Abstract
Hantaviruses are zoonotic viruses with a complex evolutionary history of virus–host coevolution and cross-species transmission. Although hantaviruses have a broad reservoir host range, virus–host relationships were previously thought to be strict, with a single virus species infecting a single host species. Here, we describe Bruges virus, a novel hantavirus harbored by the European mole (Talpa europaea), which is the well-known host of Nova virus. Phylogenetic analyses of all three genomic segments showed tree topology inconsistencies, suggesting that Bruges virus has emerged from cross-species transmission and ancient reassortment events. A high number of coinfections with Bruges and Nova viruses was detected, but no evidence was found for reassortment between these two hantaviruses. These findings highlight the complexity of hantavirus evolution and the importance of further investigation of hantavirus–reservoir relationships.
Keywords: Bruges orthohantavirus, Bunyavirales, coinfection, Talpa europaea, virus–host interaction, zoonosis
Introduction
Hantaviruses (Order Bunyavirales, Family Hantaviridae) are important zoonotic pathogens that are responsible for hantavirus diseases, which are typified by fever, thrombocytopenia, and renal and/or pulmonary injury. Hantavirus are maintained within animal reservoir populations, with humans occasionally acting as a dead-end host after inhalation of aerosols of virus-infected saliva, urine, or faeces (Maes et al. 2004). Human-to-human transmission is rare, and has been reported only for Andes virus (Wells 1997; Chaparro 1998). Since the isolation of Hantaan virus, the prototype hantavirus of hemorrhagic fever with renal syndrome, from lung tissue of the striped field mouse (Apodemus agrarius coreae), the role of rodents in the spread of pathogenic hantaviruses has been well established (Lee et al. 1978). In recent years, the host range of hantaviruses has expanded with the detection of previously undescribed hantaviruses in shrews, moles, and bats (Arai et al. 2007, 2008; Klempa et al. 2007; Sumibcay et al. 2012; Weiss et al. 2012). Although the pathogenicity of non-rodent-borne hantaviruses still warrants further investigation, shrew-borne hantavirus infections of humans have recently been reported in Africa (Heinemann et al. 2016).
Hantaviruses have a close relationship with their natural hosts. Even though spillover events can occur, hantaviruses are usually maintained by a single or a few closely related host species. Spillover infections of a single hantavirus into two or even more sympatric mammalian hosts have been documented (Schmidt-Chanasit et al. 2010; Schlegel et al. 2012) but the opposite situation where a single mammalian species serves as a reservoir host of two unique hantavirus species is less prevalent (Gu, Hejduk, et al. 2014). The most prominent example of host sharing of two hantaviruses occurs with Hantaan and Dobrava–Belgrade viruses. The striped field mouse is the reservoir of Dobrava–Belgrade virus (Kurkino and Saaremaa genotypes) in Central and Eastern Europe and Hantaan virus in Asia (Lee et al. 1978; Klempa et al. 2003). Although both Dobrava–Belgrade and Hantaan viruses have been detected in Apodemus agrarius in Russia, the geographical range of both viruses does not appear to overlap (Garanina et al. 2009; Kariwa et al. 2012). Moreover, Dobrava–Belgrade is carried by a different subspecies, A. a. agrarius present in Europe instead of A. a. coreae and other subspecies present in Asia (Kim and Park 2015).
Early observations of strict virus–host relationships and supportive phylogenetic evidence, based upon virus and host mitochondrial cytochrome b sequence data, led to an initial hypothesis of coevolution between rodent-borne hantaviruses and their hosts over millions of years (Hughes and Friedman 2000). The discovery of hantaviruses in shrews and moles has challenged those longstanding hypotheses (Guo et al. 2013). Recent phylogenetic analyses uncovered a complex evolutionary history with cross-species transmission and ancient reassortment events shaping hantavirus evolution (Bennett et al. 2014). Furthermore, ancestors of shrews and moles or bats but not rodents appear to be the natural hosts of primordial hantaviruses (Kang, Kadjo, et al. 2011; Yanagihara et al. 2014; Witkowski et al. 2016).
The complex evolution of hantaviruses is especially apparent with mole-borne hantaviruses, where multiple cases of cross-species transmission or host-switching events have occurred (Bennett et al. 2014). Thus far, five hantaviruses have been identified in moles (family Talpidae) (table 1) (Arai et al. 2008; Kang, Bennett, Dizney, et al. 2009; Kang, Bennett, Sumibcay, et al. 2009; Kang, Bennett, et al. 2011; Kang et al. 2016). Talpids are distributed throughout Eurasia and North America and 39 species have been identified to date (Wilson and Reeder 2005). More extensive screening of species of talpids will likely result in the discovery of more novel hantaviruses, and further uncover the mechanism of cross-over events that have shaped hantavirus evolution.
Table 1.
Overview of Hantaviruses Associated with Hosts of the Family Talpidae
| Hantavirus Species | Abbr. | Host Species | Subfamily | S Segment | M Segment | L Segment |
|---|---|---|---|---|---|---|
| Asama virus strain JP/N10/UT/2008/1 | ASAV | Urotrichus talpoides | Talpinae | EU929072 | EU929075 | EU929078 |
| Oxbow virus strain US/Ng1453/NG/2003/1 | OXBV | Neurotrichus gibbsii | Talpinae | FJ539166 | FJ539167 | FJ593497 |
| Rockport virus strain US/MSB57412/SA/1986/1 | RKPV | Scalopus aquaticus | Scalopinae | HM015223 | HM015219 | HM015221 |
| Dahonggou Creek virus strain CH/MSB281632/SF/1989/1 | DHCV | Scaptonyx fusicaudus | Talpinae | / | / | HQ616595 |
| Nova virus strain PL/Te34/TE/2013/1 | NVAV | Talpa europaea | Talpinae | KR072621 | KR072622 | KR072623 |
| Bruges virus strain BE/Vieux-Genappe/TE/2013/1a | BRGV | Talpa europaea | Talpinae | KX551960 | KX551961 | KX551962 |
| Bruges virus, strain DE/Wandlitz/TE/2013/1a | BRGV | Talpa europaea | Talpinae | MF683844 | MF683845 | MF683846 |
| Bruges virus, strain UK/MSB MSB48363/TE/1982/1a | BRGV | Talpa europaea | Talpinae | / | / | MF706165 |
Note.—Accession numbers of complete coding sequences are marked in bold.
Described for the first time in this article.
