Abstract
The iconic Australasian kangaroos and wallabies represent a successful marsupial radiation. However, the evolutionary relationship within the two genera, Macropus and Wallabia, is controversial: mitochondrial and nuclear genes, and morphological data have produced conflicting scenarios regarding the phylogenetic relationships, which in turn impact the classification and taxonomy. We sequenced and analyzed the genomes of 11 kangaroos to investigate the evolutionary cause of the observed phylogenetic conflict. A multilocus coalescent analysis using ∼14,900 genome fragments, each 10 kb long, significantly resolved the species relationships between and among the sister-genera Macropus and Wallabia. The phylogenomic approach reconstructed the swamp wallaby (Wallabia) as nested inside Macropus, making this genus paraphyletic. However, the phylogenomic analyses indicate multiple conflicting phylogenetic signals in the swamp wallaby genome. This is interpreted as at least one introgression event between the ancestor of the genus Wallabia and a now extinct ghost lineage outside the genus Macropus. Additional phylogenetic signals must therefore be caused by incomplete lineage sorting and/or introgression, but available statistical methods cannot convincingly disentangle the two processes. In addition, the relationships inside the Macropus subgenus M. (Notamacropus) represent a hard polytomy. Thus, the relationships between tammar, red-necked, agile, and parma wallabies remain unresolvable even with whole-genome data. Even if most methods resolve bifurcating trees from genomic data, hard polytomies, incomplete lineage sorting, and introgression complicate the interpretation of the phylogeny and thus taxonomy.
Keywords: Macropus, kangaroo, polytomy, genomics, incomplete lineage sorting
Introduction
Molecular characters are considered to be more reliable than morphological characters for phylogenetic reconstruction, because of both the sheer number of molecular characters, and the tendency for morphology to be more influenced by adaptive processes (Zou and Zhang 2016). A prominent case on how morphological characters misled taxonomical classification is found among kangaroos (Meredith et al. 2008). The genus Wallabia has been classified as the sister-group of the genus Macropus (Flannery 1989). The single living species in the genus Wallabia, the swamp wallaby falls outside Macropus in morphological analyses (Prideaux and Warburton 2010). However, when molecular data were examined the phylogeny and taxonomy of these two kangaroo genera became controversial (Kirsch et al. 1995; Meredith et al. 2008; Phillips et al. 2013). Mitochondrial (mt) genome data agreed with the morphological classification where both genera are strictly monophyletic (Phillips et al. 2013). Molecular phylogenies based on DNA hybridization (Kirsch et al. 1995), five nuclear genes (Meredith et al. 2008), and retrotransposon insertions (Dodt et al. 2017) have tended to nest Wallabia inside the genus Macropus, making the latter paraphyletic and potentially invalidating the genus name Wallabia.
Conflicting molecular phylogenetic signals, such as in the case of Macropus and Wallabia, can be explained by various types of evolutionary processes. One such process is introgression where mitochondrial and nuclear DNA can be transferred between species, and result in incongruent phylogenetic signals (Hailer et al. 2012; Toews and Brelsford 2012). Another important source of phylogenetic incongruence is incomplete lineage sorting (ILS) which is caused by nucleotide polymorphisms persisting through two or more speciation events (Nei 1987; Pamilo and Nei 1988). Finally, if too few phylogenetic characters are included, stochastic processes in phylogenetic reconstruction can lead to incongruence that does not have its origin in evolutionary processes, but as a sampling artifact. Previous sequence-based molecular phylogenetic studies of Wallabia and Macropus were based on limited data, and as such, the identified taxonomic issue of the position of Wallabia might be an artifact (Meredith et al. 2008; Phillips et al. 2013). In addition, the previously analyzed five nuclear genes (Meredith et al. 2008) exhibited a level of high phylogenetic incongruence. It has been estimated that six times as many genes would be needed to reach phylogenetic resolution (Phillips et al. 2013), further highlighting the need to analyze a larger data set. If the obtained phylogenetic signals are correct and not artifacts (Meredith et al. 2008; Phillips et al. 2013), they indicate past ILS or introgression among Macropus and Wallabia. Previous studies on species such as gibbons (Carbone 2014), equids (Jónsson et al. 2014), cats (Li et al. 2016), and bears (Kumar et al. 2017) show that a genomic approach is often needed to capture and understand the evolutionary complexity within rapidly diverged groups. Therefore, we expand the sampling of kangaroos with the complete genomes from 11 species that include most species in the two genera Macropus and Wallabia (supplementary table S1, Supplementary Material online). The taxon sampling includes the elusive black wallaroo (M. bernardus) for which genetic analyses have previously been limited to a partial mitochondrial control region sequence (Eldridge et al. 2014). The relatively recent Pliocene/Miocene divergence times of kangaroos (Meredith et al. 2008; Phillips et al. 2013) are ideal to study evolutionary processes, because fewer multiple substitutions limit the phylogenetic noise in the data set. Thus, a phylogenomic analysis allows establishing a species tree of Macropus and Wallabia and to explore phylogenetic signals and conflict.
Materials and Methods
Genome Sequencing and Assembly
For each sample, total DNA was isolated, quality controlled, and the species identity validated by sequencing a short region of the mt genome (Fumagalli et al. 1997; Sambrook and Russell 2006). The genomes for the 11 kangaroo and wallaby species (supplementary table S1, Supplementary Material online) were generated by paired-end Illumina sequencing (see Supplementary Material online). Quality trimming was done with Trimmomatic (Bolger et al. 2014). The genomes were mapped to the tammar wallaby reference genome (Macropus eugenii) (Renfree et al. 2011) with BWA-MEM (Li and Durbin 2009). Single nucleotide variations were called by FreeBayes (Garrison and Marth 2012). Duplicate reads were masked with Picard 1.136 (http://broadinstitute.github.io/picard/; last accessed November 27, 2017), and mapping statistics computed using Qualimap v2.1.1 (García-Alcalde et al. 2012). The heterozygosity for each genome was estimated by dividing the number of heterozygous sites in the consensus sequences by the sum of homozygous and heterozygous sites in 20 kb (kilo basepairs) bins (supplementary fig. S1, Supplementary Material online). Mitochondrial reads were extracted by mapping the filtered reads to the Macropus robustus mitochondrial genome (accession number Y10524). Later the mt genome sequence was assembled for each species from the trimmed paired-end mt-derived Illumina reads using MITObim 1.6 (Hahn et al. 2013).
Phylogenetic Analysis of Autosomal Loci and Mitochondrial Genomes
For phylogenetic analysis, the genome alignments were separated into Genome Fragments (GFs), assumed to be putatively in linkage equilibrium. Transposable elements and other simple repeats were removed from the analysis using the repeat-masked tammar wallaby information file (macEug2) (https://genome.ucsc.edu; last accessed November 27, 2017). The ambiguous sites (“N” and heterozygous sites) and the repeat masked regions were removed from all the 11 kangaroo genomes. A total of 14,946 nonoverlapping, 10,000 bp long GFs were extracted from the scaffolds generating 149.5 Mb Mega basepairs of sequence data. To estimate the amount of nucleotide variation in the GFs, the fraction of different base pairs between species pairs was calculated. The absolute nucleotide distance between the evolutionary closest species pair, the black and common wallaroo, as well as between the evolutionary most distant pair, the spectacled hare-wallaby and yellow-footed rock-wallaby were calculated.
