Abstract
Cells adapt to environmental changes, including fluctuations in oxygen levels, through the induction of specific gene expression programs. To identify genes regulated by hypoxia at the transcriptional level, we pulse-labeled HUVEC cells with 4-thiouridine and sequenced nascent transcripts. Then, we searched genome-wide binding profiles from the ENCODE project for factors that correlated with changes in transcription and identified binding of several components of the Sin3A co-repressor complex, including SIN3A, SAP30 and HDAC1/2, proximal to genes repressed by hypoxia. SIN3A interference revealed that it participates in the downregulation of 75% of the hypoxia-repressed genes in endothelial cells. Unexpectedly, it also blunted the induction of 47% of the upregulated genes, suggesting a role for this corepressor in gene induction. In agreement, ChIP-seq experiments showed that SIN3A preferentially localizes to the promoter region of actively transcribed genes and that SIN3A signal was enriched in hypoxia-repressed genes, prior exposure to the stimulus. Importantly, SINA3 occupancy was not altered by hypoxia in spite of changes in H3K27ac signal. In summary, our results reveal a prominent role for SIN3A in the transcriptional response to hypoxia and suggest a model where modulation of the associated histone deacetylase activity, rather than its recruitment, determines the transcriptional output.
INTRODUCTION
Organisms are constantly exposed to a wide range of environmental changes and maintenance of tissue homeostasis critically depends on transcriptional responses to different types of stresses. These responses typically involve the mobilization of hundreds of genes with roughly equal numbers of genes being up- and downregulated (1). Fluctuations in oxygen availability are a common stress that affects oxidative metabolism and the production of oxygen free radicals, thus metazoan cells constantly monitor oxygen levels. When oxygen metabolic requirement exceed its supply, a condition known as hypoxia, cells induce a transcriptional program aimed at reducing oxygen consumption and increasing its delivery. This gene expression response is mainly driven by the hypoxia inducible factors (HIFs), a family of heterodimeric transcription factors (TFs) composed of α and β subunits. The stability and transcriptional activity of HIF α subunit is regulated by oxygen, whereas HIF β subunit (Ah Receptor Nuclear Translocator, ARNT), is insensitive to hypoxia. Of the three genes encoding for HIF α subunits, HIF1A (2) and EPAS1 (also known as HIF2A) (3–5) are the best characterized members of the family. The proteins encoded by these genes share a common mechanism of regulation by hypoxia that involves the control of stability and transactivation activity through oxygen-dependent hydroxylation of specific residues (6,7). However, these genes differ in their pattern of expression. While HIF1A gene is ubiquitously expressed, EPAS1 mRNA is restricted to specific tissues and cell types; being particularly abundant in endothelial cells (3,4,6). In addition, these factors regulate partially overlapping sets of target genes (8,9). The HIF pathway is activated in a wide variety of pathological situations, including highly prevalent diseases such as cancer and cardiorespiratory diseases (10). Thus, understanding how hypoxia regulates gene expression could open up new avenues for clinical management of these diseases.
Several independent reports have investigated the regulation of gene expression in response to hypoxia from a global perspective. A consistent finding in these studies is that, although HIFs are required for both gene induction and repression (11), their DNA binding profile only correlates with gene induction; suggesting that repression is indirectly mediated by HIF (9,12–18). In keeping with this hypothesis, HIF1A directly regulates the expression of repressors such as MXI1 (19), BHLHE40 (20) and BACH1 (21), that in turn repress the expression of individual genes in response to hypoxia (21–24). However, the relative contribution of these factors to hypoxia-induced repression is unknown and the general mechanism by which hypoxia represses transcription is not clear yet. On the other hand, although the mechanism of gene upregulation is much better understood, only a relatively small fraction (∼10–20%) of the genes induced by hypoxia have a proximal HIF binding site (13,25). Although these genes could be directly regulated by HIF bound to a distant enhancer, their regulation could also be indirectly mediated by TFs acting downstream of HIF. In agreement with this possibility, HIF induces the expression of many TFs (13,26), although their role on the gene regulatory network induced by hypoxia is unclear yet. Thus, in spite of important advances, relevant questions regarding the mechanism by which hypoxia regulates transcription, and in particular gene repression, remain unanswered.
A further aspect of the transcriptional response to hypoxia where our knowledge is scarce regards to the role of cofactors (27). The role of p300/CBP as a HIF coactivator necessary for gene induction has long been established (28) and the interaction between these proteins is one of the oxygen-regulated steps in the transcriptional response to hypoxia (29). However, p300/CBP is only required for 35–50% of global HIF-1-responsive gene expression (30). Other cofactors, including CDK8 that is enriched at the regulatory regions of ∼65% of hypoxia-inducible genes (31), are thus likely to be required for a complete response. The role of corepressor complexes in the response to hypoxia has not been addressed yet, although it is known that HIF recruits histone deacetylases (HDACs) and, intriguingly, HDAC inhibitors prevent HIF-mediated transcription (reviewed in 32). Several corepressor complexes have been identified and it is generally assumed that gene repression is achieved by the combinatorial action of various enzymatic corepressor complexes that are recruited to DNA by sequence-specific TFs (repressors) that often act through enzymatic modification of histone protein tails (33). However, in spite of their role in transcription silencing, mounting evidence suggest that corepressors may have a much broader role in the regulation of gene expression including the induction of transcription (34). One of the best studied is the SIN3 complex, a conserved corepressor found from yeast to animals, that associates with HDAC1 and HDAC2 (35,36). Interestingly, SIN3A interacts with the repressors MXI1 (37) and E2F7 (38,39) that act downstream of HIF to promote gene repression in response to hypoxia.
In this work we aimed to identify factors that contribute to control the gene expression program in response to hypoxia and centered our analysis in endothelium as a relevant cell type for the induction of angiogenesis, one of the key adaptive responses to hypoxia. Through the analysis of the binding profile of almost 200 factors generated by the ENCODE project, we found that several subunits of the SIN3A complex were over-represented in the proximity of genes whose transcription is repressed by hypoxia. Further analysis demonstrated that SIN3A was required for full repression of 75–91% of genes downregulated by hypoxia. Unexpectedly, we also found that SIN3A was also required for the complete induction of 47–51% of upregulated genes. Thus, SIN3 regulates the vast majority of the transcriptional response to hypoxia. The analysis of the genome-wide binding profile of SIN3A shows that the complex is present in the promoter of hypoxia-regulated genes in normoxia and its binding to these loci is not altered by hypoxia, unlike H3K27ac signal that changes in parallel to the effects of hypoxia in gene expression. Therefore, in contrast to a simple model where repression is regulated by targeting of the corepressor complex, our analysis unveils that the recruitment of SIN3A is not required as a regulatory step in the transcriptional response to hypoxia. More generally, our results suggest that SIN3A is a general modulator of transcription that is essential to mount a complete response to environmental stresses through a mechanism that does not involve differential recruitment to target loci.
MATERIALS AND METHODS
Cell culture and treatments
Human Umbilical Vein Endothelial Cells (HUVEC) were purchased from Lonza (Lonza, C2519A) and maintained in Endothelial Cell Growth Medium (EGM-2 Bullet Kit, Lonza, CC-3162). HEK 293T were maintained in Dulbecco's modified Eagle’s medium (Gibco, 41966052) supplemented with 50 U/ml penicillin (Gibco, 15140122), 50 μg/ml streptomycin (Gibco, 15140122), 2 mM glutamine (Gibco, 25030123) and 10% (v/v) fetal bovine serum (Gibco, 10270106). All cells were grown at 37°C and 5% CO2 in a humidified incubator containing 5% CO2 and tested regularly for mycoplasma contamination. For hypoxia treatment, cells were grown at 37°C in a 1% O2, 5% CO2, 94% N2 gas mixture in a Whitley Hypoxystation H35 (Don Withley Scientific).
Metabolic labeling with 4-thiouridine and purification of newly synthesized mRNA
We used the protocol described by Dolken et al., and Schawnhausser et al. (40,41) that is represented in the Figure 1A. Briefly, exponentially growing HUVEC cells were exposed to 21 or 1% oxygen for 8 h and pulse-labeled with 4-thiouridine (400 μM, 4sU, Sigma, T4509) during the last 2 h of treatment. After treatment, total cellular RNA was isolated from cells using TRI-reagent (Ambion, AM9738). Hundred microgram of total RNA was subjected to a biotinylation reaction to label the newly transcribed RNA containing the 4sU moiety (Pierce, EZ-Link Biotin-HPDP, 21341). Then, RNA was purified using Ultrapure TM Phenol:Chloroform:Isoamylalcohol (Invitrogen, 15593–031) and labeled RNA was isolated from the total RNA by affinity chromatography using streptavidin coated magnetic beads (μMacs Streptavidin Kit; Miltenyi, 130–074-101).
