Abstract
Gene therapy by engineering patient's own blood cells to confer HIV resistance can potentially lead to a functional cure for AIDS. Toward this goal, we have previously developed an anti-HIV lentivirus vector that deploys a combination of shRNA, ribozyme and RNA decoy. To further improve this therapeutic vector against viral escape, we sought an additional reagent to target HIV integrase. Here, we report the development of a new strategy for selection and expression of aptamer for gene therapy. We developed a SELEX protocol (multi-tag SELEX) for selecting RNA aptamers against proteins with low solubility or stability, such as integrase. More importantly, we expressed these aptamers in vivo by incorporating them in the terminal loop of shRNAs. This novel strategy allowed efficient expression of the shRNA–aptamer fusions that targeted RNAs and proteins simultaneously. Expressed shRNA–aptamer fusions targeting HIV integrase or reverse transcriptase inhibited HIV replication in cell cultures. Viral inhibition was further enhanced by combining an anti-integrase aptamer with an anti-HIV Tat-Rev shRNA. This construct exhibited efficacy comparable to that of integrase inhibitor Raltegravir. Our strategy for the selection and expression of RNA aptamers can potentially extend to other gene therapy applications.
INTRODUCTION
Human immunodeficency virus (HIV) that causes acquired immunodeficiency syndrome (AIDS) is rapidly evolving. The current standard of treatment of combinatorial anti-retroviral therapy (ART) is very effective in controlling viral loads, however, ART is not curative. Patients have to endure high costs and potential side effects associated with the lifelong treatment. Gene therapy presents an attractive alternative treatment. By engineering patients’ own T cells or hematopoietic stem cells to confer HIV resistance, gene therapy can potentially lead to a functional cure for AIDS (1–3).
One of the major challenges in targeting HIV is its rapidly evolving genome that leads to drug resistance. Therefore, combinatorial strategies to simultaneously target different steps of the HIV life cycle are essential for successful gene therapy. Early attempts of combinatorial gene therapy that included RNA-based as well as protein-based strategies have proven to be very effective in cell culture and animal models (4–7). With intrinsic low immunogenic properties, multiplexing ability and small size, RNA-based reagents such as shRNAs, ribozymes and RNA decoys are excellent building blocks for combinatorial anti-HIV vectors. Most of these RNA-based therapeutics target the HIV genes or genome by base-pairing that can be attenuated by point mutation(s) in the target site. Additional RNA-based reagents that are independent of base-pairing will further enhance our combinatorial strategy for anti-HIV gene therapy.
One such class of reagents is RNA aptamers, which are short single stranded RNAs that fold into stable three-dimensional shapes. With close to unlimited potential structural conformations, aptamers are very attractive molecules for structure based targeting. Aptamers with high affinity and specificity to target proteins can be selected from complex libraries using SELEX (Selective Enrichment of Ligands by Exponential Enrichment) (8,9). Recently, aptamers with high binding affinity against HIV RT, gp120, Gag and Protease have been isolated (10–15). Currently, most therapeutic aptamers are administered exogenously to target cell surface proteins either as neutralizing reagents or as vehicles to carry therapeutic agents (14,16,17). RNA aptamers transcribed from Pol III promoter such as U6 have precise start and termination sites allowing precise prediction of length and structure of the expressed aptamers. However, Pol III driven transcripts do not contain an intrinsic nuclear export signal and predominantly reside in the nucleus (18–21), excluding their use against the proteins that function in the cytoplasm. While aptamers transcribed from Pol II promoters can be exported to the cytoplasm, the additional 5′ and 3′ sequences, the 5′ cap and the polyA tail as well as their binding proteins, may alter the structure and interfere with the aptamers’ function. Expression of functional aptamers in cells remains a major hurdle for long-term gene therapy.
Here, we report a novel strategy for the selection and stable expression of aptamer therapeutics. We developed a multi-tag SELEX method for selecting aptamers that target proteins with low solubility and/or stability, such as the HIV integrase. We took advantage of the mechanism of Exportin-5 mediated export of pre-miRNAs and designed shRNA–aptamer fusions by incorporating the aptamers as the terminal loop of shRNAs. We demonstrated that the expressed shRNA–aptamer fusions could exert prolonged inhibition of HIV replication.
MATERIALS AND METHODS
Vectors construction
To construct a maltose binding protein-integrase fusion (MBP-IN) expression vector (pJR166), an NdeI and BamHI fragment containing the IN coding region of pNL4-3 strain was released from pEt-15b-IN (22,23) and cloned into pMAL-c5X (New England Biolabs, Ispwich, MA, USA). To generate a FLAG tagged IN mammalian expression vector, a human codon optimized integrase coding sequence was synthesized according to the amino acid sequence of pEt-15b-IN and cloned into a pCDNA3 based vector (Thermofisher, Grand Island, NY, USA) that contains an N-terminal FLAG tag (pJR188). For expression of shRNAs and aptamers in mammalian cells, a plasmid (pJR255) was constructed that contained a U6 promoter for driving expression of the shRNAs, aptamers, or shRNA–aptamer fusions. It also contained an mCherry visible marker driven by CMV promoter. Two BbsI sites were inserted directly downstream of the U6 promoter to generate GTGG and TTTT overhangs upon cleavage. pJR255 was used to drive expression of shRNA and aptamers in HEK 293 cells and Ghost(3) CXCR4+/CCR5+ cells. Because CMV promoter-driven mCherry showed very weak signal in CEM cells, a similar plasmid (pJR288) was constructed that replaced the CMV promoter with the Ef1α promoter. For construction of various U6 driven shRNAs, aptamers or shRNA–aptamer fusions, 1 nmol each of oligonucleotide pair containing CACC and AAAA overhangs (Supplementary Table S1) was mixed in 1X T4 Ligase buffer (NEB), heated to 95°C for 3 min, slowly cooled to room temperature, and then ligated to the BbsI cut of either pJR255 or pJR288.
Production and purification of integrase fusion proteins
HIS tag HIV-1 integrase (HIS-IN) expression vector was a gift from Dr. Robert Craigie (NIDDK, NIH) (23). Escherichia coli strain C3016 (NEB, Ipswich, MA, USA) was transformed with the HIS-IN plasmid. A single positive colony was picked to inoculate 250 ml of SOB medium containing 100 mg/ml of ampicillin. The culture was grown overnight in a 37°C shaker (200 rpm). Induction was performed by adding 250 ml of fresh SOB containing 2 mM IPTG for another 3 hours at 22°C. HIS-IN was purified from bacterial cells with Ni-NTA agarose (Qiagen, Valencia, CA, USA.) using the manufacturer's protocol for purifying protein in native conformation. To increase the purity of the eluted protein, beads were washed first with buffer containing 300 mM NaCl and 50 mM imidazole followed by a second wash with buffer containing 500 mM of NaCl and 30 mM imidazole. Eluted HIS-IN was dialyzed overnight at 4°C with two changes of 500 ml PBS. Induction and lysis of maltose binding protein (MBP)-IN followed a similar protocol. MBP-IN was purified with pMAL protein fusion purification system (NEB, Ipswich, MA, USA). Eluted fusion protein was dialyzed against two changes of 500 ml of PBS at 4°C overnight. Purity and concentration of HIS-IN and MBP-IN fusion proteins were quantified by serial dilution and PAGE analysis using a BSA standard.
FLAG-IN and associated protein complexes were purified from pJR188-transfected HEK293 cells, 48 h post-transfection, using the M2 anti-FLAG affinity gel according to the manufacturer's protocol (Sigma-Aldrich, St. Louis, MO, USA).
Aptamer selection, identification and structure prediction
Aptamer selections basically followed Zhou et al. (10) with the following modifications. Two bacterially expressed fusions HIS-IN and MBP-IN were used alternatively in each cycle of selection and a mammalian cell expressed FLAG-IN was used in an additional cycle of enrichment. To increase the complexity of the library, primer extension with T4 polymerase instead of PCR was used to generate a double stranded template DNA from an oligonucleotide library that contained a 5′ T7 promoter sequence, a 30-nucleotide variable middle region (n), and a 3′ constant region (Supplementary Table S1). In addition, RT-PCR reactions were limited to 10 cycles of amplification. The RNA library was generated by Megashortscript T7 transcription kit (Thermofisher Scientific, Waltham, MA, USA), using the gel purified RT-PCR product as a template.