The best fitting substitution models to be used for phylogenetic reconstruction were evaluated by jModelTest (Guindon et al. 2003; Darriba et al. 2012). For the nuclear data, 100 random 10-kb GF alignments were used to estimate the best fitting substitution model. For the mt coding sequence alignment, each of the three codon positions were tested separately using an alignment of 21 species (see Supplementary Material online). RAxML version 8 (Stamatakis 2014) reconstructed Maximum Likelihood (ML) gene trees from each of the GF alignments of 10, 20, 30, and 40 kb length using the GTR + Γ+I (General Time Reversible with gamma distributed substitution rate and a fraction of invariable sites) (Tavaré 1986) substitution model. A coalescent species tree was built with ASTRAL (Mirarab et al. 2014) and support values were calculated as ASTRAL’s build-in quartet score and posterior probability (Sayyari and Mirarab 2016). An ASTRAL coalescent species tree with corresponding support values was calculated for each size fraction of GFs. The consense program (PHYLIP) (Felsenstein 2005) reconstructed a majority rule consensus tree of the ML gene trees for each size fraction.
Phylogenetic consensus networks were created using SplitsTree4 (Huson and Bryant 2006) from each size fraction of GF gene trees, using thresholds ranging from between 5% and 20%. A random set of 200 GFs each of 10 kb was concatenated to make a 2-Mb alignment. A ML tree was reconstructed with RAxML using the GTR + Γ+I model from the 2-Mb alignment. The procedure was repeated 100 times, resulting in 100 ML trees. The 100 ML trees were used to produce a majority rule consensus tree with the consense program (PHYLIP) (Felsenstein 2005). Coding sequences (CDS) were extracted from the scaffold alignments using the gene information file (gtf) from the tammar wallaby reference genome (macEug2). Exons belonging to the same gene were concatenated to create the coding sequences using custom Perl scripts. Only coding sequences fulfilling the following criteria were retained: 1) the length of the coding sequence must be divisible by 3, 2) no stop codons (TAA, TAG, or TGA) are found in any species, and 3) the number of codons that do not contain “N” or heterozygous sites in any species must be 100 or larger. The first two criteria are to reduce artifacts in annotation, and the third is to ensure enough information can be derived from each alignment. A ML tree was reconstructed using RAxML from the concatenated CDS alignment with a partitioned GTR + Γ+I model based on the three codon positions and with 200 bootstrap replicates. The assembled mtDNA sequences from the 11 sequenced kangaroo genomes, plus an additional 10 sequences from GenBank, of Macropus, Wallabia, and outgroups Lagorchestes and Petrogale, were combined into an alignment. When possible, an additional individual from the same species was included from GenBank. The mt genome of Macropus rufogriseus is a composite from two different individuals deposited in GenBank (KJ868122/KJ868121). Coding sequences from the 12 protein-coding genes were extracted from the mt alignment and concatenated. Only the 12 mt genes located on the L-strand, excluding ND6, were included. The last 48 bp from ATP8 and last 9 from ND4L were removed due to gene overlap, and all stop codons were excluded. The resulting alignment is 7,192 bp (excl. third codon positions). A partitioned ML analysis was conducted with RAxML with the GTR + Γ+I substitution model using only the first and second codon positions.
Approximately Unbiased Likelihood Analyses to Investigate Phylogenetic Signals and Topologies
To investigate if the GFs contain enough signals to discriminate between different phylogenetic topologies, the approximately unbiased (AU) likelihood test (Shimodaira 2002) values were calculated using the program CONSEL (Shimodaira and Hasegawa 2001). First a random subset of all GFs was analyzed for the support for different topologies, to determine whether the overall data could reject topologies that differ from the species tree. In a second analysis, the 146 10-kb GFs that produce a gene tree identical to the species tree topology were extracted, and trimmed into incrementally shorter sequences. The aim was to evaluate if there is enough information within a 10-kb GF to distinguish between different topologies, and as the topology supported by the GFs is known, whether there is sufficient statistical power to reject alternative topologies. In addition, the data set encompassing all 10-kb GFs was used to evaluate the phylogenetic position of the swamp wallaby and the black-gloved wallaby (see Supplementary Material online).
Average Numbers of Supporting Sites for a Branch in a Conflicting Topology
The number of parsimony informative sites was calculated for the branch supporting each of three alternative topologies for the relationship within M. (Notamacropus) (fig. 4; topologies N1–N3), or the four possible positions of the swamp wallaby (fig. 3; topologies W1–W4) from the 10-kb GFs.
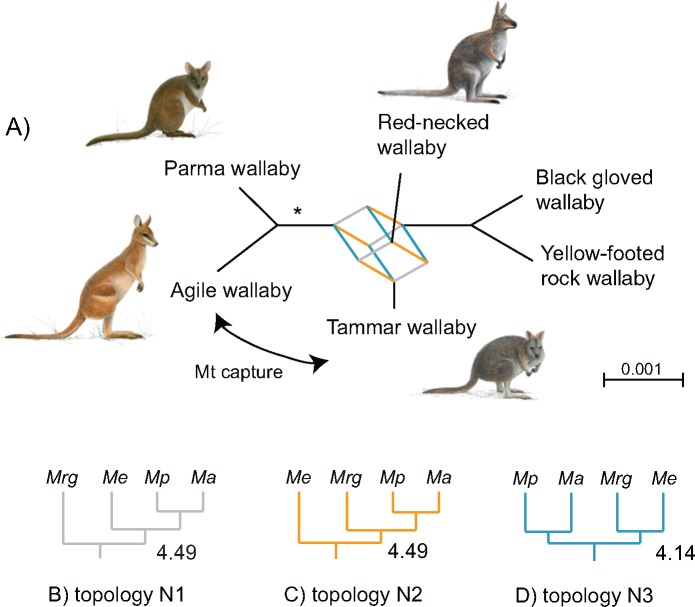
Fig. 4.
—Hard polytomy among wallabies. (A) SplitsTree network of the five species in M. (Notamacropus) showing conflict at 20% threshold. The mt capture between agile and tammar wallaby is indicated by an arrow. The * indicates that parma and agile wallabies are reconstructed as sister species by the majority of the GFs. (B) Topology N1, (C) topology N2, and (D) topology N3 (see Supplementary Material online). The number next to the supporting branch is the mean number of parsimony supporting sites (supplementary table S3, Supplementary Material online). The scale bar represents mean branch length across all gene trees. (Mrg: M. rufogriseus; Me: M. eugenii; Mp: M. parma; Ma: M. agilis)
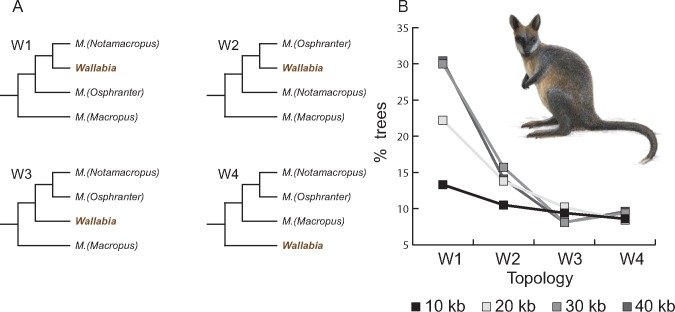
Fig. 3.
