Abstract
The RcsCDB phosphorelay system controls an extremely large regulon in Enterobacteriaceae that involves processes such as biofilm formation, flagella production, synthesis of extracellular capsules and cell division. Therefore, fine-tuning of this system is essential for virulence in pathogenic microorganisms of this group. The final master effector of the RcsCDB system is the response regulator (RR) RcsB, which activates or represses multiple genes by binding to different promoter regions. This regulatory activity of RcsB can be done alone or in combination with additional transcriptional factors in phosphorylated or dephosphorylated states. The capacity of RcsB to interact with multiple promoters and partners, either dephosphorylated or phosphorylated, suggests an extremely conformational dynamism for this RR. To shed light on the activation mechanism of RcsB and its implication on promoter recognition, we solved the crystal structure of full-length RcsB from Salmonella enterica serovar Typhimurium in the presence and absence of a phosphomimetic molecule BeF3−. These two novel structures have guided an extensive site-directed mutagenesis study at the structural and functional level that confirms RcsB conformational plasticity and dynamism. Our data allowed us to propose a β5-T switch mechanism where phosphorylation is coupled to alternative DNA binding ways and which highlights the conformational dynamism of RcsB to be so pleiotropic.
INTRODUCTION
A few bacterial regulatory proteins (CRP, FNR, IHF, FIS, ArcA, NarL and Lrp) modulate the expression of a large number of genes, being identified as global regulators (1,2). These global regulators include the response regulators (RRs) NarL and ArcA, which regulate more than 100 genes (123 for NarL and 175 for ArcA) (3). These two RR are functionally linked to sensor histidine kinases comprising the two-component system NarL/NarX and the phosphorelay system ArcA/ArcB, respectively.
The RcsCDB phosphorelay signaling system emerged as an unexpectedly complex regulatory system (4) that controls a large regulon with ∼90 genes involved in key processes related to virulence such as biofilm formation, flagella production and synthesis of extracellular capsules (5,6). The RcsCDB system is conserved in Enterobacteriaceae and responds to high osmolarity and envelope stress, among many other signals. This system is composed basically by three proteins: the hybrid histidine kinase RcsC, the phosphotransferase RcsD and the RR protein RcsB. RcsC is the sensor protein that autophosphorylates following signal detection to further transfer the phosphoryl group to RcsD, which in turn phosphorylates RcsB (4,5), which then regulates the expression of target genes.
Recent data, however, show that the Rcs phosphorelay is modulated by auxiliary proteins with capacity to influence signal detection and response. Thus, the outer membrane protein RcsF (7) and the inner membrane protein IgaA (6) modulate signal detection through RcsC (8). On the other hand, RcsB can heterodimerize with transcriptional factors to influence the final response, either in a phosphorylation-dependent manner with RcsA or phosphorylation independent with BglJ, GadE, MatA, DctR and RflM (9–14).
Therefore, RcsB is widely accepted as the master piece of the RcsCDB regulatory system controlling multiple genes by different mechanisms. Furthermore, RcsB acts as a negative or positive regulator depending on the promoter region. For example, in Escherichia coli and the intracellular bacterial pathogen Salmonella enterica serovar Typhimurium (S. Typhimurium), RcsB inhibits transcription of the flhDC flagellar master operon by binding to the promoter region P1flhDC (15) and, inversely, activates the expression of rprA, a small regulatory RNA that controls exopolysaccharide production required for biofilm formation (16). RcsB also regulates positively the operon wca—also named cps and involved in synthesis of the colonic acid capsule—through heterodimerization with the unstable co-regulator RcsA (10,17).
These multiple ways of action and varied partner proteins anticipate that RcsB should be a protein with an intrinsic high conformational dynamism and plasticity. However, protein primary sequence and structural analyses indicate that RcsB is a classical RR of the NarL/FixJ subfamily containing two domains: an N-terminal regulatory domain named receiver domain (REC), which phosphorylates a conserved catalytic Asp residue (D56 for S. Typhimurium RcsB) in the active center; and a C-terminal effector region acting as a DNA-binding domain (DBD) (18). The phosphorylated Asp at the loop connecting β3 with α3 (Lβ3α3) in the REC domains is stabilized via interactions with a catalytic Thr/Ser residue in β4, a catalytic Lys in Lβ5α5 and an essential Mg2+ ion coordinated to an Asp/Glu residue in Lβ1α1 (19). The structural characterization by X-ray crystallography of the E. coli RcsB REC domain in its unphosphorylated conformation (PDB ID: 5I4C) (20) reveals a prototypical (βα)5 organization for this sensor domain. Meanwhile, the Nuclear magnetic resonance (NMR) structure of Erwinia amylovora RcsB DBD domain (PDB ID: 1P4W) shows a prototypical helix-turn-helix (HTH) conformation (21). However, it is extremely intriguing how RcsB controls its different DNA-binding capacities in a phosphorylated or dephosphoprylated state.
To shed light in this process, we solved the structure of full length S. Typhimurium RcsB in the absence or presence of the phosphomimetic molecule BeF3− (22). These two structures provided snapshots for the RcsB conformation competent to interact with DNA in a tail-to-tail arrangement in its phosphorylated state as well as an alternative activated conformation with distinctive features to recognize the DNA, supporting the structural dynamism of this RR. We confirmed these conformations by performing in vitro, in vivo and structural studies on a battery of RcsB variants. These structures, together with the previously reported REC domain from E. coli in its unphosphorylated form (PDB ID: 5I4C) (20), allowed us to propose a switch mechanism of activation for RcsB, named β5-T coupling, which could be extended in particular to other RRs of this subfamily and in general to all the RRs.
MATERIALS AND METHODS
Cloning, mutagenesis, protein expression and purification
Cloning of RcsB from S. Typhimurium (residues 1–210) was done in vector plasmid LIC 1.1 (pETNKI-his3C-LIC-kan), provided by NKI Protein Facility (23). Site-directed mutagenesis at specific residues was performed with the Q5® Site-Directed Mutagenesis Kit (New England Biolabs, ref. E0554S). Primers used are listed in Supplementary Table S1. For protein expression, E. coli strain C43 (DE3) cells containing the appropriate vector were grown in Hyper broth (Molecular Dimensions, UK) till exponential phase (OD600 ∼0.6), then induced with 0.5 mM Isopropyl β-D-1-thiogalactopyranoside (IPTG), incubated overnight at 20°C and further centrifuged and stored at −20°C. Thawed cells were resuspended in 50 mM Tris pH 8.5, 500 mM NaCl, lysed by sonication, centrifuged (20,000 × g, 4°C) and the clarified supernatant was loaded in a His Trap HP column (GE, Healthcare) to perform affinity chromatography. Before RcsB elution, a wash with a buffer containing 50 mM Tris pH 8.5, 500 mM NaCl and 40 mM imidazole was performed to eliminate protein contaminants bound to the column. RcsB eluted in buffer containing 50 mM Tris pH 8.5, 500 mM NaCl and 200 mM imidazole. For the RcsB variant S207C, the monomeric species eluted with 200 mM imidazole while the dimeric specie containing the disulfide bond eluted with 500 mM imidazole. Purified proteins were incubated with PreScission protease at a 1:20 ratio (protein:protease) to remove the 6×His tag and then, purified again by affinity chromatography to separate digested from non-digested protein and from protease. Finally, gel filtration chromatography was run with the digested protein using a buffer containing 50 mM Tris pH 8.5, 150 mM NaCl. Collected fractions containing protein with >95% of purity were concentrated until 13 mg/ml for wild-type (WT) RcsB and 3.6 mg/ml for the dimeric species of the S207C variant. Buffer containing purified proteins were aliquoted, frozen with N2(l) and stored at −80°C in the same buffer. The RcsB variants used in the electrophoretic mobility shift assays (EMSA) experiments and gel filtration were obtained by affinity purification in batch using resin High density nickel (ABT-Agarose Beads Technologies, Spain) and the same buffer used for elution in the affinity chromatography.
Phenotypic assays to monitor activity of the S. Typhimurium RcsB variants
The activity of RcsB was tested by ectopic expression of the respective variants in a S. Typhimurium strain MD4821 (igaA1 rcsB). The allele igaA1 corresponds to an R188H mutation in IgaA causing partial loss-of-function and, as consequence, activation of the RcsCDB system (24,25). Strain MD4821 was constructed from SV4450 (igaA1) (25), which is mucoid on plates due to over-activation of the RcsCDB system. Inactivation of the rcsB gene in SV4450 was performed following the one-step inactivation procedure described by Datsenko and Wanner (26), using the oligonucleotides KO-rcsB-Fw and KO-rcsB-Rv listed in Table S1 of Supplementary Data. Strain MD4821 was subsequently transformed with plasmid pTara:500 (Addgene), which expresses T7 RNA-polymerase, to generate strain MD4822 (igaA1 rcsB pTara:500). This strain was used as recipient of the different series of plasmid LIC 1.1-derivates expressing the respective RcsB variants. This collection of strains producing distinct RcsB variants was tested for the following phenotypes: (i) motility in soft agar plates, as described (27); (ii) production of mucoid colonies on plates (25). Production of the distinct RcsB variants was verified by Coomassie staining of total protein extracts obtained from exponential cultures incubated in the presence of the inducer IPTG (0.1 mM) for 2 h.
Protein crystallization, data collection and processing
Crystallization of RcsB was achieved by the vapor diffusion method using the sitting drop technique. Crystals were obtained incubating RcsB at 13 mg/ml in the absence and presence of BeF3− (5 mM BeSO4, 30 mM NaF and 7 mM MgCl2) in a buffer containing 50 mM Tris pH 8.5 and 150 mM NaCl by mixing 0.3 μl of protein and 0.3 μl of different reservoir solutions. Crystals without BeF3− grew in 1.4 M ammonium sulfate, 8% PEG1000 and 0.1 M Hepes pH 7.5 while crystals with BeF3− grew in 29% Jeffamine ED2003, 0.1 M lithium sulfate and 0.1 M Tris pH 8.5. For data collections, just crystals without BeF3− needed cryoprotection that was achieved by replacing ammonium sulfate for lithium sulfate and addition of 12% of ethylene glycol. Crystals for S207C-RcsB were obtained by incubating the purified dimeric specie of the mutant at 3.6 mg/ml by mixing 0.8 μl of protein and 0.2 μl of different reservoir solutions. Two types of crystals were obtained, cubic ones grown in 30% polyacrylate 2100, 0.1 M sodium malonate and 0.1 M Hepes pH 7.0 and rod ones grown in 16% PEG4000, 0.2 M lithium sulfate and Tris pH 8.5. Cryoprotection for both crystal types was achieved increasing the precipitant (polyacrylate 2100 or PEG4000) to 35%. Diffraction data for RcsB crystals containing BeF3− were collected at Alba Synchrotron (Barcelona, Spain) beamline BL13-XALOC while crystals of RcsB without BeF3− and mutant S207C-RcsB were collected at Diamond light source synchrotron (Oxforshire, UK) beamline I03 and I04–1, respectively. Data integration and reduction was obtained using XDS (28) and Aimless in CCP4i suite (29). Phases were obtained by molecular replacement using Balbes and protein structure was finally obtained with cycles of tracing and refinement using the program Coot (30) and Refmac5 (31), respectively. The Ramachandran plot for refined RcsBcrossed structure showed 95.1% of residues in favored region, 4.8% in allowed region and 0.1% in generously allowed region and for refined RcsBBeF 95.3% of residues were in favored region and 4.7% in allowed region. The Ramachandran plot for S207C–RcsBcrossed structure showed 94.6% of residues in favored and 5.4% in allowed region while for S207C–RcsBAC structure showed 95.5% of residues in favored and 4.5% in allowed region. The figures were produced using PyMOL (http://www.pymol.org) and the movement analysis was performed with the Dyndom program (32).
