Abstract
In the absence of a functioning centromere, chromosome segregation becomes aberrant, leading to an increased rate of aneuploidy. The highly specific recognition of centromeres by kinetochores suggests that specific structural characteristics define this region, however, the structural details and mechanism underlying this recognition remains a matter of intense investigation. To address this, high-speed atomic force microscopy was used for direct visualization of the spontaneous dynamics of CENP-A nucleosomes at the sub-second time scale. We report that CENP-A nucleosomes change conformation spontaneously and reversibly, utilizing two major pathways: unwrapping, and looping of the DNA; enabling core transfer between neighboring DNA substrates. Along with these nucleosome dynamics we observed that CENP-A stabilizes the histone core against dissociating to histone subunits upon unwrapping DNA, unique from H3 cores which are only capable of such plasticity in the presence of remodeling factors. These findings have implications for the dynamics and integrity of nucleosomes at the centromere.
INTRODUCTION
The centromere is a specialized locus possessed by all chromosomes which aids in the segregation of sister chromatids during cell division. Failure to faithfully segregate chromosomes will result in aneuploidy, a hallmark of cancer (1). Furthermore, the presence of more than one centromere can lead to di-centric attachments, which can further result in chromosome breakage (2,3). A matter of outstanding importance is to investigate specific features of centromere derived nucleosomes which might directly aid in its role as a substrate for microtubule attachment. Numerous studies over the past two decades have shown that the characteristic feature of centromeres is the presence of centromere protein A (CENP-A); a histone in nucleosomes that replaces the H3 histone of bulk chromatin (4,5). Therefore, it is widely accepted that CENP-A nucleosomes epigenetically define the centromere and form the foundation on which the kinetochore is built.
Structural characterization of CENP-A nucleosomes by X-ray crystallographic studies revealed that the exit/entry DNA segments of the nucleosomes (13 bp) are detached from the histone core, thus shortening the DNA segment wrapped around the core by 26 bp when compared to those that contain H3 (6,7). This deficit in DNA wrapped around CENP-A nucleosomes is further supported by enzymatic digestion experiments (8). Hence, one might ask how these differences in nucleosomal DNA lengths can be translated to the structural uniqueness of CENP-A containing nucleosomes in vitro and to what degree the replacement of H3 with CENP-A affects spontaneous nucleosome dynamics.
Several recent structural studies have demonstrated the degree to which H3 and CENP-A containing histone cores can tolerate deformation. Biophysical pulling assays revealed that CENP-A is slightly more amenable to disruption than corresponding H3 nucleosomes (9); a finding that agrees well with recent all-atom molecular dynamics (MD) simulation and FRET studies (10,11). Furthermore, using high-resolution NMR spectroscopy, remodeling factors were shown to increase the plasticity of H3 histone cores, indicating that they are capable of far greater flexibility than previously supposed (12).
We recently proposed a model that explains how the shortening of nucleosomal DNA can affect the structural properties of nucleosomes, their arrays, and eventually higher order chromatin structures that define the non-canonical properties of the centromere (13). It was hypothesized that nucleosomes assembled with CENP-A follow dynamic pathways that are unique from those of bulk chromatin and likely contribute to the overall structural and dynamic properties required for a functioning centromere (10,14). Single–molecule biophysics methods capable of revealing transient states of biological systems were very beneficial for studies of nucleosomes (15). Importantly, they revealed the high inherent dynamics of isolated nucleosomes enabling their spontaneous disassembly (15–17).
In this study, we performed direct imaging of CENP-A nucleosomes by single-molecule high-speed time-lapse atomic force microscopy (AFM), enabling us to visualize dynamics of individual nucleosomes at a sub-second frame rate (13,18,19). Nucleosomes used for evaluation of DNA wrapping around the histone core were assembled on a DNA substrate containing a centrally positioned 601 motif (15–17,19). The broadly dynamic behavior of the DNA flanks was first revealed by analysis of AFM images acquired in ambient conditions. Time-lapse imaging further identified dynamic pathways unique to CENP-A nucleosomes that were not previously observed for H3. The spontaneous unwrapping of DNA flanks can be accompanied by the reversible and dynamic formation of loops with sizes equivalent to a single wrap of DNA. Furthermore, CENP-A nucleosome cores are capable of reversible translocation over the DNA substrate; a process mediated through the formation of internal DNA loops along the nucleosome core particle. Finally, the transfer of the histone core from one DNA substrate to another was visualized. The data demonstrates that CENP-A nucleosome cores have increased stability upon unwrapping of DNA, permitting it to distort freely and reversibly, which may play a critical role in centromere integrity during mitosis and replication.
MATERIALS AND METHODS
Preparation of DNA substrate and nucleosome reconstitution
The DNA substrate used in nucleosome assembly contained the 147 bp 601 strong positioning sequence (20) flanked by plasmid DNA, 154 and 122 bp in length, generated using PCR with the pGEM3Z-601 plasmid (Addgene #26656; Supplementary Figure S1; Figure 1A). Nucleosome assembly was achieved using the salt gradient dialysis method (21,22). Briefly, in 20 μl of 1× TE buffer containing 2M NaCl, recombinant human CENP-A/H4 tetramer's and H2A/H2B dimers purchased from EpiCypher (#16-0010 and #15-0311, respectively; Research Triangle Park, NC, USA) were mixed with purified DNA substrate (final concentration of 100 ng/ul) at a DNA/dimer/tetramer molar ratio of 1:2.2:1. Octamers containing H3 were purchased from EpiCypher (#16-0001) and were assembled on the 601–substrate at a ratio of 1:1 (DNA:octamer). Each assembly was loaded onto a Slide-A-Lyzer MINI dialysis unit (3500 MWCO, Thermo Fisher Scientific) and was then dialyzed at 4°C while exchanging the 2 M NaCl dialyzate from with one containing 2.5 mM NaCl (both at pH 7.5) at a rate of ∼0.5 ml/min. After 70 h, the reaction mixture was dialyzed against fresh low salt (2.5 mM NaCl) buffer for another hour. The reassembled nucleosome stock solution was stored at 4°C in the final dialysis buffer (2.5 mM NaCl, 1× TE, pH 7.5). These samples remain stable for months as confirmed using AFM imaging (17). Histone stoichiometry of assembled nucleosomes was assessed by discontinuous SDS-PAGE gel with 6% stacking and 15% separating components (Supplementary Figure S2).