—Confounding genomic signals for Wallabia. (A) Four alternative phylogenetic positions of the swamp wallaby (Wallabia) (light brown) relative to the three subgenera inside Macropus (tree W1 to W4). (B) Among each size fraction of GF alignments, the number of ML trees supporting each of the topologies W1 to W4 was extracted. The percentage of ML trees in each size fraction supporting W1 to W4 is graphically displayed in the line graph using different gray shades. The species tree W1 has the most supporting ML trees, which increases with the use of longer GF alignments.
To count numbers of parsimony sites supporting different trees, we selected three species, H1, H2, H3, and the outgroup H4. Sites that support tree ((H1, H2), H3) will have the same nucleotide for H1 and H2, but a different nucleotide for H3 and H4; that is, a “BBAA” pattern (Green et al. 2010) for these four species. For each GF alignment, the number of parsimony informative sites (grouping taxa H1 and H2) was calculated for the supporting branch using a custom Perl script. If the GF alignment supports topology N1, sites that support H1 + H2 were counted, as well as sites supporting H1 + H3 and H2 + H3. Thus, the average number of supporting sites is calculated for each topology within the GF alignment. In order to examine the number of substitutions per GF, the total number of nucleotide differences was calculated between different species pairs.
Introgression Analyses Using D-Statistics
Introgression was estimated using the D-statistic method (see Supplementary Material online). Three species were taken from the ten species of Macropus and Wallabia and phylogenetically congruent species combinations were analyzed for gene flow using D-statistics (Green et al. 2010; Durand et al. 2011). A total of 72 D-statistics tests were made using combinations of three species from the genera Macropus and Wallabia (ten species) as H1, H2, and H3. H4 is chosen as one of the closest species outside the three species, and H1, H2, and H3 are assigned so that the topology (((H1, H2), H3), H4) always agrees with the ASTRAL species tree (topology 1, fig. 1). If the (((H1, H2), H3), H4) topology disagrees with the species tree, phylogenetically informative sites will falsely be interpreted as gene flow by the D-statistics. All 96,572 scaffolds (representing 90% of genome) were involved in the analysis, and included only sites where all 12 species have a nonambiguous nucleotide.
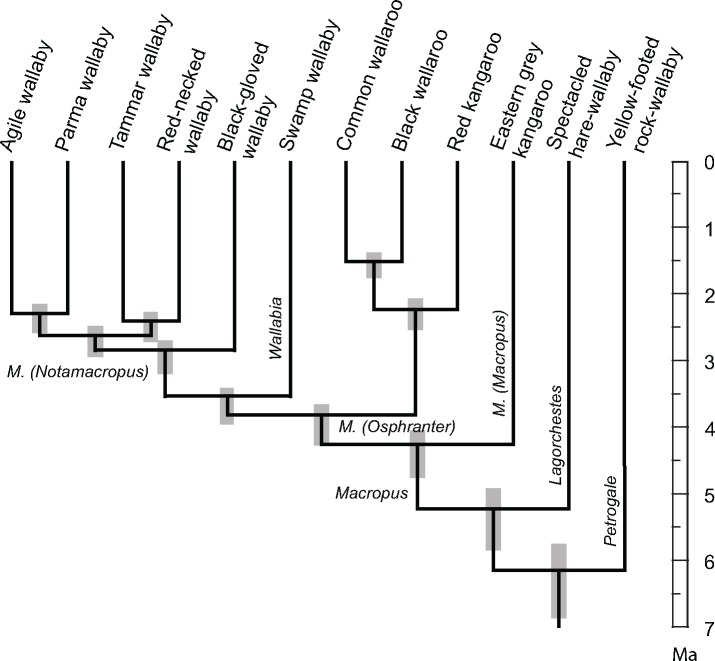
Fig. 1.
—Coalescent multilocus phylogeny. The ASTRAL multilocus coalescence species tree is based on 14,946 ML trees each reconstructed from a 10-kb long GF alignment. All branches are supported by 100% posterior probabilities. The coalescence tree reconstructs a paraphyletic genus Macropus, with the swamp wallaby (Wallabia bicolor) as the sister group to the subgenus M. (Notamacropus). The phylogeny has been scaled to divergence times. The scale bar is in million years ago (Ma).
Divergence Time Estimation
Divergence times for the genus Macropus were estimated using MCMCTREE of the PAML package (Yang 2007). Three fossil-based reference calibration priors (Meredith et al. 2008) were used:
The 3.62-Ma-old fossil, Macropus pavana, belonging to the M. (Osphranter) group (Flannery and Archer 1984; Mackness et al. 2000), used as the minimum time for split between the M. (Osphranter) and (M. (Notamacropus) + Wallabia) groups. The maximum age was set to 15.97 Ma, based on a stratigraphic bounding.
The two fossil species Macropus thor and Macropus agilis (uncertain) (Bartholomai 1975) from the Chinchilla local fauna at 3.4 Ma were used as a minimum for the split between the M. (Notamacropus) group and Wallabia, with a maximum stratigraphic bounding age of 11.61 Ma (Gradstein et al. 2004).
The 12-Myr-old sthenurine kangaroo fossil found in a middle Miocene formation in Camfield Beds in the Northern Territory (Woodburne 1985; Murray and Megirian 1992; Long et al. 2002) was used as the minimum time for the split between Macropodidae and Potoroidae. The maximum age was set to 28.4 Ma, a stratigraphic bounding (Meredith et al. 2008).
The data of three additional outgroups were included in the analyses: long-nosed potoroo (Potorous tridactylus), koala (Phascolarctos cinereus), and Tasmanian devil (Sarcophilus harrisii). CDSs were extracted from the available transcriptomes of potoroo (Udy et al. 2015) (PRJNA277745) and koala (Hobbs et al. 2014). The longest protein splice variant was taken for each CDS. For the Tasmanian devil, CDSs were extracted from the available genomic scaffolds (version sarHar1) (https://genome.ucsc.edu; last accessed November 27, 2017). After discarding the CDSs that 1) are shorter than 300 bp, 2) have a nucleotide sequence length not divisible by 3, or 3) contain an internal stop codon, 19,814 CDSs remained. We extracted a total of 7,434 protein sequences (see Supplementary Material online) from the tammar wallaby genome (macEug2), that are divisible by three, contain no in-frame stop codon and include >100 ambiguity-free codons in the 12-species alignment. Their orthology was determined with ProteinOrtho (Lechner et al. 2011) using protein sequences translated from the CDS from the three outgroups: potoroo, koala, and Tasmanian devil. The resulting orthology sets that contain one protein from each species are included. After that, all genes with stop codons in species other than M. eugenii are removed. After the orthology selection, 577 alignments remained for further analyses. Protein sequences from the four species were aligned with MAFFT (Katoh and Standley 2013) and evaluated with T-COFFEE (Notredame and Abergel 2003), from which columns with a score <7 and alignments with a total score <980 were excluded from further analysis. Corresponding sequences from the 11 new genomes were added to the alignment and the protein sequences were back-translated to nuclear CDS alignments. Finally, a data set of 182 CDSs with a length of 356 kb was concatenated and the three codon positions were partitioned. The relaxed molecular clock approach was used by implementing the “correlated rates” option. The Markov chain Monte Carlo was run for 500,000 generations and sampled every other generation, with an additional 100,000 generations that were discarded as burn-in. The species tree (fig. 1) was set as the user tree. Both fossil constraints within Macropus are used with hardened lower bound (prior tail probability set to 0.01% instead of 2.5%). All other settings were as default.
