Abstract
Cellular accumulation of repetitive RNA occurs in several dominantly-inherited genetic disorders. Expanded CUG, CCUG or GGGGCC repeats are expressed in myotonic dystrophy type 1 (DM1), myotonic dystrophy type 2 (DM2), or familial amyotrophic lateral sclerosis, respectively. Expanded repeat RNAs (ER-RNAs) exert a toxic gain-of-function and are prime therapeutic targets in these diseases. However, efforts to quantify ER-RNA levels or monitor knockdown are confounded by stable structure and heterogeneity of the ER-RNA tract and background signal from non-expanded repeats. Here, we used a thermostable group II intron reverse transcriptase (TGIRT-III) to convert ER-RNA to cDNA, followed by quantification on slot blots. We found that TGIRT-III was capable of reverse transcription (RTn) on enzymatically synthesized ER-RNAs. By using conditions that limit cDNA synthesis from off-target sequences, we observed hybridization signals on cDNA slot blots from DM1 and DM2 muscle samples but not from healthy controls. In transgenic mouse models of DM1 the cDNA slot blots accurately reflected the differences of ER-RNA expression across different transgenic lines, and showed therapeutic reductions in skeletal and cardiac muscle, accompanied by improvements of the DM1-associated splicing defects. TGIRT-III was also active on CCCCGG- and GGGGCC-repeats, suggesting that ER-RNA analysis is feasible for several repeat expansion disorders.
INTRODUCTION
More than 30 human genetic disorders are caused by genomic expansions of tandem repeats (1). At some loci, the expanded repeats cause epigenetic promoter silencing, resulting in loss of function for the expanded allele (2). At other loci the expanded repeats are expressed, causing gain-of-function by repetitive RNA, protein, or both (3–9). As a class, the disorders having RNA gain-of-function tend to have large repeat expansions in introns or in 5′- or 3′-untranslated regions (1). The mechanisms for RNA dominance are believed to involve sequestration of splicing factors, activation of RNA-sensitive signaling pathways, or repeat-associated non-AUG (RAN) translation (10–16).
Two of the leading approaches for treating repeat expansion diseases are to inhibit the transcription or accelerate the degradation of expanded repeat RNAs (ER-RNAs) [reviewed in (17)]. However, reliable methods for monitoring ER-RNA expression or knockdown are lacking, and there is uncertainty about how the disease onset or progression may depend on ER-RNA accumulation, or how much ER-RNA reduction is necessary to improve the clinical signs and symptoms. Since ER-RNA tracts do not have unique priming sites, conventional RT-PCR assays have been targeted to the 5′ or 3′ flanking sequences (18–20). However, the results may not reflect ER-RNA levels in patients, owing to cross detection of transcripts from the wild-type (WT) allele, variability of repeat length in somatic cells (21,22), and persistence of ER-RNA fragments that are disconnected from the flanking sequences. For example, the ER-RNA in DM2 cells is devoid of flanking sequences, presumably due to partial decay of the excised intron in which the repeat tract occurs (23). Detection by northern blot also has technical limitations, as it requires gel separation and transfer of large ER-RNA fragments having variable length (up to 44 000 nts in DM2) (4).
Tandem repeats are ubiquitous in the genome and transcriptome. For example, the human reference genome contains 1,055 CTG•CAG repeats of 18 or more base pairs, of which 301 lie in exons (24). Nevertheless, despite the preponderance of loci expressing non-expanded repeats, we found that total levels of CUG-repeat RNA were higher in DM1 muscle than in healthy controls, as determined by RNA slot blots hybridized with CAG-repeat probes (24-mers) (25). However, efforts to quantify levels of expanded CUG repeats (CUGexp) were confounded by the variable background in healthy controls.
Here, we examine levels of repetitive RNA by converting ER-RNA to cDNA. We show that TGIRT-III, a thermostable mobile group II intron reverse transcriptase (26), is able to reverse transcribe CUG-, CCUG-, GGGGCC and CCCCGG-repeats. By adjusting RTn conditions to suppress off-target cDNA synthesis we obtained disease-specific signals on cDNA slot blots from DM1 and DM2 patients and from (GGGGCC)160-expressing cells, enabling indirect quantification of ER-RNA.