Figure 1.
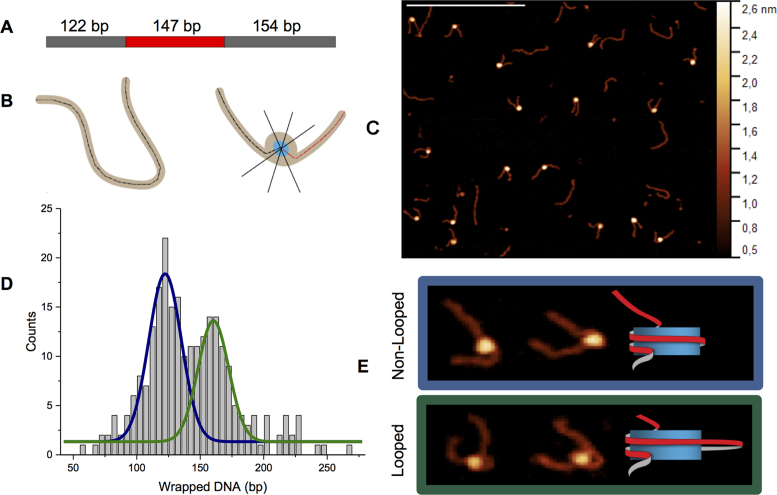
Imaging of CENP-A nucleosomes reveals the presence of looped nucleosome structure. (A) Schematic of the DNA substrate containing the 147 bp 601 positioning sequence (red) with flanking 122 and 154 bp plasmid DNA (gray). (B) Diagram of how (left) the contour length of free DNA was measured and how (right) each nucleosome ‘arm’ was measured from the end of the DNA to the center of the wrapped core (black and red dashed); the width of the particle was then measured by three height profile cross sections at FWHM (black lines through core). Half of this value was then subtracted from each ‘arm’. (C) AFM image of reconstituted CENP-A nucleosomes on an APS-mica surface (scale bar = 400 nm); each round white feature is a histone core with two DNA ‘arms’. Histograms fit with Gaussian curve of: (D) bp wrapped DNA with peaks centered at 122.3 ± 15 bp (blue) and 160.4 ± 14 bp (green; bin size = 5 bp, R2 = 0.94, 248 counts total). Blue curves correspond to non-looped nucleosomes and green curves to looped. (E) Examples of both looped and non-looped nucleosomes captured by air imaging. The cartoon to the right of the images is a demonstration of the non-looped and looped nucleosome structure.
Atomic force microscopy (imaging in air)
A 167 μM solution of 1-(3-aminopropyl)- silatrane (APS) was used to modify freshly cleaved mica for 30 minutes at RT as previously described (23,24). The nucleosome stock solution was diluted to 2 nM (based on DNA concentration) in a buffer containing 10 mM HEPES (pH 7.5) and 4 mM MgCl2. Immediately following nucleosome dilution, 7 μl of sample was deposited on chilled (4°C) APS-mica and was rinsed with 1.5 ml of ultrapure H2O after a 2-min incubation which was then followed by drying with a light flow of argon air. Prepared samples were stored under vacuum until being imaged at ambient temperature and humidity conditions on a Multimode AFM/Nanoscope IIId system using TESPA probes (Bruker Nano Inc). A typical image captured was 1.5 × 1.5 μm in size with 512 pixels/line.
High-speed atomic force microscopy
A piece of mica, ∼100 μm thick, was punched into a circle of 2 mm diameter and glued on the stage of the HS-AFM where it was functionalized with 2.5 μl of a 500 μM solution of APS for 30 minutes followed by a rinse with 20 μl of ultrapure H2O. Nucleosome stock was diluted to 2 nM in buffer containing 10 mM HEPES (pH 7.5) and 4 mM MgCl2 (imaging buffer) and 2.5 μl of the diluted sample was deposited onto the APS-mica for 2 min followed by multiple rinses with a total of 20 μl of imaging buffer. The sample surface was kept wet through the entire sample preparation process. Images were captured in the imaging buffer by high-speed AFM (RIBM) using an Olympus Micro Cantilever (BL-AC10DS- A2) that was EBD treated. Typical images acquired were 200 × 200 nm in size at a scan rate ranging from 0.2 to 0.4 s/frame.
Analysis and measurement parameters of nucleosomes imaged in air
The samples deposited on APS mica and imaged by AFM in air were analyzed using Femtoscan Online for the length of each free DNA arm. The contour lengths of DNA particles not bound by histone proteins were first measured and a histogram with a bin size of 5 nm produced a single Gaussian fit with a center at 137 nm (SD = 5.5 nm, R2 = 0.99) which was established as the mean value of free DNA contour length (Supplementary Figure S3). From this fit, a length conversion unit of 0.32 nm/bp was determined and used for subsequent calculations of DNA bp's free and wrapped around the histone core. For consistent measurements of nucleosome arm lengths, each free DNA arm was measured from the strand end to the core center as shown in Figure 1B and the full width at half maximum height (FWHM) was later subtracted from the sum of the free arms to yield the length of unwrapped DNA (Supplementary Figure S3B). The resulting value was subtracted from the statistically determined 137 nm length of the free DNA to obtain the length of wrapped DNA. This was then converted to bp using the length conversion unit established above. Gwyddion was used in obtaining height profile curves that were used to determine height, FWHM and volume measurements for nucleosome core particles. The three height and FWHM values for each nucleosome were averaged and these values were used in calculation of the nucleosome core particle volume. The averaged curves were plotted as a histogram and fit with a Gaussian curve (Supplementary Figure S3).
Analysis and measurement parameters of nucleosomes imaged by HS-AFM
Movies captured with HS-AFM were initially analyzed using the FalconViewer extension to IgorPro software. Each image was flattened using either plane or line background removal depending on the background quality. Events of interest were saved as tiffs and analyzed further in Femtoscan Online and Gwyddion as done for the dry images. A moving median was plotted along with raw data for visualization of the global dynamics taking place. Images published had an additional band pass filter applied to lower noise; this filter was not applied to images used in analysis. OriginPro 2016 was used for all graphs both in text and supplement. Nucleosome loop contour length was measured as illustrated by Supplementary Figure S10.