In each cycle of selection, RNA pools were folded in 200 μl of binding buffer (PBS pH 7.4 plus 1 mM CaCl2, 2.7 mM KCl, 2 mM MgCl2) by heating to 95°C for 3 min followed by slow cool to 37°C. The folded RNA pools were then pre-cleared by incubating with HAWP filter (0.45 μm pore size, 13 mm diameter, EMD Millipore, Concord, MA, USA) for 30 min. The tagged IN proteins were then incubated with the pre-cleared RNA pool in phosphate buffer (pH 7.4) using progressively increased NaCl concentrations in SELEX cycles (50 mM in cycle 1–2, 100 mM in cycle 3–4, 147 mM in cycle 5 and up) and incubated for an additional 15 min at 37°C. In the first selection cycle, 1.5 nmol of RNA and 0.24 nmol of protein (RNA to protein ratio of 6.5:1) were used for binding reaction. As selection cycle progressed, protein concentration was gradually decreased to 0.12 nmol. Starting from cycle 3, an increasing amount of tRNA (20 μg in cycle 3–4, 40 μg in cycle 5–6, 80 μg in cycle 7 and up) was added as nonspecific competitor. The RNA–protein complexes were isolated by passing the reaction through a HAWP filter, followed by 1 ml washes with binding buffer. Membrane bound RNAs were eluted by 200 μl of elution buffer (7 M urea and 5 mM EDTA) at 95°C for 5 min, followed by phenol/chloroform extraction. An additional round of selection was carried out with FLAG-IN complexes immuno-precipitated from 1 mg of HEK293 cell lysate with 180 μl of M2 anti-FLAG affinity gel beads. The complex was then used to bind to 60 µg of RNA with 180 µg of tRNA as nonspecific competitor.
RT-PCR products of the final three cycles of selection and the sample binding to FLAG-IN complexes were subjected to high throughput sequencing (Illumina). Sequence analysis was performed as described (24). Secondary structure predictions were carried out by Mfold web server (25).
Gel mobility shift, binding assay and northern analysis
A filter binding assay was carried out to monitor library enrichment. 5 μg of aptamer RNAs from each cycle were first treated with Antarctic phosphatase (NEB), followed by phenol extraction and ethanol precipitation. Dephosphorylated RNA pellets were resuspended in PBS and their concentrations were determined by nanodrop spectrometer. 10 pmol of aptamer RNA was then end labeled with 32P ATP with T4 polynucleotide kinase (NEB), adjusted to 200 nM with PBS then purified by G25 column (GE Life Sciences). Half of the 32P-labeled aptamer RNA was folded by heating (95°C for 3 min) and slow cooling (37°C on heat block). 1 pmol of folded RNA was incubated with 10 pmol of MBP-IN and 10 pmol of tRNA at 37°C for 20 min. The reactions were passed through nitrocellulose filters (0.45 μm pore size, 13 mm diameter, EMD Millipore, Concord, MA, USA) which were then washed with 1 ml PBS. Radioactivity of the filters containing aptamer–protein complexes was determined by scintillation counter using 1 pmol of labeled RNA as standard.
The gel mobility shift assay was carried out as described (10). RNA aptamers were transcribed from annealed oligonucleotide pairs (Supplementary Table S1) containing T7 promoter sequences using Megashortscript T7 transcription kit (Thermofisher Scientific, Waltham, MA, USA). 32P labeled and folded aptamers (final concentration of 2 nM) were incubated with HIS-IN protein (final concentration of 0, 20, 40, 80, 160 and 320 nM) at 37°C for 30 min. The complexes were then separated by 5% native polyacrylamide gel. Autoradiographs were acquired via Typhoon phosphorimaging system (GE Healtcare Life Science, Pittsburgh PA, USA). Digital images were imported and analyzed by ImageJ software (http://imagej.nih.gov/ij/index.html). 50 percent binding values were determined by the Prism 6 software (GraphPad Software Inc) using non-linear curve regression.
Northern analysis of total and fractionated RNAs was carried out as described (26) using probes listed in primer list (Supplementary Table S1). Quantification of relative band intensity was measured and calculated using ImageJ software.
Cell culture and generation of stable cell lines
Ghost(3) CXCR4+CCR5+ cells (27), HIV pNL4-3 (22) and Ba-L (28) strains were obtained from NIH AIDS Reagent Program (see acknowledgement). Ghost3 cells and HEK293 cells (ATCC CRL-1573) were grown on DMEM supplemented with 10% FBS and 1 mM glutamine. CCRF-CEM (ATCC CRM-CCL-119) cells were grown in RPMI supplemented with 10% FBS and 1 mM glutamine.
HEK293 and Ghost3 were transfected with Lipofectamine 2000 according to the manufacturer's protocol (ThermoFisher, Waltham, MA, USA). To generate stable HEK293 cell lines, 2 million cells were transfected with pJR255-based plasmids that express aptamers alone, shLuc or shLuc-aptamer fusions. Transfected cells were selected with G418 (Gold Biotechnology, St. Louis, MO, USA) for 10–14 days, followed by Fluorescent Activated Cell Sorting (FACS) to isolated mCherry positive cells. Typically, the brightest 10–30% of cells were collected. Second FACS was performed 7–10 days after the first sort to isolate stable cell populations. Cells isolated from second sort (typically 40–60% of cells) had a very stable mCherry signal and were used for lentivirus or HIV challenge experiments. Ghost3 cells were already G418 resistant, therefore, no selection step was performed. 0.5 M cells were transfected with pJR255 based plasmids that express shLuc, aptamer alone or shLuc-aptamer fusions. Transfected cells were sorted 7 days after transfection. Compared to HEK293 cells, the percentage of mCherry positive cells was much lower because no drug selection was applied. Typically, the brightest 5–10% of cells were collected. A second sort was performed 14 days later. If a decrease in mCherry signal was observed, a third sort was performed.
To generate stable CCRF-CEM cell lines, 2 million cells were transfected with pJR288-based plasmids, that express shLuc, shLuc-S1R1, shLuc-S3R1 and shLuc–S3R3, by electroporation using the Nucleofactor kit according to the manufacturer's protocol (Lonza, Basel, Switzerland). G418 selection and FACS were performed similar to that of HEK293 cells.
HIV challenge assay
For lentivirus challenge, 5 × 104 HEK293 cells stably expressing aptamer alone, shRNA alone or shRNA–aptamer fusion were transduced with HIV7-GFP (29) lentiviral particles at a MOI of 0.3 in 500 μl medium. Cells were harvested 10 days post-transduction and analyzed by FACS. For HIV challenge, 5 × 104 Ghost3 cells stably expressing shRNA alone or shRNA–aptamer fusions were infected with HIV-1 Ba-L strain at MOI of 0.02 in 500 μl medium. Viral concentration in culture was determined by p24 assay using Alliance HIV-1 p24 ELISA kit (PerkinElmer, Waltham, MA, USA).
For long term HIV challenge, 1 × 105 CCRF-CEM cells stably expressing shRNA or shRNA–aptamer fusions were infected with HIV-1 NL4-3 viruses at MOI of 0.02 in 500 μl medium. On Day 3, cells were collected by centrifugation at 200 × g for 3 min and resuspended in 1ml fresh medium. From week 1 to week 6 post-infection, 500 μl of cell cultures were collected for analysis and the continuing cultures was replenished with 500 μl fresh medium. In addition, to maintain an unsaturated cell density, 50% of cells was also replaced with fresh medium at mid-week time points.
Luciferase assay
A HEK293 cell clone stably expressing firefly luciferase was transfected with the plasmids expressing the shLuc or the shLuc-aptamer fusions. Transfected cells were harvested two or three days post-transfection and their firefly luciferase activities were determined by the Luciferase Assay System (Promega, Madison, WI, USA).
RESULTS
Multi-tag SELEX
Our previous combinatorial anti-HIV vectors target multiple steps of the HIV life cycle, including entrance, transcription and replication (4,7,30). In this work, we sought to enhance our existing vector by including an expressed aptamer that targets the HIV integrase. We used SELEX to isolate RNA aptamers that bind to HIV integrase (IN). A bacterially expressed HIS-tag mutant integrase (F185K/C280S) that retains DNA integration activity but has much improved solubility (23) was used for aptamer library enrichment. Since we intended for the aptamers to be expressed in T cells for anti-HIV gene therapy, salt concentrations of less than 147 mM of sodium chloride were used for the enrichment process. At low salt concentrations, the integrase protein became poorly soluble and very unstable especially when the tag was removed from the expressed fusion (Supplementary Figure S1A and 1B and data not shown). Therefore, the HIS-tag fusion protein was used for enrichment.