In this study, we aimed to further elucidate the role of moles in hantavirus evolution. Here, we describe Bruges virus, a novel hantavirus harbored by the European mole (Talpa europaea). This discovery marks the second hantavirus, in addition to Nova virus (Kang, Bennett, et al. 2011; Laenen et al. 2016), in the European mole. We have characterized the complete genome of Bruges virus and investigated its distribution in the European mole population. In addition, we studied the implications of the evolutionary constraints placed upon Bruges and Nova viruses for infection rates of these mole-borne hantaviruses. Our current study provides a new comprehension of the European mole as a host for mole-borne hantaviruses and gives a novel perspective on the hantavirus–host relationship.
Materials and Methods
Sample Collection
From 2013 to 2015, European moles were trapped in fields and gardens in Belgium. As moles are persecuted as a pest animal, no additional permits were required for fieldwork. Immediately after trapping, moles were stored at −20 °C until processing. Lung, kidney, heart, liver, and spleen tissue were aseptically removed and stored in RNAlater Stabilization Solution (Ambion). Samples (liver, kidney, or muscle tissue) from four European moles, captured in August 1982 in Avon County (United Kingdom), were provided by the Museum of Southwestern Biology at the University of New Mexico in Albuquerque. Lung samples from European moles captured in central Poland (Gu, Hejduk, et al. 2014) and in France (Hugot et al. 2014) were also analyzed. Moreover, lung, kidney, liver, and spleen tissue samples were collected from a single European mole found dead in the vicinity of Wandlitz village near Berlin, Germany, in March 2013 and stored at −80 °C until processing.
Hantavirus Screening
Total RNA was extracted from European mole tissue with the RNeasy Mini kit (Qiagen) according to the manufacturer’s instructions. A nested degenerate RT-PCR was performed using the OneStep RT-PCR kit (Qiagen) with primers directed at a conserved region in the polymerase gene, as described previously (Klempa et al. 2006) or primers specific for Bruges virus (supplementary table S1, Supplementary Material online). PCR amplicons were purified using ExoSAP-IT PCR Product Cleanup (Affymetrix) and sequenced according to the ddNTP chain termination method with the BigDye Terminator v3.1 cycle sequencing kit (Life Technologies) on an Applied Biosystems 3130xl Genetic Analyzer. Sequences were manually inspected using Chromas 2.4 (Technelysium) and consensus sequences were derived with Seqman 7.0 (DNAstar).
Complete Genome Sequencing
Total RNA from lung and kidney samples directed for Ion Torrent sequencing was extracted with the RNeasy Mini kit (Qiagen). Six extracts were pooled, quantified using a Qubit RNA HS assay (Life Technologies) and RNA quality was checked with an Agilent 2100 Bioanalyzer using the RNA 6000 Nano kit (Agilent). Subsequently, the RNA extract was subjected to rRNA depletion using the RiboZero kit (Epicentre) and mRNA using Dynabeads mRNA DIRECT Micro kit (Thermo Fisher Scientific) after which RNA was cleaned up with the RNeasy MinElute Clean up kit (Qiagen). Libraries were prepared using the Ion Total RNA-seq kit (Life Technologies) according to the manufacturer’s instructions. Templates were prepared with the Ion PI Hi-Q OT2 200 kit and sequencing was performed with the Ion PI Hi-Q sequencing kit. The sample was loaded on a PI chip and run on the Ion Torrent Proton platform. Initial quality assessment and FastQ generation was performed with the Torrent Suite Software 4.6 (Life Technologies). De novo assembly was initiated using CLC genomics workbench 10.0.1 (Qiagen).
For the Wandlitz strain of Bruges virus from Germany, the initial complete genome sequencing efforts were performed using Illumina NextSeq500 technology. After homogenizing the tissue using a gentleMACS dissociator (Miltenyi Biotec) we performed an ultracentrifugation-based protocol of particle-associated nucleic acids (PAN) purification (Stang et al. 2005), followed by unspecific preamplification (QuantiTect Whole Transcriptome Kit, QIAGEN). Sequencing libraries were again prepared using the Nextera Library Prep Kit (Illumina) and sequenced using paired end sequencing on an Illumina NextSeq500 system. In addition, for both samples Sanger sequencing was used in regions with low coverage.
Glycosylation Prediction and DEmARC Analysis
N-linked glycosylation sites were predicted using the NetNGlyc 1.0 Server (Gupta and Brunak 2002). Multiple sequence alignments of concatenated nucleocapsid and glycoprotein precursor proteins were constructed with MAFFT employing the iterative refinement method incorporating local pairwise alignment information and manually edited in MEGA 7.0 (Katoh et al. 2002; Kumar et al. 2016). Pairwise evolutionary distances (PED) were calculated using a WAG amino acid substitution model and maximum likelihood approach in TREE-PUZZLE (Schmidt et al. 2002). Hierarchical classification was achieved by employing the DEmARC framework in R (Lauber and Gorbalenya 2012; R Core Team 2017).
Phylogenetic Analysis
Multiple sequence alignments for nucleocapsid, glycoprotein precursor, and polymerase were constructed with MAFFT and manually edited in MEGA 7.0 (Katoh et al. 2002; Kumar et al. 2016). Bayesian phylogenetic analyses were inferred in BEAST 1.8.2 employing two independent Markov chain Monte Carlo (MCMC) runs with a chain length of 50,000,000 generations. Tree and log files of independent runs of BEAST were combined using LogCombiner 1.8.2, employing a BurnIn period of 10%. The MCMC analyses were run until effective sample sizes >200 were obtained. A consensus tree was built with TreeAnnotator 1.8.2 using the maximum clade credibility method and visualized in Figtree (Drummond and Rambaut 2007).
Results
Detection of a Novel Hantavirus in the European Mole
To characterize the relationship between mole-borne hantaviruses and their hosts, we screened renal tissue of European moles captured in Belgium for hantavirus RNA. Using a nested PCR approach with primers directed at the polymerase gene (Klempa et al. 2006), we detected a new hantavirus in a kidney sample from a European mole, captured in 2014 near Bruges, Belgium. The 347-nucleotide (nt) fragment displayed relatively low sequence similarity to other hantaviruses (73% nucleotide identity to closest neighbor Seewis virus, 75% amino acid identity to closest neighbor Bowé virus). We named this novel hantavirus Bruges virus after the origin of initial detection.