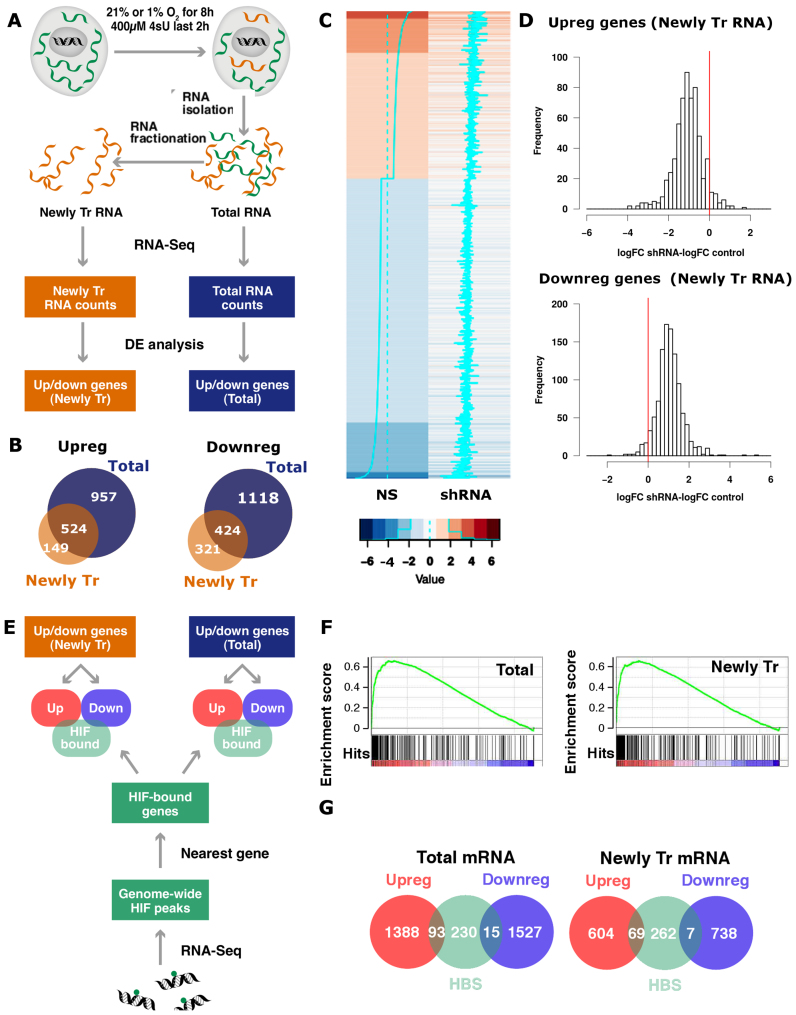
Figure 1.
Identification of the transcriptional component of the hypoxia-induced gene expression pattern. (A) Exponentially growing non-synchronized HUVEC were exposed to 21 or 1% oxygen for 8 h and pulse labeled with 4sU prior RNA extraction. An aliquot of each RNA sample was kept (Total RNA) and the rest was processed to isolate the newly transcribed (4sU-labeled) RNA pool. Poly-A RNA was purified from each of the three fractions (total, 4sU-labeled and pre-existing RNA), analyzed by high-throughput sequencing and quantified by mapping reads to the exonic regions of genes (RNAseq). The results shown derive from three independent biological replicas, each using a different pool of HUVEC donors. (B) Venn diagrams representing the number of genes significantly (FDR < 0.05) up- or downregulated shared by the total (blue circles) and newly transcribed (orange circles) fractions. (C and D) Control (NS) and EPAS1-silenced (shRNA) HUVEC were exposed to 1% oxygen for 16 h, pulse labeled with 4sU and the newly transcribed mRNA fraction from each sample was analyzed by RNAseq. The results shown derive from a single biological replica. The treatment with shRNA reduced the expression of EPAS1 to 17% of control values. (C) Heatmap representing the log-fold change (logFC) expression in hypoxia over normoxia in control (‘NS’) and EPAS1 knockdown cells. (‘shRNA’) for all transcripts affected by hypoxia (absolute logFC > 1). Each row represents a transcript in a color code that indicates their response to hypoxia (upregulated genes in red and downregulated in blue) and the magnitude of change as shown indicated in the legend below the heatmaps. The cyan lines also indicate the magnitude of the fold change for a particular transcript, and the dotted line indicates no change in expression. (D) For each gene whose expression up- (logFC > 1, top panel) or downregulated (logFC < −1, bottom panel) by hypoxia in control cells, we computed the difference between the changes induced by hypoxia in EPAS1-silenced cells and control cells, that is the logFC observed in EPAS1-silenced cells minus logFC observed in controls (‘logFC shRNA-logFC NS’). The red line indicates a difference value of zero, which would be expected in the case of lack of effect of the interference. The mean of the differences was significantly different of zero in all the cases (single sample student’s t-test, P < 0.001). (E) HUVEC were exposed to 1% oxygen for 16 h and then processed to determine ARNT and EPAS1 binding sites by ChIP-Seq. Regions bound by HIF were assigned to the nearest gene and compared to mRNAs significantly up- (‘Upreg.’) or downregulated (‘Downreg.’) by hypoxia in the total and 4sU-labeled fractions from part A. (F) The genes detected by RNA-seq in total (left graph) and newly transcribed fractions (right graph) were sorted according to their response to hypoxia (from strongly induced on the left, labeled in red color, to strongly repressed on the right, labeled in blue color) and distribution of EPAS1 binding sites across these ranked lists of genes was analyzed by Gene Set Enrichment Analysis (GSEA). HIF binding sites were significantly enriched (FDR < 0.01) in the genes whose expression was induced by hypoxia in both mRNA fractions. (G) Venn diagrams representing the number of genes significantly (FDR < 0.05) up- (red circles) or downregulated (blue circles) by hypoxia that present a HIF binding site (HBS, green circles). The same analysis was performed for genes differentially expressed in the total mRNA (left graph) and 4sU-labeled (right graph) fractions. The proportion of genes showing HIF binding sites was significantly different for the up- and downregulated categories (two-proportion test, P < 0.001).
High-throughput sequencing and bioinformatics analysis
Libraries were prepared using the standard protocol for messenger RNA sequencing (Illumina, TruSeq Stranded mRNA) and sequenced on HiSeq2000 instrument (Illumina) according to the manufacturer’s protocol. The resulting reads were aligned to the GRCh37/hg19 assembly of the human genome using TopHat (42) with the default parameters. Finally, the gene expression level was calculated as the number of reads per gene, computed using HTSeq (43) and gene features as defined in the GRCh37.75 release of the human genome (gtf file) and expressed as RPKMs. Differential gene expression analysis was performed with the Bioconductor (44) edgeR package (45) for the R statistical Software (http://www.R-project.org/). The raw reads and processed data derived from RNA-seq experiments are available at NCBI’s Gene Expression Omnibus (46,47) and are accessible through the following GEO Series accession numbers: GSE89831, GSE89838, GSE89839, GSE89840. All computations were performed using R software package (http://www.R-project.org/).
Chromatin immunoprecipitation
Chromatin immunoprecipitation was performed as previously described by Schödel et al. (14) using antibodies against HIF1A (PM14), EPAS1 (PM9) (9,48), ARNT (Novus, NB-100–110; RRID:AB_10003150), SIN3A (Abcam, ab3479; RRID:AB_303839) and a Rabbit Control IgG (Abcam, ab46540; RRID:AB_2614925).
Libraries were prepared using the Illumina ChIP-seq kit and sequenced on the Illumina GAII platform according to the manufacturer’s protocol. Sequences were mapped to the GRCh37/hg19 assembly of the human genome using ELAND software (HIF ChIPseq) or Bowtie2 (REF) (SIN3A ChIPseq). Binding peaks were determined from the aligned reads using MACS software (49) using default parameters and the pre-immune sample as a control. A single ChIP experiment was analyzed by ChIP-sequencing and Raw and mapped data are available at GEO (GSE89836 and GSE103245). The analysis of mapped reads and genomic intervals defined by the bound regions was performed with the GenomicRanges (50) and Genomation (51) Bioconductor packages.
Lentiviral production
Lentiviral vector pGIPz (Open Biosystems) was used to silence the mRNA expression of HIF1A (V2LHS_132150), EPAS1 (V2LHS_113753), SIN3A (V3LHS_343545) and MXI1 (V3LHS_408351), as well as a Non-silencing lentiviral shRNA Control (NS; RHS4346). Lentiviruses were produced and tittered in HUVEC cells as previously described (52). For transduction of target cells, lentiviruses were used at a multiplicity of infection (MOI) of 1–2 for 8 h, resulting in more than 95% transduced (GFP-positive) cells 72 h after infection.
Identification of trans-acting factors enriched in transcriptionally regulated genes
The March 2012 internal data freeze consisting in 690 ChIP-seq datasets representing 161 unique regulatory factors and generated by the ENCODE Consortium (53) was downloaded from the UCSC genome browser (54,55) and from Bioconductor using AnnotationHub version 2.4.2. We only considered peaks with -log10 FDR > 2. For TFs that had been tested in more than one cell line only the intersection of all experiments was considered. HIF1A binding sites conserved across cell types obtained from the integration of published ChIP-chip and ChIP-seq experiments data (12–15,18,56–57) and HUVEC genome-wide EPAS1 binding sites derived from our ChIP-seq were also included. Peaks of TFs binding were ascribed to the closest transcription start site (TSS). Significantly enriched binding sites for any of the TFs were determined by a Fisher’s exact test. All analyses were performed in R software package (http://www.R-project.org/).
RESULTS
Transcriptional regulation in hypoxia is indirectly mediated by HIF
HIFs are master regulators of the gene expression pattern induced by hypoxia that are required for the induction and repression of the vast majority of genes regulated by hypoxia (11). However, many of the effects of HIF on gene expression could be indirect since only ∼10–20% of the induced genes present HIF binding sites (13,25) and HIF binding does not correlate with gene repression (13,14,17,25). Therefore, we sought to identify TFs acting downstream of HIF during the adaptive response to hypoxia of HUVEC.