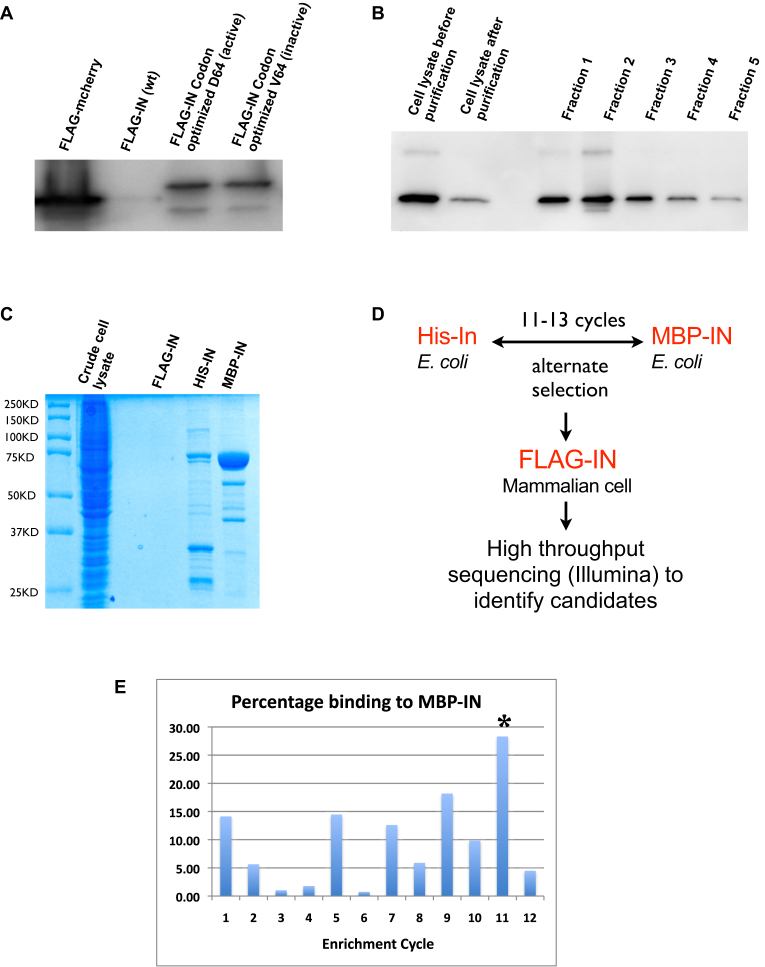
To minimize the enrichment of aptamers that bind to HIS-tag rather than integrase, we employed two additional tagged integrase fusions for aptamer selections. First, a maltose binding protein-tagged IN (MBP-IN) was expressed in E. coli. Second, a FLAG-tagged IN (FLAG-IN) was expressed in human cells in a physiologically relevant environment. Integrase interacts with a number of cellular proteins (31–34). FLAG-IN that complexed with cellular proteins presents a more native conformation that should facilitate the isolation of functional aptamers. Consistent with previous reports, we observed very low expression of wild-type integrase in mammalian cells (35). By codon optimization, we were able to increase the yield of the protein by more than ten-fold (Figure 1A). Since over-expression of the active integrase could be toxic to cells, we also generated an inactive mutant D64V. However, we obtained similar yields for both active and inactive forms from transiently transfected cells (Figure 1A) hence we used the transiently expressed active form for enrichment. A high proportion (74%) of active FLAG-IN could be purified from transfected HEK 293 cells (Figure 1B). However, the yield was still too low to allow the use of purified complexes in the early selection cycles that typically requires 10–20 μg of purified protein (Figure 1C). Therefore, bacterially expressed HIS-IN and MBP-IN (Supplementary Figure S1A and B) were used alternately in the early selection cycles while FLAG-IN complexes purified from human cells were reserved for the final round of selection (Figure 1D). We termed this strategy multi-tag SELEX.
Figure 1.
Multiple tagged integrases for SELEX. (A) Codon optimization increased the yield of an active and an inactive FLAG-IN. Western blot showing expression levels of FLAG-tagged original (wt), codon optimized active (D64) and codon-optimized inactive (V64) integrase from transiently transfected HEK293 cells. Note that yield of codon-optimized FLAG-IN was still substantially lower than FLAG-mCherry positive control. (B) FLAG-IN (D64) transiently expressed in HEK293 cells was purified by anti-FLAG M2 affinity gel (Sigma). Approximately 74% (compare lane 2 to lane 1) of FLAG-IN expressed from HEK293 cells was purified. Fraction 1 to 5 represent purified FLAG-IN protein. (C) 10% of FLAG-IN purified from two 150 cm plates of HEK 293 cells was not detectable by Coomassie staining. 0.5 μg of purified HIS-IN and MBP-IN was added for comparison. (D) Schematic of multi-tag SELEX strategy. (E) Typical change of aptamers binding during enrichment cycles monitored by the filter binding assay. Odd number cycles were enriched by MBP-IN while even number cycles were enriched by His-IN. ‘*’ marks the sample used for one additional enrichment cycle with HEK293 expressed FLAG-IN. High percentage binding at first two cycles probably represents high nonspecific binding to HIS and MBP tags. Alternate high and low binding at later cycles probably represent populations showing higher binding affinity to MBP-IN than HIS-IN.
Isolation of integrase binding aptamers
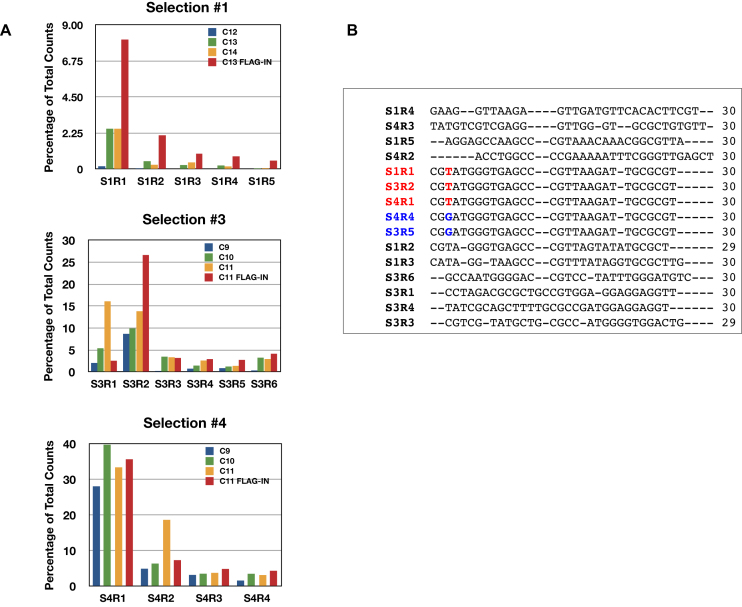
Enrichment of the library was monitored by an increase in the percentage of binding to MBP-IN (Figure 1E). Samples from cycles just reaching the plateau (typically cycle 11–13) were subjected to an additional cycle of enrichment using the cellular complexes that co-immunoprecipitated with FLAG-IN (Figure 1E). Samples of the final four cycles of selections were subjected to high throughput sequencing (Illumina). Four independent selections were performed (S1-S4). Selection 2 did not yield any candidate aptamers that were substantially enriched and was therefore rejected.
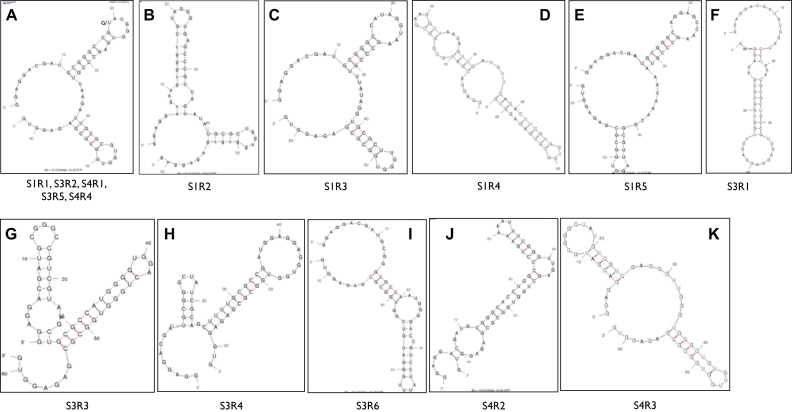
The most abundant aptamer isolated from the first selection was designated as S1R1 (Selection 1, aptameR 1). The sequence of S1R1 is identical to that of S3R2 and S4R1. This aptamer is the most abundant, representing 8.1%, 26.6% and 35.7% of total reads in the three independent selections S1, S3 and S4, respectively (Figure 2A and B). In addition, aptamers S3R5 and S4R4 differed from S1R1 by only one base and shared the same predicted secondary structure (Figures 2B and 3A). Moreover, aptamers S1R2, S1R3, S1R4, S1R5 and S4R3 also shared similarly predicted secondary structures as S1R1, having two stem-loops separated by 4–10 single-stranded spacer nucleotides (Figure 3B–E and K). Overall, aptamers with the predicted two stem–loop structure represented the majority of aptamers isolated, suggesting that aptamers with these structures might have high affinity to an exposed domain of integrase.