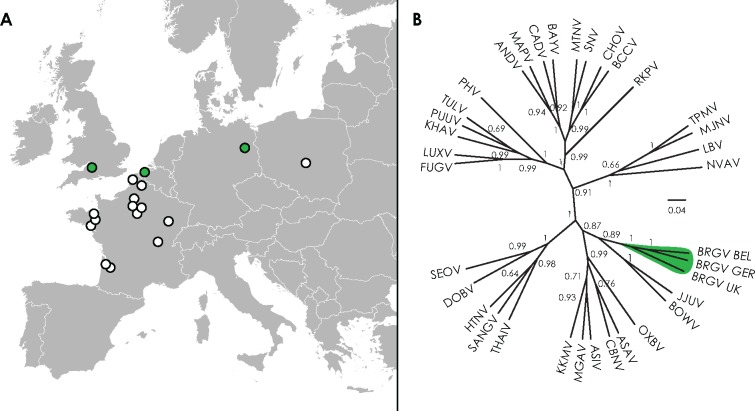
Furthermore, opportunistic testing resulted in the detection of Bruges virus in a liver sample from a European mole captured in 1982 near Avon, United Kingdom (ID number MSB48363) (1/4 European moles testing positive) and in lung tissue of a European mole found in 2013 near Wandlitz, Germany (1/1 positive) (fig. 1A). Phylogenetic analysis of a 435-nt fragment, corresponding to nucleotide positions 2535 to 2969 of the L segment of type strain Bruges virus BE/Vieux-Genappe/TE/2013/1, demonstrated that the three sequences formed a separate clade, divergent from all other hantaviruses (fig. 1B, primers available in supplementary table S1, Supplementary Material online). Although Bruges virus strains from three countries had nucleotide identities ranging from 80% to 83%, the partial L fragment was highly conserved at the amino acid (aa) level (99–100%), confirming these viruses to be strains of the same hantavirus species.
Fig. 1.
—Bruges virus (BRGV) distribution in Europe. (A) Map of Europe, showing regions in Belgium, Germany, and the United Kingdom where Bruges virus-positive European moles were captured (colored in green) and regions in France and Poland were negative European moles were captured (white circles). Samples originating from Belgium are outlined in more detail in figure 4. (B) Maximum clade credibility tree based upon the partial L segment nucleotide sequences (435 nt). Sequence alignment is available upon request. PUUV (Puumala virus), KHAV (Khabarovsk virus), PHV (Prospect Hill virus), TULV (Tula virus,), FUGV (Fugong virus), LUXV (Luxi virus), SNV (Sin Nombre virus), MTNV (Montano virus), ANDV (Andes virus), CHOV (Choclo virus), CADV (Cano Delgadito virus), BAYV (Bayou virus), BCCV (Black Creek Canal virus), MAPV (Maporal virus), RKPV (Rockport virus), TPMV (Thottapalayam virus), MJNV (Imjin virus), KKMV (Kenkeme virus), ASIV (Assikala virus), MGAV (Amga virus), CBNV (Cao Bang virus), JJUV (Jeju virus), BOWV (Bowé virus), ASAV (Asama virus), OXBV (Oxbow virus), NVAV (Nova virus), BRGV BEL (strain BE/Vieux-Genappe/TE/2013/1, Belgium), BRGV GER (strain DE/Wandlitz/TE/2013/1, Germany), BRGV UK (strain UK/Avon/TE/MSB48363/1982/1, United Kingdom), HTNV (Hantaan virus), SEOV (Seoul virus), DOBV (Dobrava–Belgrade virus), SANGV (Sangassou virus), and LBV (Laibin virus).
Additionally, European mole lung tissues from Poland (n = 13) and France (n = 119) were screened with primers targeting all hantavirus species (Klempa et al. 2006) and primers specific for Bruges virus (supplementary table S1, Supplementary Material online). However, none of the tested samples was found to be positive for Bruges virus.
Complete Genome Characterization of Bruges Virus
Two Bruges virus-positive European moles samples, originating from Belgium and Germany, were subjected to full genome sequencing. The virus strains were designated Bruges virus BE/Vieux-Genappe/TE/2013/1 and Bruges virus strain DE/Wandlitz/TE/2013/1, respectively. The complete genome sequence of Bruges virus strain BE/Vieux-Genappe/TE/2013/1 exhibited a conventional hantavirus genome organization. The 1,829-nt S segment contained a single open reading frame (ORF) (nt position 39–1331), encoding a putative nucleocapsid (N) protein of 430 aa in length. As seen also in other hantaviruses harbored by hosts of the Talpidae family, an additional open reading frame on the S segment encoding a nonstructural NSs protein was not present. The 3,641-nt M segment contained a single ORF (nt position 42–3461), encoding the glycoprotein precursor (GPC) of the Gn and Gc glycoproteins, separated by a WAVSA pentapeptide at aa positions 649–653, instead of the more commonly seen WAASA motif. N-glycosylation sites were predicted at N138, N240, N352, N404, N567, and N932, revealing the absence of the additional glycosylation site at N101, present in Nova virus strains. The 6,538-nt L segment contained a single ORF (nt position 39–6500), encoding the 2,153-aa long RNA-dependent RNA polymerase (RdRP).
Determination of the complete genome also for the DE/Wandlitz/TE/2013/1 strain from Germany enabled insights into the intraspecies variability of the new virus. The nucleotide sequence identity values were remarkably low for the complete genome (77.2%, 81.2%, and 80.9% for the S-, M-, and L-segment complete sequences, respectively), as well as coding sequences (82.2%, 81.4%, and 80.7% for the S-, M-, and L-segment coding sequences, respectively). On the other hand, the amino acid sequences were highly conserved, showing amino acid sequence identity values of 99.1%, 95.4%, and 95.9% for the N, GPC, and RdRP, respectively. The coding sequences were also of the same length. Minor insertions/deletions were observed only in the noncoding regions of the S and M segments. The WAVSA motif, instead of WAASA, and the same putative glycosylation sites were also observed in the GPC sequence of the DE/Wandlitz/TE/2013/1 strain.