EMSA experiments
EMSA experiments with WT and mutant RcsB proteins were run in 10% acrylamide gel using 0.5× Tris/Borate/EDTA (TBE) buffer, containing 4 mM MgCl2, either for gel preparation or running buffer. To perform the EMSAs, dsDNA for rcsA and P1flhDC regions was prepared by equimolecular hybridization with their corresponding oligos (Supplementary Table S1) resuspended in water and incubated during 10 min at 85°C followed by cooled down to room temperature overnight. A total of 20 μM of RcsB was incubated with 0.5 μM of hybridized DNA (P1flhDC sequence 5′- CGTCGAATTAGGAAAAATCTTAGGCA -3′ and rcsA sequence 5′- CCTGTTTTACTAAGGTTTATCCGAAAATA -3′) and 0.025 mg/ml of poly d(I-C) (Roche, ref 10108812001) in 50 mM Tris pH 8.5, 50 mM MgCl2 and 10% glycerol in absence/presence of 50 mM acetylphosphate (AcP). When appropriate, 0.3 mM copper phenanthroline (CuPh) was added to the mixture in order to promote disulfide bond formation at mutant S207C. Gels were stored for 3 h in the cold room and pre-run at 150 V during 1 h at 4°C before loading and running the samples. Each gel was stained by the addition of 6 μl of GelRed nucleic acid gel stain (Biotium, Inc. Fremont, CA, USA) in 20 ml of 0.5× TBE buffer and incubated for 30 min. Gel staining was visualized with ultraviolet light.
Synthesis of radioactive AcP
Radioactive [32P]-AcP used in the RcsB phosphorylation assay was obtained incubating for 2 h at room temperature 1.5 U of acetate kinase with 100 μCi/μl of [γ-32P] adenosine triphosphate (3000 Ci/mmol Perkin Elmer) in 2.5 mM Tris pH 8.0, 6 mM potassium acetate and 1 mM MgCl2 buffer. The [32P]-AcP was freed from acetate kinase by filtering using Microcon-10 kDa Centrifugal Filter Unit (GE Healthcare). [32P]-AcP was stored at −20°C.
Phosphorylation experiments with [32P]-AcP
Phosphorylation of RcsB WT and catalytic mutants was performed using 1 mg ml−1 (40 nM) of protein incubated with 12.5 mM [32P]-AcP in a solution containing 50 mM Tris–HCl pH 8.5, 500 mM KCl, 5 mM MgCl2, 20 mM dithiothreitol (DTT) and 1 mM ethylenediaminetetraaceticacid (EDTA). Phosphorylation was stopped at different incubation times, 5, 10, 20, 40 and 60 min adding loading buffer containing 4% sodium dodecyl sulphate (SDS) and 50 mM EDTA. Then, the samples were subjected to SDS-polyacrylamide gel electrophoresis (PAGE) on 15% gel and run at 150 V at room temperature. Phosphorylated proteins were visualized by phosphorimaging using a Fluoro Image Analyzer FLA-5000 (Fuji) and evaluated with the MultiGauge software (Fuji). Data were normalized against the WT. Phosphorylation of additional RcsB mutants involved in DNA recognition was performed using 0.5 mg ml−1 (20 nM) of protein and incubating during 10, 30 and 37 min at 37°C followed by the same protocol described above.
Phosphorylation of RcsB using native PAGE
Phosphorylation of RcsB was achieved incubating 26 μM of protein with a buffer containing 50 mM Tris pH 8.5, 150 mM NaCl, 25 mM AcP and 10 mM MgCl2. At different time points (1, 5, 15, 30 and 60 min) phosphorylation was stopped by the addition of loading buffer containing 50 mM EDTA. The samples were loaded in a 12% native gel, which was run at 150 V in the cold room and stained with coomassie brilliant blue solution.
Phostag experiments
RcsB phosphorylation was assessed by using Phos-tag acrylamide in SDS-PAGE using a 10% gel containing 75 μM Phos-tag and 150 μM MnCl2. RcsB at 87 μM was phosphorylated with 50 mM AcP at 37°C in a buffer containing 50 mM Tris pH 8.5, 150 mM NaCl and 10 mM MgCl2. At different time points (10, 30 and 60 min.) RcsB phosphorylation was stopped by adding SDS-loading buffer and heating 5 min at 95°C. Gel was run at 4°C at 150 V and stained with coomassie brilliant blue solution.
Gel filtration analysis of RcsB phosphorylation
Gel filtration chromatography was carried out in a Superdex 200 increase 10/300GL column (GE Healthcare). The sample (200 μl) containing 200 μg of protein in 50 mM Tris pH 8.5 and 200 mM NaCl buffer was incubated with or without 50 mM AcP and 50 mM of MgCl2 at 37°C for 1 h and was filtered with a 0.45 μm cellulose acetate centrifuge tube filter. Then the sample was individually applied to the column equilibrated with the same buffer and run at a flow rate of 0.7 ml min−1.
RESULTS
The active phosphorylated S. Typhimurium RcsB form is an asymmetric dimer
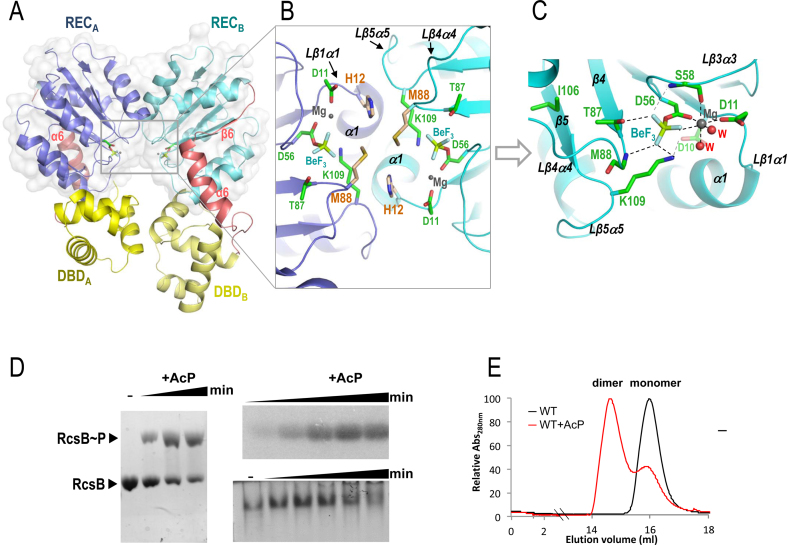
To understand the activation mechanism through phosphorylation in S. Typhimurium RcsB, we solved the crystal structure of the full length protein in the presence of the phosphomimetic BeF3− (RcsBBeF) (Table 1). The asymmetric unit of the crystal contains a dimer of RcsB where each subunit presents a molecule of BeF3− bound to the active center (Figure 1A–C). As it was anticipated by sequence similarities, each REC domain (1–124) of S. Typhimurium RcsB showed the canonical (βα)5 fold (β1–β5 and α1–α5) (Supplementary Figure S1) as the crystal structure of the isolated REC domain of E. coli RcsB (PDB ID: 5I4C; root-mean-square deviation (RMSD) of 0.92 Å for 121 residues), which share 100% sequence identity. Meanwhile, each C-terminal DBD domain (residues 153–210) presents the expected HTH fold (α7–α10) (Figure 1A and Supplementary Figure S1) similar to the NMR structure of the isolated DBD of RcsB from E. amylovora (PDB ID: 1P4W; RMSD 1.13 Å for 59 residues). The REC and DBD domains are connected by a flexible region of 24 residues, designated as REC–DBD connector, which involves an alpha helix (α6; residues 132–144) and two longs loops connecting this helix with the REC (Lα5α6; residues 125–131) and the DBD (Lα6α7; residues 145–152) domains (Figure 1A and Supplementary Figure S1). Interestingly, the Lα5α6 contains a small β6 (residues 128–130) that expands the beta sheet in the REC domain from five to six strands (Figure 1A and Supplementary Figure S2A). Although both REC and DBD domains in the RcsBBeF dimer structure have almost identical structure (RMSD for REC 0.47 Å in 122 residues and for DBD 1.6 Å in 59 residues), the RcsB dimer shows high asymmetry. This asymmetry is due to a different relative orientation of REC and DBD domains in each subunit (Figure 1A). Thus, if the REC domains are superposed, the DBD domains present a relative displacement corresponding to 79° of rotation and −4.2 Å of translation (calculated by Dyndom (32), using the REC–DBD connector as bending region (Supplementary Figure S3). This asymmetry exposes the conformational dynamism of RcsB and the role of the REC–DBD connector as a bending structural element, which is reflected in the low or absent electron density for residues 127–130 and 144–149 in one subunit that precluded their modeling.
Table 1. Crystallograhic data and refinement statistics.
| Crystal containing | RcsBBeF | RcsBcrossed | S207C–RcsBcrossed | S207C–RcsBAC |
|---|---|---|---|---|
| Data collection | ||||
| Space group | P 1 | P412121 | P6322 | P212121 |
| Cell dimensions | 37.4, 54.3 | 108.83, 108.83 | 92.95, 92.95 | 69.62, 74.22 |
| a, b, c (Å) | 55.0 | 306.54 | 120.72 | 72.21 |
| α, β, γ (°) | 62.6, 81.6, 80.0 | 90, 90, 90 | 90, 90, 120 | 90, 90, 90 |
| Resolution (Å)a | 48.7–2.1 | 102.6–2.256 | 80.5–2.6 | 75.2–2.5 |
| (2.16–2.10) | (2.264–2.256) | (2.72–2.60) | (2.6–2.5) | |
| Rmerge (%)* | 7.0 (85.1) | 17.5 (288.2) | 6.1 (124.1) | 6.3 (59.0) |
| Rpim (%)* | 3.1 (37.0) | 3.5 (65.2) | 1.4 (30.7) | 2.7 (29.5) |
| Mean I/σI | 16.4 (2.3) | 17.9 (1.7) | 31.8 (2.6) | 17.7 (2.7) |
| Completeness (%) | 97.8 (96.4) | 100 (92.2) | 96.4 (81.0) | 100 (100) |
| Redundancy | 7.1 (7.2) | 25.4 (20.0) | 20.2 (17.8) | 6.8 (5.9) |
| Refinement | ||||
| Resolution (Å) | 2.1 | 2.3 | 2.6 | 2.5 |
| Reflections | 152127/21487 | 2227190/87626 | 195331/9679 | 96094/14033 |
| total/unique | (12541/1736) | (16942/845) | (17096/958) | (9069/1541) |
| R work/Rfree (%) | 0.21/0.26 | 0.22/0.25 | 0.22/0.25 | 0.22/0.27 |
| No. atoms | ||||
| Protein | 3062 | 9375 | 1566 | 3052 |
| Ligand/ion | 8/2 | 48/50 | 10 | |
| Water | 88 | 111 | 12 | 31 |
| B-factors (Å2) | ||||
| Protein | 49.1 | 52.1 | 77.9 | 59.2 |
| Ligand/ion | 28.1/31.5 | 62.6/71.9 | 77.6 | |
| Water | 45.3 | 46.3 | 68.5 | 50.7 |
| R.m.s. deviations | ||||
| Bond lengths (Å) | 0.01 | 0.008 | 0.006 | 0.006 |
| Bond angles (°) | 1.37 | 1.19 | 1.02 | 1.1 |
*A single crystal was used for each structure.
aValues in parentheses are for highest-resolution shell.
Figure 1.