Figure 2.
Similar aptamers were isolated in three independent selections. (A) Change of abundance of aptamers during the last four cycles of enrichment. In most cases, further enrichment of aptamers was achieved by FLAG-IN. One aptamer (S1R1, S3R2 and S4R1 with identical sequence) dominated in all three selections. S3 and S4 used a steeper slope of increasing stringency in selection cycles so plateau was reached at cycle 11, two cycles earlier than S1. (B) Alignment of the most abundant 15 aptamers. Only sequences of the variable region of the aptamers are shown. Aptamers with identical sequences were highlighted (red or blue) and the single base difference between S1R1 and S3R5 (red T and blue G) is shown.
Figure 3.
Predicted secondary structures of the 15 most abundant aptamers. Structure in panel (A) is shared with the most number of aptamers. S1R1 and S3R5 differed only by a single base at (position 19) the loop of the first stem-loop structure.
Functional assay for integrase binding aptamers
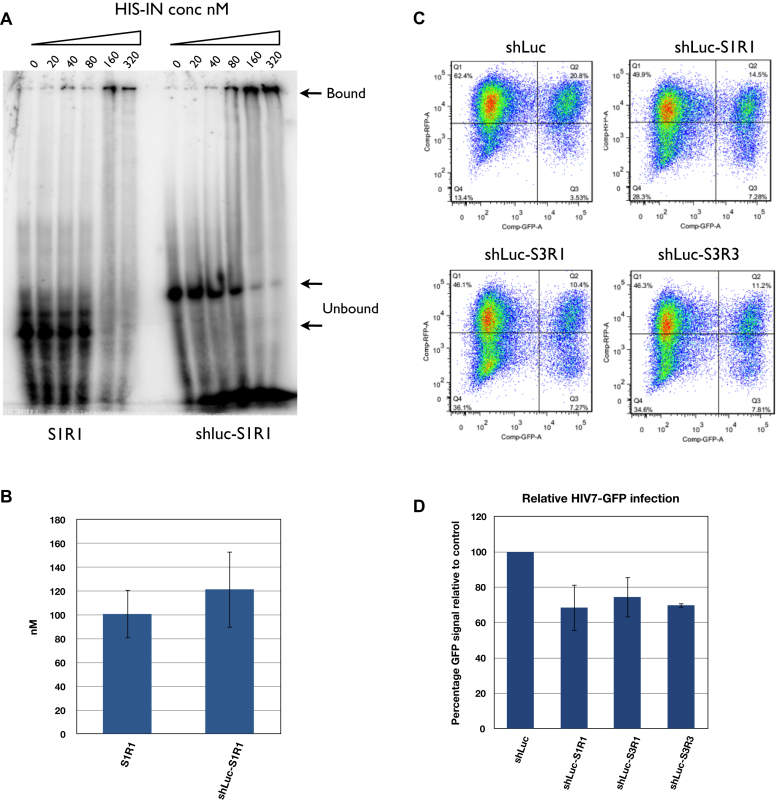
We intended to express the aptamers in T cells or hematopoietic stem cells (HSC) for combinatorial gene therapy. Therefore, we sought a direct functional assay to test the inhibitory effect of the aptamer candidates. To this end, we constructed plasmids (pJR255) consisting of (i) a U6 promoter for driving the expression of the aptamers, (ii) a G418-resistant cassette for selection and (iii) an mCherry marker for cell purification and functional analysis (Supplementary Figure S2). RNAs expressed from U6 (pol III) promoter lack nucleus export signals and stay in the nucleus (18–21). Integrase functions primarily in the nucleus. These nuclear aptamers may be effective against its functions. We first tested the aptamers with a distinct secondary structure, namely S1R1, S3R1 and S3R3 for resistance to infection of the self-inactivating (Sin) lentiviral vector (HIV-7-GFP) (29). Stable HEK293 lines were established by transfecting an empty plasmid backbone (negative control) or plasmids expressing S1R1, S3R1 or S3R3, selecting for G418 resistance and sorting for mCherry positive signals. Each population of cells was than transduced with lentiviral particles at an MOI of 0.3. The transduced cells were analyzed by FACS 10 days after transduction to minimize the effects of GFP signals from nonintegrated viral DNA. If an aptamer exerts any inhibition of HIV integrase, the percentage of GFP and mCherry double-positive cells should be reduced compared to the control. By this measure, FACS data showed that all three aptamers expressed directly from the U6 promoter lacked any inhibition of the lentivirus infection, suggesting that integrase aptamers expressed directly from U6 promoter might not be effective against lentivirus infection (Supplementary Figure S3A and 3B).
Aptamers incorporated into shRNA loops behave like canonical shRNAs
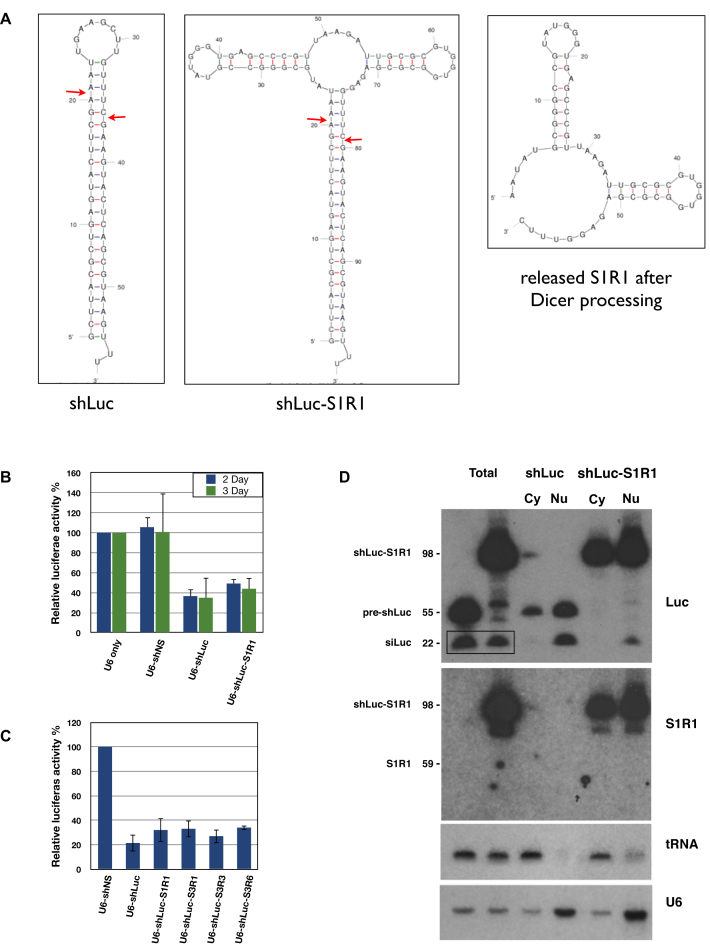
Pre-miRNAs are exported to the cytoplasm mainly by Exportin-5 that recognizes the 3′ two base overhangs of the stem-loop structures (36,37). Some pre-miRNAs contain large loops; nonetheless, these pre-miRNAs are still efficiently exported to the cytoplasm, cleaved by Dicer and loaded into RNA Induced Silencing Complex (RISC) to mediate silencing of target mRNAs (38,39). We took advantage of these properties to design a chimeric RNA therapeutic that incorporated the aptamer into the terminal loop of an shRNA. This shRNA–aptamer fusion may be exported to the cytoplasm and processed by Dicer to release the siRNA. In principle, either the released aptamer or the unprocessed fusion could contribute to HIV inhibition.
First, we used a luciferase assay to test if the shRNA–aptamer fusion could behave like a canonical shRNA that is exported into the cytoplasm and processed by Dicer. A HEK293 clone that stably expressed a firefly luciferase gene was transfected with plasmids containing (i) only the U6 promoter, (ii) U6 driving a nonspecific shRNA (shNS), (iii) U6 driving an shRNA targeting luciferase with an artificial 10-base loop (shLuc) (40) and (iv) shLuc with the aptamer S1R1 as the loop (shLuc-S1R1) (Figure 4A). The shLuc-S1R1 showed substantial knockdown of luciferase activity, albeit slightly weaker than the shLuc with 10-base loop (Figure 4B). Similar results were observed with the shLuc incorporated with three other aptamers S3R1, S3R3 and S3R6 (Figure 4C). These results indicated that the shLuc–aptamer fusions behaved like a canonical shRNA that could be processed by Dicer to release the siLuc RNA, resulting in knockdown of the luciferase target.