Phylogenetic Analyses and Taxonomic Placement
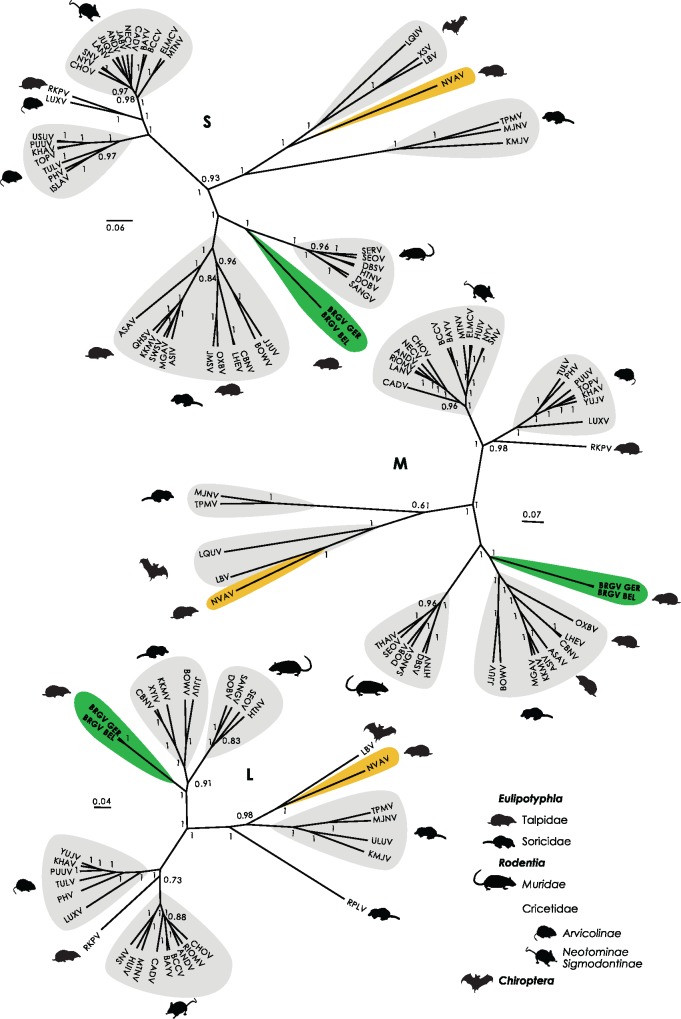
Bayesian phylogenetic inference of the complete coding sequence of the S segment revealed that even though Bruges and Nova viruses infected the same reservoir host, they were highly divergent viruses (fig. 2). Although Nova virus formed a monophyletic clade with bat-borne hantaviruses, the S segment of Bruges virus appeared to be divergent with closest relationship to hantaviruses associated with hosts from the Muridae family. A phylogenetic tree of the complete coding sequence of the M segment confirmed the high divergence between Bruges and Nova viruses. Furthermore, analysis of the M segment uncovered a closer relationship between Bruges virus and hantaviruses harbored by Eulipotyphla hosts. Bayesian inference of the amino acid sequence of the complete L segment showed that Bruges virus clustered closer to the root, forming a separate monophyletic group. These results indicate that Bruges virus appeared to be highly divergent from other hantaviruses. Inconsistencies in tree topologies suggest that Bruges virus may have emerged from ancient reassortment events.
Fig. 2.
—Bayesian phylogenetic analysis of the nucleocapsid (S), glycoprotein precursor (M), and RNA-dependent RNA polymerase (L) amino acid sequences. Maximum clade credibility trees display posterior probability values from 0.5. Virus clustering by host species is marked according to the legend.
To determine the taxonomic position of Bruges virus within the family Hantaviridae, the concatenated N protein and GPC sequences of the Belgian and German strains of Bruges virus were included in multiple sequence alignments with all other hantavirus species approved by the International Committee on Taxonomy of Viruses (ICTV). In accordance with the most recent taxonomy report (Adams et al. 2017), hierarchical clustering was implemented using the DEmARC framework (Lauber and Gorbalenya 2012). PED ranges with highest threshold support measure (TSM) values were determined for species and genus groups. For each taxonomy level, a threshold with an optimal clustering cost of zero could be further specified. Hierarchical clustering using DEmARC demonstrated that Bruges virus was a distinct species, as likewise recognized in the current taxonomy report (fig. 3).
Fig. 3.
—DEmARC analysis of the concatenated nucleocapsid and glycoprotein precursor of the family Hantaviridae. Frequency distribution graphs of PED values show the intragroup genetic divergence. For each classification level, box-and-whisker plots are used to plot level-specific PED frequency distributions. A Bayesian phylogenetic tree is used for vertical grouping of hantavirus species. The two hierarchical classification levels are colored orange and purple.
Prevalence of Bruges Virus Infection in the European Mole Population in Belgium
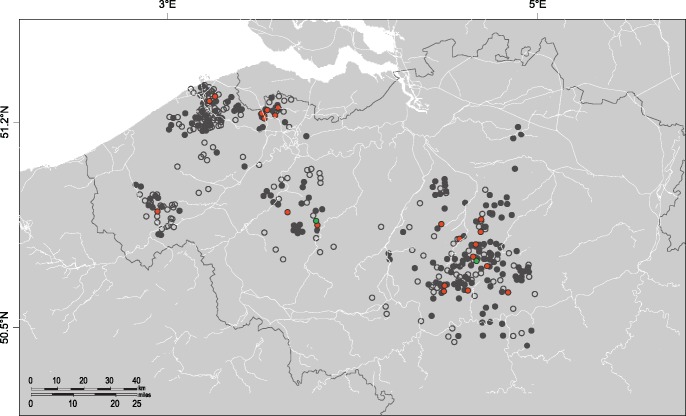
Nova virus has been shown to be highly adapted to infection of the European mole, resulting in high RNA positivity rates (Gu, Hejduk, et al. 2014). We previously reported that Nova virus had a high prevalence and widespread distribution in the European mole population in Belgium (Laenen et al. 2016). In analyzing kidney tissues from 479 European moles captured in Belgium for Nova virus RNA by RT-PCR, 255 of 479 (53.2%) tested positive, suggesting efficient transmission. By contrast, in using primers directed at the S segment of Bruges virus (supplementary table S1, Supplementary Material online), only 22 of the 479 (4.6%) samples were positive. Bruges virus-positive samples, however, were widely distributed across the entire sampling region, indicating a widespread distribution (fig. 4).
Fig. 4.
—Distribution of Bruges and Nova viruses in the European mole in Belgium. Negative samples are denoted by open circles. Samples uniquely positive for Nova virus are marked in gray. Samples uniquely positive for Bruges virus are marked in green. Samples that are coinfected with Bruges and Nova viruses are marked in red.
Coinfections with Bruges and Nova Viruses
Remarkably, 20 of 22 (90.9%) Bruges virus-positive European moles were coinfected (fig. 4), as evidenced by partial S-segment sequences of both Bruges virus and Nova virus in kidney tissues. This represents the first time coinfection with two hantavirus species have been detected in nature. Bruges virus-positive samples from the United Kingdom and Germany were negative for Nova virus, although Nova virus may circulate there as well. Using Ion Torrent and Sanger sequencing, the complete S-, M-, and L-genomic segments of both Bruges virus (BRGV BE/Vieux-Genappe/TE/2013/1) and Nova virus (NVAV BE/Vieux-Genappe/TE/2013/2) were recovered from a dually infected European mole from Belgium, indicating that both viruses were present. Moreover, both Nova virus and Bruges virus were detected in kidney, lung, heart, liver, and spleen tissue, denoting a broad tissue distribution.