RESULTS
DNA wrapping of CENP-A nucleosomes characterized by AFM
Mononucleosomes containing human CENP-A were reconstituted on a 423 bp DNA substrate containing the 147 bp Widom 601 positioning sequence (20,21) between 122 and 154 bp flanks of non-specific plasmid DNA (Figure 1A and Supplementary Figure S1). Static imaging by AFM at ambient temperature and humidity conditions revealed centrally positioned nucleosomes, which are distinguishable from ‘naked’ DNA by the wrapped histone core which appears as a bright round feature flanked by two arms of unbound DNA (Figure 1B). The contour lengths of all free DNA molecules were measured and plotted as a histogram (Supplementary Figure S3A). Fitting a Gaussian curve to the histogram produced an average at 423 ± 17 bp. To determine the length of DNA wrapped in a nucleosome, the contour length of each free DNA arm was measured and subtracted from the 423 bp length of the ‘naked’ DNA. Each measurement was made from the end of the strand to the center of the core for consistent measurement and FWHM of three cross section height profiles of each core particle was subtracted from the sum of its respective arms (Figure 1B). A histogram of the wrapped DNA yielded a bimodal Gaussian distribution with the first peak centered at 122.3 ± 15 bp and a second peak with a center at 160.4 ± 14 bp (Figure 1D) which is unique from the single peak distribution previously reported for similar studies of H3 nucleosomes (17). The population with a peak centered at 122.3 ± 15 bp agrees with the value previously reported (7). A histogram of nucleosome heights produced a broad distribution with a single peak centered at 2.4 ± 0.3 nm (Supplementary Figure S3C). Broad, single peak distributions were also obtained for the volume and FWHM of the nucleosome population (Supplementary Figure S3D and E). Strikingly, another unexpected observation was that a significant population of the CENP-A nucleosomes contain DNA looped out of the particle (Figure 1E) which was not previously observed for H3 nucleosomes in air (17). To ensure that the bimodal distribution of wrapped DNA and the presence of looped structures was not a sample preparation artifact, mononucleosomes containing H3 were assembled on the same 601 substrate and were prepared for imaging under the same conditions as CENP-A (Supplementary Figure S4A). Consistent with previous studies, the DNA wrapped by these particles was found to have a single peak distribution centered at 143.9 ± 20 bp (Supplementary Figure S4B) with a broad, single peak height distribution like that of CENP-A (Supplementary Figure S4C) Additionally, the H3 mononucleosome control had no visible looped structures. The contrast in dynamics between CENP-A and H3 nucleosomes, including the looping of the former, is further established below.
Dynamics of CENP-A nucleosomes visualized by time-lapse AFM
The rather broad distributions of all parameters (wrapped DNA, height, volume) of CENP-A nucleosomes on the 601 substrate obtained at ambient conditions suggest dynamic behavior of CENP-A nucleosomes. To directly characterize the dynamic events, we turned to time-lapse AFM, in which nucleosome samples are imaged in aqueous solution without drying of the sample. Moreover, we used high-speed AFM (HS-AFM), which allows for direct observation of the nucleosomes at a sub-second image acquisition rate (18,19,25,26). As a control for sample preparation and imaging conditions, H3 mononucleosomes assembled on the 601 substrate were imaged. Each of the H3 control nucleosomes were found to exhibit the same dynamic unwrapping behavior as was commonly observed in previously studies (15,16,19,23,26). Movie snapshots from Movies S1 and S2 and analysis of two of these particles are presented as Supplementary Figure S5. Also consistent with previous studies, the histone core of the H3 nucleosomes rapidly breaks apart into smaller histone subunits upon full unwrapping of DNA which is visible in frame 4 of Supplementary Figure S5A.
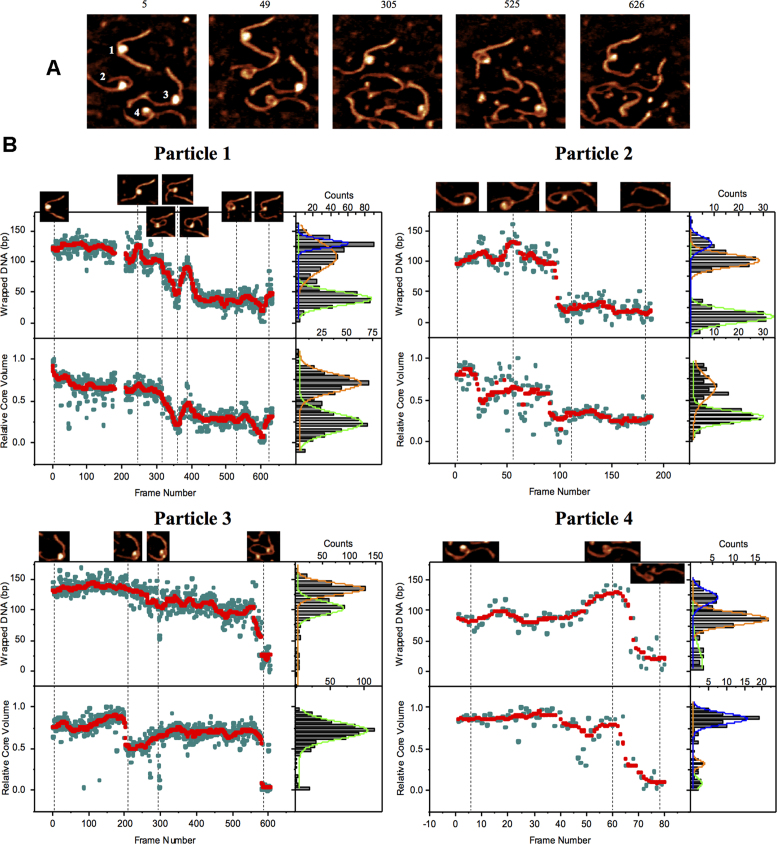
With imaging conditions established, CENP-A mono-nucleosomes were next imaged. Figure 2A shows a few frames out of the 632 consecutive frames assembled as Movie S3. The selected area contained four CENP-A nucleosomes; the dynamics of which were captured simultaneously at a rate of 3.3 frames/s. Every nucleosome in this dataset spontaneously unwraps and the dynamics were characterized by measuring the lengths of DNA arms. These measurements were made for each of the four particles across all 632 frames as presented in Figure 2B and Supplementary Figure S5–S7. The two shortest-lived particles, particles 2 and 4, are initially wrapped less than the 121 bp mean value (Figure 1D), with 100 ± 11 and 86 ± 13 bp, respectively. Next, the partially unwrapped nucleosomes begin a wrapping process, achieving wrapped values of 126 ± 15 bp and 121 ± 11 bp for particles 2 and 4, respectively. The transient wrapping is followed by unwrapping of both particles until they become fully unwrapped after ∼100 frames (Figure 2B; particles 2 and 4).