Results
Genome Sequencing and Heterozygosity of Australian Kangaroos
The genomes of 11 kangaroos from four different genera were sequenced to genome coverages between 6.7× (red kangaroo) and 17.2× (black wallaroo) (supplementary tables S1 and S2, Supplementary Material online). The heterozygosity among the investigated species ranges between 0.1% and 0.22% (supplementary fig. S1, Supplementary Material online), which is typical for natural populations (Li et al. 2010; Yim et al. 2014). After removing repetitive, ambiguous, heterozygous sites, and missing information from the 12-species multiple alignment (see Supplementary Material online), 14,946 GFs each with a length of 10 kb were extracted. In addition, 20 (2,418), 30 (540), and 40 kb (135) GFs were extracted from the tammar wallaby genome (see Supplementary Material online). However, using larger GFs violates the underlying assumption of coalescence approaches using unlinked loci. The observed substitutions for the 10-kb GFs between the closest species pair (black wallaroo and common wallaroo) were 48.3 ± 14.8 (observed p distance [p] = 0.005) and that between the most distant species pair 184.2 ± 26.4 (P = 0.018) (yellow-footed rock-wallaby and spectacled hare wallaby), and indicate sufficient information for phylogenetic analyses (supplementary fig. S2, Supplementary Material online). Statistical analyses demonstrate that 10 kb long GFs sequences already contain on an average, sufficient phylogenetic signal to reject alternative ML topologies (supplementary fig. S3A–C, Supplementary Material online).
Coalescent Analysis Reconstructs a Paraphyletic Macropus
Individual ML trees were calculated from each of the 14,946 GF alignments using the best fitting substitution model GTR + Γ+I (see Supplementary Material online) in RAxML (Stamatakis 2014). The ML trees were used to reconstruct a multilocus coalescent species tree (fig. 1) using ASTRAL (Mirarab et al. 2014; Edwards 2016). Posterior probability support values were calculated with ASTRAL for each 10, 20, 30, and 40 kb GF data set (Sayyari and Mirarab 2016). The posterior probability support was 100% for all nodes, except for the node joining tammar wallaby and red-necked wallaby (58% [20 kb], 18.8% [30 kb], or 27% [40 kb]). The coalescent species trees agree with a nested phylogenetic position of Wallabia inside the genus Macropus as the sistergroup to M. (Notamacropus) (Meredith et al. 2008), but differs in that the tammar wallaby is the sister group to the red-necked wallaby instead of to the agile wallaby. As Meredith et al. (2008) suggested, the subgenera M. (Osphranter) and M. (Notamacropus)+Wallabia are sistergroups. The black wallaroo is placed inside M. (Osphranter) as the sistergroup to the common wallaroo (M. robustus).
Mitochondrial Analysis Indicates Mitochondrial Capture across Species
A ML tree based on complete mitochondrial genomes (supplementary fig. S4, Supplementary Material online) agrees with the previously published tree (Phillips et al. 2013; Mitchell et al. 2014) except for the phylogenetic position of the black-gloved wallaby, which we find to be the deepest diverging member of M. (Notamacropus), as is different from previous analyses based on a limited mt data set (Phillips et al. 2013). Within M. (Notamacropus), the tammar wallaby (M. eugenii) is the sister species to the agile wallaby (M. agilis), different from the nuclear coalescent species tree (fig. 1). The mt phylogenetic placement of Wallabia as the sister-group to the genus Macropus is strongly supported.
Phylogenetic Analysis of Concatenated Genome Fragments and Coding Sequences
For comparison to the multilocus analyses, analyses of alignments from 200 random, concatenated 10 kb GF alignments (2 Mb) reconstructed a topology consistent with the coalescent species tree (fig. 1), except for the relationship inside M. (Notamacropus) (supplementary fig. S5A, Supplementary Material online). All nodes, except inside M. (Notamacropus) (44%) receive 100% support, suggesting that the majority of the individual ML trees do not differ topologically. The consensus tree based on concatenated GFs places the tammar wallaby as the sister group to agile, parma, and red-necked wallabies. Phylogenetic analysis of 5,017 concatenated CDS (7.49 Mb), is consistent with the coalescent species tree, but resolves the phylogeny inside M. (Notamacropus) differently (supplementary fig. S5B, Supplementary Material online), as the red-necked wallaby is the sister species to the agile, parma, and tammar wallabies. Thus, coalescent analysis of ML trees, and concatenation of GFs and CDS reconstruct the same internal topology of Macropus and Wallabia, but differ in the relationships within M. (Notamacropus).
Network Analyses Show Strong Phylogenetic Conflict among Kangaroos
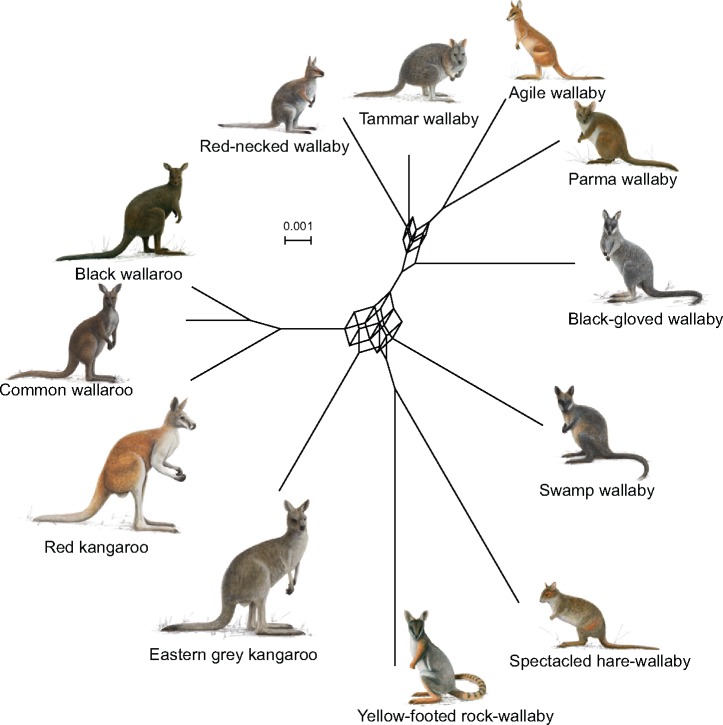
A SplitsTree consensus network of all 10 kb long GF alignments depicts the complex relationships of branches, which occur in at least 10% of the GF ML-trees (fig. 2). The cuboid structures in the network reveal that numerous GFs contain conflicting phylogenetic signals. The network analyses show the intricate evolutionary processes for the deeper relationships inside Macropus. Both the M. (Notamacropus) and M. (Osphranter) clades are monophyletic in the network at the 10% threshold. Increasing the threshold in the network to branches that are supported by at least 20% of the GF ML-trees, shows only conflict among the four species in core-M. (Notamacropus) (excluding black-gloved wallaby) (supplementary fig. S6, Supplementary Material online). In the network analysis, the relationships within M. (Osphranter) are stable and show no conflict until reducing the threshold to 5% (supplementary fig. S6, Supplementary Material online). SplitsTree networks reconstructed for GF alignments of 20- to 40-kb lengths reconstruct nearly identical networks to that of the 10-kb GFs (supplementary fig. S6, Supplementary Material online).