Figure 4.
shRNA–aptamer fusions behave like canonical shRNAs. (A) Predicted secondary structures of shLuc, shLuc-S1R1 and the released S1R1. Secondary structures were predicted by M-fold (18). Potential Dicer cleavage sites were marked by red arrows. (B) Percentage knockdown of firefly luciferase by shNS (NonSpecific), shLuc and shLuc-S1R1 fusion at day 2 or day 3 after transfection. Average and standard deviation of three independent experiments. (C) Knockdown of other shLuc-aptamer fusions at day 3 after transfection. Average and standard deviation of two independent experiments. (D) Northern blot showing processing, distribution and stability of shLuc and shLuc-S1R1 fusion. Probes are listed to the right of the panels. RNAs and their corresponding sizes were listed on the left of the panels. Box in top panel highlights mature siRNA against luciferase. tRNA and U6 RNA were used as quality control for fractionation.
To further assess the distribution and processing of the shLuc-aptamer fusion, we performed Northern blot analyses on fractionated samples. When detecting the luciferase guide strand, both shLuc and shLuc-S1R1 showed similar steady state levels (Figure 4D, top panel). Moreover, both shLuc and shLuc-S1R1 showed similar distributions between the nucleus (nu) and the cytoplasm (cy), indicating that the large S1R1 loop neither destabilized the shRNA–aptamer fusion nor hindered its nuclear export. Consistent with the observed 20% weaker knockdown of luciferase activity, 25% less processed siRNA was detected from shLuc-S1R1 than from shLuc (Figure 4D top panel box), indicating that the large S1R1 loop moderately inhibited Dicer processing.
To further determine the fate of the aptamer S1R1 released from the shLuc-S1R1 fusion, we probed the same blot with an S1R1 probe. When detected by the S1R1 probe, the shLuc-S1R1 fusion was distributed in both the nucleus and the cytoplasm, similar to that detected by the luciferase probe. However, the released S1R1 aptamer was not detectable by the S1R1 probe (Figure 4D middle panel), suggesting that the S1R1 aptamer released by Dicer processing was rapidly degraded, similar to the terminal loops and the passenger strand of miRNAs. These results showed that aptamers incorporated into the terminal loop of an shRNA could be exported to the cytoplasm and the shRNA–aptamer fusion could be maintained at a high steady state level. In contrast, the released aptamer did not accumulate to detectable levels.
shRNA–aptamer fusions inhibit lentivirus replication
Despite an undetectable steady state level of released aptamer from the shRNA–aptamer fusion, the high level of shLuc-aptamer fusion may still bind to and inhibit integrase function. To test this possibility, we first tested if the stem of the shRNA affected the binding affinity of the aptamer using a gel mobility assay (Figure 5A). The binding affinity of S1R1 to HIS-IN was 101 nM ± 20 while that of shLuc-S1R1 was not clearly distinguishable at 121 nM ± 32 (P = 0.8536) (Figure 5B), suggesting that the addition of the shLuc stem did not appreciably alter the binding of the aptamer S1R1 to integrase. We next tested if the shLuc-S1R1 and other fusions contained any anti-HIV activity in a functional assay. Stable HEK293 cell lines expressing the shLuc alone, fusions of shLuc-S1R1, shLuc-S3R1 and shLuc–S3R3 were generated. When infected by lentiviral particle HIV-7-GFP, all three shLuc-aptamers fusions showed moderate but consistent inhibition of GFP expression, ranging from 20% to 30% (Figure 5C and D). This was an improvement compared to aptamer alone driven by the same U6 promoter (Supplementary Figure S3A and 3B). These results indicated that when expressed from U6 promoter, the shRNA–aptamer fusions have higher anti-HIV activity than the aptamers alone.
Figure 5.
shLuc-aptamer fusion showed weaker binding to HIS-IN but stronger inhibition of lentivirus infection than aptamer alone. (A) Mobility shift assay comparing aptamer S1R1 alone and shLuc-S1R1 fusion. Unbound aptamers were indicated by lower arrows (59 nucleotides long for S1R1 and 96 nucleotides long for shLuc-S1R1). Bound aptamers appeared to form very large complexes with IN and retained in the wells (upper arrow). Signals from lower bands were used to calculate the 50% binding value. (B) Quantification of affinity of S1R1 and shLuc-S1R1 to HIS-IN. Concentrations at which 50% of probe shift were shown. Binding of S1R1 was the average of three independent assays while that of shLuc-S1R1 was the average of two independent assays. (C) FACS analysis of HEK293 cells expressing shLuc alone or shLuc-aptamer fusions infected by HIV7-GFP lentivirus at MOI of 0.3. (D) Quantification of FACS analysis. Percentage of double-positive (Q2) over total GFP-positive (Q2+Q3) was compared to shLuc control (100%). Averages and standard deviations of three independent assays are shown.
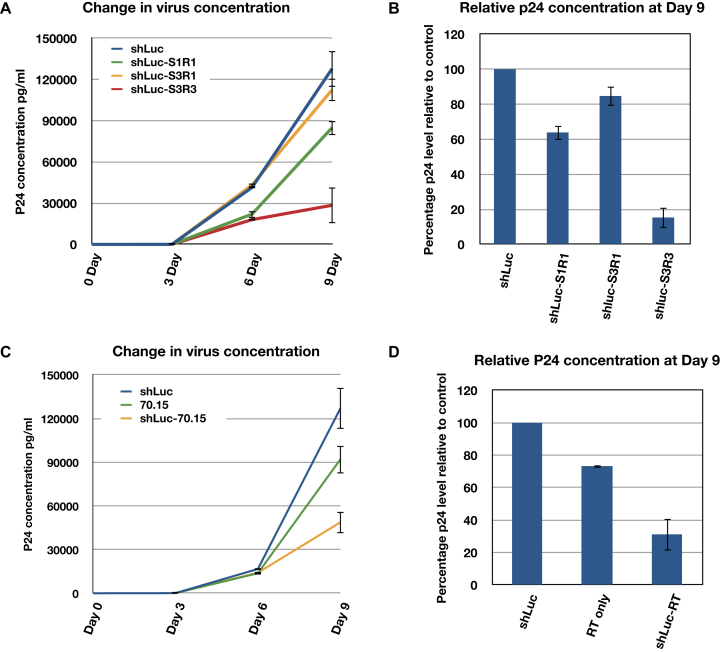
To test if the shRNA-fusions can exert inhibition during subsequent infection cycles, we repeated our assays using replication competent HIV. We generated stable cell lines expressing the shLuc alone and the shLuc-S1R1, shLuc-S3R1 and shLuc–S3R3 fusions in Ghost3 + CXCR4 + CCR5 cells (abbreviated as Ghost3 cells in following text) (27). We then infected the Ghost3 lines with the M-tropic HIV-1 Ba-L strain at a MOI of 0.02. The viral concentration in cultures was monitored by p24 assay for 9 days. While shLuc-S1R1 and shLuc-S3R1 showed similar inhibition level as single cycle infection at 20–30% inhibition, shLuc–S3R3 consistently showed 85% inhibition towards replication competent HIV viruses (Figure 6A and B) suggesting that the shRNA–aptamer fusions could be more effective in multiple infection cycles.
Figure 6.
Expressed shRNA–aptamer fusions exhibited stronger inhibition of HIV replication in multiple cycle infection. (A) shLuc integrase aptamer fusions (shLuc-S1R1, shLuc-S3R1, shLuc–S3R3) inhibited Ba-L strain virus replication in Ghost3 cells to different extents. (B) Inhibition of HIV replication at day 9 post-infection. C. Expressed shLuc RT aptamer fusion (shLuc-70.15) exhibited stronger inhibition than expressed RT aptamer (70.15) alone. D. Inhibition of HIV replication at day 9 post-infection. In A and C, single representative experiment with triplicate samples are shown. In B and D, percentage inhibition is represented by relative p24 concentration to shLuc control. Averages and standard deviations of two (D) or three (B) independent biological assays are shown.
To further test if this strategy could be applied to aptamers targeting proteins primarily function in the cytoplasm, a fusion of shLuc and an RT aptamer (shLuc-70.15) was constructed (41). Stable Ghost3 cells expressing shLuc, 70.15 or shLuc-70.15 fusion were challenged by Ba-L strain HIV and the p24 concentration in cultures were monitored for 9 days. Like the results found in IN aptamers, expressed shLuc-70.15 fusion showed stronger HIV inhibition than the RT aptamer alone (Figure 6C and D), suggesting that fusing an shRNA to an aptamer could enhance its efficacy and that this strategy could be used to target cytoplasmic proteins.