Figure 2.
Visualization of CENP-A nucleosomes by HS-AFM reveals increased dynamic behaviors. (A) HS-AFM images representing ‘snapshots’ of the diverse dynamics of four nucleosomes across 632 consecutive frames captured at a a rate of 3.3 frames/sec as can be seen in Movie S3. The frame number corresponding to the time at which each frame was captured is above each frame and the four particles are referenced according to their assigned number in the first image. Each image is 200 nm × 200 nm. (B) Analysis of the wrapped DNA (bp) and relative core volume (normalized 0 to 1) as a function of time. Raw data is shown as cyan circles. Moving median (i±5; red) of the raw data makes the overall dynamic trends clear. The raw data from each particle was plotted as a histogram and fit by a Gaussian curve with centers at the following values where error is represented by std. dev. of the curve: Particle 1; wrapped DNA = 125 ± 15, 105 ± 26, 38 ± 12 bp (R2 = 0.99), volume = 0.67 ± 0.1, 0.29 ± 0.1 (R2 = 0.95), each histogram total count = 598. Particle 2; wrapped DNA = 126 ± 15, 100 ± 11.4, 21 ±12.5 bp (R2 = 0.98), volume = 0.64 ± 0.15, 0.30 ± 0.08 (R2 = 0.90), each histogram total count = 188. Particle 3; wrapped DNA = 134 ± 11, 103 ± 12.5 bp (R2 = 0.99), volume = 0.72 ± 0.11 (R2 = 0.96), each histogram total count = 605. Particle 4; wrapped DNA = 121.4 ± 11, 86.7 ± 13, 17.1 ± 15 bp (R2 = 0.99), volume = 0.87 ± 0.06, 0.32 ± 0.04, 0.09 ± 0.06 (R2 = 0.88), each histogram total count = 79. Bin size for wrapped DNA = 10 bp and for relative core volume = 0.05. Total measurements made for analysis of these particles exceeds 15 000.
Particles 1 and 3 (Figure 2B) demonstrate a dynamic behavior different from that of 2 and 4. Initially both nucleosome cores are wrapped to values close to the expected 121 bp and DNA dissociates from each of the cores gradually over the ∼600 frames. Particle 1 begins with 125 ± 15 bp of wrapped DNA for ∼200 frames until it unwraps to 105 ± 26 bp for ∼200 frames. This is a process with multiples unwinding-rewinding step as is evident from the large width of the Gaussians approximated from the experimental histograms (see Figure 2B). Unique from the other four nucleosomes, particle 1 never fully unwraps, instead rewrapping 38 ± 12 bp around a smaller histone complex, where it remains stable for over 200 frames. Particle 3 begins wrapped with 134 ± 11 bp, where it remains for over 400 frames, until partially unwrapping ∼30 bp of DNA to 103 ± 12.5 bp, where it remains until eventually fully unwrapping (Figure 2B).
Dynamic formation of DNA loops
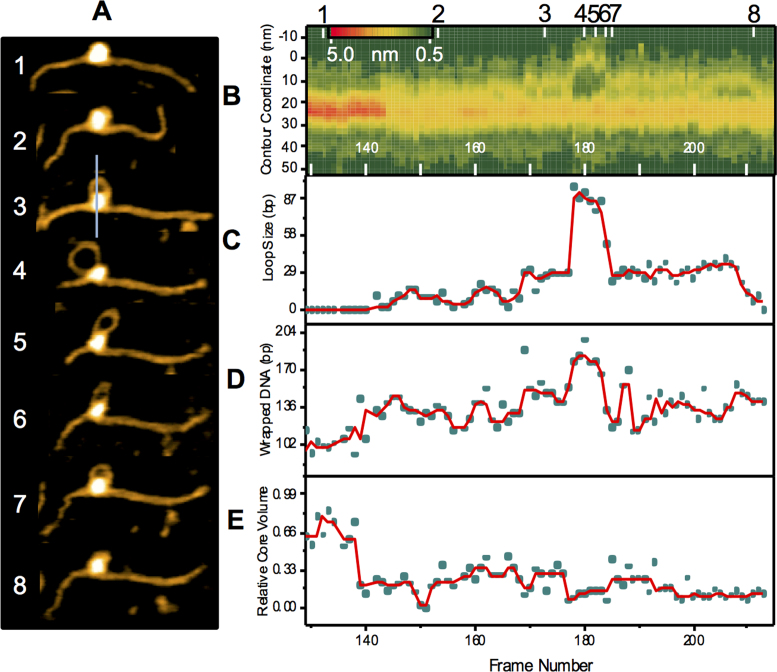
As discussed above, AFM images of dry nucleosome samples with looped out nucleosomal DNA were identified (Figure 1E). Given that looping out of DNA in chromatin is an issue of great biological importance (27–29), we next focused our attention on dynamics of this specific behavior. A few frames representing snapshots of this looping process were selected from Movie S4 and are shown in Figure 3A with additional examples of nucleosomes displaying this behavior assembled as Supplementary Figure S9. The images in Figure 3A visually illustrate that the loop grows gradually (frames 1 through 3), until it reaches a size of ∼90 bp, as seen in frame 4. Next, the loop shrinks (frames 5 though 7) becoming again small (∼20 bp) in frame 8. To characterize this process, images of all 100 frames were analyzed. This was done by contour length measurements of the looped DNA, height profile analysis of a line bisecting the apex of the loop and core, relative volume of the nucleosome core particle, and by changes in the DNA wrapped around the core via nucleosome arm contour measurements (Supplementary Figure S10). The resulting analysis is plotted in Figure 3B–E. Contour length analysis of the loop (Figure 3B and C) shows that its formation between these frames is reversible, as it dramatically changes size five times as seen on the loop cross sections and the loop size graph. The first two times the loop is formed, it grows to be ∼15 bp until retightening around the core. Shortly after, the formation of the first of these two loops was found to be accompanied by a more than two-fold reduction in relative core volume where it remained for the subsequent looping events (Figure 3C and E). This reduction in volume can be attributed to an approximately two-fold reduction in relative height of the nucleosome (Supplementary Figure S11), while the width of the particle is seen to slightly increase. This reduction in height can be explained in part by the loss of DNA from the histone core due to looping, but is likely also due to rearrangement of the histone core as the looping process begins. The potential dissociation of H2A/H2B was not observed during the loop formation events. A height reduction of this magnitude which is never reached again despite shrinking of the loop, suggests that upon looping of the DNA, an internal rearrangement of the histone core has occurred. Unlike the first two loops formed, the third grows to a size of ∼30 bp where it remains stable for eight frames until growing to ∼90 bp. After five frames, the ∼90 bp loop shrinks back to ∼30 bp where it remains stable for over 20 frames.