Fig. 2.
—Network analysis of kangaroo relationships. A SplitsTree network analysis based on 14,946 GF ML trees, at the 10% threshold level, depicts the complex phylogenetic signal in the kangaroo genomes. Increasing the length of the GFs does not notably affect the complexity of the network. Networks at additional threshold levels for the 10-kb GF networks are shown in supplementary figure S6, Supplementary Material online. The scale bar represents mean branch length across all gene trees. Kangaroo paintings have been provided by Jon Baldur Hlidberg.
Uncertain Position of the Swamp Wallaby Inside Macropus
A consensus analysis of the ML trees reconstructed from 10-kb GF alignments supports the extent of phylogenetic conflict in the phylogenetic network (supplementary fig. S7, Supplementary Material online). There is a trend for increased support using longer GFs for some nodes in the tree (supplementary fig. S7, Supplementary Material online), however support for nodes with high amount of conflict are unaffected by increased GF sequence lengths. The two nodes for M. (Osphranter) are well supported and occur in 90–99% of the GF ML-trees regardless of the GF length. The support for joining Macropus and Wallabia increases from 68% (10 kb) to 97% (40 kb), and similar gains in support are seen for other nodes (supplementary fig. S7, Supplementary Material online). The swamp wallaby is recovered as sister to M. (Notamacropus) with low support (36% for 10 kb) but this rises to 55% for 40 kb GFs. Alternative positions for swamp wallaby are shown in figure 3, and though the conflict remains, it reduces as the longer GFs are analyzed (supplementary fig. S7, Supplementary Material online). The amount of parsimony informative substitutions for the internal branch supporting the swamp wallaby placement in each alternative topology is 8.7, 8.4, 8.9, and 9.5, respectively (supplementary table S3, Supplementary Material online), indicating that there are adequate supporting sites per 10 kb.
Whole Genome Analyses Identify a Hard Polytomy Inside M. (Notamacropus)
Consense and network analyses (figs. 2 and 4;supplementary fig. S7, Supplementary Material online) of individual 10-kb GF ML trees find nearly equal numbers of supporting GFs for each of the possible topologies for the four-taxon complex in core-M. (Notamacropus). Support for agile and parma wallabies increases with the use of 40-kb GFs, from 55% to 79%. About 1,377 of the 10-kb GFs reconstruct tammar wallaby as the closest sister-species to agile and parma (topology N1, fig. 4), whereas 1,556 GFs favor red-necked wallaby as the sister group (topology N2, fig. 4). Instead, 1,442 GFs support a sistergroup relationship of red-necked wallaby and tammar wallaby (topology N3, fig. 4). The average numbers of parsimony informative substitutions for the supporting internal branch for each of the three topologies is 4.14, 4.49, and 4.49 (supplementary table S3, Supplementary Material online). Increasing the GF length does not alter the relative number of supporting GFs for each topology, indicating that massive phylogenetic conflict in the genomes of the core-M. (Notamacropus) species confounds phylogenetic analyses (supplementary fig. S7, Supplementary Material online).
The AU likelihood test (Shimodaira 2002) of the sequence data shows that two of the alternative possibilities for the swamp wallaby, as the sister group to M. (Osphranter) or to (M. (Notamacropus) + M. (Osphranter)) cannot be statistically rejected (supplementary table S4 and fig. S8, Supplementary Material online). By contrast, sister group relationships between the swamp wallaby and M. (Macropus) or Macropus as a whole can be rejected with the 10-kb GF sequences. Alternative positions of the black-gloved wallaby, placed either as the sistergroup to M. (Osphranter), inside M. (Osphranter) or as sister to M. (Osphranter) and M. (Notamacropus) were rejected. Although the posterior probability support for the coalescent topology was significant for the deeper branches (fig. 1), the relationship among the three subgenera M. (Notamacropus), (M. (Osphranter), (M. (Macropus) cannot be distinguished by genomic data (supplementary fig. S9, Supplementary Material online).
Complex Introgression Signals in Macropus Genomes
The extent of introgression among the analyzed kangaroo species was evaluated using D-statistics (Green et al. 2010) for 72 combinations involving 10 species congruent with the species tree (fig. 1 and see Supplementary Material online). The analyses of the subgenus M. (Notamacropus) detects signal for gene flow between the common ancestor of agile and parma wallabies and the black-gloved wallaby (supplementary table S5A, Supplementary Material online). However, there is some evidence for gene flow among all species of M. (Notamacropus). The D-statistics analyses identified two major introgression events of similar magnitude, from M. (Notamacropus) to both M. (Osphranter) and M. (Macropus). These introgression events can be more parsimoniously interpreted as a single introgression of ancestral alleles from a ghost lineage to the swamp wallaby genome (supplementary fig. S10D and table S5E, Supplementary Material online). An introgression signal between the common ancestors to the core-M. (Notamacropus) and the swamp wallaby was also identified (supplementary fig. S10B and table S5C, Supplementary Material online), whereas no introgression signals between the swamp wallaby and the black-gloved wallaby were found. Evidence for introgression between swamp wallaby and the common ancestor of M. (Osphranter) was also found (supplementary fig. S10C and table S5D, Supplementary Material online). Finally, gene flow between black wallaroo and red kangaroo was detected (supplementary fig. S10A and table S5B, Supplementary Material online).
Divergence Times Suggest a Climate-Triggered Radiation of Kangaroos
The divergence times of the genus Macropus and Wallabia were estimated based on a concatenated CDS alignment including three outgroups, the long-nosed potoroo (Potorous tridactylus), the koala (Phascolarctos cinereus), and the Tasmanian devil (Sarcophilus harrisii) using three fossil-based calibration priors (supplementary table S6, Supplementary Material online). The deepest divergence inside Macropus is estimated to 4.3 (4.04–4.75) Ma, which is somewhat younger than previous estimates (Meredith et al. 2008; Phillips et al. 2013). The divergence coincides with the Pliocene expansion of Australian grasslands (Martin 2006; Strömberg 2011). The divergences between the three Macropus sub genera took place within a narrow time span of 0.5 million years (4.3–3.8 Ma). The origins of the swamp wallaby and the black-gloved wallaby are estimated at 3.6 and 2.9 Ma, respectively. The deepest splits inside the core-M. (Notamacropus) clade occurred at 2.6 Ma, and the four species radiated within a time span of 200,000-300,000 years, coinciding with a rapid expansion of Australian grass lands (Martin 2006; Strömberg 2011).