Aptamer S3R3 in combination with an shRNA targeting Tat-Rev has similar potency as the integrase inhibitor Raltegravir
We used the same assay to further test the efficacy of three other candidates S3R4, S3R6 and S4R2. S3R3 remained the most effective in inhibiting proliferation of HIV-1 in Ghost3 cells, consistently showing 80–85% p24 reduction compare to the control cells expressing shLuc (Supplementary Figure S4A and 4B). Moreover, S3R3 showed a higher affinity (47nM ± 3) toward HIS-IN than S1R1 (101 nM ± 20) (Supplementary Figure S5A and 5B). Therefore, we focused on S3R3 for further studies.
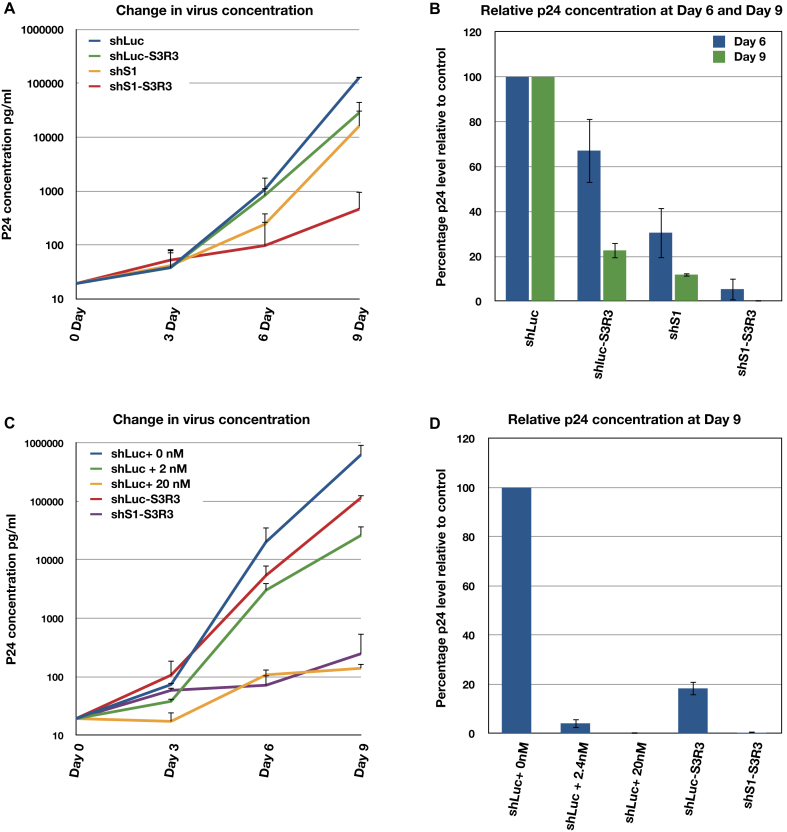
First, we compared the anti-HIV activities of S3R3 to that of an shRNA directed against the HIV Tat-Rev region (shS1) (42). Stable Ghost3 cells expressing shS1 were generated. These cells were then infected with the HIV Ba-L strain at a MOI of 0.02 for 9 days. p24 levels were compared to those from cells expressing the shLuc control and shLuc–S3R3. We found that shS1 cells showed two-fold stronger inhibition than shLuc–S3R3 at day 6. However, the difference became less at day 9 when shLuc–S3R3 showed 78% inhibition while shS1 showed 88% inhibition (Figure 7A and B). To test if shS1 and S3R3 exhibited synergy in inhibiting HIV propagation, a similar plasmid expressing a fusion RNA of shS1 and S3R3 (shS1–S3R3) was constructed. Stable Ghost3 cells lines were generated and then challenged with HIV-1 Ba-L strain. Although each individual RNA provided 80–90% inhibition of HIV replication, the combined shS1–S3R3 fusion RNA resulted in a 100-fold stronger inhibition than the single RNA at day 9 (Figure 7A and B).
Figure 7.
shS1–S3R3 fusion has comparable efficacy as Integrase inhibitor Raltegravir. (A and B) shS1 and aptamer S3R3 (shS1–S3R3) showed synergistic inhibition against Ba-L HIV replication in Ghost3 cells. (A) Changes in virus concentration in Ghost3 cells infected with Ba-L virus were monitored by p24 assays. (B) Percentage inhibition represented by relative p24 concentration to shLuc control is shown at day 6 and day 9 post-infection. (C and D) shS1–S3R3 showed efficacy comparable to integrase inhibitor Raltegravir. (C) Changed in virus concentration in Ghost3 cells infected with Ba-L virus. (D) Inhibition of HIV replication at day 9 post-infection. In A and C, single representative experiment with triplicate samples are shown. Y-axis uses semi-log scale so only positive errors are shown. In B and D, averages and standard deviations of two independent assays are shown.
To further assess the potency of the aptamer S3R3, we compared the efficacy of shLuc–S3R3 and the shS1–S3R3 to the FDA approved anti-integrase drug Raltegravir. We monitored HIV Ba-L proliferation in Ghost3 cells expressing shLuc–S3R3 to that of control cells expressing shLuc in the presence or absence of Raltegravir. We tested 2nM and 20nM concentration of Raltegravir, corresponding to IC50 and IC95 in 10% FBS cell culture (43,44). As observed previously, shLuc–S3R3 showed 80–85% inhibition at day 9 compared to 96% inhibition for 2nM and >99% inhibition for 20 nM Raltegravir (Figure 7C and D). These results showed that S3R3 alone possessed anti-HIV activity that was weaker than a low dose of Raltegravir. On the other hand, the combination of shS1 and S3R3 (shS1–S3R3) showed much stronger inhibition with a p24 level comparable to those treated with a high dose of Raltegravir (Figure 7C and D). These results demonstrated that combining anti-HIV shRNA and anti-integrase aptamers could be a very effective gene therapy strategy against HIV.
Aptamer S3R3 is effective in long-term inhibition of HIV replication
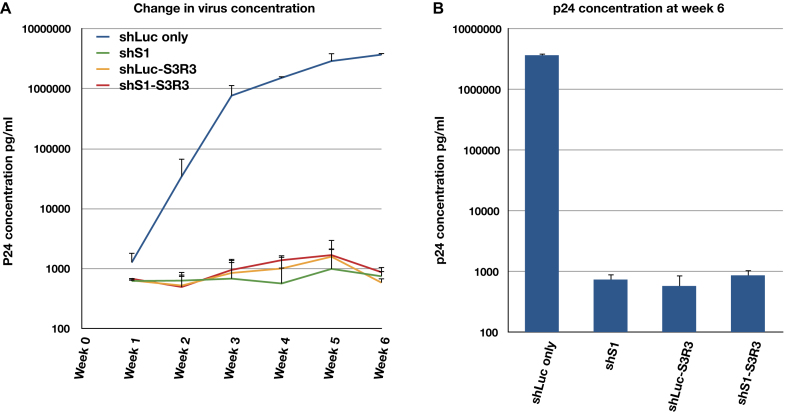
We further tested the long-term efficacy of shRNA–aptamer fusions in CD4 positive T cell line. Stable CEM cell lines expressing shLuc, shLuc–S3R3, shS1 or shS1–S3R3 were generated (see Material and Method). 1 × 105 stable cells were then infected with T-tropic HIV pNL4-3 at a MOI of 0.01. Viral concentrations were monitored for 6 weeks by p24 assay (Figure 8). Surprisingly, aptamer alone (shLuc–S3R3) and shRNA alone (shS1) showed similar efficacy as the combination (shS1–S3R3). In all cases, the viral concentration was more than three orders of magnitude lower than that of control (shLuc) and did not increase over the observed 6 weeks. This inhibition was much stronger than that observed in Ghost3 cells (compare Figure 8 to Figure 7). Aptamer S3R3 was selected against integrase derived from pNL4-3 strain. This might explain a much stronger inhibition in this assay than in the short-term assays that challenged by the Ba-L strain. Alternatively, the fusions might exhibit higher potency in its natural host of T cells than in the engineered Ghost3 cell. In summary, anti-integrase aptamer expressed as an shRNA–aptamer fusion can confer long term resistance to HIV-1 replication in T cells. This strategy of expressing an aptamer into the terminal loop of an shRNA can be applicable for gene therapy against HIV and can potentially be adopted to treat other diseases.
Figure 8.
shRNA–aptamer fusions confer long-term resistance to HIV replication in T cells. (A) Changes in virus concentration in CEM cells infected with pNL4-3 virus were monitored by p24 assays for 6 weeks. (B) p24 concentration at the end of week 6 post infection. In panel A, a single representative experiment with triplicate samples is shown. Y-axis uses semi-log scale so only positive errors are shown. In panel B, the averages and standard deviations of two independent assays are shown.