Figure 3.
Spontaneous looping of CENP-A nucleosomes visualized by high speed AFM. (A) Images representing stages of the looping of DNA from the nucleosome complex were selected from Movie S4; numbers (1–8) correspond to the location of the frames (A) in the time trajectory. The line in image three shows how height profile cross sections were measured to obtain plot (B); this represents the cross sections as a function of frame number where the maximum height of the core is aligned for each cross section. Deviations in the upper yellow line make clear the dynamic nature of this looped complex. The reduction in height shown as a transition from red to yellow immediately prior to loop formation at approx. frame 140 suggests a rearrangement of the histone core as the DNA loosens around it for loop formation (C) A plot of loop size as a function of time shows the dynamic growth and shrinking of the loop. (D) Wrapped DNA (bp) was determined by arm length measurements which revealed that changes in arm do contribute to the formation of the loop. (E) The core volume was found to decrease by two times initially which is largely due to a reduction in height. Raw data is represented as cyan circles, and the moving median by a red line. In total, looping was observed for a total of 10 nucleosomes from the 52 particles imaged.
Measurements of the DNA arm contour lengths lend insight to the mechanism by which the loop is forming; either by sliding of the DNA around the particle, or by falling away of the DNA from the core particle. As each of these loops forms, one of the DNA arms remains relatively stable, while the other is seen to decrease in size in unison with loop growth, suggesting that the loop is formed through the sliding of one of the DNA arms around the histone core. Since these arm lengths are used in the calculation of the wrapped DNA, a decrease in the length of the arm produces an artificially high value for bp of wrapped DNA, as was observed in AFM images in ambient conditions (Figure 1). Additional looping events are shown in Supplementary Figure S9.
Translocation and transfer of nucleosomes
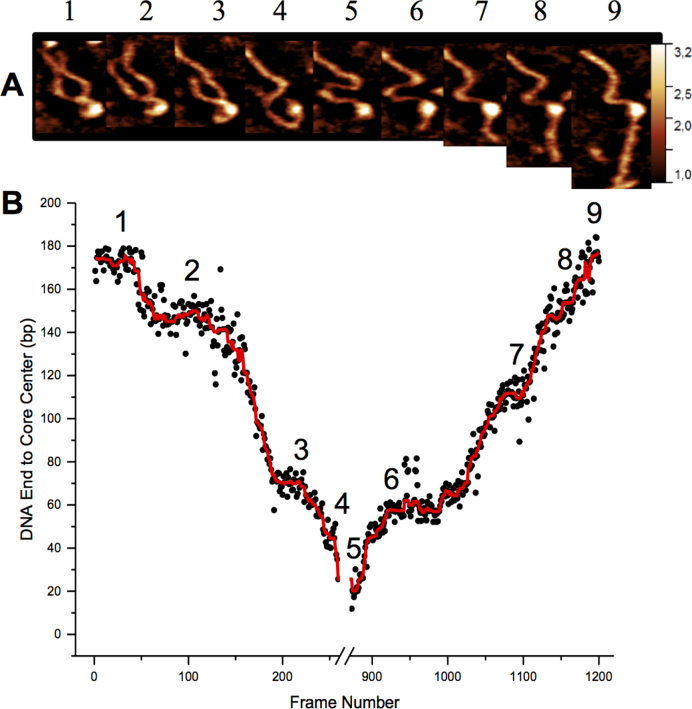
Previous studies have illustrated that the spontaneous unwrapping of nucleosomes is the major DNA dissociation event for H3 containing chromatin subunits; as was also observed for H3 nucleosome control assembled in this study (Movies S1, S2 and Supplementary Figure S5). However, potential translocation (or, hopping), disruption of protein-protein contacts, or sliding of the histone core were not taken into consideration. In our previous time-lapse experiments we did visualize sliding, although it was limited to a relatively short range (19). Long-range sliding was only previously observed in the presence of the detergent, 3-[(3-cholamidopropyl)dimethylammonio]-l-propanesulfonate (CHAPS), suggesting that such long range translocation of canonical nucleosomes is only possible in the presence of CHAPS (17,19). In contrast, CENP-A nucleosomes behave rather differently and are capable of long-range translocation. One of such events is shown in Figure 4; this set of images (out of >500 assembled as Movie's S5 and S6) demonstrates that translocation takes place over 180 bp and that the process is bi-directional. The initial translocation takes place while the nucleosome is partly unwrapped, and the DNA moves in a corkscrew like motion around the core until it is at the end of the DNA substrate (Figure 4A and Movie S5). The contour length of the ‘shrinking’ DNA arm was measured from its end, to the center of the core particle for 250 frames until the core had moved to the end of the DNA substrate. This ‘forward’ translocation moves the DNA a total of ∼180 bp but then appears to stall twice along the way. The first time is after the DNA moves ∼25 bp and the second after it moves ∼75 bp. Lastly, it moves ∼70 bp to the end of the DNA strand where it stays for few hundred frames (Figure 4A and B). Next, the DNA begins a reverse translocation of ∼180 bp, back to its starting position (Figure 4B and Movie S6). Unlike the partly wrapped, corkscrew-like translocation, the reverse propagation stalls four times after moving the following distances in order of the translocation: ∼60, ∼52, ∼40, ∼25 bp (Figure 4B). Similar translocation events were clearly observed for seven nucleosome particles.
Figure 4.