Discussion
Phylogenomic analyses of most of the large kangaroos revealed a surprisingly complex evolutionary history, despite distinct morphological appearances and different ecologies. The extant species in Macropus and Wallabia originated ∼4.3 Ma, during a time when Australia’s climate became cooler and drier, leading to an expansion of grasslands across the continent (Strömberg 2011) and opening up new ecological niches. The phylogenomic multilocus coalescence analysis reconstructed a well-supported species tree of Macropus and Wallabia. The results of our phylogenomic analyses of the kangaroo relationships are mostly congruent with a five-nuclear gene study based on 6 kb (Meredith et al. 2008), except for that study grouping the agile and tammar wallabies. Dodt et al. (2017) found that this difference may stem from misidentification of Meredith et al.’s (2008) tammar wallaby sequences, which are near identical to M. agilis, but differ from the tammar wallaby reference genome. Our phylogeny differs more substantially from mitochondrial analyses (Phillips et al. 2013; Mitchell et al. 2014). The disagreement is possibly caused by two mitochondrial capture events, from an extinct species to the swamp wallaby, and from the agile wallaby to the tammar wallaby, although the latter placement is less well supported. Nevertheless, mitochondrial capture is not uncommon in mammals and emphasizes the need to involve nuclear markers in phylogenetic analysis (Hailer et al. 2012; Toews and Brelsford 2012). Haldane’s Rule (Haldane 1922), a phenomenon where female hybrids remain fertile but male hybrids are sterile has been observed in kangaroos, between the eastern and western gray kangaroos in M. (Macropus) (Watson and Demuth 2012) and could have facilitated mt captures (Li et al. 2016).
The evolutionary history of the swamp wallaby remains unresolved as a bifurcating tree even by analyses of genome data. Although the swamp wallaby is the sister group to the genus Macropus in mt analyses (supplementary fig. S4, Supplementary Material online) (Phillips et al. 2013; Mitchell et al. 2014), the phylogenetic position of the swamp wallaby inside the genus Macropus based on autosomal GF alignments is uncertain. The strongest phylogenetic signal from genome data places the swamp wallaby as the sister-group to M. (Notamacropus). However, there are phylogenetic signals that place the swamp wallaby as the sistergroup to M. (Osphranter), or to M. (Notamacropus) and M. (Osphranter). Some autosomal GFs even favor a phylogeny where the swamp wallaby is the sistergroup to Macropus, identical to the signal from mt genome analyses (Phillips et al. 2013). The evolutionary inconsistencies are easiest explained by the three lineages (M.(Osphranter), M.(Notamacropus), Wallabia) diverging nearly simultaneously potentially triggered by climate changes (Strömberg 2011). Divergence time estimation, analyses of parsimony informative sites and the results from the AU likelihood test support an emergence of the three lineages in rapid succession. After the initial rapid radiation, the mt genome introgressed from a now extinct species (“ghost lineage”) into the ancestral swamp wallaby causing the deviant mt phylogeny. Other, less parsimonious scenarios are possible but involve processes less frequently observed in nature. A second potential scenario is a mitochondrial capture from the ancestor of Macropus to a “proto-Wallabia” hybrid species that emerged from interbreeding of ancestral M. (Notamacropus) and M. (Osphranter) species. A third scenario is “genomic swamping,” whereby the nuclear genome of that Macropus ancestor was swamped via hybridization by male M. (Notamacropus), and perhaps to a lesser extent, M. (Osphranter). The nuclear genome of the ancestral Wallabia population could be eroded, but the maternally transferred mt genome maintained. Male-mediated genome swamping had been observed in some primate species (Zinner et al. 2009; Jiang et al. 2016). Interestingly, the swamp wallaby is one of only three extant kangaroos displaying an unequal chromosome number between males and females (2n = 10♀/11♂) (Westerman et al. 2010). Most species of Macropus have a stable karyotype with 2n = 16 (Westerman et al. 2010). Hybridization and introgression can lead to compromised chromosomal stability, as observed in sterile tammar and red-necked wallaby hybrids (Metcalfe et al. 2007) and sterile red-necked and swamp wallaby hybrids (O’Neill et al. 1998, 2002). In the light of the genome analyses, the swamp wallaby’s unusual chromosomal arrangement might be caused by one or several ancient introgression events. The swamp wallaby’s chromosomal arrangement might have led to a reproductive barrier to its parent species, causing genetic isolation.
Our genome analyses confirm the suggestion of Meredith et al. (2008) that Macropus is paraphyletic, and thus not a valid genus name as currently constituted. The required revision could follow Meredith et al. (2008) and subsume Wallabia into Macropus in its own subgenus, Macropus (Wallabia) bicolor or instead follow Jackson and Groves (2015) and maintain Wallabia, but elevate each of the three Macropus subgenera to genera. A third option is to raise both the Osphranter and Macropus subgenera to genera, and place the Notamacropus subgenus within Wallabia (which has taxonomic priority). Morphological, ecological, and palaeontological considerations may also help to guide this taxonomic decision.
Phylogenomic analyses leave the relationship among the three Macropus subgenera unresolved. Although the coalescent and consensus trees favor a sister-group relationship between M. (Osphranter) and M. (Notamacropus), alternative topologies cannot be excluded using the AU likelihood test, and are supported by a large fraction of GF ML trees. Divergence time estimates show that the three subgenera evolved within <500,000 years (4.3–3.8 Ma), thus leaving little time to fix phylogenetically informative substitutions and random signal from incomplete lineage sorting dominates the analysis. Relationships within M. (Notamacropus) are also problematic due to massive conflicting signals. This subgenus radiated quickly at 2.6 Ma (2.5–2.9), possibly triggered by the rapid expansion of grasslands in Australia (Martin 2006; Strömberg 2011). The phylogenomic analyses of five of the seven living species suggest that the deepest divergence among the four core-M. (Notamacropus) species is best presented as a hard polytomy. It remains to be seen how inclusion of the remaining species M. parryi and M. dorsalis will affect the result. In the case of a hard polytomy, alternative topologies occur at near-equal frequencies due to the lack of phylogenetic information. This is a consequence of rapid, more, or less simultaneous divergences that leave too little time for accumulation and fixation of genetic differences. A good example of a hard polytomy is the basal radiation among Neoaves (Suh 2016). Such divergences should not be presented as bifurcating trees, but are better represented as phylogenetic networks.
To what extent introgression has influenced the phylogenetic inconsistencies among Macropus is difficult to estimate with the current data set, and is problematic for gene flow analyses due to the unresolved phylogeny. To differentiate gene flow from ILS with current statistical methods, it is fundamental to have a resolved phylogeny, and is currently impossible for rapidly evolving groups such as core-M. (Notamacropus) even on the basis of genome data. Ongoing introgression between Macropus subgenera is unlikely, although numerous kangaroo hybrids are known, these are infertile (Close and Lowry 1989; Metcalfe et al. 2007). Not even the closely related members of M. (Notamacropus) produce fertile offspring (Close and Lowry 1989). The two species in the subgenus M. (Macropus) are the only species where fertile hybrids have been identified, but population genetic screening found only minimal signs of introgression in the wild (Zenger et al. 2003).
The relationships inside Macropus are highly complex with large numbers of conflicting loci. The swamp wallaby is a so-called phylogenetically “rogue” taxon, because different loci in its genome support placing it at different phylogenetic positions. The term “rogue taxon” is generally used for species that cannot be solidly placed in an evolutionary context, because of missing data, elevated substitutions rates or homoplasy (Sanderson and Shaffer 2002). The analysis of kangaroo evolution show that ILS and introgression can play an equally strong part in generating rogue taxa, and omitting such species to improve support values, removes important evolutionary information. Interpretation of morphological character change over time, or molecular changes in the Macropus + Wallabia complex is fraught with problems due to their network-like evolution. Thus, whole genome studies are necessary to gain an understanding of rapidly diverging groups and the results may not always lead to a bifurcating tree (Hallström and Janke 2010).