Discussion
Selected species of rodent-borne hantaviruses in Eurasia and the Americas cause mild to life-threatening diseases in humans, characterized by renal and/or cardiopulmonary insufficiency or failure (Kruger et al. 2015). Thus, their impact on human health underscores the importance of understanding the reservoir host range and transmission dynamics of hantaviruses. However, the characteristics of hantavirus infection in rodents, shrews, moles, and bats have not been fully elucidated. Here, we report the detection and genomic characterization of a new hantavirus, named Bruges virus, in the European mole, previously recognized as the reservoir of Nova virus (Kang, Bennett, Sumibcay, et al. 2009). The European mole has a broad geographic range throughout much of continental Europe (Amori et al. 2017). From detection of Bruges virus in European moles from Belgium, Germany, and the United Kingdom, we can infer that a widespread dispersal of Bruges virus mirroring the wide host range is plausible. On the other hand, no Bruges virus RNA was found in European moles from Poland and France, but this may be due to the sample size and low prevalence of Bruges virus infection, as observed in Belgium. Bruges virus strains, originating from Belgium, Germany, or the United Kingdom, showed considerable nucleotide sequence variability while sharing high amino-acid similarities, indicating long-term circulation of the virus in the regions of their detection. It is therefore likely that such geographic variants/lineages occur elsewhere across Europe.
Complete genome characterization demonstrated that Bruges virus has an expected genome organization encoding an N protein, GPC, and RdRP. The GPC contained a WAVSA pentapeptide sequence at the position of the WAASA cleaving site. Although the WAASA site is usually considered to be well conserved, a WAVSA sequence was previously seen in Asama virus, a hantavirus harbored by the Japanese shrew mole (Urotrichus talpoides) (Arai et al. 2008), Qian Hu Shan virus in the greater striped-back shrew (Sorex cylindricauda) (Zuo et al. 2014) and several New World hantaviruses (e.g., Castelo dos Sonhos virus, Maciel virus, Pergamino virus, and Araraquara virus) (Firth et al. 2012). The pentapeptide sequence, part of the C region at the carboxyterminal end of the putative signal peptide should have a small uncharged amino acid at position −3 of the cleavage site for efficient recognition by a signal peptidase, a requirement that is fulfilled by the WAVSA motif (Lober et al. 2001).
Bayesian phylogenetic inference of the amino acid sequences of the N protein, GPC, and RdRP of Bruges virus demonstrated that Bruges virus was highly divergent from Nova virus and all other hantaviruses. These findings confirm that while for most other hantaviruses close virus–host associations are apparent through phylogenetic analysis, mole-borne hantaviruses are scattered across the hantavirus phylogenetic tree. It suggests that they were likely involved in several cross-species transmission events. The probability of spill-over of hantaviruses to moles could depend on behavioral factors influencing virus exposure and host susceptibility factors (e.g., innate immune response, receptor compatibility) (Plowright et al. 2017). Unfortunately, little is known about the immunology and genetics of talpid species. Complete genome sequencing of moles could possibly provide a foundation for better characterization of hantavirus–host interactions.
Furthermore, the genome segment-specific inconsistencies in the tree topologies suggest that Bruges virus might have emerged from ancient reassortment events, with the S segment more closely related to Muridae-associated hantaviruses, whereas the M segment was closer to hantaviruses hosted by shrews and moles and the L segment formed a monophyletic clade closer to the root. Phylogenetic inference indicated that Bruges virus may be the result of a complicated evolutionary process likely involving cross-species transmission and reassortment events. However, alternative explanations for these observations cannot be completely ruled out. High sequence divergence of Bruges virus and/or poor taxon sampling could bias phylogenetic estimation. Moreover, the high evolutionary distance of Bruges virus from other taxa could lead to long-branch attraction, thereby incorrectly grouping divergent taxa together. The detection and whole-genome sequencing of other hantaviruses in a broad range of hosts could be pivotal to further elucidation of this complex process.
Hantavirus prevalence in the natural host population relies on myriad factors (e.g., host evolutionary history, environmental factors, host population dynamics, virus characteristics) (Kallio et al. 2006; Clement et al. 2010; Voutilainen et al. 2016; Drewes et al. 2017; Tian et al. 2017). Bruges virus represents the second hantavirus, apart from Nova virus, detected in the European mole. The high prevalence of Nova virus infection in European moles has been extensively studied in France and Poland (Gu, Dormion, et al. 2014; Gu, Hejduk, et al. 2014). Recently, the high positivity rate of 53.2% (255 of 479 positive) and widespread distribution of Nova virus were confirmed in European moles captured in Belgium (Laenen et al. 2016). Here, we had the opportunity to investigate the dispersal of Bruges virus in the Belgian mole population under identical conditions. A significantly lower positivity rate of 4.6% (22 of 479 positive) was observed for Bruges virus, suggesting the possibility of a lower transmission efficiency of Bruges virus in the European mole population and raising the question of whether the European mole is the preferential host of Bruges virus. Each hantavirus is generally associated with a single, or a few closely related, host species, with examples of spillover events to sympatric hosts (Hjelle and Yates 2001; Schmidt et al. 2016). Although Bruges virus was demonstrated to have a broad tissue distribution in the European mole, we cannot definitely exclude the possibility of a spillover infection. Experimental infections of bank voles, rats, and deer mice have contributed to a better understanding of hantavirus host persistence mechanisms (Yanagihara et al. 1985; Botten et al. 2002, 2003; Easterbrook et al. 2007; Schountz et al. 2012). Unfortunately, the European mole does not thrive under laboratory conditions, excluding the possibility of long-term monitoring of virus excretion. Notwithstanding a less efficient transmission among European moles than Nova virus, Bruges virus was found to be competently infecting European moles across Belgium. Moreover, independent detection of Bruges virus in European moles from Belgium, Germany, and the United Kingdom further strongly indicates that European mole is a genuine reservoir host of Bruges virus and not only a randomly infected dead-end host sharing the habitat with the authentic reservoir host.
This study represents the first report of dual infections of the same host with two genetically distinct hantaviruses. These findings clearly indicate that infection with a hantavirus does not prevent a secondary infection with another hantavirus species. Studies of the natural host immune response and viral persistence have been challenging because of a lack of suitable reagents for most host species (Schountz and Prescott 2014). Experimental infections of rats with Seoul virus and deer mice with Sin Nombre virus have demonstrated that regulatory T cells contribute to hantavirus persistence, despite the presence of neutralizing antibodies (Easterbrook et al. 2007; Schountz et al. 2007). Some degree of cross-neutralizing activity against other hantavirus species was seen after Hantaan or Andes virus DNA vaccination (Hooper et al. 1999, 2006). A lack of a protective response against a secondary hantavirus infection in European moles could be explained by the higher degree of antigenic variability between Bruges virus and Nova virus and/or lower titers of neutralizing antibodies that are presumably produced during viral persistence.