Nucleosome translocation is reversible along a DNA substrate. (A) Images of the forward (1–4) and reverse (5–9) translocation of a nucleosome core particle. Forward movement of the complex appears to be achieved via a corkscrew motion of the DNA as best visualized in Movie S5. The reverse movement is of a less wrapped complex and ends up at about the same position on the DNA substrate as where it began (Movie S6). The white arrows in images 1 and 6 point to the arm length for which the contour length measurements are shown in (B) where the distance was measured from the end of that arm to the center of the core particle for every frame of the video. Frames between ∼250 and ∼850 were excluded because the core remained at the end of the DNA substrate (as seen in image 5) for this time and no translocation took place. Black circles represent the data points and the red line is a moving median. Sliding was observed for a total of 6 particles from the 52 particles imaged.
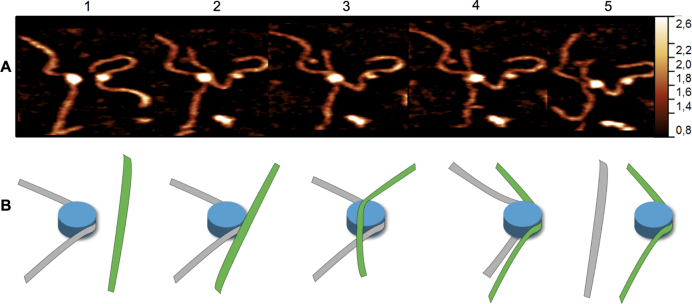
Following the reverse translocation, the core particle was seen to transfer from one DNA molecule to another. A set of a few AFM images is shown as Figure 5A; the full set of data can be seen in Movie S7. Initially, two nucleosomes are shown in the same scanning area. One nucleosome unwraps leaving a free DNA substrate (frame 2) which is later approached by the second nucleosome core particle (frame 3). This nucleosome core particle becomes associated simultaneously with two DNA molecules (frames 3 and 4), eventually transferring its core to the free DNA substrate. Schematically this process is shown in Figure 5B.
Figure 5.
The stable CENP-A histone core is capable of spontaneous inter-strand transfer. (A) HS-AFM images depicting the transfer of a partially wrapped CENP-A containing histone core from one strand to another as seen in Movie S7 and depicted in schematic (B). Image one shows the parent strand (grey) on the left is partly wrapped with the histone core (blue) free from interaction with the acceptor strand (green) on the right (image and schematic 1). Interaction of the acceptor strand (image and schematic 2) soon leads to a histone/DNA complex containing both parent and acceptor strands (image and schematic 3). Within a second of this dual substrate complex, the acceptor strand took full control of the core with the parent strand only partly interacting (image and schematic 4) until the acceptor/core complex is completely free from interaction with the parent substrate (image and schematic 5).
DISCUSSION
In this study, we identified the dynamics of CENP-A nucleosomes by AFM techniques including high-speed time-lapse imaging which revealed that CENP-A nucleosomes are capable of spontaneous unwrapping. As was reported previously for H3 nucleosomes and observed for the H3 control sample in this study (15,17,19), the spontaneous unwrapping of CENP-A nucleosomes was the eventual fate for all particles imaged. The simultaneous visualization of four nucleosomes as they unwrap (Figure 2 and Movie S3) illustrates that the phenomenon is common and that spontaneous dynamics are an intrinsic feature of all nucleosomes. Unlike H3 containing nucleosomes however, the unwrapping process/pathway for each CENP-A nucleosome is different; some of them unwrap rapidly like H3 (e.g. particles 2 and 4), while others take a longer time via dynamic pathways (e.g. particles 1 and 3). While a similar process was observed for canonical nucleosomes, we identified a number of dynamic behaviors unique to CENP-A nucleosome that are not found in the canonical nucleosome (16,19). Details of these features are discussed below.
Nucleosome unwrapping via loop formation
A distinct feature of CENP-A nucleosomes is the bimodal distribution of wrapped DNA as shown in Figure 1D. The major peak is associated with wrapping ∼120 bp DNA which makes ∼1.5 turns around the CENP-A core compared with ∼1.7 turns for canonical particles. This value is in line with recent crystallographic and Cryo EM studies (7,30). This difference was discussed and highlighted in our recent paper in which different models of nucleosomal arrays based on these differences in turn number were proposed, as supported by several studies (7,13,30). However, we identified a second population of nucleosomes with much shorter DNA flanks that were determined to have 160 bp of wrapped DNA which corresponds to as many as two full turns of DNA around the core. This population is unique to CENP-A nucleosomes as such a bimodal distribution is not observed with canonical nucleosomes which have been shown both in this (Supplementary Figure S4B) and previous studies to have a single population with ∼1.7 DNA turns (15,31). High-resolution images in Figure 1E revealed that the arm length deficit for a portion of the CENP-A population is not due to the elevated wrapping of DNA but rather to large segments of DNA that are looped out from the core. These looped nucleosomes are formed in such a way that the non-looped DNA, in contact with the core, consists of both 601 sequence and an adjacent segment of non-specific DNA. The mechanism by which such loops assemble was next probed using time-lapse imaging in which (Figure 3) we observed small loops are formed initially which continue to grow over time. Two processes contribute to the looping-out of DNA: First, a segment of DNA dissociates from the histone core, as evidenced by decrease the particle size (compare frames 1 and 4 in Figure 3A). Second, a segment of the left flank moves around the core. Importantly, the segment remained bound to the histone core forming a stably existing loop. As it is seen from frames 5 through 8, the loop shrinks and this process is primarily due to moving of the left flank of the DNA substrate. Supplementary Figure S9 shows additional looping events that generally follow the same pattern.
Translocation of histone core
The spontaneous translocation of CENP-A histone cores is another dynamic property of CENP-A nucleosomes as illustrated in Figure 4. In this example, the core moves over ∼180 bp and then eventually returns to the initial position. It stops at the end of the DNA substrate and has a few pauses along the translocation path. A similar pattern is observed for the reverse translocation. We used arm length measurements to estimate the DNA wrapping efficiency of the histone core during the translocation. Initially (frame 1 in Figure 4A.), 104 bp of DNA are seen to wrap around the histone core, which is 16 bp shorter than for the mean value for CENP-A nucleosomes estimated in Figure 1D. However, only a portion (∼40 bp) of the DNA is in direct contact with the core, as the rest forms a looped structure. As translocation progresses, this loop structure changes in size, which results in fluctuations of the DNA segment in contact with the core. Once at the end of the DNA substrate, the loop unwraps, leaving ∼40 bp of DNA still bound to the core. Spontaneously the distal end of DNA moved beyond the scan size, so determination of nucleosome size was further made through volume measurements. The results shown in Supplementary Figure S12 demonstrate that the nucleosome volume fluctuates, but generally remains constant, suggesting that during translocation the CENP-A core remains associated with ∼40 bp. This is confirmed when the distal DNA end later reenters the frame and arm measurements provided the expected DNA contact value of ∼40 bp.