The gene profiling studies published to date were based on the analysis of steady state mRNA levels, and thus cannot differentiate between effects on transcription and decay rates. To address this caveat, we pulse-treated HUVEC for 2 h with 4sU to label nascent RNA and then characterized the pattern of newly transcribed mRNAs by affinity capture of the labeled transcripts followed by high-throughput sequencing (Figure 1A, top). Incorporation of 4sU into mRNA after a short pulse can be used as a proxy for transcription rate (40,41) and comparison of changes in this nascent fraction to those in total RNA allowed us to identify and focus on those genes whose regulation by hypoxia occurs at the transcriptional level (Figure 1A). This analysis identified 1481 and 673 upregulated genes (FDR < 0.05) in the total and newly transcribed mRNA fractions respectively and 1542 and 745 downregulated genes in these same fractions (Figure 1B and Supplementary Table S1). Comparison of the genes differentially expressed in each of these fractions revealed that only 35.4% of the genes whose expression was induced by hypoxia showed statistically significant changes in their transcription rate. Likewise, only 27.5% of downregulated genes had significantly repressed their transcription. Thus, although transcription is the major determinant of the gene expression changes induced by hypoxia (Tiana et al., manuscript in preparation), we only found statistically significant changes in transcription rate for a subset of the genes showing altered steady state expression levels.
We next decided to investigate the role of HIF on the changes in transcription induced by hypoxia and focussed on EPAS1 which is the predominant HIF isoform expressed in HUVEC (Supplementary Figure S1). Figure 1C represents the effect of hypoxia on transcription (logarithm of the ratio of nascent mRNA levels in hypoxia over normoxia) in control (‘NS’) and EPAS1 silenced cells (‘shEPAS1’, knock-down to 15–19.8% of control values) as a heat map where each row represents an individual gene and the color code indicates the magnitude and direction of the changes induced by hypoxia (log-fold change hypoxia over normoxia). To quantify the magnitude of the effect, we first classified genes as upregulated (log-fold change > 1) or downregulated (log-fold change < −1) attending to the effect of hypoxia in their transcription rate in control cells. Then, for each gene, we calculated the difference of log-fold induction values in EPAS1-silenced (Figure 1D, ‘shRNA’) minus control cells (Figure 1D, ‘NS’). Of the 617 genes whose transcription was induced by hypoxia in control HUVEC, 581 (94%) showed reduced expression in knock-down cells (Figure 1D top panel; mean of differences = −1.05, 95% CI [−1.11, −0.99]). Similarly, EPAS1-silencing attenuated the transcriptional repression of 1021 out of 1067 genes (96%), resulting in positive values of fold induction differences (Figure 1D bottom panel; mean of differences = +1.04, 95% CI [+1.01, +1.08]).
These experiments indicate that EPAS1 globally mediates the effects of hypoxia on transcription in HUVEC, affecting both induction and repression to a similar extent. Next, to investigate if the effects on transcription were directly mediated by HIF we determined the genome-wide binding patterns of EPAS1 and ARNT in HUVEC by chromatin immunoprecipitation followed by high-throughput sequencing (ChIP-seq) (Figure 1E). To increase the stringency in the annotation we only considered genomic segments bound by both EPAS1 and ARNT, as proposed in previous studies (14). This analysis yielded a total of 392 regions bound by ARNT/EPAS1 in HUVEC (Supplementary Table S2), a value of similar magnitude to that found in other cell types (12,13,17). In order to compare pan-genomic binding data with the effects of hypoxia on gene expression, EPAS1 binding regions were assigned to the nearest gene locus, resulting in a total of 338 genes bound by this factor (Supplementary Table S2). EPAS1 bound genes strongly clustered with genes upregulated by hypoxia in a Gene Set Enrichment Analysis (GSEA) (58) in both total (Figure 1F, left graph) and newly transcribed (Figure 1F, right graph) mRNA fractions; strongly supporting that HIF binding increases transcription rate. However, only 6% (93 out of 1481) and 10% (69 out of 673) of loci bound by EPAS1 were significantly induced by hypoxia in the total and newly transcribed fractions respectively (Figure 1G). In the case of gene repression, 1% of the genes with reduced expression in total (15 out of 1527) or newly transcribed (7 out of 745) fractions were bound by EPAS1 (Figure 1G). This proportion is significantly lower than the 6–10% observed for upregulated genes (normal approximation of proportions, Student’s t-test, P < 0.01). Thus, almost all the transcriptional repression and, at least part, of the induction are indirectly mediated by HIF in HUVEC.
Identification of candidate DNA-binding proteins contributing to gene regulation in hypoxia
In order to identify the DNA-binding proteins that could mediate the indirect effects of HIF on transcription, we employed motif finding algorithms (59–61) to search for factors over-represented in the vicinity of genes whose transcription was significantly affected by hypoxia. However, this approach is limited by design to sequence-specific TFs and shows low specificity (62).
To overcome these limitations and expand our search to coregulatory proteins binding DNA indirectly, we exploited the hundreds of ChIP-seq experiments from the ENCODE Consortium (53). Specifically, we searched for trans-acting factors over-represented in the regulatory regions of genes whose transcription is regulated by hypoxia (Figure 2A). Since the number of factors experimentally determined in HUVEC is small, we used binding sites of all TFs analyzed by ENCODE in any cell line as a proxy for the genome-wide regulatory landscape. To exclude cell-type specific binding events and maximize the selection of sites likely to be present in HUVEC, for each TF we only considered binding sites that were conserved (i.e. overlapping) in all cell lines studied by the ENCODE consortium (Figure 2A). These stringent criteria resulted in a list of binding sites expected to be conserved across cell lines. In addition to the factors analyzed by ENCODE, we also included a data set describing HIF1A, EPAS1 and ARNT binding sites derived from the integration of published ChIP-chip and ChIP-seq experiments (63). Finally, we also included into this analysis pipeline the genome-wide EPAS1 binding sites derived from the ChIP-seq experiment described herein. For each DNA-interacting factor, we ascribed binding sites to the nearest TSS and compared the distribution of binding sites in genes induced or repressed by hypoxia with those whose transcription remained constant (Figure 2A).
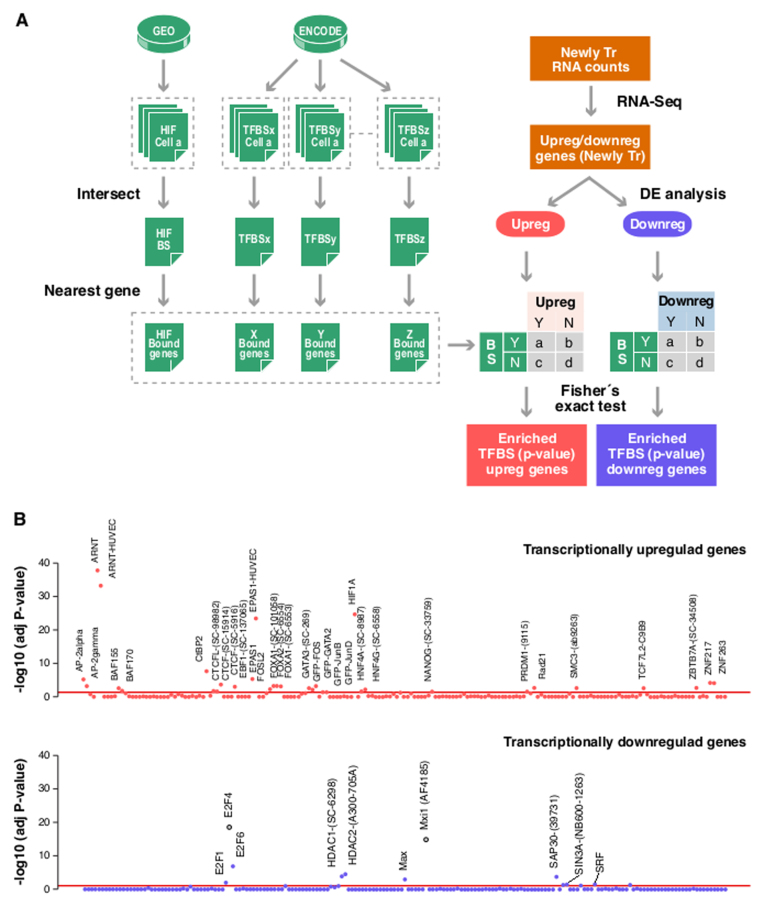
Figure 2.
TF binding sites over-represented in genes regulated by hypoxia. The presence of binding sites for each one of the 166 factors (‘TFBSx’, ‘TFBSy’,…‘TFBSz’) assayed by the ENCODE project and HIF binding sites determined elsewhere (‘HIF’) as determined for each of the genes whose transcription is significantly modulated by hypoxia in HUVEC cells (‘Newly Tr. RNA counts’). For factors assayed in more than one cell line by ENCODE, we only considered those peaks present in all cell types. For each TF we assigned binding sites to the nearest gene. For association studies, genes were categorized according to their response to hypoxia into up- or downregulated sets and the distribution of binding sites for each factor in each set was compared to that expected by chance by Fisher exact test. The resulting P-values were adjusted for multiple testing. (A) Schematic diagram depicting the analysis strategy. (B) The figure shows the association between binding for each of the analyzed factors (x-axis) and the induction (upper graph) or repression (bottom graph) of the nearest genes. Each dot represents the FDR-adjusted P-value obtained in the Fisher’s for a single factor. Red line correspond to an adjusted P-value of 0.05. The identity of the factors significantly over-represented is indicated.
As expected, this analysis revealed that the distribution of HIF1A, EPAS1 and ARNT binding sites was strongly biased toward genes whose transcription was induced by hypoxia (Figure 2B, upper graph, ‘transcriptionally upregulated genes’; Fisher’s exact test, FDR-adjusted P-value < 0.0001). We also found enrichment for other factors, albeit showing a much less skewed distribution toward upregulated genes (Figure 2B, upper graph). Thus, these additional TFs could contribute to gene induction in hypoxia acting cooperatively with HIF or downstream of it.