Dimerization of full-length RcsB bound to BeF3−. (A) Cartoon representation of the dimerization surface of RcsB bound to the phosphomimetic BeF3− (labeled as BeF) through the REC domain (blue and cyan) and DBD domains (yellow shades). Surface of the REC domains is shown. The REC–DBD connector is colored in salmon. (B) A framed detail of the dimerization surface and catalytic center in the dimer showing relevant residues, BeF3−, Mg+2 ion (Mg). (C) Detail of the active site in one of the subunits showing interactions by relevant residues, BeF3− (BeF) and Mg+2 (Mg) ion and water molecules (W) as dashed lines. (D) Autophosphorylation of RcsB with 50 mM AcP is visualized using different techniques (Phostag-acrylamide, autoradiography and native-PAGE). (E) RcsB dimer formation upon phosphorylation is visualized by gel filtration with 200 μg of purified protein using a Superdex200 increased 10/300GL (GE, Healthcare)
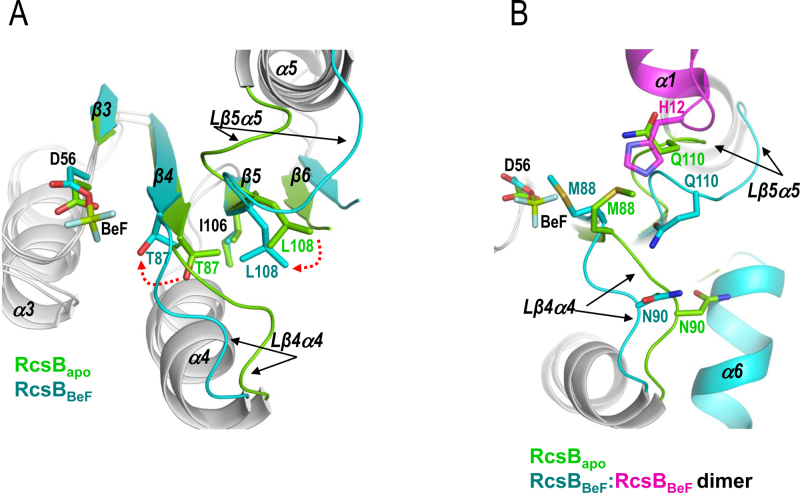
The dimer structure of RcsBBeF is generated by the interaction of the REC domains through the interface provided by α1 and the loops Lβ1α1 and Lβ5α5 (Figure 1B and Supplementary Table S2), similarly to the α1–α5 dimerization surface observed in other activated RRs of NarL/FixJ subfamily such as VraR from Staphyloccocus aureus (PDB ID: 4IF4) (33), Spr1814 from Streptococcys pneumoniae (PDB ID: 4ZMR) (34) and DesR from Bacillus subtilis (PDB ID: 4LDZ) (35) (Supplementary Figure S2B and Table S3). This type of α1–α5 dimerization differs from the α4β5α5 dimerization observed for the OmpR/PhoB subfamily of RRs, which represents approximately one-third of all RRs (36). The presence of the small β6 in RcsB, VraR, DesR (Supplementary Figure S2A) and Spr1814 impairs dimerization through α4β5α5 as it would be sandwiched into the dimers generating clashes, thus, explaining the acquisition for an alternative dimerization surface (Supplementary Figure S2C). Additionally, the Lβ4α4 also contributes to REC dimerization through interactions with the same loop in the other subunit between residues M88-M88, or with Lβ1α1 of the other subunit, between M88 and H12 (Figure 1B and Supplementary Table S2). Dimerization of RcsBBeF is not only produced through the REC domain but also through the DBD domains (Supplementary Table S2), which dimerize similarly as the NarL DBD domains bound to its own promoters, nirB and narG (37,38). Similar DBD dimerization has been also observed in VraR although in this case each DBD involves a different dimer in a tetrameric complex and not within the same dimer as observed in RcsBBeF (Supplementary Figure S4). Indeed, according to the PISA server (39), the total interface area of the dimer was 1180 Å2, where 800 Å2 was provided by the REC domains and 381 Å2 by the DBDs. The asymmetry observed in the RcsBBeF dimer is reflected by a different interaction between REC and DBD at each subunit of the dimer (Figure 1A and Supplementary Table S4). In subunit A, residues from Lβ3α3 mainly interact with α7 (with a salt bridge between Asp66 and Arg160) and from Lβ4α4 with Lα10 while in subunit B residues from α3 and α4 mainly interact with α10—with a salt bridge between Asp100 and Arg150—(Supplementary Table S4 and Figure S5A).
The asymmetric dimerization observed for RcsBBeF was induced by the presence of the phosphomimetic BeF3− (Figure 1A), which promoted an arrangement at the active site commonly observed in other RR structures (Figure 1C). In this way, BeF3− was stabilized in the active center by interactions with the side chain of the catalytic conserved residue D56, side chain of the conserved catalytic residues T87 at β4 and K109 at Lβ5α5, main chain of S58 at Lβ3α3 and main chain nitrogen of M88 at Lβ4α4, the Mg2+ ion and two water molecules. Additionally, K109 was also salt bridged with D10 at the end of β1. Finally, the Mg2+ sphere of hexa-coordination was completed by the interaction with D56, D11 at Lβ1α1, main chain oxygen of S58 and two water molecules (Figure 1C and Supplementary Figure S5B). This arrangement at the active site confirmed that RcsBBeF shows the phosphorylated conformation.
Phosphorylation of RcsB could also be observed in vitro upon incubation with the small phosphodonor AcP and visualized either in Phostag-acrylamide gels, native gels or by autoradiography (Figure 1D). Meanwhile, RcsB dimerization driven by phosphorylation could also be followed by gel filtration, demonstrating that phosphorylated RcsB either in the crystal structure or in solution, forms stable dimers (Figure 1E).
The conformation of phosphorylated RcsB is competent to interact with DNA
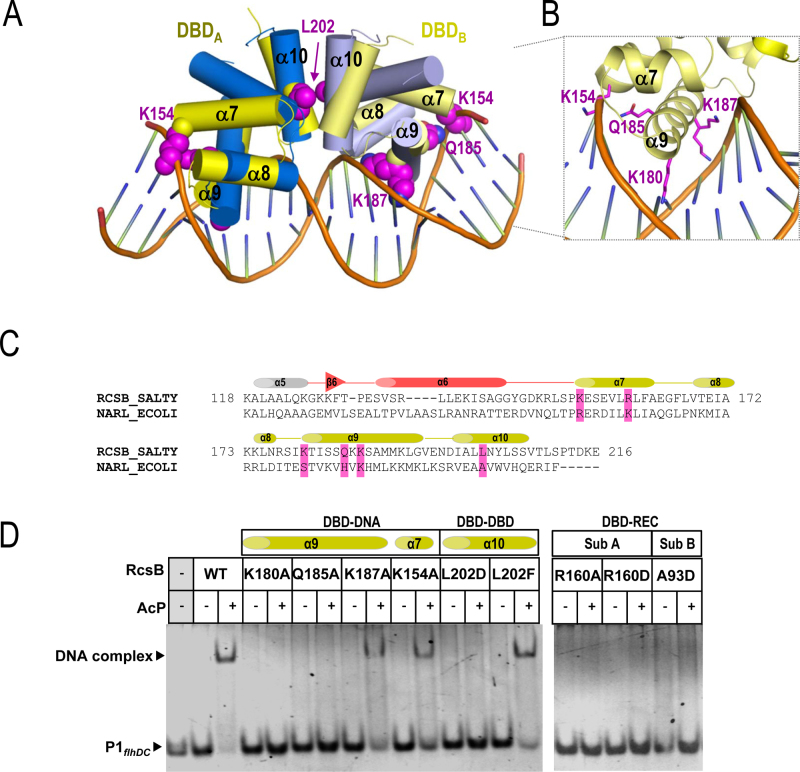
As mentioned before, dimerization of the DBDs in RcsBBeF is similar to other RRs that belong to the NarL/FixJ subfamily. The superposition of DBDs dimer from RcsBBeF with the two DBDs from NarL recognizing the promoter nirB in a tail-to-tail arrangement (PDB ID: 1ZG1) (37,38) showed high structural alignment (RMSD of 2Å for 120 residues, 60 residues for each domain). Such alignment supports a DNA binding competent conformation for RcsBBeF. This superposition enabled us to propose structural elements and residues involved in DNA recognition (Figure 2A). Thus, RcsB α9 helix was inserted in the major groove, as it is expected for a DNA recognition helix, while α7, the support helix for α8 and α9, also contributed to DNA interaction. In this model, the residues K180, T181, S184, Q185 and K187, in α9 would be projected toward the major grove interacting with DNA bases, supporting their recognition role while residue R154 in α7 interact with the DNA backbone stabilizing the DNA–RcsB complex. To test this recognition model, we designed the RcsB mutants K180A, Q185A, K187A in α9 and K154A present in α7 (Figure 2B and C). These variants were analyzed for their capacity to interact with bona fide promoters recognized by RcsB (Figure 2D and Supplementary Figure S6). As a previous step, we tested in vitro by EMSA assays the binding of phosphorylated WT RcsB to promoters P1flhDC (−228 to −203) in the flhDC operon (15) and rcsA (−275 to −247) in the rcsA gene (10) (Figure 2D; Supplementary Figure S6A and B). We selected these regions since P1flhDC accounts for the RcsB box while a similar consensus sequence is observed in thercsA promoter, which is autoregulated by RcsA and RcsB in the so named RcsAB box (4,40) (Supplementary Figure S6B). Binding of RcsB to both promoters was achieved in the presence of AcP, supporting that phosphorylation induced the competent conformation required to bind these promoters (Figure 2D and Supplementary Figure S6A). However, binding of RcsB∼P to P1flhDC caused a higher band shift compared to the rcsA promoter, which could account for higher affinity to the former promoter. Since RcsA assists the binding of RcsB to the rcsA promoter (10), the absence of this protein would explain the weaker binding of RcsB alone to the rcsA promoter. We then analyzed the DNA-binding capacity of the mutants at the recognition residues proposed by our RcsB–DNA model by additional EMSA assays (Figure 2D and Supplementary Figure S6A). K180A and Q185A mutations abolished interaction of RcsB with DNA in both promoters; however, K187A still allowed binding of the RcsB variant to P1flhDC although with lower affinity than the WT (Figure 2D and Supplementary Figure S6C). K180 was previously reported to play a critical role in DNA-binding to the flhDC promoter in E. coli RcsB (41). Our data therefore confirmed the participation of α9 helix in the process of DNA recognition and binding. The K154A mutation resulted in lower binding affinity of RcsB for both P1flhDC and rcsA promoters but did not impair DNA binding (Figure 2D and Supplementary Figure S6). This observation suggested a secondary role for K154 in DNA interaction by stabilizing the DNA–RcsB complex through contacts with the DNA backbone, as it was observed for the corresponding residue (K159) in NarL (37). It has also been suggested that K154 acetylation inhibits the ability of E. coli RcsB to activate rprA transcription in vivo (42).
Figure 2.
Model of the interaction of RcsB with DNA. (A) Superposition of the DBD domains in RcsBBeF structure (in yellow) with the DBD NarL structure bound to DNA (PDB ID: 1ZG1) (in blue). (B) Side chain for residues Lys154, Lys180, Gln185 and Lys187 are shown as sphere in magenta. A framed detailed view of same residues in RcsB interacting with DNA is shown (C) Sequence alignment of RcsB and NarL comprising REC–DBD connector (in salmon) and DBD (yellow). Mutated residues at the DBD in RcsB involved in interactions are highlighted in magenta. (D) EMSA assays with RcsB WT and mutants in the absence and presence of phosphorylation (with 50 mM AcP) and P1flhDC site (−228 to −203) in the flhDC operon.
To further confirm our recognition model, we analyzed the relevance of the relative DBD–DBD arrangement as it was observed in the RcsBBeF structure for DNA recognition. L202D and L202F mutations were introduced in α10 since L202 interacts with itself via hydrophobic contacts to maintain the DBD dimer (Figure 2A and C). EMSA assays showed that the L202D RcsB variant did not bind DNA while the L202F mutation restored DNA binding (Figure 2D and Supplementary Figure S6A). Interestingly, both mutants could dimerize upon phosphorylation (Supplementary Figure S7A), confirming the phosphorylation induced dimerization mediated by REC domains, but the strength of the DBD–DBD interaction is crucial to adopt a DNA-binding competent conformation. These results also confirm our DNA-recognition model (Figure 2A).