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
We thank Dion Wedd at the Territory Wildlife Park and Hannah Maloney (QUT) for the collection of the black wallaroo sample. In addition, we extend our gratitude to the veterinarians at the Antwerp and Hannover zoos for DNA samples. Dr Rachel O’Neill kindly provided DNA from Petrogale xanthopus, as did Mike Bunce from Macropus irma. Jon Baldur Hlidberg (www.fauna.is) provided the kangaroo paintings. We thank the two anonymous reviewers for their helpful criticism. This study was supported by Hesse's funding program LOEWE (Landes-Offensive zur Entwicklung Wissenschaftlich-ökonomischer Exzellenz) and the Leibniz Association.
Literature Cited
- Bartholomai A. 1975. The genus Macropus Shaw (Marsupialia; Macropodidae) in the upper Cainozoic deposits of Queensland. Mem Qld Museum 17:195–235. [Google Scholar]
- Bolger AM, Lohse M, Usadel B.. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30(15):2114–2120.http://dx.doi.org/10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbone L. 2014. Gibbon genome and the fast karyotype evolution of small apes. Nature 513(7517):195–201.http://dx.doi.org/10.1038/nature13679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Close RL, Lowry PS.. 1989. Hybrids in marsupial research. Aust J Zool. 37(3):259–267.http://dx.doi.org/10.1071/ZO9890259 [Google Scholar]
- Darriba D, Taboada GL, Doallo R, Posada D.. 2012. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods 9(8):772..http://dx.doi.org/10.1038/nmeth.2109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodt WG, Gallus S, Phillips MJ, Nilsson MA. 2017. Resolving kangaroo phylogeny and overcoming retrotransposon ascertainment bias. Sci Rep. 7 (1):16811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand EY, Patterson N, Reich D, Slatkin M.. 2011. Testing for ancient admixture between closely related populations. Mol Biol Evol. 28(8):2239–2252.http://dx.doi.org/10.1093/molbev/msr048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards SV. 2016. Implementing and testing the multispecies coalescent model: a valuable paradigm for phylogenomics. Mol Phylogenet Evol. 94(Pt A):447–462. [DOI] [PubMed] [Google Scholar]
- Eldridge MD, Potter S, Johnson CN, Ritchie EG.. 2014. Differing impact of a major biogeographic barrier on genetic structure in two large kangaroos from the monsoon tropics of Northern Australia. Ecol Evol. 4(5):554–567.http://dx.doi.org/10.1002/ece3.954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. 2005. PHYLIP (Phylogeny Inference Package) version 3.6. Seattle (WA): University of Washington.
- Flannery T. 1989. Phylogeny of the Macropodoidea; a study in convergence In: Kangaroos, wallabies and rat-kangaroos. Chipping Norton (Australia: ): Surrey Beatty & Sons; p. 1–46. [Google Scholar]
- Flannery TF, Archer M.. 1984. The macropodoids (Marsupialia) of the early Pliocene Bow local fauna, central-eastern New South Wales. Aust Zool. 21:357–384. [Google Scholar]
- Fumagalli L, Pope LC, Taberlet P, Moritz C.. 1997. Versatile primers for the amplification of the mitochondrial DNA control region in marsupials. Mol Ecol. 6(12):1199–1201.http://dx.doi.org/10.1046/j.1365-294X.1997.00298.x [DOI] [PubMed] [Google Scholar]
- García-Alcalde F, et al. 2012. Qualimap: evaluating next-generation sequencing alignment data. Bioinformatics 28(20):2678–2679. [DOI] [PubMed] [Google Scholar]
- Garrison E, Marth G.. 2012. Haplotype-based variant detection from short-read sequencing. arXiv preprint arXiv: 1207.3907.
- Gradstein FM, Ogg JG, Smith AG, Bleeker W, Lourens LJ.. 2004. A new geologic time scale, with special reference to Precambrian and Neogene. Episodes 27:83–100. [Google Scholar]
- Green RE, et al. 2010. A draft sequence of the Neandertal genome. Science 328(5979):710–722.http://dx.doi.org/10.1126/science.1188021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, Gascuel O, Rannala B.. 2003. A simple, fast and accurate method to estimate large phylogenies by maximum-likelihood. Syst Biol. 52(5):696–704.http://dx.doi.org/10.1080/10635150390235520 [DOI] [PubMed] [Google Scholar]
- Hahn C, Bachmann L, Chevreux B.. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads – a baiting and iterative mapping approach. Nucleic Acids Res. 41(13):e129.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hailer F, et al. 2012. Nuclear genomic sequences reveal that polar bears are an old and distinct bear lineage. Science 336(6079):344–347.http://dx.doi.org/10.1126/science.1216424 [DOI] [PubMed] [Google Scholar]
- Haldane JBS. 1922. Sex ratio and unisexual sterility in hybrid animals. J Genet. 12(2):101–109.http://dx.doi.org/10.1007/BF02983075 [Google Scholar]
- Hallström BM, Janke A.. 2010. Mammalian evolution may not be strictly bifurcating. Mol Biol Evol. 27(12):2804–2816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobbs M, et al. 2014. A transcriptome resource for the koala (Phascolarctos cinereus): insights into koala retrovirus transcription and sequence diversity. BMC Genomics 15(1):1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huson DH, Bryant D.. 2006. Application of phylogenetic networks in evolutionary studies. Mol Biol Evol. 23(2):254–267.http://dx.doi.org/10.1093/molbev/msj030 [DOI] [PubMed] [Google Scholar]
- Jackson S, Groves C.. 2015. Taxonomy of Australian mammals. Clayton South (Australia: ): CSIRO Publishing. [Google Scholar]
- Jiang J, et al. 2016. Mitochondrial genome and nuclear markers provide new insight into the evolutionary history of macaques. PLoS One 11(5):e0154665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jónsson H, et al. 2014. Speciation with gene flow in equids despite extensive chromosomal plasticity. Proc Natl Acad Sci U S A. 111(52):18655–18660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM.. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.http://dx.doi.org/10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirsch JAW, Lapointe FJ, Foeste A.. 1995. Resolution of portions of the kangaroo phylogeny (Marsupialia: Macropodidae) using DNA hybridization. Biol J Linn Soc. 55(4):309–328.http://dx.doi.org/10.1111/j.1095-8312.1995.tb01068.x [Google Scholar]
- Kumar V, et al. 2017. The evolutionary history of bears is shaped by gene flow across species. Sci Rep. 7:46487.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechner M, et al. 2011. Proteinortho: detection of (co-) orthologs in large-scale analysis. BMC Bioinformatics 12:124.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Davis BW, Eizirik E, Murphy WJ.. 2016. Phylogenomic evidence for ancient hybridization in the genomes of living cats (Felidae). Genome Res. 26(1):1–11.http://dx.doi.org/10.1101/gr.186668.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R.. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25(14):1754–1760.http://dx.doi.org/10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, et al. 2010. The sequence and de novo assembly of the giant panda genome. Nature 463(7279):311–317.http://dx.doi.org/10.1038/nature08696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long J, Archer M, Flannery TF, Hand SJ.. 2002. Prehistoric mammals of Australia and New Guinea: one hundred million years of evolution. Sydney (Australia: ): University of New South Wales Press. [Google Scholar]