Importantly, the analysis of genes whose transcription is repressed under hypoxia also revealed a set of DNA-binding factors that were significantly over-represented (Figure 2B, bottom graph, ‘transcriptionally downregulated genes’). Among them was the transcriptional repressor MXI1, a factor known to act downstream of HIF to repress specific genes (19), the MXI binding partner MAX, and members of the E2F family, particularly the transcriptional repressors E2F4 and E2F6. In addition to these sequence-specific TFs, we also found significant enrichment for HDAC1 and HDAC2 and the proteins SIN3 Transcription Regulator Family Member A (SIN3A) and Sin3A Associated Protein 30 kDa (SAP30). Notably, HDAC1/2, SAP30 and SIN3A are components of the HDAC co-repressor complex Sin3A which is required for transcriptional repression by MXI1 (37) and E2Fs (38,39).
Taken together, these results suggest that the Sin3A co-repressor complex could mediate the repression of transcription observed under hypoxia.
Depletion of SIN3A disrupts the hypoxic gene expression pattern
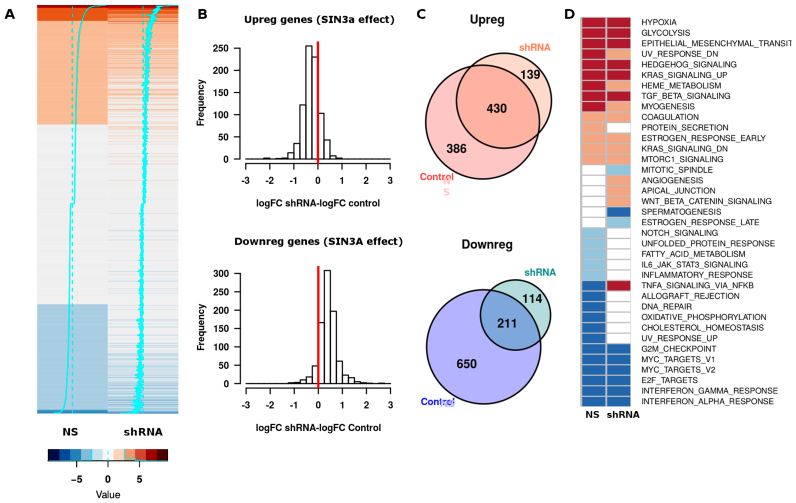
In the view of these results, we decided to focus our attention on the role of the SIN3A complex on the transcriptional response to hypoxia. To investigate the functional role of this complex, we analyzed the effect of SIN3A knock-down on the hypoxic transcriptome by means of RNA-sequencing (Figure 3 and Supplementary Table S3). Figure 3A represents the effect of hypoxia on transcription in control (‘NS’) and SIN3A silenced cells (‘shRNA’) as a heat map where each row represents an individual gene and the color code indicates the magnitude and direction of the changes induced by hypoxia (log-fold change hypoxia over normoxia). SIN3A knock-down to 36% of control levels had a profound impact on the transcriptional response to hypoxia and, unexpectedly, it affected not only gene repression but also impaired gene induction. To quantify the magnitude of the effect, we first classified genes as upregulated (FDR < 0.001 and log-fold change > 0) or downregulated (FDR < 0.001 and log-fold change < 0) attending to the effect of hypoxia in the mRNA levels in control cells. Then, for each gene, we calculated the difference of log-fold induction values (log2 hypoxic over normoxic mRNA values) in SIN3A-silenced minus their value in control cells. SIN3A RNA interference attenuated the repressive effect of hypoxia for 779 out of the 861 genes (91%) that were downregulated in control cells (Figure 3B, bottom graph; mean of differences = +0.41, 95% CI [+0.38,+0.43]). In agreement, <25% (211 out of 861) of the genes significantly repressed by hypoxia in control conditions were still found significantly repressed in knockdown cells (Figure 4C, bottom graph). Gene induction was also affected by SIN3A interference, with 661 out of 816 upregulated genes showing reduced induction in knock-down cells (Figure 3B, upper graph). This effect was of smaller magnitude compared to the effect on repressed genes (mean of differences = −0.27, 95% CI [−0.29, −0.25]), but statistically significant (single sample Student’s t-test, P < 0.001). Accordingly, 53% (430 out of 816) of the genes induced in control cells were still significantly upregulated upon SIN3A interference (Figure 3C, upper graph). In agreement with these results, analysis of the ontology terms associated to the genes regulated by hypoxia revealed that SIN3A is required for a wide range of biological pathways regulated by hypoxia (Figure 3D). In particular, SIN3A interference affected metabolic functions repressed by hypoxia such as oxidative phosphorylation and fatty acid metabolism (Figure 3D).
Figure 3.
Effect of SIN3A knock-down on hypoxia-induced gene regulation. Exponentially growing non-synchronized HUVEC were transduced with lentiviral particles encoding for shRNA against SIN3A mRNA (‘shRNA’) or for a non-specific shRNA (‘NS’); 72 h after infection cells were exposed to 21 or 1% oxygen for 16 h, poly-A RNA was purified and analyzed by RNAseq. The results from two independent biological replicates, each using a different pool of HUVEC donors, were used to compute gene expression values. The treatment with shRNA reduced the expression of SIN3A to 34% (replicate 1) and 38% (replicate 2) of control values. The effect of hypoxia on each gene was calculated as the log-fold change of hypoxic over normoxic values (‘logFC’). (A) Heatmap representing the changes in expression, as log2 fold changes of hypoxic compared with normoxic conditions (logFC), in control (‘NS’) and SIN3A-silenced cells (‘shRNA’). The graph represents all transcripts differentially expressed in control cells (FDR < 0.01) as was described in Figure 2. (B) For each gene whose expression was significantly (FDR < 0.01) up- (upper graph) or downregulated (bottom graph) by hypoxia in control cells, we computed the difference between the changes induced by hypoxia in SIN3A-silenced cells and control cells (‘logFC shRNA-logFC NS’). The red line indicates a difference value of zero, which would be expected in the case of lack of effect of the interference. The mean of the differences was significantly different of zero in all the cases (single sample student’s t-test, P < 0.001). (C) Venn diagrams representing the number of genes significantly up- (upper graph) or downregulated (bottom graph) in control and SIN3A (‘shRNA’) interfered cells. (D) The heatmap represents the biological functions associated to genes differentially induced (represented in red) and repressed (blue) in control (‘NS’) and silenced cells (‘ShRNA’) as determined by GSEA against the ‘Hallmarks’ gene sets. Light colors represent adjusted P-value < 0.05 and dark colors adjusted P-value < 0.01.
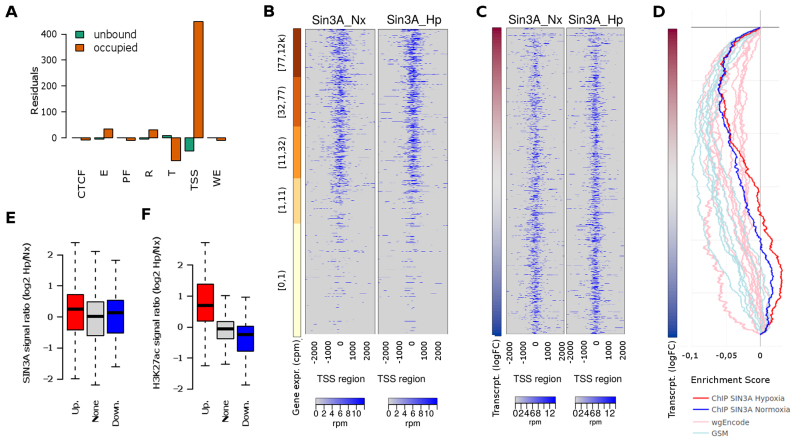
Figure 4.
Genome-wide SIN3A binding profile. Exponentially growing non-synchronized HUVEC were exposed to 1% oxygen for 16 h and then processed to determine SIN3A binding profile by ChIP-Seq. (A) The distribution of the normoxic SIN3A binding regions across the functional regions of the HUVEC genome was studied and compared to that expected by chance using a chi-squared goodness of fit test (Chi-square6 = 213 740, P < 10−15). The graph represents the Pearson residuals resulting from the test. The functional regions (chromatin states as defined by the ENCODE consortium) are: ‘TSS’, predicted promoter region including TSS; ‘PF’, Predicted promoter flanking region; ‘E’, Predicted enhancer; ‘WE’, Predicted weak enhancer or open chromatin cis regulatory element; ‘CTCF’, CTCF enriched element; ‘T’, Predicted transcribed region; ‘R’, Predicted Repressed or Low Activity region. (B and C) Heatmaps showing the enrichment of SIN3A in a 4 Kb region centered in the TSS. Each row represent a gene and rows are sorted according to gene expression level as determined from RNA-seq (B) or log fold change in transcription (Newly transcribed fraction) induced by hypoxia (C). In the latter case, only genes with detectable expression (cpm > 1 in at least two samples) are represented. (D) GSEA graph the distribution of the normoxic SIN3A (blue) and hypoxic (red) SIN3A binding sites from this study (ChIP SIN3A) and those determined by the ENCODE project (‘wgEncode’, ligth red) and individual research groups (‘GSM’, light blue) across genes expressed in HUVEC sorted according to their transcriptional response to hypoxia as in C. The information about each SIN3A data set together with ES and P-values for echrichment is shown in Supplementary Table S4. (E) The log-ratio of the normalized SIN3A signal in hypoxia over normoxia was computed for all promoters. The boxplot represent the distribution of values obtained for genes upregulated (‘Up.’, logFC > 1 and adjusted P-values < 0.001), downregulated (‘Down.’, logFC < −1 and adjusted P-values < 0.001) and not effected by hypoxia (‘None’). The differences between groups were not statistically significant for α = 0.01 (ANOVA F29 053 = 3.707, P = 0.0246). (F) Wig files from reference (56) were downloaded from the NCBI GEO repository (GSE50144). The log-ratio of the normalized H3K27ac signal in hypoxia over normoxia was computed for all promoters and represented as in (E). The differences between mean signals in each group were statistically significant (ANOVA F29 053 = 247.9, P < 2e-16) and the signal in both up- and downregulated groups was significantly different to that in control genes (post-hoc Tukey HDS, adjusted P-value < 2e-16).