Translocation has previously been observed for H3 nucleosomes, but it was over very short distance ∼40 bp (19). Ranges exceeding 100 bp were only achieved in the presence of CHAPS detergent, which like other detergents stabilizes the histone core from dissociation in dilute solutions of mononucleosomes (17,19). In this study, we revealed that CENP-A is capable of reversible long range translocation of ∼180 bp in the absence of stabilizing factors/conditions. Critical to this ability is the stability of the CENP-A histone core upon loosening or unwrapping of DNA. Despite losing two-thirds of its contacts with DNA prior to translocation, the core remains whole and continues spontaneous interactions with and along the DNA substrate (Supplementary Figure S13A); differs from cores containing H3 which rapidly break apart to smaller subunits upon unwrapping (Supplementary Figure S13B). As CENP-C has been shown to stabilize fully wrapped CENP-A nucleosomes, our results suggest that the unwrapped core is also stable and capable of maintaining dynamic contacts with DNA (11).
Interstrand transfer of CENP-A core
Transfer of the nucleosome core from one DNA substrate to another, as illustrated in Figure 5, is another characteristic of CENP-A nucleosomes that has not been directly visualized before. Evidence of interstrand nucleosome transfer was presented and discussed in landmark studies (32,33) and was recently demonstrated in a magnetic tweezers study (34). Furthermore, our results directly support such an ability. An interesting feature of the transfer process, as Figure 5 illustrates, is that the transfer takes place between two DNA substrates. Note that exchange between the two DNA strands takes place with the CENP-A nucleosome core in contact with ∼40 bp, as schematically shown in the cartoon (Figure 5B). We would like to emphasize two important features of the CENP-A nucleosome core that makes such a transfer possible: the retained integrity of the histone core, and the long lifetime of the core-DNA complex after unwrapping. Again, these properties are unique to the stable CENP-A core and were not previously observed for canonical nucleosomes (19).
Overall, the experiments presented here reveal the highly dynamic properties of CENP-A nucleosomes that contribute to centromeric chromatin remodeling. Nucleosomes spontaneously undergo the unfolding process utilizing two major pathways: looping, and translocation along DNA, which enables the transfer of nucleosomes from one DNA to another. These spontaneous processes can be modulated by environmental conditions. Nucleosome remodeling can occur by different pathways with the necessary pathway for a specific genetic process (repair, replication, transcription) being promoted through recruitment of a specialized remodeling factor (35); for example, Chd1 can be used for sliding of nucleosomes (36). Our discoveries of the nanoscale dynamics intrinsic to CENP-A nucleosomes suggest that dynamic rearrangements of centromeric chromatin can occur in the absence of remodeling factors and at the same time facilitate dynamics of chromatin catalyzed by the remodeling factors. In addition to CENP-A nucleosome dynamics, we observed that CENP-A stabilizes nucleosome core particles against complete dissociation upon loosening or unwrapping of DNA. We speculate that this novel property of CENP-A might permit it to stay associated with H2A, H2B and H4, thereby permitting rapid nucleosome re-assembly following mitosis, transcription or replication induced eviction. Furthermore, the stabilization of fully wrapped CENP-A nucleosomes by CENP-C (11) along with the intrinsic stability of unwrapped CENP-A cores, may contribute to its longevity on the chromatin fiber, thus contributing to the cell cycle independent ‘memory’ of centromeric chromatin over several cell cycles (37).
Supplementary Material
ACKNOWLEDGEMENTS
We thank Yamini Dalal for the gift of recombinant histones, training, enthusiastic feedback, and critical review of the manuscript. We acknowledge Daniel Melters (Dalal lab) for thoughtful discussions and critical review of the manuscript.
Author contributions: Y.L.L. and M.S.D. designed the project; M.S.D., S.B., M.H., Z.S. performed AFM experiments and data analyses. All authors wrote and edited the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Science Foundation [MCB 1515346 to Y.L.L.]; National Institutes of Health [GM096039 and GM118006 both to Y.L.L.]; M.H. was partially supported by the Bukey Memorial Fellowship. Funding for open access charge: National Science Foundation [MCB 1515346 to Y.L.L.].
Conflict of interest statement. None declared.