The consequences of multiple infections on hantavirus fitness have not been studied. An important question to ask is whether two coinfecting hantaviruses will be in competition or will cooperate for more efficient host exploitation. It has been reported that viruses that have evolved under conditions where both single and multiple infections occur, exhibit adaptive phenotypic plasticity in response to coinfections with important implications for virus epidemiology and virulence (Leggett, Benmayor, et al. 2013). Viruses can evolve to be better adapted to coinfections by faster replication or interaction with the host immune system (Leggett, Benmayor, et al. 2013; Leggett, Buckling, et al. 2013; Bose et al. 2016).
The presence of coinfection also raises questions concerning possible reassortment of both hantaviruses. Reassortment between two hantavirus species can lead to the generation of virus progeny with new characteristics than can differ from the two parental viruses. Although reassortment events within a single hantavirus species have been reported, interspecies reassortment seems less likely, mainly because situations where two hantavirus species infect the same host are thought to rarely occur (Klempa et al. 2003; Razzauti et al. 2008). In vitro, reassortment between two hantavirus species led to the generation of a new variant with new properties and a higher replication efficiency (Rizvanov et al. 2004; Handke et al. 2010; Kirsanovs et al. 2010). Although Bruges virus and Nova virus are genetically highly divergent viruses, the possibility of historic or future reassortment events should be kept in mind. The design of our study did not allow us to distinguish reassortment from a simple coinfection. Simultaneous detection of all six virus segments (three segments of both viruses) in the same tissue specimen does not necessarily rule out the possibility of a reassortment event. Only through the rarely successful virus isolations and subsequent clonal purification of the new isolates could one show that the two viruses “coexist” in the same host without reassortment.
Furthermore, recent detection of hantaviruses in moles and findings of a single hantavirus species circulating in multiple hosts raises the question of how frequently have host-switching events have occurred during hantavirus evolution. Preferential host-jumping and local adaption could resemble what is perceived as virus–host coevolution (Ramsden et al. 2009). Geoghegan and coworkers recently highlighted the role of cross-species transmission during evolution. They reported that virus–host codivergence occurs less frequently than previously expected, whereas all studied virus families had the potential to jump species, indicating that cross-species transmission could play a more central role in virus evolution (Geoghegan et al. 2017).
In this study, we describe Bruges virus, a hantavirus able to coinfect its host, the European mole, together with Nova virus as another mole-borne hantavirus. This raises questions of host specificity and hantavirus–host codivergence. More research is warranted to gain insights into hantavirus ecology, transmission dynamics and virus–host evolution to elucidate these important questions.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
This research has been supported by KU Leuven projects IDO/07/005 and CREA/11/027 and the Slovak Research and Development Agency under the contract No. APVV-15-0232 and in part by U.S. Public Health Service grant R01AI075057 from the National Institute of Allergy and Infectious Diseases and grant P20GM103516 from the National Institute of General Medical Sciences, National Institutes of Health.
Literature Cited
- Adams MJ, et al. 2017. Changes to taxonomy and the International Code of Virus Classification and Nomenclature ratified by the International Committee on Taxonomy of Viruses (2017). Arch Virol. 162(8):2505–2538. [DOI] [PubMed] [Google Scholar]
- Amori G, et al. 2017. Talpa europaea. The IUCN Red List of Threatened Species 2017: e.T41481A22320754. http://dx.doi.org/10.2305/IUCN.UK.2017-2.RLTS.T41481A22320754.en, last accessed August 21, 2017. [Google Scholar]
- Arai S, et al. 2007. Hantavirus in northern short-tailed shrew, United States. Emerg Infect Dis. 13(9):1420–1423.http://dx.doi.org/10.3201/eid1309.070484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arai S, et al. 2008. Molecular phylogeny of a newfound hantavirus in the Japanese shrew mole (Urotrichus talpoides). Proc Natl Acad Sci U S A. 105(42):16296–16301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett SN, Gu SH, Kang HJ, Arai S, Yanagihara R.. 2014. Reconstructing the evolutionary origins and phylogeography of hantaviruses. Trends Microbiol. 22(8):473–482.http://dx.doi.org/10.1016/j.tim.2014.04.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bose J, Kloesener MH, Schulte RD.. 2016. Multiple-genotype infections and their complex effect on virulence. Zoology (Jena) 119(4):339–349.http://dx.doi.org/10.1016/j.zool.2016.06.003 [DOI] [PubMed] [Google Scholar]
- Botten J, et al. 2002. Shedding and intracage transmission of Sin Nombre hantavirus in the deer mouse (Peromyscus maniculatus) model. J Virol. 76(15):7587–7594.http://dx.doi.org/10.1128/JVI.76.15.7587-7594.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botten J, et al. 2003. Persistent Sin Nombre virus infection in the deer mouse (Peromyscus maniculatus) model: sites of replication and strand-specific expression. J Virol. 77(2):1540–1550.http://dx.doi.org/10.1128/JVI.77.2.1540-1550.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaparro J. 1998. Assessment of person-to-person transmission of hantavirus pulmonary syndrome in a Chilean hospital setting. J Hosp Infect. 40(4):281–285.http://dx.doi.org/10.1016/S0195-6701(98)90304-8 [DOI] [PubMed] [Google Scholar]
- Clement J, et al. 2010. Beechnuts and outbreaks of nephropathia epidemica (NE): of mast, mice and men. Nephrol Dial Transplant. 25(6):1740–1746.http://dx.doi.org/10.1093/ndt/gfq122 [DOI] [PubMed] [Google Scholar]