SIN3A binding profile is not altered by hypoxia
The effect of SIN3A on gene induction was unanticipated given its established function as a corepressor and the observed association with genes repressed under hypoxia (Figure 2B). In addition, hypoxia did not alter the expression of SIN3A nor affected its subcellular localization (Supplementary Figure S2). For these reasons, we next determined the genome-wide binding pattern of SIN3A in HUVEC. Sequencing of the chromatin immunoprecipitated with SIN3A revealed 16 875 regions (FDR < 5%) bound by this factor in normoxic HUVEC (Supplementary Table S4A), which is similar to the number of sites described by the ENCODE project in other cell lines (mean 11 617 peaks, range 6024–21 309 peaks, n = 9 datasets). Interestingly, the analysis of the distribution of these regions across the different functional elements defined by the genome-wide segmentation of HUVEC (57), showed a strong enrichment of SIN3A in active promoter regions (Figure 4A), with a distribution that was significantly different to that expected by chance (Chi-square6 = 213 740, P < 10−15). The correlation between SIN3A signal and gene expression, is further analyzed in Figure 4B, which represents the SIN3A signal in a region of 4 Kb centered at the TSS across all human genes (each row represents a gene) sorted according to their expression in HUVEC. This representation shows that SIN3A signal is prominent on the promoters of genes actively transcribed in normoxic HUVEC but faint on genes with very low (1–10 counts per million of reads) or not detectable expression (<1 count per million of reads) (Figure 4B). The same pattern was found when we analyzed the 11 948 regions (FDR < 5%) bound by SIN3A in hypoxic conditions (Figure 4B and Supplementary Table S4B). Although paradoxical, this result concurs with previous reports describing the presence of SIN3A in active promoters (64) and the fact that HDACs are highly recruited to actively transcribed genes (65). Next, we studied whether the distribution of SIN3A correlated with the changes on transcription induced by hypoxia. To this end we selected genes expressed in HUVEC (those with cpm > 1 in at least two samples of the RNA-seq experiments) and represented SIN3A signal across genes sorted by their response to hypoxia (log-fold change of the ratio hypoxia over normoxia) (Figure 4C). Although we detected SIN3A in the promoters of the majority of expressed genes, the distribution of SIN3A binding sites was slightly skewed toward genes repressed by hypoxia (Figure 4C). The bias in distribution of SIN3A signal with respect to hypoxia response is more clearly observed when SIN3A binding loci were examined for enrichment among hypoxia-regulated genes using GSEA (Figure 4D and Supplementary Table S5). Interestingly, the skewed distribution was observed in normoxic cells prior to the exposure to the stimuli (Figure 4C, left panel and blue line in Figure 4D), in agreement with the enrichment of SIN3A proximal to genes repressed by hypoxia found in the analysis of the ENCODE datasets (Figures 2B and 4D). Moreover, the binding pattern of SIN3A under hypoxia (Figure 4C, right panel, and red line in Figure 4D) was very similar to the one observed under normoxic conditions. In agreement, we did not observe statistically significant differences in the recruitment of SIN3A to hypoxia-responsive genes (Figure 4E). These results strongly argue against the regulation of SIN3A recruitment and point to modulation of the activity of the pre-assembled complex in response to hypoxia. Consistent with this possibility, the transcription of yeast genes INO1 and CHO in response to nutrients was found to be mediated by modulation of the associated HDAC activity rather than recruitment of the SIN3A complex (64). To test this possibility, we analyzed publicly available datasets (GSE50144) of H3K27 acetylation in HUVEC (66) and found that, in response to hypoxia, the signal of this histone modification was increased in the promoters of induced genes and reduced in repressed genes (Figure 4F). Altogether these results suggest that the activity of the complex, rather than its recruitment to target genes, could be regulated in response to hypoxia to modulate the transcription of the hypoxia-responsive genes.
DISCUSSION
In this work, we aimed to provide novel insights into the transcriptional response to hypoxia, a type of stress commonly found in a wide variety of physiological and pathological situations. Specifically, our objective was to identify novel TFs involved in this response.
To this end we employed a technique that, in contrast with previous studies, allowed us to determine the effect of hypoxia on transcription, avoiding the confounding effect that RNA decay rates might have on the gene expression pattern. This experimental approach identified a set of genes whose expression was regulated by hypoxia at the level of transcription and thus optimal for the identification of TFs that could cooperate with HIF or mediate its effects. Interestingly, the overlap between the genes found significantly regulated by hypoxia (FDR < 0.05) in the total and newly transcribed mRNA fractions was only partial, with only about a third of the changes in total mRNA being also detected at the level of transcription. However, the variability between triplicates was slightly higher in the newly transcribed than in total mRNA fraction, probably due to the additional processing steps required to isolate the 4sU-fraction, which prevented reaching statistical significance when the effect of hypoxia on transcription rates was modest. Thus, these numbers most likely underestimate the true overlap between the changes in expression and transcription. In agreement, when we focused our analysis in genes showing robust changes in expression (logFC > 1 or logFC < −1), the fraction of genes showing concordant changes in the total and newly transcribed fraction increased up to 60% approximately.
Next, in order to identify factors enriched in the proximity of hypoxia-regulated targets, we exploited datasets of genome-wide binding profiles generated by ENCODE. Although ENCODE experiments were performed under normoxic conditions, we hypothesized that any hypoxia-regulated factor is likely to be present to some extent under normoxia and hence to be detected on its target genes. The comparison of the fraction of hypoxia-regulated and control genes bound by each factor led us to the identification of sets of proteins significantly enriched in genes transcriptionally induced or repressed by hypoxia. In addition to HIF1A, EPAS1 and ARNT, we identified 24 additional factors over-represented in upregulated genes. Among them, we found FOS and its binding partners JUND and JUNB, in agreement with previous reports suggesting a cooperation between HIF and AP-1 in the induction of hypoxia-regulated genes (9,56,67). In addition, some of these factors might be acting downstream of HIF to control the hypoxic gene expression pattern. In support to this possibility, we found that only 10% of the genes transcriptionally upregulated by hypoxia were bound by HIF. Although the uncertainty associated with the assignation of binding sites to the nearest gene could contribute to the reduced correlation between binding and gene expression, this result strongly suggests that most transcriptional changes are indirectly induced by HIF, as previously suggested (13).
In the case of downregulated genes, we found enrichment for ten proteins, including several repressors of the E2F family, MXI1 and its dimerization partner MAX, as well as several components of the SIN3A HDAC corepressor complex. These findings are particularly relevant as the mechanisms that mediate gene repression by hypoxia are poorly understood and the role of corepressors in the transcriptional response to hypoxia has not been studied before. For these reasons we decided to further investigate the role of the SIN3A corepressor.
Knock-down of SIN3A had a profound impact on the transcriptional response to hypoxia, affecting to 75% of the genes downregulated by hypoxia and 47% of upregulated genes. However, it is important to note that in most cases SIN3A depletion did not fully prevent, but rather attenuated the effect of hypoxia on gene expression. This could be a consequence of incomplete SIN3A interference as we could only reduce its expression to 36% of control levels on average. However, it is well known that removal of corepressor function results in gene expression changes that tend to be subtle, suggesting that they function to fine-tune transcript levels rather than turn transcription on and off (34). Given the role of SIN3A as a corepressor, the effect of its interference on hypoxia-triggered gene induction was puzzling. In some cases, the attenuated induction was a consequence of increased basal gene expression upon SIN3A interference, providing a potential explanation for this effect that is in agreement with the function of SIN3A as a corepressor. However, this was not the case for the majority of genes (360 out of 386) showing attenuated induction, suggesting that SIN3A may be required for full induction of some genes. Although these results might seem paradoxical, SIN3A was initially described in yeast as a protein with dual functions as activator and repressor (68,69) and recent works are putting forward its role as an activator of specific genes (reviewed in 36). Of particular relevance to our work, previous works suggest that the SIN3A HDAC complex is required for the induction of genes in response to environmental insults such as heat or osmotic stress (64,65) in yeast and xenobiotics in mammalian cells (70). Thus, additionally to its undisputed role as corepressor, SIN3A seems to be also playing a role on transcriptional activation. In fact, the role of corepressors in transcriptional upregulation is not unique to SIN3A. Loss of different types of corepressor proteins usually results in similar numbers of genes decreasing and increasing their expression (34), so that emerging models propose that recruitment of both corepressor and coactivator complexes is needed for gene induction (33). Importantly, the requirement of SIN3A for hypoxia-triggered gene induction could explain, at least in part, why HDAC inhibitors, that in most cases promote gene expression, in the context of several HIF target promoters prevent transcription (30,71).