MATERIALS AND METHODS
Tandem repeat plasmids and in vitro transcription
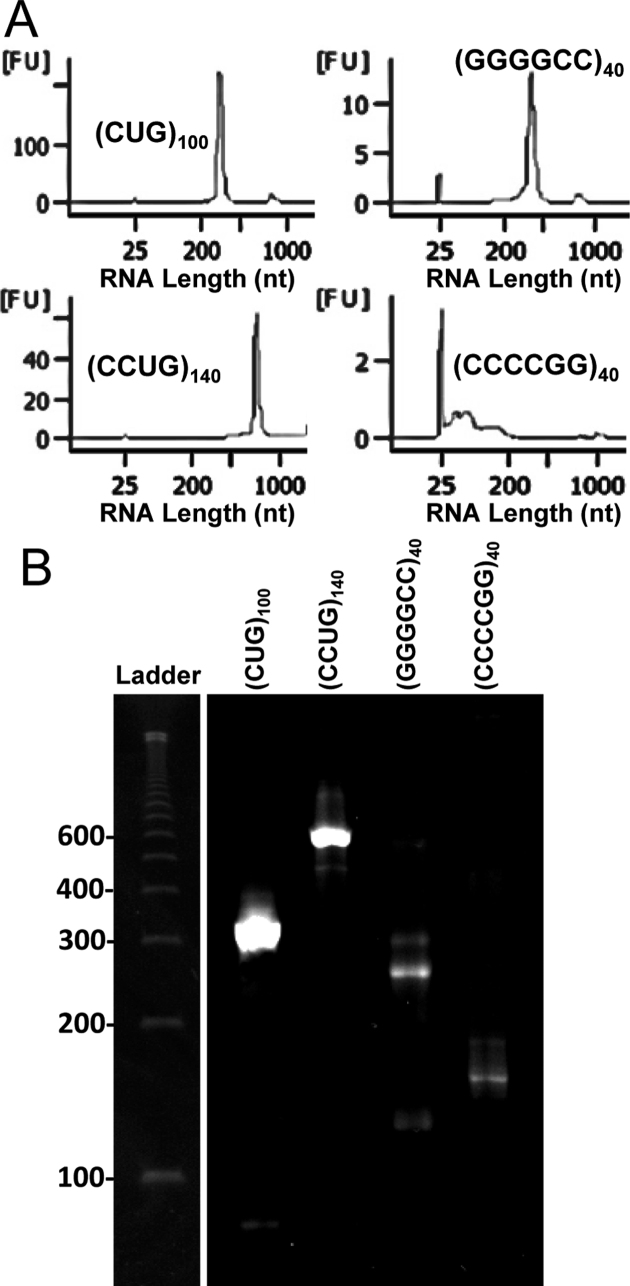
(CTG)100, (CCTG)140 and (GGGGCC)40 tracts [(repeat sequence)# of repeats] were generated and cloned in plasmid pDWD, as previously described (27). A promoter for T7 RNA polymerase was inserted 37 bp upstream of the repeat tract, and 20 bp downstream in the case of (GGGGCC)40 to produce r(GGCCCC)40. Length and purity of each repeat tract was confirmed by Sanger sequencing. In vitro transcription of XbaI-linearized plasmid was performed using HiScribe T7 Quick High Yield RNA Synthesis Kit (New England Biolabs). Template was removed using DNase I followed by ethanol precipitation of RNA. The in vitro transcribed ER-RNAs were assessed by capillary and polyacrylamide TBE-urea gel electrophoresis (see below).