- Drewes S, et al. 2017. Host-associated absence of human Puumala virus infections in Northern and Eastern Germany. Emerg Infect Dis. 23(1):83–86.http://dx.doi.org/10.3201/eid2301.160224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond AJ, Rambaut A.. 2007. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol. 7:214.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Easterbrook JD, Zink MC, Klein SL.. 2007. Regulatory T cells enhance persistence of the zoonotic pathogen Seoul virus in its reservoir host. Proc Natl Acad Sci U S A. 104(39):15502–15507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firth C, et al. 2012. Diversity and distribution of hantaviruses in South America. J Virol. 86(24):13756–13766.http://dx.doi.org/10.1128/JVI.02341-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garanina SB, et al. 2009. Genetic diversity and geographic distribution of hantaviruses in Russia. Zoonoses Public Health 56(6–7):297–309.http://dx.doi.org/10.1111/j.1863-2378.2008.01210.x [DOI] [PubMed] [Google Scholar]
- Geoghegan JL, Duchene S, Holmes EC.. 2017. Comparative analysis estimates the relative frequencies of co-divergence and cross-species transmission within viral families. PLoS Pathog. 13(2):e1006215.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu SH, Dormion J, Hugot JP, Yanagihara R.. 2014. High prevalence of Nova hantavirus infection in the European mole (Talpa europaea) in France. Epidemiol Infect. 142(6):1167–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu SH, Hejduk J, et al. 2014. Co-circulation of soricid- and talpid-borne hantaviruses in Poland. Infect Genet Evol. 28:296–303.http://dx.doi.org/10.1016/j.meegid.2014.10.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo WP, et al. 2013. Phylogeny and origins of hantaviruses harbored by bats, insectivores, and rodents. PLoS Pathog. 9(2):e1003159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta R, Brunak S.. 2002. Prediction of glycosylation across the human proteome and the correlation to protein function. Pac Symp Biocomput. 7:310–322. [PubMed] [Google Scholar]
- Handke W, et al. 2010. Generation and characterization of genetic reassortants between Puumala and Prospect Hill hantavirus in vitro. J Gen Virol. 91(Pt 9):2351–2359. [DOI] [PubMed] [Google Scholar]
- Heinemann P, et al. 2016. Human infections by non-rodent-associated hantaviruses in Africa. J Infect Dis. 214(10):1507–1511. [DOI] [PubMed] [Google Scholar]
- Hjelle B, Yates T.. 2001. Modeling hantavirus maintenance and transmission in rodent communities. Curr Top Microbiol Immunol. 256:77–90. [DOI] [PubMed] [Google Scholar]
- Hooper JW, Custer DM, Smith J, Wahl-Jensen V.. 2006. Hantaan/Andes virus DNA vaccine elicits a broadly cross-reactive neutralizing antibody response in nonhuman primates. Virology 347(1):208–216.http://dx.doi.org/10.1016/j.virol.2005.11.035 [DOI] [PubMed] [Google Scholar]
- Hooper JW, Kamrud KI, Elgh F, Custer D, Schmaljohn CS.. 1999. DNA vaccination with hantavirus M segment elicits neutralizing antibodies and protects against seoul virus infection. Virology 255(2):269–278. [DOI] [PubMed] [Google Scholar]
- Hughes AL, Friedman R.. 2000. Evolutionary diversification of protein-coding genes of hantaviruses. Mol Biol Evol. 17(10):1558–1568.http://dx.doi.org/10.1093/oxfordjournals.molbev.a026254 [DOI] [PubMed] [Google Scholar]
- Hugot JP, et al. 2014. Genetic diversity of Talpa europaea and Nova hanta virus (NVAV) in France. Bull Acad Vet Fr. 167(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallio ER, et al. 2006. Prolonged survival of Puumala hantavirus outside the host: evidence for indirect transmission via the environment. J Gen Virol. 87(Pt 8):2127–2134. [DOI] [PubMed] [Google Scholar]
- Kang HJ, Bennett SN, Hope AG, Cook JA, Yanagihara R.. 2011. Shared ancestry between a newfound mole-borne hantavirus and hantaviruses harbored by cricetid rodents. J Virol. 85(15):7496–7503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang HJ, Bennett SN, Dizney L, et al. 2009. Host switch during evolution of a genetically distinct hantavirus in the American shrew mole (Neurotrichus gibbsii). Virology 388(1):8–14.http://dx.doi.org/10.1016/j.virol.2009.03.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang HJ, Bennett SN, Sumibcay L, et al. 2009. Evolutionary insights from a genetically divergent hantavirus harbored by the European common mole (Talpa europaea). PLoS One 4(7):e6149.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang HJ, Gu SH, Cook JA, Yanagihara R.. 2016. Dahonggou Creek virus, a divergent lineage of hantavirus harbored by the long-tailed mole (Scaptonyx fusicaudus). Trop Med Health 44:16.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang HJ, Kadjo B, Dubey S, Jacquet F, Yanagihara R.. 2011. Molecular evolution of Azagny virus, a newfound hantavirus harbored by the West African pygmy shrew (Crocidura obscurior) in Cote d’Ivoire. Virol J. 8(1):373.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kariwa H, et al. 2012. Isolation and characterization of hantaviruses in Far East Russia and etiology of hemorrhagic fever with renal syndrome in the region. Am J Trop Med Hyg. 86(3):545–553.http://dx.doi.org/10.4269/ajtmh.2012.11-0297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Misawa K, Kuma K, Miyata T.. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30(14):3059–3066.http://dx.doi.org/10.1093/nar/gkf436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HR, Park YC.. 2015. Genetic diversity and genetic structure of the striped field mouse Apodemus agrarius coreae (Muridae, Rodentia) in Korea. Gene 572(2):292–297.http://dx.doi.org/10.1016/j.gene.2015.08.014 [DOI] [PubMed] [Google Scholar]
- Kirsanovs S, et al. 2010. Genetic reassortment between high-virulent and low-virulent Dobrava-Belgrade virus strains. Virus Genes 41(3):319–328.http://dx.doi.org/10.1007/s11262-010-0523-2 [DOI] [PubMed] [Google Scholar]
- Klempa B, et al. 2003. Genetic interaction between distinct Dobrava hantavirus subtypes in Apodemus agrarius and A. flavicollis in nature. J Virol. 77(1):804–809.http://dx.doi.org/10.1128/JVI.77.1.804-809.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klempa B, et al. 2006. Hantavirus in African wood mouse, Guinea. Emerg Infect Dis. 12(5):838–840.http://dx.doi.org/10.3201/eid1205.051487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klempa B, et al. 2007. Novel hantavirus sequences in Shrew, Guinea. Emerg Infect Dis. 13(3):520–522.http://dx.doi.org/10.3201/eid1303.061198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruger DH, Figueiredo LT, Song JW, Klempa B.. 2015. Hantaviruses–globally emerging pathogens. J Clin Virol. 64:128–136.http://dx.doi.org/10.1016/j.jcv.2014.08.033 [DOI] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Tamura K.. 2016. MEGA7: molecular evolutionary genetics analysis Version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874.http://dx.doi.org/10.1093/molbev/msw054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laenen L, et al. 2016. Spatio-temporal analysis of Nova virus, a divergent hantavirus circulating in the European mole in Belgium. Mol Ecol. 25(23):5994–6008.http://dx.doi.org/10.1111/mec.13887 [DOI] [PubMed] [Google Scholar]