Aiming to understand the molecular mechanism by which SIN3A affects the transcriptional response to hypoxia we determined its genome-wide binding profile. Interestingly, we found that SIN3A is preferentially found at the promoter region of actively transcribed genes, in agreement with a recent report showing that SIN3A and NANOG co-occupy active promoters (72). Although, SIN3A signal was found in most active promoters, it was absent from about 30% of the hypoxia-inducible genes. In contrast, it was absent from 5% of repressed genes only; resulting in a distribution of SIN3A binding sites slightly skewed toward hypoxia-repressed genes. This distribution was highly reproducible as it was also observed in other SIN3A binding datasets, including those from ENCODE and individual GEO submissions (Figure 4D and Supplementary Table S5). A question arising from these results is whether this pattern is shared by other corepressor complexes. In addition to SIN3A, ENCODE also determined the binding profiles of COREST, CTBP2, MTA3, SMARCA4, SMARCB1, SMARCC1, SMARCC2, MBD4, CHD1, CHD2, KDM5A, KDM5B and HDAC6 that are subunits of different corepressor and chromatin remodeling complexes such as CoREST, NURD and SWI/SNF. However, none of these factors were enriched in genes downregulated by hypoxia (Figure 2). On the other hand, analysis of datasets generated by individual groups and deposited in GEO confirmed an over-representation of binding of components of the SIN3A complex but not subunits of the NCor, SMRT, NURD or CoREST corepressors in repressed genes (supplementary Tables S5 and 6). Thus, the skewed distribution of SIN3A binding sites is not observed for other corepressors. Interestingly, the comparison of normoxic and hypoxic SIN3A binding profiles revealed very similar patterns suggesting that, in spite of the effect of SIN3A interference on gene expression, hypoxia did not regulate SIN3A recruitment. In agreement, although we found individual cases where SIN3A is recruited by hypoxia to repressed genes (Supplementary Figure S3), as a general rule we did not find significant changes in the SIN3A signal associated to promoters of upregulated nor downregulated genes (Figure 4). In contrast, H3K27ac signal on the promoter of repressed genes was significantly reduced in response to hypoxia. Although we can not rule alternative mechanisms, our data support a model where hypoxia regulates the activity of SIN3A complexes constitutively located at target genes, rather that its recruitment. Supporting this possibility, a recent work demonstrates that yeast Sin3 can be detected at target promoters INO1 and CHO2 and contributes to the induction or repression of these genes through its interaction with activators or repressors that are recruited to these promoters under different conditions (64). Thus, a plausible explanation is that HIFs induce the expression of sequence-specific repressors that in turn activate the HDAC activity of the SIN3A complex already bound to target genes. In this regard, the SIN3A complex interacts to MXI1 to silence gene expression (73) and we found enrichment of MXI1 binding to downregulated genes (Figure 3). Moreover, MXI1 mRNA is robustly induced by hypoxia in HUVEC cells (Supplementary Table S1) in a HIF-dependent manner (data not shown). To investigate the possibility of MXI1 acting downstream of HIF to regulate SIN3A located at target genes, we studied the effect of MXI1 interference on hypoxia induced gene repression (Supplementary Figure S4). Although silencing of MXI1 attenuated the repression of 245 out of 409 genes found significantly repressed in control HUVEC cells (Supplementary Figure S3), the effect was of very small magnitude (mean of differences = +0.06, 95% CI [+0.02, +0.09]). Accordingly, the number and identity of genes significantly repressed by hypoxia remained unaltered in silenced cells. This result is in agreement with other reports (74) and suggests functional redundancy with other transcriptional repressors in the MAD family, such as MXD4, that are also induced by hypoxia in HUVEC cells. Interestingly, the SIN3A complex also binds to the repressor members of the E2F family (38,39) including E2F4 and E2F6 that were also found enriched in downregulated genes (Figure 3). E2F4 and E2F6 mRNAs were not induced by hypoxia in HUVEC (Supplementary Table S1), but both of them have been implicated in the repression of BRCA1 and RAD51 in hypoxia (75,76). In addition, a recent report indicates that HIF1A physically interacts with E2F7 to form an active repressor complex that prevents transcription (77). Thus, it is feasible that a similar mechanism is operative in HUVEC cells whereby HIF may directly interact with E2F4/6 and regulate the activity of SIN3A on repressed genes. Future work will address this hypothesis and the role of Mad and E2F family members on hypoxia-induced gene repression. Besides MXI1 and E2Fs we found other factors that, although enriched in downregulated genes, did not pass the stringent threshold of adjusted P-value < 0.005. Among them is bHLHe40 (adjusted P-value = 0.0107, Fisher’s exact test for the association with downregulated genes). As discussed before, bHLHe40 has been shown to mediate the repression of specific genes downstream of HIF.
Regardless the specific mechanism, our results demonstrate that SIN3A is required for a complete transcriptional response to hypoxia. SIN3A interference not only affected individual genes but also altered paradigmatic hypoxia-regulated processes, such as suppression of oxidative phosphorylation and fatty acid oxidation (Figure 3). Interestingly, recent reports in Saccharomyces and Drosophila (78) demonstrate that loss of SIN3 affect mitochondrial activity, suggesting an evolutionarily conserved role for SIN3 in the control of cellular energy production. Further studies will be required to test the role of SIN3A in the changes induced by hypoxia on mitochondrial function.
Altogether, our results delineate an unanticipated role for the SIN3A corepressor complex in the transcriptional response to hypoxia, highlighting that the regulation of gene expression by environmental oxygen is substantially more complex than has previously been considered. More generally, our findings strongly support a prominent role for SIN3A complex in the up- and downregulation of transcription in response to a wide array of environmental stresses.
AVAILABILITY
All datasets generated as part of this study are available at NCBI’s Gene Expression Omnibus (GEO) (46,47) and are accessible through the following GEO Series accession numbers:
RNA-Seq 4SU-labeled HUVEC three biological replicates three RNA fractions: GSE89831
RNA-Seq HUVEC shRNA Sin3A two biological replicate total RNA fractions: GSE89840
RNA-Seq 4SU-labeled HUVEC shRNA EPAS1 one biological replicate two RNA fractions: GSE89838
RNA-Seq HUVEC shRNA EPAS1/HIF1A/MXI1 one biological replicate total RNA fractions: GSE89839
ChIP-Seq HUVEC EPAS1/HIF1A/ARNT one biological replicate: GSE89836
ChIP-Seq HUVEC SIN3A one biological replicate: GSE103245
The reference series for all the experiments in this publication is GSE89841
Supplementary Material
ACKNOWLEDGEMENTS
We thank Diego Villar for critical review of the manuscript and suggestions. We also like to thank Ramon Diaz-Uriarte for his advice on statistical analysis and for lending us computing power to perform the analysis of high-throughput data.
Footnotes
Present address: Rebecca Worsley-Hunt, AG Ohler-Computational Regulatory Genomics, Max Delbrück Center for Molecular Medicine. Berlin Inst. for Medical Systems Biology, Berlin, Germany.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Ministerio de Ciencia e Innovación (Spanish Ministry of Science and Innovation, MICINN) [SAF2011_24225 to L.d.P., SAF2014–53819-R to L.d.P., B.J.]; Canadian Institutes of Health Research (CIHR) [MOP-82875 to W.W.).]; Natural Sciences and Engineering Research Council of Canada (NSERC) [RGPIN355532–10 to W.W.W.]; National Institutes of Health [1R01GM084875 to W.W.W.]; CIHR Fellowship (to R.W.H.); Michael Smith Foundation for Health Research Fellowship (to R.W.H.); Caja Madrid Foundation for Visiting Professor Fellowship (to L.d.P). Funding for open access charge: Spanish Ministry of Science and Innovation, MICINN, [SAF2014-53819-R].
Conflict of interest statement. None declared.