DISCUSSION
In this study, we developed a strategy to select and stably express RNA aptamers against the HIV integrase and reverse transcriptase. By incorporating the aptamers into the terminal loop of an shRNA, we achieved long-term inhibition of HIV replication in a cell culture system. The SELEX method has been useful for isolating RNA aptamers against specific protein targets. Typically, aptamers with high binding affinity are selected by binding to a single soluble protein. Applying this approach to unstable or low solubility proteins such as the HIV integrase has been technically challenging. Tags such as MBP, HIS and FLAG can improve solubility and/or stability of the tagged proteins and thus facilitate purification and selection. Varying the capture method is a longstanding approach to reduce background. We employed multiple tagged proteins in alternate selection cycles in a modified SELEX protocol, we termed multi-tag SELEX. This method allows the selection of aptamers against low solubility or unstable proteins while minimize non-specific binding to the tags. In most cases, target proteins expressed in mammalian cells is preferable over bacterially expressed ones. HIV integrase undergoes various post-translational modifications such as phosphorylation, acetylation and sumoylation in mammalian cells (45). This increase the likelihood of obtaining functional aptamers against modified IN protein. Furthermore, the integrase should be in its native conformation and therefore, be more likely to form complexes with other cellular proteins (31–34). This increases the chance of obtaining aptamers against IN protein epitopes available in host cells. Ideally, selection should be alternated among all three tagged proteins. However, the limited yield of FLAG-IN restricted the use of FLAG-tagged integrase to only one selection cycle. Nevertheless, using this strategy, we have successfully isolated the first RNA aptamers that target HIV integrase under physiological salt concentrations. This strategy should see general applications for other difficult and specific targets, such as a functional domain of a protein that may have low solubility. Importantly, his method also allows selection of more physiologically relevant aptamers.
Another hurdle for using aptamers in gene therapy is the lack of sustained expression. Currently, most nucleic acid aptamers are administered extracellularly to block interaction of surface receptor and viral proteins. Some aptamers can be internalized together with the receptors. However, this method only allows a limited level of delivery and is subject to cyclical variation. A sustained high level of RNA aptamer can be achieved by Pol III promoter driven expression. However, without an intrinsic nuclear export signal, Pol III transcripts stay in the nucleus, thereby limiting their application for targeting nuclear proteins, such as transcription factors (18,20,21,46–48). By incorporating the aptamer into the terminal loop of an shRNA, the U6 promoter driven shRNA–aptamer fusions persisted at a high level in the nucleus and the cytoplasm. A portion of the fusion is processed by Dicer, resulting in knockdown of target genes. We could not detect the released aptamer moiety in a Northern blot assay, consistent with the rapid degradation of uncapped and unprotected RNA in the cytoplasm. However, the shRNA–aptamer fusions targeting either IN or RT exhibited a stronger inhibition than the aptamer alone. The mechanism of this enhancement is not clear. In principle, several factors or a combination of them could contribute to a stronger inhibition. The shRNAs facilitated the export of the fusions to the cytoplasm where newly translated viral polyproteins might be more susceptible to aptamer binding. The stronger inhibition observed in multiple infection cycle than in single infection cycle suggested that this might be one of the mechanism. The stem structure of an shRNA is very stable. It could have stabilized the active aptamer structures and/or increased the steady-state levels of the primary transcript. Importantly, the integrase aptamer S3R3 showed a strong synergy with an shRNA targeting the tat-rev region and together the shS1–S3R3 fusion exhibited a very strong and prolonged inhibition of HIV replication in cell cultures.
This strategy of combining shRNAs and aptamers allows both sequence-based and structure-based targeting by one shRNA–aptamer fusion. The shRNA and aptamer combination allows flexibility in targeting either the same gene (protein and mRNA), two genes (one protein and one mRNA) or one protein plus one non-coding RNA. Moreover, by using multiplexed vectors (7), multiple shRNA–aptamer fusions can be expressed from a single transcript. This allows inhibition of multiple targets at once and will be particularly useful to combat the ever-evolving HIV.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Chris Gandhi and Arin Nam for critical reading and commenting on this manuscript. Research reported in this publication included work performed in the Analytical Cytometry Core, Integrative Genomics Core and Drug Discovery & Structural Biology Core supported by the National Cancer Institute of the National Institutes of Health under award number P30CA033572. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
The following reagent was obtained through the NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH: Ghost(3)X4/R5 from Dr. Vineet N. KewalRamani and Dr. Dan R. Littman; pNL4-3 from Dr. Malcolm Martin; and HIV-1Ba-L from Dr. Suzanne Gartner, Dr. Mikulas Popovic and Dr. Robert Gallo. HIS-IN expression vector pET-15b-IN (F185H/C280S) plasmid was a gift from Dr. Robert Craigie NIDDK, NIH.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institute of Health [RO1 AI042552 and AI029329 to J.J.R.] Funding for open access charge: National Institute of Health [RO1 AI042552].
Conflict of interest statement. None declared.
REFERENCES
- 1. Leibman R.S., Riley J.L.. Engineering T cells to functionally cure HIV-1 infection. Mol. Ther. 2015; 23:1149–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. DiGiusto D.L. Stem cell gene therapy for HIV: strategies to inhibit viral entry and replication. Curr. HIV/AIDS Rep. 2015; 12:79–87. [DOI] [PubMed] [Google Scholar]
- 3. Pernet O., Yadav S.S., An D.S.. Stem cell-based therapies for HIV/AIDS. Adv. Drug Deliv. Rev. 2016; 103:187–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Li M.J., Kim J., Li S., Zaia J., Yee J.K., Anderson J., Akkina R., Rossi J.J.. Long-term inhibition of HIV-1 infection in primary hematopoietic cells by lentiviral vector delivery of a triple combination of anti-HIV shRNA, anti-CCR5 ribozyme, and a nucleolar-localizing TAR decoy. Mol. Ther. 2005; 12:900–909. [DOI] [PubMed] [Google Scholar]
- 5. Asparuhova M.B., Barde I., Trono D., Schranz K., Schümperli D.. Development and characterization of a triple combination gene therapy vector inhibiting HIV-1 multiplication. J. Gene Med. 2008; 10:1059–1070. [DOI] [PubMed] [Google Scholar]
- 6. Anderson J.S., Javien J., Nolta J.A., Bauer G.. Preintegration HIV-1 inhibition by a combination lentiviral vector containing a chimeric TRIM5α protein, a CCR5 shRNA, and a TAR decoy. Mol. Ther. 2009; 17:2103–2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chung J., Scherer L.J., Gu A., Gardner A.M., Torres-Coronado M., Epps E.W., Digiusto D.L., Rossi J.J.. Optimized lentiviral vectors for HIV gene therapy: multiplexed expression of small RNAs and inclusion of MGMT(P140K) drug resistance gene. Mol. Ther. 2014; 22:952–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tuerk C., Gold L.. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990; 249:505–510. [DOI] [PubMed] [Google Scholar]
- 9. Ellington A.D., Szostak J.W.. Selection in vitro of single-stranded DNA molecules that fold into specific ligand-binding structures. Nature. 1992; 355:850–852. [DOI] [PubMed] [Google Scholar]