As RcsBBeF showed asymmetry, the REC–DBD interaction surface was different for each subunit of the dimer. Thus, we evaluated the impact of this asymmetry for DNA recognition by introducing mutations at specific residues promoting specific interactions at each subunit (Supplementary Figure S5A). In subunit A, we generated R160A and R160D mutations (Figure 2C) since this R160 residue makes a salt bridge with D66 at Lβ3α3 (Supplementary Figure S5A). EMSA assays showed that both mutations—R160A and R160D—impaired DNA binding (Figure 2D and Supplementary Figure S6A) supporting that the proper orientation REC–DBD is required for DNA binding and the important role of R160 (and its counterpart D66) in maintaining this orientation. In subunit B, we generated the A93D mutation since A93 is in close proximity to Lα9α10 (Supplementary Figure S5A). EMSA assays showed that the RcsB A93D variant was impaired to bind both promoters (Figure 2D and Supplementary Figure S6A), possibly due to electrostatic clashes between the new aspartic residue and Lα9α10. Altogether, the data support that the asymmetric conformation of RcsB, having different REC–DBD relative disposition, is required to bind to these promoters. Finally, we confirmed that the lack of DNA-binding capacity of K180A, Q185A, L202D, R160A, R160D and A93D mutants was specific and not due to improper protein folding. We analyzed the phosphorylation capacity of these mutants, confirming that all of them showed similar phosphorylation ability as the WT protein (Supplementary Figure S7B). Overall, the mutational analysis supports that the conformation observed for phosphorylated RcsBBeF corresponds to the one competent for DNA binding, as the structural analysis predicted. Furthermore, it also reveals that RcsB binds the P1flhDC and rcsA promoters regions in a tail-to-tail arrangement.
The structure of RcsB in the absence of BeF3− shows alternative oligomeric organization
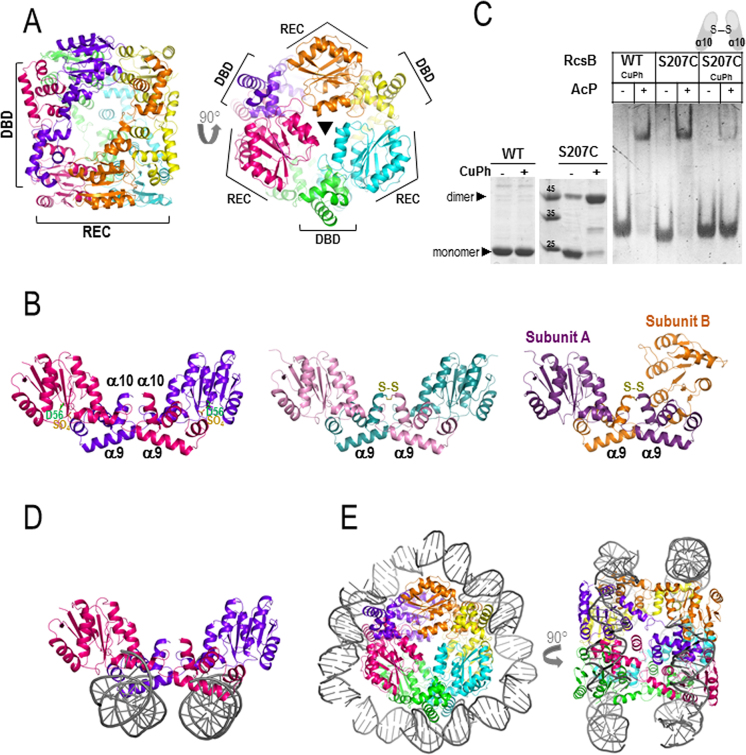
To provide clues about the mechanism that drives RcsB phosphorylation, we sought to solve the structure of its unphosphorylated form. Crystals of RcsB full-length in the absence of BeF3− showed an asymmetric unit containing six molecules of RcsB, which form three dimers arranged in a hexameric structure that resembled a cylinder (∼60 × 70 Å) (Table 1 and Figure 3A). The bases at each side of this cylinder were generated by the interaction of three REC domains (each dimer provides a REC domain to each of the bases), related by a 3-fold axis (Figure 3A). Meanwhile, the DBD domains were placed at the central part of the cylinder (Figure 3A). These three dimers are structurally similar (RMSD ∼1 Å) as well as the individual subunits (RMSD <1 Å) (Supplementary Tables S5 and 6) with a bit higher deviations at Lβ5α5 (RMSD ∼1.4 between 109 and 114 residues) (Supplementary Figure S8). However, these dimers show a different quaternary arrangement than the dimers obtained for RcsBBeF. While RcsBBeF dimerize mainly through the interaction of REC domains with a secondary contribution of DBD–DBD interactions (Figure 1A), RcsB in the absence of BeF3− dimerizes by the interaction of the REC from one subunit with the DBD from the other subunit as the DBDs were crossed or interchanged between subunits in the dimer (Figure 3B). This ‘crossed’ conformation (RcsBcrossed) is achieved due to the separation between REC and DBD with a complete absence of interactions between these domains within a subunit. Although this domain separation is observed in other members of the NarL/FixJ subfamily (34,35), none of them shows a similar type of ‘crossed dimerization’. In addition to the intersubunit REC–DBD interaction in RcsBcrossed, both DBDs also interact through Lα9α10 and α10. The total interface area of RcsBcrossed was of 1591 Å2, around 30% larger (400 Å2) that the observed for RcsBBeF dimer suggesting that should be stable in solution.
Figure 3.
Structure of RcsB in the absence of BeF3−. (A) RcsB hexamer structure, in the absence of BeF3−, composed by three dimers related by a 3-fold axis resembling a cylinder. The bases of the cylinder are generated by the REC domains and DBDs are at the central part of the cylinder. (B) Dimer crossed conformation of RcsB (RcsBcrossed; in dark pink and purple) with a sulfate ion (SO42− labeled as SO4) bound at the active site (on the left) shows dimerization mainly through REC and DBD from different subunits. Dimer structures of the S207C-RcsB mutant in the crossed conformation at the middle (S207C–RcsBcrossed; in pink and blue) and in an alternative asymmetric crossed dimer conformation on the right (S207C–RcsBAC; in violet and orange) (C) Formation of RcsBcrossed dimers with mutant S207C in the absence and presence of 0.1 mM of CuPh is observed in SDS–PAGE (left panel). EMSA experiments with WT and mutant S207C in absence and presence of 50 mM AcP and 0.1 mM CuPh (right panel) (D) In this conformation, RcsB can bind two separate dsDNA fragments (in gray) (E) Model of the RcsB hexamer wrapped by two separate dsDNA fragments.
Comparison between RcsBcrossed and RcsBBeF revealed that the individual domains are almost identical (RMSD for REC ∼0.7 for 122 residues and DBD ∼0.8 for 59 residues) with low deviations at α1 and Lβ3α3 and higher deviations at Lβ5α5, as observed for the individual subunits in RcsBcrossed (Supplementary Figure S8). However, the disposition between REC and DBD domains are different in both RcsBcrossed and RcsBBeF structures due to the flexibility provided by the REC–DBD connector (Supplementary Figure S3). In RcsBcrossed, the small β6 in the REC–DBD connector shows a similar conformation to subunit B in RcsBBeF, however, Lα6α7 is reoriented, allowing DBD to rotate 101.9° and translate 8.2 Å with respect to subunit B of RcsBBeF (Supplementary Figure S3). This displacement moves away the DBD domain in such a way that REC and DBD do not interact anymore. Comparison between subunits A and B at RcsBBeF with RcsBcrossed reveals that the different relative orientations of REC and DBD are acquired by a gradual opening from the closest REC–DBD conformation presented at subunit A of RcsBBeF (Supplementary Figure S3 and Table S7). From this orientation, the DBD could move away and rotate ∼80° to acquire the disposition observed in subunit B of RcsBBeF and even ∼100° more to adopt the RcsBcrossed conformation (Supplementary Figure S3). Despite the fact that the REC–DBD connector is the flexible region allowing these movements, Lα6α7 plays a major role as bending region in these movements.
To validate RcsBcrossed conformation, we generated the RcsB mutant S207C at α10. If RcsB adopts the crossed conformation, this cysteine residue could be in an appropriate distance to generate a disulfide bond that could lock the dimer in this conformation. SDS-PAGE in absence of reducing agents showed that purified protein of this mutant presented a consistent amount of preformed covalent dimer (Figure 3C), supporting that this type of dimerization is present in the E. coli expression system. Furthermore, we were able to purify the dimer species of S207C and crystallization assays produced two types of crystals that contained S207C-RcsB tied through the expected disulfide bond at DBD (Table 1, Figure 3B and Supplementary Figure S9A). One type of crystal contained a S207C-RcsB dimer in the crossed conformation (S207C–RcsBcrossed), as observed in the RcsBcrossed structure (RMSD 1.27 Å for 410 residues), which was able to form the hexameric structure by crystallographic symmetry similar to the WT (RMSD 1.8 Å for 1197 residues) (Figure 3B and Supplementary Figure S9B). The other type of crystal contained a dimer resembling the crossed conformation but the REC domain for one of the subunits (subunit B) had moved 84° and 1.6 Å preventing the formation of the hexameric structure, thus producing an alternative asymmetric crossed dimer structure (S207C–RcsBAC) (Figure 3B and Supplementary Figure S9C). In this way, the conformation between the subunits in RcsBcrossed, S207C–RcsBcrossed and subunit A in S207C–RcsBAC were identical (Supplementary Figures S8, 9C and 10) while the conformation of the subunit B in S207C–RcsBAC seemed to be halfway between RcsBcrossed and subunit B of RcsBBeF (Supplementary Figures S8 and 11). Importantly, the S207C–RcsB structures support that the crossed conformation and its macromolecular arrangement as homohexamers might be a biological conformation relevant for RcsB. In addition, these structures are consistent with a conformational dynamism in RcsB despite being trapped by the disulfide bond at the DBD. Comparison of the individual domains in RcsBcrossed with S207C–RcsBcrossed and S207C–RcsBAC shows also deviations at α1 and Lβ3α3 with higher deviations at Lβ5α5. However, S207C–RcsBAC subunit B shows additional deviations at Lβ4α4 and α4 (Supplementary Figure S8) pointing to this region as promoting the alternative conformation observed in this subunit.
Interestingly, in the RcsBcrossed structure and S207C–RcsB structures, the helix α9 is exposed to the solvent in such a way that could be competent to bind DNA (Figure 3B). However, the relative disposition of the DBD domains is different to the observed in RcsBBeF and therefore should not be competent to bind tail-to-tail to the promoters previously analyzed. To test this hypothesis, we analyzed by EMSA the capacity of phosphorylated RcsB S207C mutant to bind the promoter region P1flhDC in the absence and presence of CuPh. The phosphorylated WT RcsB could bind DNA either in the absence or presence of CuPh, however, DNA binding of phosphorylated RcsB S207C was severely impaired in the presence of CuPh, suggesting than the crossed conformation was not adequate to bind this promoter region (Figure 3C). Indeed, EMSA assays with the purified S207C dimer species confirmed that the crossed conformation showed much lower binding affinity to P1flhDC despite high concentrations used (Supplementary Figure S12A). However, the superposition of the RcsBcrossed DBD domains with the corresponding one of NarL in the NarL–nirB complex structure proposed an alternative DNA-binding model for this RcsB conformation. This model showed that RcsBcrossed could bind to two separate dsDNA fragments in an almost parallel disposition and having different orientation (Figure 3D). If we extend this superposition to the hexamer, the two separate dsDNA fragments would generate a supercoiled structure of ∼140 bp that would wrap the hexamer (Figure 3E). The biological relevance of this type of supercoiling structure remains to be explored.