REFERENCES
- 1. de Nadal E., Ammerer G., Posas F.. Controlling gene expression in response to stress. Nat. Rev. Genet. 2011; 12:833–845. [DOI] [PubMed] [Google Scholar]
- 2. Wang G.L., Semenza G.L.. Characterization of hypoxia-inducible factor 1 and regulation of DNA binding activity by hypoxia. J. Biol. Chem. 1993; 268:21513–21518. [PubMed] [Google Scholar]
- 3. Tian H., McKnight S.L., Russell D.W.. Endothelial PAS domain protein 1 (EPAS1), a transcription factor selectively expressed in endothelial cells. Genes Dev. 1997; 11:72–82. [DOI] [PubMed] [Google Scholar]
- 4. Ema M., Taya S., Yokotani N., Sogawa K., Matsuda Y., Fujii-Kuriyama Y.. A novel bHLH-PAS factor with close sequence similarity to hypoxia-inducible factor 1alpha regulates the VEGF expression and is potentially involved in lung and vascular development. Proc. Natl. Acad. Sci. U.S.A. 1997; 94:4273–4278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hogenesch J.B., Chan W.K., Jackiw V.H., Brown R.C., Gu Y.Z., Pray-Grant M., Perdew G.H., Bradfield C.A.. Characterization of a subset of the basic-helix-loop-helix-PAS superfamily that interacts with components of the dioxin signaling pathway. J. Biol. Chem. 1997; 272:8581–8593. [DOI] [PubMed] [Google Scholar]
- 6. Wiesener M.S., Turley H., Allen W.E., Willam C., Eckardt K.U., Talks K.L., Wood S.M., Gatter K.C., Harris A.L., Pugh C.W. et al. . Induction of endothelial PAS domain protein-1 by hypoxia: characterization and comparison with hypoxia-inducible factor-1alpha. Blood. 1998; 92:2260–2268. [PubMed] [Google Scholar]
- 7. Kaelin W.G., Ratcliffe P.J.. Oxygen sensing by metazoans: the central role of the HIF hydroxylase pathway. Mol. Cell. 2008; 30:393–402. [DOI] [PubMed] [Google Scholar]
- 8. Hu C.-J., Wang L.-Y., Chodosh L.A., Keith B., Simon M.C.. Differential roles of hypoxia-inducible factor 1alpha (HIF-1alpha) and HIF-2alpha in hypoxic gene regulation. Mol. Cell. Biol. 2003; 23:9361–9374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Salama R., Masson N., Simpson P., Sciesielski L.K., Sun M., Tian Y.-M., Ratcliffe P.J., Mole D.R.. Heterogeneous effects of direct hypoxia pathway activation in kidney cancer. PLoS One. 2015; 10:e0134645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Semenza G.L. Hypoxia-inducible factors in physiology and medicine. Cell. 2012; 148:399–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Elvidge G.P., Glenny L., Appelhoff R.J., Ratcliffe P.J., Ragoussis J., Gleadle J.M.. Concordant regulation of gene expression by hypoxia and 2-oxoglutarate-dependent dioxygenase inhibition: the role of HIF-1alpha, HIF-2alpha, and other pathways. J. Biol. Chem. 2006; 281:15215–15226. [DOI] [PubMed] [Google Scholar]
- 12. Mole D.R., Blancher C., Copley R.R., Pollard P.J., Gleadle J.M., Ragoussis J., Ratcliffe P.J.. Genome-wide association of hypoxia-inducible factor (HIF)-1alpha and HIF-2alpha DNA binding with expression profiling of hypoxia-inducible transcripts. J. Biol. Chem. 2009; 284:16767–16775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Xia X., Lemieux M.E., Li W., Carroll J.S., Brown M., Liu X.S., Kung A.L.. Integrative analysis of HIF binding and transactivation reveals its role in maintaining histone methylation homeostasis. Proc. Natl. Acad. Sci. U.S.A. 2009; 106:4260–4265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Schödel J., Oikonomopoulos S., Ragoussis J., Pugh C.W., Ratcliffe P.J., Mole D.R.. High-resolution genome-wide mapping of HIF-binding sites by ChIP-seq. Blood. 2011; 117:e207–e217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Xia X., Kung A.L.. Preferential binding of HIF-1 to transcriptionally active loci determines cell-type specific response to hypoxia. Genome Biol. 2009; 10:R113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Choudhry H., Scho’del J., Oikonomopoulos S., Camps C., Grampp S., Harris A.L., Ratcliffe P.J., Ragoussis J., Mole D.R.. Extensive regulation of the non-coding transcriptome by hypoxia: Role of HIF in releasing paused RNApol2. EMBO Rep. 2014; 15:70–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ortiz-Barahona A., Villar D., Pescador N., Amigo J., del Peso L.. Genome-wide identification of hypoxia-inducible factor binding sites and target genes by a probabilistic model integrating transcription-profiling data and in silico binding site prediction. Nucleic Acids Res. 2010; 38:2332–2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Schödel J., Bardella C., Sciesielski L.K., Brown J.M., Pugh C.W., Buckle V., Tomlinson I.P., Ratcliffe P.J., Mole D.R.. Common genetic variants at the 11q13.3 renal cancer susceptibility locus influence binding of HIF to an enhancer of cyclin D1 expression. Nat. Genet. 2012; 44:420–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhang H., Gao P., Fukuda R., Kumar G., Krishnamachary B., Zeller K.I., Dang C.V., Semenza G.L.. HIF-1 inhibits mitochondrial biogenesis and cellular respiration in VHL-deficient renal cell carcinoma by repression of C-MYC activity. Cancer Cell. 2007; 11:407–420. [DOI] [PubMed] [Google Scholar]
- 20. Miyazaki K., Kawamoto T., Tanimoto K., Nishiyama M., Honda H., Kato Y.. Identification of functional hypoxia response elements in the promoter region of the DEC1 and DEC2 genes. J. Biol. Chem. 2002; 277:47014–47021. [DOI] [PubMed] [Google Scholar]
- 21. Kitamuro T., Takahashi K., Ogawa K., Udono-Fujimori R., Takeda K., Furuyama K., Nakayama M., Sun J., Fujita H., Hida W. et al. . Bach1 functions as a hypoxia-inducible repressor for the heme oxygenase-1 gene in human cells. J. Biol. Chem. 2003; 278:9125–9133. [DOI] [PubMed] [Google Scholar]
- 22. Yun Z., Maecker H.L., Johnson R.S., Giaccia A.J.. Inhibition of PPAR gamma 2 gene expression by the HIF-1-regulated gene DEC1/Stra13: a mechanism for regulation of adipogenesis by hypoxia. Dev. Cell. 2002; 2:331–341. [DOI] [PubMed] [Google Scholar]
- 23. Choi S.M., Cho H.-J., Cho H., Kim K.H., Kim J.B., Park H.. Stra13/DEC1 and DEC2 inhibit sterol regulatory element binding protein-1c in a hypoxia-inducible factor-dependent mechanism. Nucleic Acids Res. 2008; 36:6372–6385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ivanov S. V, Salnikow K., Ivanova A.V., Bai L., Lerman M.I.. Hypoxic repression of STAT1 and its downstream genes by a pVHL/HIF-1 target DEC1/STRA13. Oncogene. 2007; 26:802–812. [DOI] [PubMed] [Google Scholar]
- 25. Mole D.R., Blancher C., Copley R.R., Pollard P.J., Gleadle J.M., Ragoussis J., Ratcliffe P.J.. Genome-wide association of hypoxia-inducible factor (HIF)-1alpha and HIF-2alpha DNA binding with expression profiling of hypoxia-inducible transcripts. J. Biol. Chem. 2009; 284:16767–16775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tsai Y.-P., Wu K.-J.. Hypoxia-regulated target genes implicated in tumor metastasis. J. Biomed. Sci. 2012; 19:102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Dengler V.L., Galbraith M.D., Espinosa J.M.. Transcriptional regulation by hypoxia inducible factors. Crit. Rev. Biochem. Mol. Biol. 2015; 49:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Arany Z., Huang L.E., Eckner R., Bhattacharya S., Jiang C., Goldberg M.A., Bunn H.F., Livingston D.M.. An essential role for p300/CBP in the cellular response to hypoxia. Proc. Natl. Acad. Sci. U.S.A. 1996; 93:12969–12973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lando D., Peet D.J., Gorman J.J., Whelan D.A., Whitelaw M.L., Bruick R.K.. FIH-1 is an asparaginyl hydroxylase enzyme that regulates the transcriptional activity of hypoxia-inducible factor. Genes Dev. 2002; 16:1466–1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kasper L.H., Boussouar F., Boyd K., Xu W., Biesen M., Rehg J., Baudino T.A., Cleveland J.L., Brindle P.K., Alarcon J. et al. . Two transactivation mechanisms cooperate for the bulk of HIF-1-responsive gene expression. EMBO J. 2005; 24:3846–3858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Galbraith M.D., Allen M.A., Bensard C.L., Wang X., Schwinn M.K., Qin B., Long H.W., Daniels D.L., Hahn W.C., Dowell R.D. et al. . HIF1A employs CDK8-mediator to stimulate RNAPII elongation in response to hypoxia. Cell. 2013; 153:1327–1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Perez-Perri J.I., Acevedo J.M., Wappner P.. Epigenetics: new questions on the response to hypoxia. Int. J. Mol. Sci. 2011; 12:4705–4721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Perissi V., Jepsen K., Glass C.K., Rosenfeld M.G.. Deconstructing repression: evolving models of co-repressor action. Nat. Rev. Genet. 2010; 11:109–123. [DOI] [PubMed] [Google Scholar]
- 34. Reynolds N., O’Shaughnessy A., Hendrich B.. Transcriptional repressors: multifaceted regulators of gene expression. Development. 2013; 140:505–512. [DOI] [PubMed] [Google Scholar]
- 35. Grzenda A., Lomberk G., Zhang J.S., Urrutia R.. Sin3: Master scaffold and transcriptional corepressor. Biochim. Biophys. Acta. 2009; 1789:443–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kadamb R., Mittal S., Bansal N., Batra H., Saluja D.. Sin3: insight into its transcription regulatory functions. Eur. J. Cell Biol. 2013; 92:237–246. [DOI] [PubMed] [Google Scholar]
- 37. Rao G., Alland L., Guida P., Schreiber-Agus N., Chen K., Chin L., Rochelle J.M., Seldin M.F., Skoultchi A.I., DePinho R.A.. Mouse Sin3A interacts with and can functionally substitute for the amino-terminal repression of the Myc antagonist Mxi1. Oncogene. 1996; 12:1165–1172. [PubMed] [Google Scholar]
- 38. Lai A., Kennedy B.K., Barbie D.A., Bertos N.R., Yang X.J., Theberge M.C., Tsai S.C., Seto E., Zhang Y., Kuzmichev A. et al. . RBP1 recruits the mSIN3-histone deacetylase complex to the pocket of retinoblastoma tumor suppressor family proteins found in limited discrete regions of the nucleus at growth arrest. Mol. Cell. Biol. 2001; 21:2918–2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. David G., Grandinetti K.B., Finnerty P.M., Simpson N., Chu G.C., DePinho R.A.. Specific requirement of the chromatin modifier mSin3B in cell cycle exit and cellular differentiation. Proc. Natl. Acad. Sci. U.S.A. 2008; 105:4168–4172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Schwanhäusser B., Busse D., Li N., Dittmar G., Schuchhardt J., Wolf J., Chen W., Selbach M.. Global quantification of mammalian gene expression control. Nature. 2011; 473:337–342. [DOI] [PubMed] [Google Scholar]