- 10. Zhou J., Swiderski P., Li H., Zhang J., Neff C.P., Akkina R., Rossi J.J.. Selection, characterization and application of new RNA HIV gp 120 aptamers for facile delivery of Dicer substrate siRNAs into HIV infected cells. Nucleic Acids Res. 2009; 37:3094–3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ramalingam D., Duclair S., Datta S.A., Ellington A., Rein A., Prasad V.R.. RNA aptamers directed to human immunodeficiency virus type 1 Gag polyprotein bind to the matrix and nucleocapsid domains and inhibit virus production. J. Virol. 2011; 85:305–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ditzler M.A., Bose D., Shkriabai N., Marchand B., Sarafianos S.G., Kvaratskhelia M., Burke D.H.. Broad-spectrum aptamer inhibitors of HIV reverse transcriptase closely mimic natural substrates. Nucleic Acids Res. 2011; 39:8237–8247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Whatley A.S., Ditzler M.A., Lange M.J., Biondi E., Sawyer A.W., Chang J.L., Franken J.D., Burke D.H.. Potent inhibition of HIV-1 reverse transcriptase and replication by nonpseudoknot, “UCAA-motif” RNA Aptamers. Mol. Ther. Nucleic Acids. 2013; 2:e71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Shum K.T., Zhou J., Rossi J.J.. Aptamer-based therapeutics: new approaches to combat human viral diseases. Pharmaceuticals (Basel). 2013; 6:1507–1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Duclair S., Gautam A., Ellington A., Prasad V.R.. High-affinity RNA aptamers against the HIV-1 protease inhibit both in vitro protease activity and late events of viral replication. Mol. Ther. Nucleic Acids. 2015; 4:e228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kruspe S., Mittelberger F., Szameit K., Hahn U.. Aptamers as drug delivery vehicles. ChemMedChem. 2014; 9:1998–2011. [DOI] [PubMed] [Google Scholar]
- 17. Zhou J., Rossi J.. Aptamers as targeted therapeutics: current potential and challenges. Nat. Rev. Drug Discov. 2017; 16:181–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Good P.D., Krikos A.J., Li S.X., Bertrand E., Lee N.S., Giver L., Ellington A., Zaia J.A., Rossi J.J., Engelke D.R.. Expression of small, therapeutic RNAs in human cell nuclei. Gene Ther. 1997; 4:45–54. [DOI] [PubMed] [Google Scholar]
- 19. Bertrand E., Castanotto D., Zhou C., Carbonnelle C., Lee N.S., Good P., Chatterjee S., Grange T., Pictet R. et al. . The expression cassette determines the functional activity of ribozymes in mammalian cells by controlling their intracellular localization. RNA. 1997; 3:75–88. [PMC free article] [PubMed] [Google Scholar]
- 20. Paul C.P., Good P.D., Li S.X., Kleihauer A., Rossi J.J., Engelke D.R.. Localized expression of small RNA inhibitors in human cells. Mo.l Ther. 2003; 7:237–247. [DOI] [PubMed] [Google Scholar]
- 21. Lee N.S., Kim D.H., Alluin J., Robbins M., Gu S., Li H., Kim J., Salvaterra P.M., Rossi J.J.. Functional and intracellular localization properties of U6 promoter-expressed siRNAs, shRNAs, and chimeric VA1 shRNAs in mammalian cells. RNA. 2008; 14:1823–1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Adachi A., Gendelman H.E., Koenig S., Folks T., Willey R., Rabson A., Martin M.A.. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J. Virol. 1986; 59:284–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jenkins T.M., Engelman A., Ghirlando R., Craigie R.. A soluble active mutant of HIV-1 integrase: involvement of both the core and carboxyl-terminal domains in multimerization. J. Biol. Chem. 1996; 271:7712–7718. [DOI] [PubMed] [Google Scholar]
- 24. Zhou J., Satheesan S., Li H., Weinberg M.S., Morris K.V., Burnett J.C., Rossi J.J.. Cell-specific RNA aptamer against human CCR5 specifically targets HIV-1 susceptible cells and inhibits HIV-1 infectivity. Chem. Biol. 2015; 22:379–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003; 31:3406–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Castanotto D., Lingeman R., Riggs A.D., Rossi J.J.. CRM1 mediates nuclear-cytoplasmic shuttling of mature microRNAs. Proc. Natl. Acad. Sci. U.S.A. 2009; 106:21655–21659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mörner A., Björndal A., Albert J., Kewalramani V.N., Littman D.R., Inoue R., Thorstensson R., Fenyö E.M., Björling E.. Primary human immunodeficiency virus type 2 (HIV-2) isolates, like HIV-1 isolates, frequently use CCR5 but show promiscuity in coreceptor usage. J. Virol. 1999; 73:2343–2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gartner S., Markovits P., Markovitz D.M., Kaplan M.H., Gallo R.C., Popovic M.. The role of mononuclear phagocytes in HTLV-III/LAV infection. Science. 1986; 233:215–219. [DOI] [PubMed] [Google Scholar]
- 29. Yam P.Y., Li S., Wu J., Hu J., Zaia J.A., Yee J.K.. Design of HIV vectors for efficient gene delivery into human hematopoietic cells. Mol. Ther. 2002; 5:479–484. [DOI] [PubMed] [Google Scholar]
- 30. DiGiusto D.L., Krishnan A., Li L., Li H., Li S., Rao A., Mi S., Yam P., Stinson S. et al. . RNA-based gene therapy for HIV with lentiviral vector-modified CD34(+) cells in patients undergoing transplantation for AIDS-related lymphoma. Sci. Transl. Med. 2010; 2:36ra43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Cherepanov P., Maertens G., Proost P., Devreese B., Van Beeumen J., Engelborghs Y., De Clercq E., Debyser Z.. HIV-1 integrase forms stable tetramers and associates with LEDGF/p75 protein in human cells. J. Biol. Chem. 2003; 278:372–381. [DOI] [PubMed] [Google Scholar]
- 32. Llano M., Vanegas M., Hutchins N., Thompson D., Delgado S., Poeschla E.M.. Identification and characterization of the chromatin-binding domains of the HIV-1 integrase interactor LEDGF/p75. J. Mol. Biol. 2006; 360:760–773. [DOI] [PubMed] [Google Scholar]
- 33. Zheng Y., Ao Z., Wang B., Jayappa K.D., Yao X.. Host protein Ku70 binds and protects HIV-1 integrase from proteasomal degradation and is required for HIV replication. J. Biol. Chem. 2011; 286:17722–17735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Arhel N., Kirchhoff F.. Host proteins involved in HIV infection: new therapeutic targets. Biochim. Biophys. Acta. 2010; 1802:313–321. [DOI] [PubMed] [Google Scholar]
- 35. Cherepanov P., Pluymers W., Claeys A., Proost P., De Clercq E., Debyser Z.. High-level expression of active HIV-1 integrase from a synthetic gene in human cells. FASEB J. 2000; 14:1389–1399. [DOI] [PubMed] [Google Scholar]
- 36. Okada C., Yamashita E., Lee S.J., Shibata S., Katahira J., Nakagawa A., Yoneda Y., Tsukihara T.. A high-resolution structure of the pre-microRNA nuclear export machinery. Science. 2009; 326:1275–1279. [DOI] [PubMed] [Google Scholar]
- 37. Lee S.J., Jiko C., Yamashita E., Tsukihara T.. Selective nuclear export mechanism of small RNAs. Curr. Opin. Struct. Biol. 2011; 21:101–108. [DOI] [PubMed] [Google Scholar]
- 38. Feng Y., Zhang X., Graves P., Zeng Y.. A comprehensive analysis of precursor microRNA cleavage by human Dicer. RNA. 2012; 18:2083–2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Winter J., Link S., Witzigmann D., Hildenbrand C., Previti C., Diederichs S.. Loop-miRs: active microRNAs generated from single-stranded loop regions. Nucleic Acids Res. 2013; 41:5503–5512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Li M.J., Rossi J.J.. Lentiviral vector delivery of recombinant small interfering RNA expression cassettes. Methods Enzymol. 2005; 392:218–226. [DOI] [PubMed] [Google Scholar]
- 41. Lange M.J., Sharma T.K., Whatley A.S., Landon L.A., Tempesta M.A., Johnson M.C., Burke D.H.. Robust suppression of HIV replication by intracellularly expressed reverse transcriptase aptamers is independent of ribozyme processing. Mol. Ther. 2012; 20:2304–2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Aagaard L.A., Zhang J., von Eije K.J., Li H., Saetrom P., Amarzguioui M., Rossi J.J.. Engineering and optimization of the miR-106b cluster for ectopic expression of multiplexed anti-HIV RNAs. Gene Ther. 2008; 15:1536–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Summa V., Petrocchi A., Bonelli F., Crescenzi B., Donghi M., Ferrara M., Fiore F., Gardelli C., Gonzalez Paz O. et al. . Discovery of raltegravir, a potent, selective orally bioavailable HIV-integrase inhibitor for the treatment of HIV-AIDS infection. J. Med. Chem. 2008; 51:5843–5855. [DOI] [PubMed] [Google Scholar]
- 44. Temesgen Z., Siraj D.S.. Raltegravir: first in class HIV integrase inhibitor. Ther. Clin. Risk Manag. 2008; 4:493–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zheng Y., Yao X.. Posttranslational modifications of HIV-1 integrase by various cellular proteins during viral replication. Viruses. 2013; 5:1787–1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Mi J., Zhang X., Rabbani Z.N., Liu Y., Su Z., Vujaskovic Z., Kontos C.D., Sullenger B.A., Clary B.M.. H1 RNA polymerase III promoter-driven expression of an RNA aptamer leads to high-level inhibition of intracellular protein activity. Nucleic Acids Res. 2006; 34:3577–3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Choi Y.S., Hur J., Lee H.K., Jeong S.. The RNA aptamer disrupts protein-protein interaction between beta-catenin and nuclear factor-kappaB p50 and regulates the expression of C-reactive protein. FEBS Lett. 2009; 583:1415–1421. [DOI] [PubMed] [Google Scholar]
- 48. Salamanca H.H., Antonyak M.A., Cerione R.A., Shi H., Lis J.T.. Inhibiting heat shock factor 1 in human cancer cells with a potent RNA aptamer. PLoS One. 2014; 9:e96330. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.