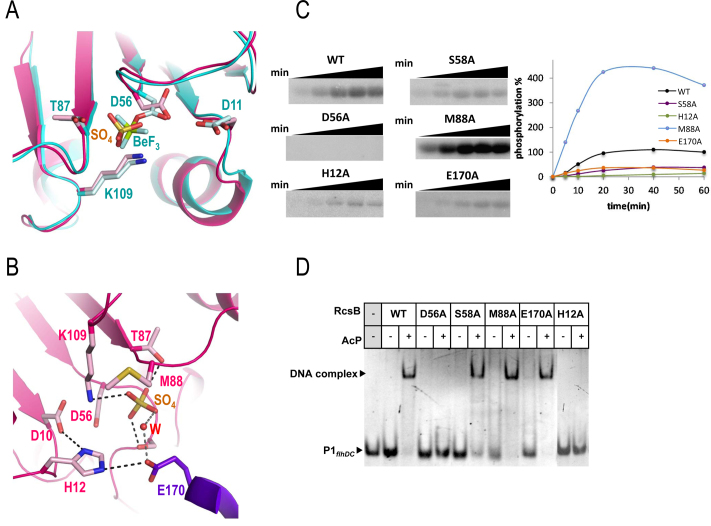
The crossed RcsB dimers present a phosphorylated conformation stabilized by the DBD domains
Due to the conformation of RcsBcrossed, the active site of each subunit is facing at opposite directions on the dimer, pointing toward the solvent and is not locked by the α1–α5 dimerization surface as in the RcsBBeF conformation (Figure 3B). A closer look to the active site in the RcsBcrossed dimers showed the presence of a sulfate ion located at a similar position than the BeF3− (Figure 4A). Comparison with the active site of RcsBBeF showed that the sulfate ion maintained the conserved contacts expected for a phosphomimetic molecule which involved interaction with the side chains of T87 and the catalytic K109 (Figure 4A). Similarly, the structural elements Lβ4α4 and Lβ5α5 showed the characteristic disposition of the phosphorylated state, although Lβ5α5 showed certain flexibility (comparing the subunits of RcsBcrossed) probably due to lack of interactions with the other subunit of the dimer as is observed in RcsBBeF. All these structural features correlate with RcsBcrossed having a phosphorylated conformation. Interestingly, in the active site, the sulfate ion has additional interactions than BeF3−, contacting the side chain of S58 that, in turn, interacts with the side chain of E170 in the DBD of the other subunit (Figure 4B). In some subunits of the hexamer, E170 can contact the sulfate ion directly or through a water molecule and can also interact with H12, a residue that contributes to the dimerization surface in the RcsBBeF dimers (Figure 1B). Thus, the interacting network S58-E170-H12 might contribute to the stabilization of the sulfate ion at the active site and favor the crossed conformation through intermolecular REC–DBD interactions (Figure 4B). Interestingly, in the active site of RcsBcrossed the side chain of M88 shows flexibility and adopts alternative orientations in different subunits, in contrast to its fixed conformation observed in RcsBBeF structure where it contributes to lock the phosphomimetic BeF3− at the active site (Figure 1B). These alternative orientations could modulate the access to bind the phosphate at the active site implying that M88 could work as a ‘phosphoryl gate’. Furthermore, M88 is also involved in RcsB oligomerization since stabilizes α1–α5 dimerization in RcsBBeF through interactions with M88 and H12 of the other subunit (Figure 1A). To better understand the role of these residues in RcsB activity, we generated H12A, S58A, M88A and E170A mutations to test the phosphorylation capacity of these variants in relation with the WT and the non-phosphorylatable mutant D56A as negative control (Figure 4C). As expected, D56A mutant did not show phosphorylation and the RcsB variants H12A, S58A and E170A showed reduced levels of phosphorylation in comparison with the WT (Figure 4C). In contrast, the M88A variant exhibited higher phosphorylation level than the WT. An estimation of the initial phosphorylation rate of these mutants accounted for a reduction of the phosphorylation level around ∼90, 75 and 60% for H12A, S58A and E170A, respectively, when compared to the WT. Meanwhile the M88A mutation increased the rate of phosphorylation to almost 5× higher than WT (Figure 4C). As it has been observed in DesR, homodimerization favors autophosphorylation (35); thus, the reduced phosphorylation observed for H12A might be due to its reduced contribution for the dimer formation in the RcsBBeF confirming its role promoting dimerization. Meanwhile, the decreased level of phosphorylation observed for S58A and E170A confirms their role stabilizing the sulfate ion as it is observed in the RcsBcrossed structure (Figure 4B). The increased phosphorylation observed for M88A could be linked to its putative function as a phosphoryl gate regulating the access to the active site, highlighting an important role for this residue controlling phosphorylation levels. The previous analysis in vivo of RcsB M88A mutation in E. coli showed that this mutation induces a phenotype compatible with constitutive phosphorylated form, supporting the hyperphosphorylation observed in vitro (9).
Figure 4.
Active site and functional studies on catalytic residues in RcsB. (A) Detail of the active site for RcsBcrossed (in pink) superposed with RcsBBeF (in cyan). Catalytic residues as well as the sulfate ion (SO42− labeled as SO4) and BeF3−(labeled as BeF) are shown as sticks. (B) Another view of the active site in RcsBcrossed (pink for one subunit and purple for the other). Residue E170 (in purple) from the DBD of the other subunit contributes to the active site. Catalytic and relevant residues are shown as sticks together with the sulfate ion (SO4) and a water molecule (W). (C) Phosphorylation assays of WT and mutants of RcsB with AcP32. Phosphorylation was followed at 5, 10, 20, 40 and 60 min and was evaluated with the MultiGauge software (Fuji). (D) EMSAs of RcsB WT and mutant forms with P1flhDC were performed in the absence and presence of 50 mM of AcP.
We also checked by EMSA experiments whether the decreased or enhanced phosphorylation levels in some of the RcsB variants impacted binding to the promoter region P1flhDC (Figure 4D). As expected, D56A could not bind DNA due to its inability to be phosphorylated. EMSA assays with S58A, E170A and M88A variants showed binding to DNA as the WT (Figure 4D). However, a concentration-dependent assay demonstrated lower binding affinity for S58A and E170A in a clear correlation with their reduced phosphorylation level (Supplementary Figure S12B). Gel filtration analysis of E170A showed a slight reduction in dimer formation induced by AcP (Supplementary Figure S13), confirming the correlation among phosphorylation, dimerization and DNA binding. Indeed, the H12A variant did not show binding to P1flhDC, consistent with a role of H12 stabilizing the dimer in the conformation observed in RcsBBeF necessary to bind this promoter region, as it was confirmed by gel filtration (Supplementary Figure S13). Although M88A showed a higher affinity for the P1flhDC promoter, this increment is not correlated with the increase in the phosphorylation level observed for this mutant (Supplementary Figure S12B). This result indicates that the increment in phosphorylation of M88 is not directly proportional to an increase level of dimerization, as it was confirmed by gel filtration (Supplementary Figure S13), which supports a dual role of this M88 residue in oligomerization and phosphorylation.
In contrast to RcsBcrossed, the S207C–RcsBcrossed and S207C–RcsBAC structures did not contain a sulfate ion at the active site but their REC domains, except for the subunit B of S207C–RcsBAC, displayed a characteristic activated conformation (see below), similar to that of RcsBcrossed and RcsBBeF (Supplementary Figure S14). This fact supports that RcsB crossed conformation corresponds to a phosphorylated state, induced by the disulfide bond in S207C–RcsB structures.
Effect of RcsB mutations in colanic capsule formation and motility
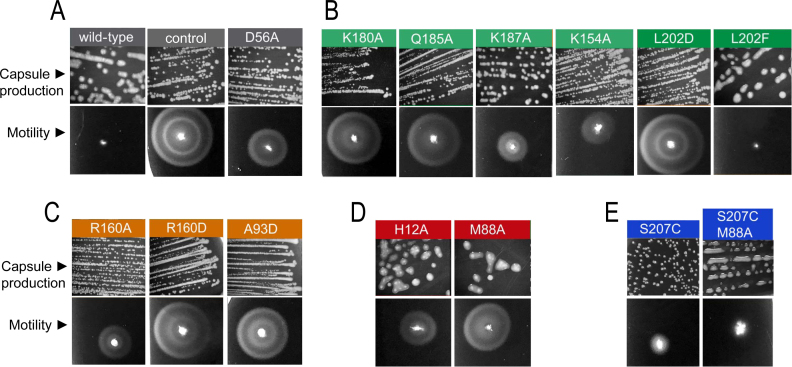
To assess the biological impact of the conformations observed in the structures of S. Typhimurium RcsB, we tested in vivo the production of colanic acid capsule and motility in S. Typhimurium MD4821 strain lacking endogenous RcsB and expressing the different RcsB variants previously characterized in vitro (see ‘Materials and Methods’ section for details). This strain, MD4821, also produces an IgaA repressor protein partially deficient for function, which facilitates monitoring of phenotypes linked to activity of the RcsCDB phosphorelay (25). To note that the RcsA–RcsB heterodimer is required to reach optimal production levels of colanic acid capsule (wca) operon whereas the RcsB homodimer is sufficient to represses transcription of flhDC, encoding the flagella master regulator (10,15). As expected, the absence of RcsB (empty plasmid as a control) yielded a phenotype of no capsule production but high motility consistent with lack of wca expression but production of the FlhDC master regulators (Figure 5A). In contrast, expression of WT RcsB in the genetic background of MD4821 strain resulted in colonic capsule production (mucoid colonies) and motility inhibition (Figure 5A). Expression of the RcsB variant D56A, unable to phosphorylate, yielded a phenotype closer to the control with absence of capsule and a high reduction in motility, confirming that both processes may require phosphorylated RcsB (Figure 5A). The reduced motility observed for the D56A mutant could be explained by the equilibrium between conformations of RRs independently of the phosphorylation, which would produce a small pool of active RcsB when is overexpressed. Expression of the mutants generated in residues of the DBD that interact with DNA, K180A and Q185A, showed a similar phenotype to the control, that was, normal motility but no capsule production (Figure 5B). However, K187A and K154A showed a phenotype closer to D56A (Figure 5B), consistent with their low-binding capacity to the P1flhDC promoter in the EMSA experiments. Interestingly, L202, a residue that interacts with L202 in the other subunit to maintain the DBD dimer, has an opposite effect when mutated to an aspartic residue (L202D)—showing no capsule production but motility—compared to the capsule production but no motility observed following production of the L202F variant (Figure 5B). This opposite effect explains the EMSA experiments that showed no binding to P1flhDC promoter for L202D and binding for L202F variant. This observation, therefore, confirms the importance of DBD–DBD interaction to generate the tail-to-tail arrangement required to bind rcsA/wca and flhDC promoters. Expression of RcsB variants with mutations R160A, R160D and A93D, residues involved in interactions between REC and DBD at each subunit in the RcsBBeF structure, showed no capsule production but motility confirming the impairment of these RcsB variants to bind to DNA. Nonetheless, a partial effect on motility was observed for R160A, clearly correlating the promoter binding capacity with the severity of the mutation (Figure 5C). The production of the H12A and M88A variants resulted in high capsule production but some motility with higher motility for M88A (Figure 5D). This finding indicated that the H12A and M88A mutations could affect at different extent the binding of RcsB to distinct promoters. Our in vitro experiments showed antagonistic behaviors for these two mutants since H12A phosphorylates a 60% less than the WT and does not show dimer formation (Supplementary Figure S13) unable to bind P1flhDC promoter while M88A hyperphosphorylates, dimerizes and binds to P1flhDC promoter. These apparent contradictory results for these variants in vitro and the in vivo phenotypic experiments could be explained by the participation of these residues in the conformational dynamism of RcsB and in the process of homo- and hetero-dimerization, thus, having different impact in the binding to those promoters regulated only by RcsB or those requiring auxiliary regulators. Finally, expression of the RcsB variant S207C did not result in capsule production while motility was reduced in a great extent (Figure 5E), indicating that the S207C mutation affects specifically the binding of RcsB to the rcsA/wcA promoters. Therefore, the in vivo tests support our model involving a crossed conformation for RcsB acquired through a disulfide bond and produced even though the reduced environment in E. coli cytoplasm. This crossed conformation could have evolved to modulate the regulatory capacity of RcsB. Indeed, expression of a double mutant S207C/M88A recovered the phenotype of capsule production observed for M88A but, similarly to S207C, showed absence of motility (Figure 5E) restoring this double mutant the WT phenotype. The characterization of these RcsB variants therefore confirms the conformational dynamism of RcsB, which facilitates different molecular arrangements either by homo- or heterodimerization with other proteins, as a main feature to modulate its transcriptional regulatory capacity.