- 41. Dölken L., Ruzsics Z., Rädle B., Friedel C.C., Zimmer R., Mages J., Hoffmann R., Dickinson P., Forster T., Ghazal P. et al. . High-resolution gene expression profiling for simultaneous kinetic parameter analysis of RNA synthesis and decay. RNA. 2008; 14:1959–1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Trapnell C., Pachter L., Salzberg S.L.. TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics. 2009; 25:1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Anders S., Pyl P.T., Huber W.. HTSeq-A Python framework to work with high-throughput sequencing data. Bioinformatics. 2015; 31:166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Gentleman R., Carey V., Bates D., Bolstad B., Dettling M., Dudoit S., Ellis B., Gautier L., Ge Y., Gentry J. et al. . Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004; 5:R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Robinson M.D., McCarthy D.J., Smyth G.K.. edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2009; 26:139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Edgar R., Domrachev M., Lash A.E.. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002; 30:207–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Barrett T., Wilhite S.E., Ledoux P., Evangelista C., Kim I.F., Tomashevsky M., Marshall K.A., Phillippy K.H., Sherman P.M., Holko M. et al. . NCBI GEO: archive for functional genomics data sets–update. Nucleic Acids Res. 2013; 41:D991–D995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wiesener M.S., Jürgensen J.S., Rosenberger C., Scholze C.K., Hörstrup J.H., Warnecke C., Mandriota S., Bechmann I., Frei U.A., Pugh C.W. et al. . Widespread hypoxia-inducible expression of HIF-2alpha in distinct cell populations of different organs. FASEB J. 2003; 17:271–273. [DOI] [PubMed] [Google Scholar]
- 49. Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W. et al. . Model-based analysis of ChIP-Seq (MACS). Genome Biol. 2008; 9:R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Lawrence M., Huber W., Pagès H., Aboyoun P., Carlson M., Gentleman R., Morgan M.T., Carey V.J.. Software for computing and annotating genomic ranges. PLoS Comput. Biol. 2013; 9:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Akalin A., Franke V., Vlahoviček K., Mason C.E., Schübeler D.. Genomation: a toolkit to summarize, annotate and visualize genomic intervals. Bioinformatics. 2015; 31:1127–1129. [DOI] [PubMed] [Google Scholar]
- 52. Orgaz J.L., Ladhani O., Hoek K.S., Fernández-Barral A., Mihic D., Aguilera O., Seftor E.A., Bernad A., Rodríguez-Peralto J.L., Hendrix M.J.C. et al. . ‘Loss of pigment epithelium-derived factor enables migration, invasion and metastatic spread of human melanoma’. Oncogene. 2009; 28:4147–4161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. ENCODE Project Consortium An integrated encyclopedia of DNA elements in the human genome. Nature. 2012; 489:57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Speir M.L., Zweig A.S., Rosenbloom K.R., Raney B.J., Paten B., Nejad P., Lee B.T., Learned K., Karolchik D., Hinrichs A.S. et al. . The UCSC Genome Browser database: 2016 update. Nucleic Acids Res. 2016; 44:D717–D725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Rosenbloom K.R., Sloan C.A., Malladi V.S., Dreszer T.R., Learned K., Kirkup V.M., Wong M.C., Maddren M., Fang R., Heitner S.G. et al. . ENCODE data in the UCSC Genome Browser: year 5 update. Nucleic Acids Res. 2013; 41:D56–D63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Villar D., Ortiz-Barahona A., Gómez-Maldonado L., Pescador N., Sánchez-Cabo F., Hackl H., Rodriguez B.A.T.B. A.T., Trajanoski Z., Dopazo A., Huang T.H.M.T.H.M. et al. . Cooperativity of stress-responsive transcription factors in core hypoxia-inducible factor binding regions. PLoS One. 2012; 7:e45708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Hoffman M.M., Ernst J., Wilder S.P., Kundaje A., Harris R.S., Libbrecht M., Giardine B., Ellenbogen P.M., Bilmes J.A., Birney E. et al. . Integrative annotation of chromatin elements from ENCODE data. Nucleic Acids Res. 2013; 41:827–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S. et al. . Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 2005; 102:15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Ho Sui S.J., Mortimer J.R., Arenillas D.J., Brumm J., Walsh C.J., Kennedy B.P., Wasserman W.W.. oPOSSUM: Identification of over-represented transcription factor binding sites in co-expressed genes. Nucleic Acids Res. 2005; 33:3154–3164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Kwon A.T., Arenillas D.J., Worsley Hunt R., Wasserman W.W.. oPOSSUM-3: advanced analysis of regulatory motif over-representation across genes or ChIP-Seq datasets. G3 (Bethesda). 2012; 2:987–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Frith M.C., Fu Y., Yu L., Chen J.F., Hansen U., Weng Z.. Detection of functional DNA motifs via statistical over-representation. Nucleic Acids Res. 2004; 32:1372–1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Zambelli F., Pesole G., Pavesi G.. Motif discovery and transcription factor binding sites before and after the next-generation sequencing era. Brief. Bioinform. 2013; 14:225–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Roche O., Deguiz M.L.M.L., Tiana M.M., Galiana-Ribote C., Martinez-Alcazar D., Rey-Serra C., Ranz-Ribeiro B., Casitas R., Galera R., Fernández-Navarro I. et al. . Identification of non-coding genetic variants in samples from hypoxemic respiratory disease patients that affect the transcriptional response to hypoxia. Nucleic Acids Res. 2016; 44:9315–9330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Kliewe F., Engelhardt M., Aref R., Schüller H.-J.. Promoter recruitment of corepressors Sin3 and Cyc8 by activator proteins of the yeast Saccharomyces cerevisiae. Curr. Genet. 2017; 63:739–750. [DOI] [PubMed] [Google Scholar]
- 65. Wang Z., Zang C., Cui K., Schones D.E., Barski A., Peng W., Zhao K.. Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes. Cell. 2009; 138:1019–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Inoue T., Kohro T., Tanaka T., Kanki Y., Li G., Poh H.-M., Mimura I., Kobayashi M., Taguchi A., Maejima T. et al. . Cross-enhancement of ANGPTL4 transcription by HIF1 alpha and PPAR beta/delta is the result of the conformational proximity of two response elements. Genome Biol. 2014; 15:R63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Yamashita K., Discher D.J., Hu J., Bishopric N.H., Webster K.A.. Molecular regulation of the endothelin-1 gene by hypoxia. Contributions of hypoxia-inducible factor-1, activator protein-1, GATA-2, AND p300/CBP. J. Biol. Chem. 2001; 276:12645–12653. [DOI] [PubMed] [Google Scholar]
- 68. Vidal M., Gaber R.F.. RPD3 encodes a second factor required to achieve maximum positive and negative transcriptional states in Saccharomyces cerevisiae. Mol. Cell. Biol. 1991; 11:6317–6327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Vidal M., Strich R., Esposito R.E., Gaber R.F.. RPD1 (SIN3/UME4) is required for maximal activation and repression of diverse yeast genes. Mol. Cell. Biol. 1991; 11:6306–6316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Solaimani P., Wang F., Hankinson O.. SIN3A, generally regarded as a transcriptional repressor, is required for induction of gene transcription by the aryl hydrocarbon receptor. J. Biol. Chem. 2014; 289:33655–33662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Fath D.M., Kong X., Liang D., Lin Z., Chou A., Jiang Y., Fang J., Caro J., Sang N.. Histone deacetylase inhibitors repress the transactivation potential of hypoxia-inducible factors independently of direct acetylation of HIF- alpha. J. Biol. Chem. 2006; 281:13612–13619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Saunders A., Huang X., Fidalgo M., Reimer M.H., Faiola F., Ding J., Sànchez-Priego C., Guallar D., Sàenz C., Li D. et al. . The SIN3A/HDAC corepressor complex functionally cooperates with NANOG to promote pluripotency. Cell Rep. 2017; 18:1713–1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Schreiber-Agus N., Chin L., Chen K., Torres R., Rao G., Guida P., Skoultchi A.I., DePinho R.A.. An amino-terminal domain of Mxi1 mediates anti-myc oncogenic activity and interacts with a homolog of the Yeast Transcriptional Repressor SIN3. Cell. 1995; 80:777–786. [DOI] [PubMed] [Google Scholar]
- 74. Löfstedt T., Fredlund E., Noguera R., Navarro S., Holmquist-Mengelbier L., Beckman S., Påhlman S., Axelson H.. HIF-1α induces MXI1 by alternate promoter usage in human neuroblastoma cells. Exp. Cell Res. 2009; 315:1924–1936. [DOI] [PubMed] [Google Scholar]
- 75. Bindra R.S., Gibson S.L., Meng A., Westermark U., Jasin M., Pierce A.J., Bristow R.G., Classon M.K., Glazer P.M.. Hypoxia-induced down-regulation of BRCA1 expression by E2Fs. Cancer Res. 2005; 65:11597–11604. [DOI] [PubMed] [Google Scholar]
- 76. Bindra R.S., Glazer P.M.. Repression of RAD51 gene expression by E2F4/p130 complexes in hypoxia. Oncogene. 2007; 26:2048–2057. [DOI] [PubMed] [Google Scholar]
- 77. de Bruin A., Cornelissen P.W.A., Kirchmaier B.C., Mokry M., Iich E., Nirmala E., Liang K., D Végh A.M., Scholman K.T., Groot Koerkamp M.J. et al. . Genome-wide analysis reveals NRP1 as a direct HIF1α-E2F7 target in the regulation of motorneuron guidance in vivo. Nucleic Acids Res. 2016; 44:3549–3566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Barnes V.L., Strunk B.S., Lee I., Hüttemann M., Pile L.L.A., Scarpulla R., Scarpulla R., Hughes T., Marton M., Jones A. et al. . Loss of the SIN3 transcriptional corepressor results in aberrant mitochondrial function. BMC Biochem. 2010; 11:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.