RNA isolation and reverse transcription (RTn)
RNA was isolated from tissue or cells using Tri-Reagent (Sigma-Aldrich) and treated with DNase I (Qiagen), as previously described (28,29). RTn was performed using 0.5 μg of in vitro transcript or 2 ug of total cellular RNA and primed with CAG-, CAGG-, GGCCCC- or GGGGCC-repeat primers listed in Table 1 (1.0 μM primer for in vitro ER-RNA transcripts, 0.5 μM for total cellular RNA). Where indicated the repeat primers were RNA/DNA chimeric oligonucleotides (‘hybrid primers’) comprised of RNA except for 4 nt of DNA at the 3′ end. Hybrid primers were used to reduce both primer carry through and detection of off-target cDNA extension in slot blots, as described below. Alternatively, RTn of in vitro transcripts was primed using a non-repeat primer that anneals 3′ of the repeat tract (0.1 μM). Where indicated the primers were 5′ end-labeled with HEX or IRD-700 fluorophores. For Superscript III (SS-III) (ThermoFisher) the RTn was performed in 20 μl reactions containing 20 mM Tris–HCl pH 8.4, 50 mM KCl, 10 mM MgCl2 and 0.5 mM dNTPs (dCTP, dATP and dGTP). dTTP was omitted and 0.05 mM dideoxy-UTP (ddUTP) (TriLink Biotechnologies) or FAM-ddUTP (Enzo Life Sciences) was included to terminate extension beyond the repeat tract. dATP was also omitted for RTn of GGGGCC- and GGCCCC-repeats. Components were brought to 80°C (90°C for GGGGCC templates) for 2 min then ramp cooled over 45 min to 25°C (GeneAmp PCR system 9700, Applied Biosystems) before addition of 10 mM DTT and 200 U SS-III enzyme. Extension was performed at 55°C for 20 min. The RNA template and RNA component of hybrid primer were then removed by digestion with RNase H and RNase A (Qiagen) for 15 min at 37°C.
Table 1. Primer and probe sequences.
| Name | Type | Sequence (5′→3′) |
|---|---|---|
| pDWD-specific *, ‡ | DNA RT primer | CGGGTGGGTTACCGAAGAC |
| pDWD-specific hybrid | rna/DNA RT primer | cgggtgggttaccgaAGAC |
| (CAG)6+1 | DNA RT primer | CAGCAGCAGCAGCAGCAGC |
| (CAG)6+1 hybrid | rna/DNA RT primer | cagcagcagcagcagCAGC |
| (CAGG)4+2 * | DNA RT primer | CAGGCAGGCAGGCAGGCA |
| (CAGG)4+2 hybrid | rna/DNA RT primer | caggcaggcaggcaGGCA |
| (GGCCCC)3+3 * | DNA RT primer | GGCCCCGGCCCCGGCCCCGGC |
| (GGCCCC)3+3 hybrid | rna/DNA RT primer | ggccccggccccggcccCGGC |
| (GGGGCC)3+3 * | DNA RT primer | CGGGGCCGGGGCCGGGGCCGG |
| (GGGGCC)3+3 hybrid | rna/DNA RT primer | cggggccggggccggggCCGG |
| (CTG)7 LNA Δ | Digoxigenin LNA Probe | CTGCTGCTGCTGCTGCTGCTG |
| (CCTG)6 LNA Δ | Digoxigenin LNA Probe | CCTGCCTGCCTGCCTGCCTGC |
| (GGGGCC) LNA Δ | Digoxigenin LNA Probe | GGGGCCGGGGCCGGGGCC |
| hACTA1-Ex1F | qPCR forward primer | CCAGGCCGGAGCCATT |
| hACTA1-Ex2R | qPCR reverse primer | GTAGCTACCCCGCCCAGAAACT |
| hACTA1-probe | qPCR probe | ACCACCGCCCTCGTGTGCG |
| hDMPK 3′UTR-F | qPCR forward primer | CTATCGTTGGTTCGCAAAGTG |
| hDMPK 3′UTR-R | qPCR reverse primer | GCAAATTTCCCGAGTAAGCAG |
| hDMPK 3′UTR-probe | qPCR probe | AAGCTTTCTTGTGCATGACGCCC |
| mSerca1 Ex22-F † | RT-PCR splicing primer | CTCATGGTCCTCAAGATCTCAC |
| mSerca1 Ex22-R | RT-PCR splicing primer | GGGTCAGTGCCTCAGCTTTG |
| mClcn1 Ex7A-F † | RT-PCR splicing primer | TGAAGGAATACCTCACACTCAAGG |
| mClcn1 Ex7A-R | RT-PCR splicing primer | CACGGAACACAAAGGCACTG |
| mTmem63b Ex5-F † | RT-PCR splicing primer | CTGGCTCTGGACTTCATGTGCTTTC |
| mTmem63b Ex5-R | RT-PCR splicing primer | GAGACGGAGGTGAGACGCTCATACC |
| mCacna1s Ex29-F † | RT-PCR splicing primer | GAGATCCTTGGAATGTGTTTGACTTCCT |
| mCacna1s Ex29-R | RT-PCR splicing primer | GGTTCAGCAGCTTGACCAGTCTCAT |
Key: N = DNA, n = RNA, N = LNA. RT primers 5′-end-labeled with HEX (*), FAM (†), IRD-700 (‡), or Digoxigenin (Δ).