Figure 5.
Effect of RcsB mutations in capsule formation and motility. Overexpression of RcsB mutants in the Salmonella Typhimurium strain MD4821 (igaA1 rcsB) to monitor effect on colanic capsule production and motility in vivo. (A) Expression of the WT, unphosphorylatable mutant D56A and empty vector (as control). (B) Expression of mutants at residues present in the DBD domain that either interact with DNA according to the model RcsBBeF–DNA or contribute to DBD–DBD dimerization (L202). (C) Expression of mutants at residues that contribute to specific interactions between REC and DBD at each subunit. (D) Expression of mutants at relevant residues H12 and M88 that contribute to RcsB dimerization and catalysis. (E) Expression of mutants at residue S207C that traps DBD–DBD in the crossed conformation due to disulfide bond formation. A double mutant (S207C/M88A) containing an additional mutation M88A is also shown.
A switch between unphosphorylated and phosphorylated conformations in RcsB
Extensive studies have proposed the Y-T coupling mechanism for RR activation through phosphorylation (43). In this mechanism, the phosphorylation of the Asp residue at the REC domain induces a conformational rearrangement at the active site, which is propagated along the central β-sheet to produce a change in the α4β5α5 surface that allows RR dimerization or interaction with target proteins through this surface (19). A Thr/Ser in β4, which coordinates with the phosphoryl group, and Tyr/Phe in β5, reorients their side chains to inward conformations, propagating the signal from the active site to the α4β5α5 surface and so designated the mechanism as Y-T coupling. Since NarL/FixJ subfamily of RR uses the surface α1–α5 to dimerize after phosphorylation, we sought to determine whether this RR uses the Y-T coupling mechanism to shift from the unphosphorylated conformation (usually depicted as inactive) to the phosphorylated conformation (usually depicted as active) or an alternative one. Structural comparison of the unphosphorylated conformation (PDB ID: 5I4C) and phosphorylated conformation (RcsBBeF) of RcsB REC domains gives clues for the mechanism of activation by phosphorylation for this type of RRs (Figure 6A and Supplementary Figure S15A). Phosphorylation of D56 induces the movement of T87, characteristic of the Y-T mechanism, so its hydroxyl group forms a direct hydrogen bond with the BeF3− or the sulfate ion (Figure 6A). On the contrary, I106, which would corresponds to the Tyr position, does not undergo any type of conformational change, suggesting that this type of RR follows only partially the Y-T mechanism (Figure 6A). Indeed, other members of the NarL/FixJ subfamily also lack the conformational change at the Tyr position due to movement of the Thr since the residue in this position shows a constitutive inward conformation (34,35). Interestingly, the restriction of movement at Tyr position is also conditioned by the presence of the β6, which expands the central β-sheet and clamps the central and N-terminal part of β5 where Tyr is placed. In addition, β6 also induces the inward conformation of the residue at Tyr position, even in the unphosphorylated state (Figure 6A and Supplementary Figure S15A). Therefore, RcsB and other members of NarL/FixJ subfamily do not follow strictly a classical Y-T coupling mechanism, which makes sense since the presence of the β6 in this group of RRs blocks α4β5α5 surface, impairing its use to dimerize or to interact with target proteins. Alternatively, movement of T87 helps L108 to occupy the hydrophobic pocket left by T87 (Figure 6A and Supplementary Figure S15A). L108 is a conserved residue in the NarL/FixJ but not in the OmpR/PhoB subfamily of RRs and is placed at the end of β5 and two residues upstream of Tyr position (Supplementary Figure S1). In this way, the orientation of T87 and L108 is key to define the inactive or active state in RRs of NarL/FixJ subfamily. Moreover, the displacement of L108 causes the movement of Lβ5α5, which is just after L108, to approach slightly the catalytic K109 to interact at the active site with the phosphate (Figure 6A). The Lβ5α5 movement, which is the largest conformational change observed in the REC domain between unphosphorylated and phosphorylated conformations (Figure 6 and Supplementary Figure S15A), opens a groove where α1 and Lβ1α1 from the second REC domain is accommodated during the dimerization process (Figure 6B). In this way, movement of Lβ5α5 allows H12 insertion into the groove for dimerization, otherwise, it would clash with Q110 at Lβ5α5, a residue that interacts with Lβ4α4 stabilizing its conformation (Figure 6B). Oppositely, α1 does not show any important movements, except for alternative conformations of side chains, thus, the dimerization process should be controlled by the opening of Lβ5α5 induced by the phosphorylation. Additionally, movement of Lβ4α4 is also necessary in order to avoid clashes during dimerization between M88 and the H12 coming from the other subunit, as well as between N90 and the helix α6 at the REC–DBD connector (Figure 6B). Inspection of the key residues T87 and L108 in RcsBcrossed confirms the active conformation for this structure as they show a similar orientation to RcsBBeF (Supplementary Figure S15B). Furthermore, a look at the structures of S207C–RcsBcrossed and S207C–RcsBAC shows that the orientation of T87 and L108 in S207C–RcsBcrossed and in subunit A of S207C–RcsBAC is similar to RcsBBeF and RcsBcrossed standing for an active conformation (Supplementary Figure S16A), even though the active centers are empty in these structures. By contrast, subunit B in S207C–RcsBAC is between the inactive–active conformations due to movement of Lβ4α4 at N90 and to Lβ5α5 at Q110 as in the unphosphorylated REC structure of RcsB (PDB ID: 5I4C) (Supplementary Figure S16). Altogether, the switch between inactive and active conformations in RcsB seems to be a variation of the Y-T coupling mechanism where the effect of Thr movement induced by the phosphorylation is transduced to a different direction by the constraints of β5 movement imposed by the presence of β6. This mechanism seems to be general for the NarL/FixJ subfamily since residues T87 and L108 and the structural element β6 are conserved.
Figure 6.
Switch mechanism (β5-T coupling) of RcsB between the inactive and active conformation. (A) Comparison between the unphosphorylated (PDB ID: 5I4C in green) and phosphorylated conformation (RcsBBeF in cyan) reveals movements, induced by the presence of β6, in the switch residues T87 and L108 while I106 shows the similar inward conformation. (A and B) These movements produce conformational changes, mainly in Lβ4α4 and Lα5β5, that allow α1 and Lβ1α1 from the other subunit (in magenta) to insert H12 for RcsB dimerization. (B) The movements of Lβ4α4 and Lβ5α5 are necessary to avoid clashes between H12 (in magenta), Q110 at Lβ5α5 and M88 at Lβ4α4. Additionally, movement of Lβ4α4 avoids clashes between N90 and α6 at the REC–DBD connector.
DISCUSSION
RcsB is a pleiotropic RR that is involved in transcription of a variety of genes related with bacterial virulence either alone or in complex with additional co-regulators in a fashion dependent or independent of phosphorylation. These characteristics anticipate that RcsB could adopt alternative conformations to conduct its different modes of regulation. Indeed, we present here four structures of full-length RcsB mainly in the active conformation, which are stabilized in some cases by the phosphomimetic BeF3− or a sulfate ion. These structures clearly demonstrate the pronounced conformational dynamism of the DBD domain with respect to the REC domain. This conformational dynamism allows different relative disposition of both domains by the REC–DBD connector. This structural flexibility seems to be intrinsic of NarL/FixJ subfamily, since the different structures known for members of this subfamily display multiple alternative dispositions of DBD domain with respect to REC domains in the active state, meanwhile the REC domain always adopt a similar dimeric disposition. The crystal structure of RcsB in the presence of the phosphomimetic BeF3− has allowed us to visualize, for the first time, the conformation competent to bind DNA for a full-length dimeric RR of the NarL/FixJ subfamily. This is an asymmetric dimer conformation acquired thanks to the dimerization driven by the REC domains through interaction with the structural elements α1–α5 surface, which produce a face-to-face disposition of REC domains that lock the phosphoryl groups at the active site, reducing in this manner the possibility of dephosphorylation. In addition, the dimer is reinforced by the interaction of the DBD domains, mainly through α10. To acquire this asymmetric conformation the DBD domains present different conformational disposition with respect to the REC domains by moving away from this domain. Although the structure of full-length RcsB in the unphosphorylated conformation has not been solved, the available structure of other members of Narl/FixJ subfamily in this state showed a monomeric closed compact conformation where REC and DBD domains present a high number of intramolecular contacts (33,44). Therefore, the phosphorylation induces the dissociation of REC and DBD domains, allowing alternative relative dispositions of both domains thanks to the flexible REC–DBD connector. Indeed, the relaxation of DBD domain from REC mediated by phosphorylation is observed in other subfamilies of RRs (33–35). In this phosphorylated asymmetric conformation, the DBD acquires an organization competent to bind promoters in a tail-to-tail arrangement, as we have confirmed in vitro by checking the binding of different mutants to P1flhDC and the rcsA promoters, similar to the organization observed in the complex structure of DBD–NarL with its nirB promoter (38). Furthermore, the in vivo analysis using RcsB variants corroborate this asymmetric conformation as competent to recognize and bind the bona fide RcsB-regulated promoters. On the other hand, the structure of RcsB containing a sulfate ion at the active site showed an interesting new type of dimerization where the REC domain from one subunit interacts with the DBD domain from the other subunit but any intramolecular REC–DBD contact is observed. This conformation, which we have named ‘crossed’, seems to be a further step in the process of REC–DBD relaxation mediated by phosphorylation which is possible due to the REC–DBD connector. Although similar crossed dimers have not been reported yet, the extended conformation of RR, where the REC and DBD domains have not intramolecular contacts, is characteristic of the active state of RR from NarL/FixJ subfamily (34,35). Surprisingly, in the crossed conformation, the α9 DNA recognition helix of RcsB is exposed to the solvent in a disposition able to bind DNA. However, the relative arrangement of α9 helices in this crossed conformation would not allow the recognition of two consecutive sequences within the same DNA strands but, instead, two independent sites in two DNA strands which would have an almost parallel orientation. According to the crystal structure, the crossed dimer can acquire a hexameric quaternary structure. In this hexameric conformation, the two DNA chains that bind each RcsB dimer would be connected, wrapping the RcsB hexamer and generating a ‘nucleosome-like structure’ surrounded by a DNA of ∼140 bp. It could be reasonably to assume that this hexameric structure is induced by crystallographic contacts and, consequently, that it lacks of biological relevance. However, the stabilization of this arrangement by a disulfide bond (S207C variant) confirms that the RcsB dimer adopts the crossed conformation in vivo. Despite the reducing character of the E. coli cytoplasm, a high proportion of expressed RcsB S207C is present as a dimer and the three dimensional-structure of this dimer confirms the crossed conformation. Furthermore, the S207C-RcsBcrossed dimer acquires a hexameric organization identical to the observed for the WT RcsBcrossed, supporting that this arrangement is not induced by crystal contacts since both proteins crystallized in different space groups. Finally, the in vivo analysis of this mutant showed that S207C-RcsB is able to inhibit flhDC transcription as the WT protein but, unlike WT RcsB, was unable to promote capsule formation. Altogether, in vitro, in vivo and crystallographic data supports that the RcsBcrossed dimer or hexamer structure are biologically relevant. We do not know yet which regions of DNA might be recognized by this conformation, which elucidation will be the objective of future work, but it is well known that transcription and DNA topology are mutually linked, thus we cannot exclude a regulatory role for the crossed RcsB species. In this way, it has been proposed that DNA supercoiling can work as a regulator of gene expression, in order to fine tuning differential expression of multiple operons, modulated by nutrient and environmental signals (45). Binding to two dsDNA has been also observed for the transcriptional factor Ets-1 (PDB ID: 3RI4) involved in activation or repression of numerous genes (46). As RcsB regulates transcription of many genes, it is tempting to speculate that RcsB in a crossed conformation could bind to alternative promoter regions. In this conformation, RcsB would function as a repressor of gene transcription or even as a transcriptional enhancer to activate transcription over a large distance, as it has been observed for the RR NtrC (47).