REFERENCES
- 1. Gordon D.J., Resio B., Pellman D.. Causes and consequences of aneuploidy in cancer. Nat. Rev. Genet. 2012; 13:189–203. [DOI] [PubMed] [Google Scholar]
- 2. Kops G.J., Weaver B.A., Cleveland D.W.. On the road to cancer: aneuploidy and the mitotic checkpoint. Nat. Rev. Cancer. 2005; 5:773–785. [DOI] [PubMed] [Google Scholar]
- 3. Panigrahi A.K., Pati D.. Road to the crossroads of life and death: Linking sister chromatid cohesion and separation to aneuploidy, apoptosis and cancer. Crit. Rev. Oncol./Hematol. 2009; 72:181–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Black B.E., Jansen L.E.T., Maddox P.S., Foltz D.R., Desai A.B., Shah J.V., Cleveland D.W.. Centromere identity maintained by nucleosomes assembled with histone H3 containing the CENP—a targeting domain. Mol. Cell. 2007; 25:309–322. [DOI] [PubMed] [Google Scholar]
- 5. McKinley K.L., Cheeseman I.M.. The molecular basis for centromere identity and function. Nat. Rev. Mol. Cell. Biol. 2016; 17:16–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Tachiwana H., Kagawa W., Kurumizaka H.. Comparison between the CENP-A and histone H3 structures in nucleosomes. Nucleus. 2012; 3:6–11. [DOI] [PubMed] [Google Scholar]
- 7. Tachiwana H., Kagawa W., Shiga T., Osakabe A., Miya Y., Saito K., Hayashi-Takanaka Y., Oda T., Sato M., Park S.-Y. et al. Crystal structure of the human centromeric nucleosome containing CENP-A. Nature. 2011; 476:232–235. [DOI] [PubMed] [Google Scholar]
- 8. Yoda K., Ando S., Morishita S., Houmura K., Hashimoto K., Takeyasu K., Okazaki T.. Human centromere protein A (CENP-A) can replace histone H3 in nucleosome reconstitution in vitro. Proc. Natl. Acad. Sci. U.S.A. 2000; 97:7266–7271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kim S.H., Vlijm R., van der Torre J., Dalal Y., Dekker C.. CENP-A and H3 nucleosomes display a similar stability to force-mediated disassembly. PLOS ONE. 2016; 11:e0165078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Winogradoff D., Zhao H., Dalal Y., Papoian G.A.. Shearing of the CENP-A dimerization interface mediates plasticity in the octameric centromeric nucleosome. Scientific Rep. 2015; 5:17038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Falk S.J., Lee J., Sekulic N., Sennett M.A., Lee T.-H., Black B.E.. CENP-C directs a structural transition of CENP-A nucleosomes mainly through sliding of DNA gyres. Nat. Struct. Mol. Biol. 2016; 23:204–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sinha K.K., Gross J.D., Narlikar G.J.. Distortion of histone octamer core promotes nucleosome mobilization by a chromatin remodeler. Science. 2017; 355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lyubchenko Y.L. Centromere chromatin: a loose grip on the nucleosome?. Nat. Struct. Mol. Biol. 2014; 21:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Melters D.P., Nye J., Zhao H., Dalal Y.. Chromatin dynamics in vivo: a game of musical chairs. Genes. 2015; 6:751–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lyubchenko Y.L. Nanoscale nucleosome dynamics assessed with time-lapse AFM. Biophys. Rev. 2014; 6:181–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Shlyakhtenko L.S., Lushnikov A.Y., Lyubchenko Y.L.. Dynamics of nucleosomes revealed by time-lapse atomic force microscopy. Biochemistry. 2009; 48:7842–7848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Menshikova I., Menshikov E., Filenko N., Lyubchenko Y.L.. Nucleosomes structure and dynamics: effect of CHAPS. Int. J. Biochem. Mol. Biol. 2011; 2:129–137. [PMC free article] [PubMed] [Google Scholar]
- 18. Uchihashi T., Ando T.. Braga PC, Ricci D. Atomic Force Microscopy in Biomedical Research: Methods and Protocols. 2011; Totowa: Humana Press; 285–300. [Google Scholar]
- 19. Miyagi A., Ando T., Lyubchenko Y.L.. Dynamics of nucleosomes assessed with time-lapse high-speed atomic force microscopy. Biochemistry. 2011; 50:7901–7908. [DOI] [PubMed] [Google Scholar]
- 20. Lowary P.T., Widom J.. New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning1. J. Mol. Biol. 1998; 276:19–42. [DOI] [PubMed] [Google Scholar]
- 21. Guse A., Fuller C.J., Straight A.F.. A cell free system for functional centromere and kinetochore assembly. Nat. Protoc. 2012; 7:1847–1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Luger K., Rechsteiner T.J., Richmond T.J.. Preparation of nucleosome core particle from recombinant histones. Methods Enzymol. 1999; 304:3–19. [DOI] [PubMed] [Google Scholar]
- 23. Lyubchenko Y.L. Preparation of DNA and nucleoprotein samples for AFM imaging. Micron. 2011; 42:196–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Shlyakhtenko L.S., Gall A.A., Filonov A., Cerovac Z., Lushnikov A., Lyubchenko Y.L.. Silatrane-based surface chemistry for immobilization of DNA, protein-DNA complexes and other biological materials. Ultramicroscopy. 2003; 97:279–287. [DOI] [PubMed] [Google Scholar]
- 25. Katan A.J., Vlijm R., Lusser A., Dekker C.. Dynamics of nucleosomal structures measured by high-speed atomic force microscopy. Small. 2015; 11:976–984. [DOI] [PubMed] [Google Scholar]
- 26. Lyubchenko Y.L., Shlyakhtenko L.S., Ando T.. Imaging of nucleic acids with atomic force microscopy. Methods (San Diego, Calif.). 2011; 54:274–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kulić I.M., Schiessel H.. Nucleosome repositioning via loop formation. Biophys. J. 2003; 84:3197–3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Pasi M., Lavery R.. Structure and dynamics of DNA loops on nucleosomes studied with atomistic, microsecond-scale molecular dynamics. Nucleic Acids Res. 2016; 44:5450–5456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bazett-Jones D.P., Côté J., Landel C.C., Peterson C.L., Workman J.L.. The SWI/SNF complex creates loop domains in DNA and polynucleosome arrays and can disrupt DNA-histone contacts within these domains. Mol. Cell. Biol. 1999; 19:1470–1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Roulland Y., Ouararhni K., Naidenov M., Ramos L., Shuaib M., Syed S.H., Lone I.N., Boopathi R., Fontaine E., Papai G. et al. The flexible ends of CENP-A nucleosome are required for mitotic fidelity. Mol. Cell. 2016; 63:674–685. [DOI] [PubMed] [Google Scholar]
- 31. Luger K., Mäder A.W., Richmond R.K., Sargent D.F., Richmond T.J.. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature. 1997; 389:251–260. [DOI] [PubMed] [Google Scholar]
- 32. Beard P. Mobility of histones on the chromosome of simian virus 40. Cell. 1978; 15:955–967. [DOI] [PubMed] [Google Scholar]
- 33. Stacks P.C., Schumaker V.N.. Nucleosome dissociation and transfer in concentrated salt solutions. Nucleic Acids Res. 1979; 7:2457–2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Brennan L.D., Forties R.A., Patel S.S., Wang M.D.. DNA looping mediates nucleosome transfer. Nat. Commun. 2016; 7:13337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Clapier C.R., Cairns B.R.. The biology of chromatin remodeling complexes. Annu. Rev. Biochem. 2009; 78:273–304. [DOI] [PubMed] [Google Scholar]
- 36. McKnight J.N., Tsukiyama T., Bowman G.D.. Sequence-targeted nucleosome sliding in vivo by a hybrid Chd1 chromatin remodeler. Genome Res. 2016; 26:693–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ross J.E., Woodlief K.S., Sullivan B.A.. Inheritance of the CENP-A chromatin domain is spatially and temporally constrained at human centromeres. Epigenet. Chromatin. 2016; 9:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.