genome Res. 2014; 24:422–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Artieri C.G., Fraser H.B.. Evolution at two levels of gene expression in yeast. Genome Res. 2014; 24:411–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Albert F.W., Muzzey D., Weissman J.S., Kruglyak L.. Genetic influences on translation in yeast. PLoS Genet. 2014; 10:e1004692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Cenik C., Cenik E.S., Byeon G.W., Grubert F., Candille S.I., Spacek D., Alsallakh B., Tilgner H., Araya C.L., Tang H. et al. . Integrative analysis of RNA, translation, and protein levels reveals distinct regulatory variation across humans. Genome Res. 2015; 25:1610–1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Laurent J.M., Vogel C., Kwon T., Craig S.A., Boutz D.R., Huse H.K., Nozue K., Walia H., Whiteley M., Ronald P.C. et al. . Protein abundances are more conserved than mRNA abundances across diverse taxa. Proteomics. 2010; 10:4209–4212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Schrimpf S.P., Weiss M., Reiter L., Ahrens C.H., Jovanovic M., Malmstrom J., Brunner E., Mohanty S., Lercher M.J., Hunziker P.E. et al. . Comparative functional analysis of the Caenorhabditis elegans and Drosophila melanogaster proteomes. PLoS Biol. 2009; 7:e48. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.