The structural and functional data suggested that the crossed conformation of RcsB would represent an active conformation with regulatory capacity, although additional experimental data would be required to confirm this point. In addition, the active site of RcsB in the crossed conformation is exposed to the solvent favoring the released of the phosphoryl group, unlike in the RcsBBeF structure where the active site is locked. The presence of a sulfate ion bound in this exposed active site could support the crossed dimer as an intermediate step previous for RcsB dephosphorylation, since sulfates tend to mimic phosphoryl groups where the additional oxygen represents the water molecule in place to attack the phosphate (48). This notion is supported by the structure of S207C–RcsBAC, where the dimer shows a REC domain (at subunit A) in the active conformation while the second REC domain (subunit B) adopts a conformation near the inactive state. Concomitant with this difference in the activation state of each subunit in S207C–RcsBAC, the inactive REC domain (subunit B) acquires an alternative disposition that precludes the crossed hexameric organization, suggesting that once a subunit is dephosphorylated, the hexamer is disassembled.
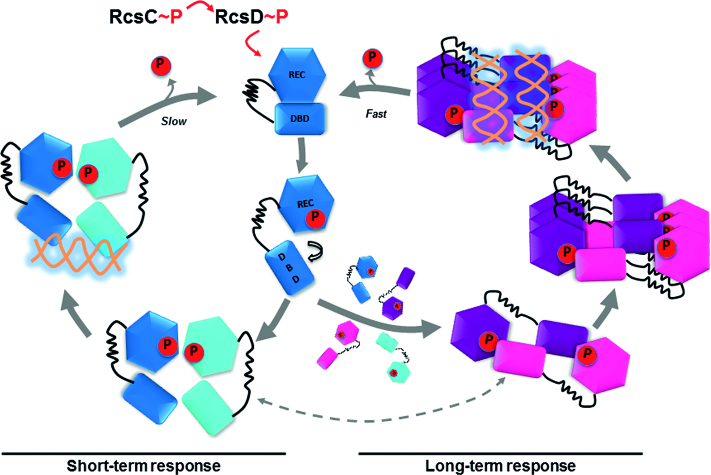
Since RcsB is a pleotropic regulator, as other members of NarL/FixJ subfamily, a conceptually attractive working model for how RcsB promotes transcriptional regulation of multiple genes could be based on its high conformational dynamism. In our working model, the initial activation of the Rcs phosphorelay by different signals triggers the RcsB phosphorylation (Figure 7). This phosphorylation induces relaxation of DBD from the REC domain and, as a consequence, the generation of a more dynamic RcsB conformation that dimerizes through the interface provided by α1–α5 surface characteristic of NarL/FixJ subfamily. This dimer presents an asymmetric conformation able to bind with high-affinity to DNA sequences with two half-sites, mainly with tail-to-tail arrangement, as in P1flhDC, wca and rcsA promoters, producing a rapid short-term response. The high dynamism of phosphorylated RcsB could allow to adopt alternative conformations, including the crossed dimer. The crossed conformation is able to bind a second pool of DNA sequence with different features as a dimer or by acquiring the hexameric architecture and inducing DNA supercoiling. The crossed dimers and hexamers could be responsible of a second wave of response by acting either as transcriptional repressors or enhancers. The acquisition of different conformations in a phosphorylation dependent manner could be the basis of sequential gene expression observed in the regulation mediated by several RRs. Indeed, it has been observed for the RR PhoP that the transcription timing is dependent on the amount of phosphorylated PhoP protein and the amount of PhoP boxes present in the promoter regions (49). Finally, in the crossed conformation the active site is solvent exposed favoring RcsB desphosphorylation and, consequently, the acquisition of the closed unphosphorylated conformation once the signal stops. Since the number of RR exceeds the number of sequences with two consecutives half-sites to recognize (50) it is tentative speculate a relationship between the RcsB regulon expression timing and the sequential acquisition of the different conformations. The prototypical asymmetric conformation would appear first, recognizing the classical two consecutives half-site and generating a short-time response. If signal persists, an excess of unbound phosphorylated RcsB molecules will be present in the cell, allowing the acquisition of the crossed conformation due to its high dynamism. The crossed RcsB dimers or hexamers would recognize a second pool of DNA sequences and would generate a long-term response. Obviously, this is an oversimplified working model since auxiliary transcription regulators, such as RcsA, GadE, BglJ, MatA and Rflm, also participates in the RcsB complex regulatory system. However, we anticipate that features of this working model could be applied to other members of the NarL/FixJ subfamily since the conformational dynamism observed in RcsB is common for many of them.
Figure 7.
Model for RcsB DNA-binding regulation mediated by phosphorylation. The initial activation of the Rcs phosphorelay triggers RcsB phosphorylation, which dimerizes through the α1–α5 surface. This asymmetric dimer is able to bind, with high-affinity, DNA sequences with two half-sites, mainly with tail-to-tail arrangement, producing a fast short-term response. If signal persists, an excess of highly dynamic phosphorylated RcsB molecules could form crossed dimers able to bind a second pool of DNA sequence with different features and acquire the hexameric architecture inducing DNA supercoiling, producing a long-term response. We suggest that the asymmetric dimer could show a slow dephosphorylation as dimerization locks the active site hindering the release of the phosphoryl group while the crossed dimer could dephosphorylate faster, as the active site is more solvent exposed. We cannot exclude that RcsB could fluctuate between the asymmetric and crossed conformations (represented by dashed gray lines), during its regulation, reflecting its conformational dynamism.
The structural and functional information accumulated during the last two decades, mainly in the OmpR/PhoB subfamily, have been vital to propose a mechanism of activation by phosphorylation in RRs depicted as Y-T coupling (43). This mechanism explains how Asp phosphorylation induces conformational changes at specific residues or structural elements that are propagated to the α4β5α5 surface, which is used by the OmpR/PhoB family to dimerize. However, this mechanism seems not be universal for RRs, since alternative mechanisms deferring for the Y-T coupling has been proposed (51,52). This would be in line with the observation of alternative dimerization surfaces in other RRs, suggesting that these RRs should not follow a canonical Y-T coupling mechanism, with final output effect involving modification of the α4β5α5 surface. This is the case of RcsB and other RRs of the NarL/FixJ subfamily that dimerize using α1–α5 surface and also does not follow completely the Y-T mechanism, as the residue that occupies the position of Tyr at β5 adopts a constitutive active inward position. The structural analysis of RcsB and other NarL/FixJ members in the inactive and active conformation has allowed us to propose a variant of this mechanism where the phosphorylation induces a movement of the Thr which is not coupled with a conformational change at the Tyr but with a conformational change in a Leu conserved in NarL/FixJ subfamily placed two positions upstream. This shift in the effect of the Thr movement toward the C-terminal part of β5 is due to the presence of β6 that clamps the N-terminal part of β5 and induces the constitutive inward conformation of the Tyr. Thus, the phosphorylation is coupled with strong conformational changes in the Lβ5α5 instead of α4β5α5 surface, allowing the dimerization by the α1–α5 surface. The conservation in the NarL/FixJ subfamily of β6, the constitutive Tyr inward conformation and the presence of a conserved Leu two residues upstream of the Tyr supports that this mechanism should be common for the members of this subfamily of RRs. We refer to this mechanism of activation as ‘β5-T coupling’ and we propose it encompasses the classical Y-T mechanism since, in both cases, the movement of the Thr residue is coupled with residues in β5 (Tyr or Leu positions), redirecting the conformational changes toward alternative surfaces. Given the conservation of the Thr, we hypothesize that similar coupling mechanisms could be present in the RRs with alternative forms of dimerization. In this way the Thr inward conformation induced by the Asp phosphorylation could be coupled with the movement of residues at positions other than Tyr or Leu, which would transduce the phosphorylation effect toward different directions, generating alternative surfaces of dimerization.
ACCESSION NUMBERS
Atomic coordinates and structure factors for the reported crystal structures have been deposited at the Protein Data bank under accession number 5O8Z for RcsBBeF, 5O8Y for RcsBcrossed, 6E02 for S207C–RcsBcrossed and 6E03 for S207C–RcsBAC.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank the NKI Protein Facility for provision of LIC vector(s) and to the IBV-CSIC Crystallogenesis Facility for protein crystallization screenings. The experimental results reported in this article derive from measurements made at the synchrotrons Diamond Light Source (DLS, Didcot, UK) and ALBA (Cerdanyola del Valles, Spain). Data collection experiments for the best crystals of RcsB structures were taken respectively at ALBA synchrotron beamline BL13 (Xaloc) and at Diamond Light source DLS I03 and I04–1 beamlines. We thank the staff of the beamlines used at the synchrotrons for assistance in the measurement of the crystals.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Spanish Government (Ministry of Economy and Competitiveness) Grants [BIO2013–42619-P, BIO2016–78571-P to A. M, BFU2016–78606-P to P.C, BIO2016–77639-P to F.G.dP]; Valencian Government Grant Prometeo [II/2014/029 to A.M.]; Ministry of Economy and Competitiveness Ramón y Cajal contract (to P.C.); Diamond Light Source block allocation group (BAG) Proposal [MX10121]; Spanish Synchrotron Radiation Facility ALBA Proposal [2015071314]. Funding for open access charge: Spanish Government (Ministry of Economy and Competitiveness) Grants [BIO2013-42619-P, BIO2016-78571-P to A. M, BIO2016-77639-P to F.G.dP, BFU2016-78606-P to P.C.]; Valencian Government grant Prometeo [II/2014/029 to A.M.].
Conflict of interest statement. None declared.