- Mackness BS, Whitehead PW, McNamara GC.. 2000. New potassium-argon basalt date in relation to the Pliocene Bluff Downs Local Fauna, northern Australia. Aust. J Earth Sci. 47(4):807–811. [Google Scholar]
- Martin HA. 2006. Cenozoic climatic change and the development of the arid vegetation in Australia. J Arid Environ. 66(3):533–563.http://dx.doi.org/10.1016/j.jaridenv.2006.01.009 [Google Scholar]
- Meredith RW, Westerman M, Springer MS.. 2008. A phylogeny and timescale for the living genera of kangaroos and kin (Macropodiformes: Marsupialia) based on nuclear DNA sequences. Aust J Zool. 56(6):395–410.http://dx.doi.org/10.1071/ZO08044 [Google Scholar]
- Metcalfe CJ, et al. 2007. Genomic instability within centromeres of interspecific marsupial hybrids. Genetics 177(4):2507–2517.http://dx.doi.org/10.1534/genetics.107.082313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirarab S, et al. 2014. ASTRAL: genome-scale coalescent-based species tree estimation. Bioinformatics 30(17):i541–i548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell KJ, et al. 2014. Molecular phylogeny, biogeography, and habitat preference evolution of marsupials. Mol Biol Evol. 31(9):2322–2330.http://dx.doi.org/10.1093/molbev/msu176 [DOI] [PubMed] [Google Scholar]
- Murray PF, Megirian D.. 1992. Continuity and contrast in middle and late Miocene vertebrate communities from the Northern Territory. Vol. 9.Beagle: Records of the Museums and Art Galleries of the Northern Territory. p. 195–218. [Google Scholar]
- Nei M. 1987. Molecular evolutionary genetics. New York: Columbia University Press. [Google Scholar]
- Notredame C, Abergel C.. 2003. Using multiple alignment methods to assess the quality of genomic data analysis In: Andrade MA, editor.Bioinformatics and genomes: current Perspectives. Wymondham, England: Horizon Scientific Press; p. 30–50. [Google Scholar]
- O’Neill RJ, O’Neill MJ, Graves JA.. 1998. Undermethylation associated with retroelement activation and chromosome remodelling in an interspecific mammalian hybrid. Nature 393:68–72. [DOI] [PubMed] [Google Scholar]
- O’Neill RJ, O’Neill MJ, Graves JA.. 2002. Corrigendum: undermethylation associated with retroelement activation and chromosome remodelling in an interspecific mammalian hybrid. Nature 420:106. [DOI] [PubMed] [Google Scholar]
- Pamilo P, Nei M.. 1988. Relationships between gene trees and species trees. Mol Biol Evol. 5(5):568–583. [DOI] [PubMed] [Google Scholar]
- Phillips MJ, Haouchar D, Pratt RC, Gibb GC, Bunce M.. 2013. Inferring kangaroo phylogeny from incongruent nuclear and mitochondrial genes. PLoS One 8(2):e57745.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prideaux G, Warburton N.. 2010. An osteology-based appraisal of the phylogeny and evolution of kangaroos and wallabies (Macropodidae: Marsupialia). Zool J Linn Soc. 159(4):954–987. [Google Scholar]
- Renfree MB, et al. 2011. Genome sequence of an Australian kangaroo, Macropus eugenii, provides insight into the evolution of mammalian reproduction and development. Genome Biol. 12(8):R81.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Russell DW.. 2006. Isolation of high-molecular-weight DNA from mammalian cells using Proteinase K and phenol. CSH Protoc. 2006(1):pdb-prot4036.. [DOI] [PubMed] [Google Scholar]
- Sanderson M, Shaffer H.. 2002. Troubleshooting molecular phylogenetic analyses. Annu Rev Ecol Syst. 33(1):49–72.http://dx.doi.org/10.1146/annurev.ecolsys.33.010802.150509 [Google Scholar]
- Sayyari E, Mirarab S.. 2016. Fast coalescent-based computation of local branch support from quartet frequencies. Mol Biol Evol. 33(7):1654–1668.http://dx.doi.org/10.1093/molbev/msw079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimodaira H. 2002. An approximately unbiased test of phylogenetic tree selection. Syst Biol. 51(3):492–508.http://dx.doi.org/10.1080/10635150290069913 [DOI] [PubMed] [Google Scholar]
- Shimodaira H, Hasegawa M.. 2001. CONSEL: for assessing the confidence of phylogenetic tree selection. Bioinformatics 17(12):1246–1247.http://dx.doi.org/10.1093/bioinformatics/17.12.1246 [DOI] [PubMed] [Google Scholar]
- Stamatakis A. 2014. RAxML Version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30(9):1312–1313.http://dx.doi.org/10.1093/bioinformatics/btu033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strömberg CAE. 2011. Evolution of grasses and grassland ecosystems. Annu Rev Earth Planet Sci. 39(1):517–544. [Google Scholar]
- Suh A. 2016. The phylogenomic forest of bird trees contains a hard polytomy at the root of Neoaves. Zool Scripta 45:50–62.http://dx.doi.org/10.1111/zsc.12213 [Google Scholar]
- Tavaré S. 1986. Some probabilistic and statistical problems in the analysis of DNA sequences. Lect Math Life Sci. 17:57–86. [Google Scholar]
- Toews DP, Brelsford A.. 2012. The biogeography of mitochondrial and nuclear discordance in animals. Mol Ecol. 21(16):3907–3930.http://dx.doi.org/10.1111/j.1365-294X.2012.05664.x [DOI] [PubMed] [Google Scholar]
- Udy DB, Voorhies M, Chan PP, Lowe TM, Dumont S.. 2015. Draft de novo transcriptome of the rat kangaroo Potorous tridactylus as a tool for cell biology. PLoS One 10(8):e0134738.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson ET, Demuth JP.. 2012. Haldane’s Rule in marsupials: what happens when both sexes are functionally hemizygous? J Hered. 103(3):453–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westerman M, Meredith RW, Springer MS.. 2010. Cytogenetics meets phylogenetics: a review of karyotype evolution in diprotodontian marsupials. J Hered. 101(6):690–702. [DOI] [PubMed] [Google Scholar]
- Woodburne MO, Turnbull WD, Plane M, Lundelius L.. 1985. Biochronology of the continental mammal record of Australia and New Guinea. Vol. 5. Special Publication. South Australia Department of Mines and Energy, Adelaide. p. 347–364. [Google Scholar]
- Yang Z. 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 24(8):1586–1591.http://dx.doi.org/10.1093/molbev/msm088 [DOI] [PubMed] [Google Scholar]
- Yim H-S, et al. 2014. Minke whale genome and aquatic adaptation in cetaceans. Nat Genet. 46(1):88–92.http://dx.doi.org/10.1038/ng.2835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenger KR, Eldridge MD, Cooper DW.. 2003. Intraspecific variation, sex-biased dispersal and phylogeography of the eastern grey kangaroo (Macropus giganteus). Heredity 91(2):153–162.http://dx.doi.org/10.1038/sj.hdy.6800293 [DOI] [PubMed] [Google Scholar]
- Zinner D, Groeneveld LF, Keller C, Roos C.. 2009. Mitochondrial phylogeography of baboons (Papio spp.): indication for introgressive hybridization? BMC Evol Biol. 9:83.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou Z, Zhang J.. 2016. Morphological and molecular convergences in mammalian phylogenetics. Nat Commun. 7:12758.. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.