- Lauber C, Gorbalenya AE.. 2012. Partitioning the genetic diversity of a virus family: approach and evaluation through a case study of picornaviruses. J Virol. 86(7):3890–3904.http://dx.doi.org/10.1128/JVI.07173-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HW, Lee PW, Johnson KM.. 1978. Isolation of the etiologic agent of Korean Hemorrhagic fever. J Infect Dis. 137(3):298–308. [DOI] [PubMed] [Google Scholar]
- Leggett HC, Benmayor R, Hodgson DJ, Buckling A.. 2013. Experimental evolution of adaptive phenotypic plasticity in a parasite. Curr Biol. 23(2):139–142. [DOI] [PubMed] [Google Scholar]
- Leggett HC, Buckling A, Long GH, Boots M.. 2013. Generalism and the evolution of parasite virulence. Trends Ecol Evol. 28(10):592–596.http://dx.doi.org/10.1016/j.tree.2013.07.002 [DOI] [PubMed] [Google Scholar]
- Lober C, Anheier B, Lindow S, Klenk HD, Feldmann H.. 2001. The Hantaan virus glycoprotein precursor is cleaved at the conserved pentapeptide WAASA. Virology 289(2):224–229. [DOI] [PubMed] [Google Scholar]
- Maes P, Clement J, Gavrilovskaya I, Van Ranst M.. 2004. Hantaviruses: immunology, treatment, and prevention. Viral Immunol. 17(4):481–497.http://dx.doi.org/10.1089/vim.2004.17.481 [DOI] [PubMed] [Google Scholar]
- Plowright RK, et al. 2017. Pathways to zoonotic spillover. Nat Rev Microbiol. 15(8):502–510.http://dx.doi.org/10.1038/nrmicro.2017.45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. 2017. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing. [Google Scholar]
- Ramsden C, Holmes EC, Charleston MA.. 2009. Hantavirus evolution in relation to its rodent and insectivore hosts: no evidence for codivergence. Mol Biol Evol. 26(1):143–153. [DOI] [PubMed] [Google Scholar]
- Razzauti M, Plyusnina A, Henttonen H, Plyusnin A.. 2008. Accumulation of point mutations and reassortment of genomic RNA segments are involved in the microevolution of Puumala hantavirus in a bank vole (Myodes glareolus) population. J Gen Virol. 89(7):1649–1660. [DOI] [PubMed] [Google Scholar]
- Rizvanov AA, Khaiboullina SF, St Jeor S.. 2004. Development of reassortant viruses between pathogenic hantavirus strains. Virology 327(2):225–232.http://dx.doi.org/10.1016/j.virol.2004.07.012 [DOI] [PubMed] [Google Scholar]
- Schlegel M, et al. 2012. Tula virus infections in the Eurasian water vole in Central Europe. Vector Borne Zoonotic Dis. 12(6):503–513.http://dx.doi.org/10.1089/vbz.2011.0784 [DOI] [PubMed] [Google Scholar]
- Schmidt HA, Strimmer K, Vingron M, von Haeseler A.. 2002. TREE-PUZZLE: maximum likelihood phylogenetic analysis using quartets and parallel computing. Bioinformatics 18(3):502–504.http://dx.doi.org/10.1093/bioinformatics/18.3.502 [DOI] [PubMed] [Google Scholar]
- Schmidt S, et al. 2016. High genetic structuring of Tula hantavirus. Arch Virol. 161(5):1135–1149. [DOI] [PubMed] [Google Scholar]
- Schmidt-Chanasit J, et al. 2010. Extensive host sharing of central European Tula virus. J Virol. 84(1):459–474.http://dx.doi.org/10.1128/JVI.01226-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schountz T, et al. 2007. Regulatory T cell-like responses in deer mice persistently infected with Sin Nombre virus. Proc Natl Acad Sci U S A. 104(39):15496–15501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schountz T, et al. 2012. Kinetics of immune responses in deer mice experimentally infected with Sin Nombre virus. J Virol. 86(18):10015–10027.http://dx.doi.org/10.1128/JVI.06875-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schountz T, Prescott J.. 2014. Hantavirus immunology of rodent reservoirs: current status and future directions. Viruses 6(3):1317–1335.http://dx.doi.org/10.3390/v6031317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stang A, Korn K, Wildner O, Uberla K.. 2005. Characterization of virus isolates by particle-associated nucleic acid PCR. J Clin Microbiol. 43(2):716–720.http://dx.doi.org/10.1128/JCM.43.2.716-720.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumibcay L, et al. 2012. Divergent lineage of a novel hantavirus in the banana pipistrelle (Neoromicia nanus) in Cote d’Ivoire. Virol J. 9:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian H, et al. 2017. Anthropogenically driven environmental changes shift the ecological dynamics of hemorrhagic fever with renal syndrome. PLoS Pathog. 13(1):e1006198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voutilainen L, Kallio ER, Niemimaa J, Vapalahti O, Henttonen H.. 2016. Temporal dynamics of Puumala hantavirus infection in cyclic populations of bank voles. Sci Rep. 6:21323.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss S, et al. 2012. Hantavirus in bat, Sierra Leone. Emerg Infect Dis. 18(1):159–161.http://dx.doi.org/10.3201/eid1801.111026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells RM. 1997. An unusual hantavirus outbreak in southern Argentina: person-to-person transmission? Hantavirus Pulmonary Syndrome Study Group for Patagonia. Emerg Infect Dis. 3(2):171–174.http://dx.doi.org/10.3201/eid0302.970210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson DE, Reeder DM.. 2005. Mammal species of the world: a taxonomic and geographic reference. Baltimore, Maryland: Johns Hopkins University Press. [Google Scholar]
- Witkowski PT, et al. 2016. Phylogenetic analysis of a newfound bat-borne hantavirus supports a laurasiatherian host association for ancestral mammalian hantaviruses. Infect Genet Evol. 41:113–119.http://dx.doi.org/10.1016/j.meegid.2016.03.036 [DOI] [PubMed] [Google Scholar]
- Yanagihara R, Amyx HL, Gajdusek DC.. 1985. Experimental infection with Puumala virus, the etiologic agent of nephropathia epidemica, in bank voles (Clethrionomys glareolus). J Virol. 55(1):34–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagihara R, Gu SH, Arai S, Kang HJ, Song JW.. 2014. Hantaviruses: rediscovery and new beginnings. Virus Res. 187:6–14.http://dx.doi.org/10.1016/j.virusres.2013.12.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo SQ, et al. 2014. A new hantavirus from the stripe-backed shrew (Sorex cylindricauda) in the People’s Republic of China. Virus Res. 184:82–86. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.