REFERENCES
- 1. Martinez-Antonio A., Collado-Vides J.. Identifying global regulators in transcriptional regulatory networks in bacteria. Curr. Opin. Microbiol. 2003; 6:482–489. [DOI] [PubMed] [Google Scholar]
- 2. Gottesman S. Bacterial regulation: global regulatory networks. Annu. Rev. Genet. 1984; 18:415–441. [DOI] [PubMed] [Google Scholar]
- 3. Gama-Castro S., Salgado H., Santos-Zavaleta A., Ledezma-Tejeida D., Muniz-Rascado L., Garcia-Sotelo J.S., Alquicira-Hernandez K., Martinez-Flores I., Pannier L., Castro-Mondragon J.A. et al. . RegulonDB version 9.0: high-level integration of gene regulation, coexpression, motif clustering and beyond. Nucleic Acids Res. 2016; 44:D133–D143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Majdalani N., Gottesman S.. The Rcs phosphorelay: a complex signal transduction system. Annu. Rev. Microbiol. 2005; 59:379–405. [DOI] [PubMed] [Google Scholar]
- 5. Clarke D.J. The Rcs phosphorelay: more than just a two-component pathway. Future Microbiol. 2010; 5:1173–1184. [DOI] [PubMed] [Google Scholar]
- 6. Mariscotti J.F., Garcia-del Portillo F.. Genome expression analyses revealing the modulation of the Salmonella Rcs regulon by the attenuator IgaA. J. Bacteriol. 2009; 191:1855–1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Farris C., Sanowar S., Bader M.W., Pfuetzner R., Miller S.I.. Antimicrobial peptides activate the Rcs regulon through the outer membrane lipoprotein RcsF. J. Bacteriol. 2010; 192:4894–4903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cho S.H., Szewczyk J., Pesavento C., Zietek M., Banzhaf M., Roszczenko P., Asmar A., Laloux G., Hov A.K., Leverrier P. et al. . Detecting envelope stress by monitoring beta-barrel assembly. Cell. 2014; 159:1652–1664. [DOI] [PubMed] [Google Scholar]
- 9. Pannen D., Fabisch M., Gausling L., Schnetz K.. Interaction of the RcsB response regulator with auxiliary transcription regulators in Escherichia coli. J. Biol. Chem. 2016; 291:2357–2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wehland M., Bernhard F.. The RcsAB box. Characterization of a new operator essential for the regulation of exopolysaccharide biosynthesis in enteric bacteria. J. Biol. Chem. 2000; 275:7013–7020. [DOI] [PubMed] [Google Scholar]
- 11. Stratmann T., Pul U., Wurm R., Wagner R., Schnetz K.. RcsB-BglJ activates the Escherichia coli leuO gene, encoding an H-NS antagonist and pleiotropic regulator of virulence determinants. Mol. Microbiol. 2012; 83:1109–1123. [DOI] [PubMed] [Google Scholar]
- 12. Castanie-Cornet M.P., Cam K., Bastiat B., Cros A., Bordes P., Gutierrez C.. Acid stress response in Escherichia coli: mechanism of regulation of gadA transcription by RcsB and GadE. Nucleic Acids Res. 2010; 38:3546–3554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Venkatesh G.R., Kembou Koungni F.C., Paukner A., Stratmann T., Blissenbach B., Schnetz K.. BglJ-RcsB heterodimers relieve repression of the Escherichia coli bgl operon by H-NS. J. Bacteriol. 2010; 192:6456–6464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kuhne C., Singer H.M., Grabisch E., Codutti L., Carlomagno T., Scrima A., Erhardt M.. RflM mediates target specificity of the RcsCDB phosphorelay system for transcriptional repression of flagellar synthesis in Salmonella enterica. Mol. Microbiol. 2016; 101:841–855. [DOI] [PubMed] [Google Scholar]
- 15. Mouslim C., Hughes K.T.. The effect of cell growth phase on the regulatory cross-talk between flagellar and Spi1 virulence gene expression. PLoS Pathog. 2014; 10:e1003987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Latasa C., Garcia B., Echeverz M., Toledo-Arana A., Valle J., Campoy S., Garcia-del Portillo F., Solano C., Lasa I.. Salmonella biofilm development depends on the phosphorylation status of RcsB. J. Bacteriol. 2012; 194:3708–3722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wang Q., Harshey R.M.. Rcs signalling-activated transcription of rcsA induces strong anti-sense transcription of upstream fliPQR flagellar genes from a weak intergenic promoter: regulatory roles for the anti-sense transcript in virulence and motility. Mol. Microbiol. 2009; 74:71–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Stout V., Gottesman S.. RcsB and RcsC: a two-component regulator of capsule synthesis in Escherichia coli. J. Bacteriol. 1990; 172:659–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gao R., Stock A.M.. Biological insights from structures of two-component proteins. Annu. Rev. Microbiol. 2009; 63:133–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Filippova E.V., Wawrzak Z., Ruan J., Pshenychnyi S., Schultz R.M., Wolfe A.J., Anderson W.F.. Crystal structure of nonphosphorylated receiver domain of the stress response regulator RcsB from Escherichia coli. Protein Sci. 2016; 25:2216–2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pristovsek P., Sengupta K., Lohr F., Schafer B., von Trebra M.W., Ruterjans H., Bernhard F.. Structural analysis of the DNA-binding domain of the Erwinia amylovora RcsB protein and its interaction with the RcsAB box. J. Biol. Chem. 2003; 278:17752–17759. [DOI] [PubMed] [Google Scholar]
- 22. Wemmer D.E., Kern D.. Beryllofluoride binding mimics phosphorylation of aspartate in response regulators. J. Bacteriol. 2005; 187:8229–8230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Luna-Vargas M.P., Christodoulou E., Alfieri A., van Dijk W.J., Stadnik M., Hibbert R.G., Sahtoe D.D., Clerici M., Marco V.D., Littler D. et al. . Enabling high-throughput ligation-independent cloning and protein expression for the family of ubiquitin specific proteases. J. Struct. Biol. 2011; 175:113–119. [DOI] [PubMed] [Google Scholar]
- 24. Dominguez-Bernal G., Pucciarelli M.G., Ramos-Morales F., Garcia-Quintanilla M., Cano D.A., Casadesus J., Garcia-del Portillo F.. Repression of the RcsC-YojN-RcsB phosphorelay by the IgaA protein is a requisite for Salmonella virulence. Mol. Microbiol. 2004; 53:1437–1449. [DOI] [PubMed] [Google Scholar]
- 25. Cano D.A., Dominguez-Bernal G., Tierrez A., Garcia-Del Portillo F., Casadesus J.. Regulation of capsule synthesis and cell motility in Salmonella enterica by the essential gene igaA. Genetics. 2002; 162:1513–1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Datsenko K.A., Wanner B.L.. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U.S.A. 2000; 97:6640–6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rosu V., Chevance F.F., Karlinsey J.E., Hirano T., Hughes K.T.. Translation inhibition of the Salmonella fliC gene by the fliC 5′ untranslated region, fliC coding sequences, and FlgM. J. Bacteriol. 2006; 188:4497–4507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kabsch W. Xds. Acta Crystallogr. D Biol. Crystallogr. 2010; 66:125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Winn M.D., Ballard C.C., Cowtan K.D., Dodson E.J., Emsley P., Evans P.R., Keegan R.M., Krissinel E.B., Leslie A.G., McCoy A. et al. . Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 2011; 67:235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Emsley P., Lohkamp B., Scott W.G., Cowtan K.. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010; 66:486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Murshudov G.N., Skubak P., Lebedev A.A., Pannu N.S., Steiner R.A., Nicholls R.A., Winn M.D., Long F., Vagin A.A.. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D Biol. Crystallogr. 2011; 67:355–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Taylor D., Cawley G., Hayward S.. Quantitative method for the assignment of hinge and shear mechanism in protein domain movements. Bioinformatics. 2014; 30:3189–3196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Leonard P.G., Golemi-Kotra D., Stock A.M.. Phosphorylation-dependent conformational changes and domain rearrangements in Staphylococcus aureus VraR activation. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:8525–8530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Park A.K., Lee J.H., Chi Y.M., Park H.. Structural characterization of the full-length response regulator spr1814 in complex with a phosphate analogue reveals a novel conformational plasticity of the linker region. Biochem. Biophys. Res. Commun. 2016; 473:625–629. [DOI] [PubMed] [Google Scholar]
- 35. Trajtenberg F., Albanesi D., Ruetalo N., Botti H., Mechaly A.E., Nieves M., Aguilar P.S., Cybulski L., Larrieux N., de Mendoza D. et al. . Allosteric activation of bacterial response regulators: the role of the cognate histidine kinase beyond phosphorylation. MBio. 2014; 5:e02105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Gao R., Mack T.R., Stock A.M.. Bacterial response regulators: versatile regulatory strategies from common domains. Trends Biochem. Sci. 2007; 32:225–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Maris A.E., Kaczor-Grzeskowiak M., Ma Z., Kopka M.L., Gunsalus R.P., Dickerson R.E.. Primary and secondary modes of DNA recognition by the NarL two-component response regulator. Biochemistry. 2005; 44:14538–14552. [DOI] [PubMed] [Google Scholar]
- 38. Maris A.E., Sawaya M.R., Kaczor-Grzeskowiak M., Jarvis M.R., Bearson S.M., Kopka M.L., Schroder I., Gunsalus R.P., Dickerson R.E.. Dimerization allows DNA target site recognition by the NarL response regulator. Nat. Struct. Biol. 2002; 9:771–778. [DOI] [PubMed] [Google Scholar]
- 39. Krissinel E., Henrick K.. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007; 372:774–797. [DOI] [PubMed] [Google Scholar]
- 40. Francez-Charlot A., Laugel B., Van Gemert A., Dubarry N., Wiorowski F., Castanie-Cornet M.P., Gutierrez C., Cam K.. RcsCDB His-Asp phosphorelay system negatively regulates the flhDC operon in Escherichia coli. Mol. Microbiol. 2003; 49:823–832. [DOI] [PubMed] [Google Scholar]
- 41. Thao S., Chen C.S., Zhu H., Escalante-Semerena J.C.. Nepsilon-lysine acetylation of a bacterial transcription factor inhibits Its DNA-binding activity. PLoS One. 2010; 5:e15123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hu L.I., Chi B.K., Kuhn M.L., Filippova E.V., Walker-Peddakotla A.J., Basell K., Becher D., Anderson W.F., Antelmann H., Wolfe A.J.. Acetylation of the response regulator RcsB controls transcription from a small RNA promoter. J. Bacteriol. 2013; 195:4174–4186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Stock A.M., Guhaniyogi J.. A new perspective on response regulator activation. J. Bacteriol. 2006; 188:7328–7330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Baikalov I., Schroder I., Kaczor-Grzeskowiak M., Grzeskowiak K., Gunsalus R.P., Dickerson R.E.. Structure of the Escherichia coli response regulator NarL. Biochemistry. 1996; 35:11053–11061. [DOI] [PubMed] [Google Scholar]
- 45. Hatfield G.W., Benham C.J.. DNA topology-mediated control of global gene expression in Escherichia coli. Annu. Rev. Genet. 2002; 36:175–203. [DOI] [PubMed] [Google Scholar]
- 46. Babayeva N.D., Baranovskaya O.I., Tahirov T.H.. Structural basis of Ets1 cooperative binding to widely separated sites on promoter DNA. PLoS One. 2012; 7:e33698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Liu Y., Bondarenko V., Ninfa A., Studitsky V.M.. DNA supercoiling allows enhancer action over a large distance. Proc. Natl. Acad. Sci. U.S.A. 2001; 98:14883–14888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Casino P., Rubio V., Marina A.. Structural insight into partner specificity and phosphoryl transfer in two-component signal transduction. Cell. 2009; 139:325–336. [DOI] [PubMed] [Google Scholar]
- 49. Zwir I., Yeo W.S., Shin D., Latifi T., Huang H., Groisman E.A.. Bacterial nucleoid-associated protein uncouples transcription levels from transcription timing. MBio. 2014; 5:doi:10.1128/mBio.01485-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Cai S.J., Inouye M.. EnvZ-OmpR interaction and osmoregulation in Escherichia coli. J. Biol. Chem. 2002; 277:24155–24161. [DOI] [PubMed] [Google Scholar]
- 51. Fernandez I., Otero L.H., Klinke S., Carrica Mdel C., Goldbaum F.A.. Snapshots of conformational changes shed light into the NtrX receiver domain signal transduction mechanism. J. Mol. Biol. 2015; 427:3258–3272. [DOI] [PubMed] [Google Scholar]
- 52. Villali J., Pontiggia F., Clarkson M.W., Hagan M.F., Kern D.. Evidence against the “Y-T coupling” mechanism of activation in the response regulator NtrC. J. Mol. Biol. 2014; 426:1554–1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.