Purified recombinant TGIRT-III was prepared as previously described (26) or obtained from Ingex, St. Louis, MO, USA. The template RNA was mixed with indicated primer in 450 mM NaCl, 5 mM MgCl2, 20 mM Tris–HCl pH 7.5, brought to 80°C (or 90°C for GGGGCC templates) for 2 min, ramp cooled over 45 min to 25°C, and then placed on ice. After addition of 5 mM DTT and 560 U TGIRT-III, reactions were held on ice for 30 min. dNTPs and dideoxy-UTP were then added as described above. RTn mix was brought to 50°C, then increased by 2°C increments every 2 min to 60°C, and then held at 60°C for a total extension time of 20 min. RNA was then removed using RNase H and RNase A as described above.
Analysis of cDNA products by capillary or gel electrophoresis
Residual primers were removed on silica columns (Zymo Research). cDNAs generated with HEX-labeled primers or FAM-ddUTP were analyzed by capillary electrophoresis (Genewiz). cDNAs generated with IRD-700-labeled primers were separated on 6% polyacrylamide urea gels and analyzed by infrared laser scanning (Odyssey, LI-COR).
cDNA slot blots
After RNase A/H treatment, cDNA samples were mixed with 400 μL of denaturing buffer (0.5 M NaOH and 10 mM EDTA) then applied to Hybond-N+ membrane (GE Healthcare) using a Bio-Dot SF slot blot manifold (Bio-Rad). Prior to sample application the membrane was soaked in denaturing buffer. Following application of sample, the slots were rinsed with 400 μl denaturing buffer. The membrane was removed from the manifold, rinsed with 2× saline–sodium citrate (SSC) for 2 min, baked at 120°C for 20 min, then soaked for 30 min at 70°C in pre-hybridization buffer [2× SSC, 0.1% N-laurosyl sarcosine, 0.2% sodium dodecyl sulfate (SDS), 10% blocking reagent (Roche; Cat #: 11096176001), and 1% dimethyl sulfoxide (DMSO)]. Digoxigenin-labeled locked nucleic acid (LNA) probe having the indicated repeat sequence (10 pmol/ml; Table 1) was added and hybridized overnight at 70°C (or 75°C for GGGGCC probe). The membrane was then washed twice in low stringency buffer (2× SSC, 0.1% SDS) for 5 min at room temperature, followed by two 15-min incubations in pre-heated higher stringency buffer (0.5× SSC, 0.1% SDS) at 70°C. The membrane was rinsed in 50 ml wash buffer (0.1 M maleic acid, 0.15 M NaCl, 0.3% Tween-20, pH to 7.5 with NaOH) at room temperature for 2 min, then in 25 ml blocking buffer (0.1 M maleic acid, 0.15 M NaCl, pH 7.5, 10% blocking solution (Roche)) for 30 min with shaking. Anti-Digoxigenin-AP Fab antibody fragment was added to blocking buffer (1:20 000; Roche Cat#: 11093274910) and held for 30 min at room temperature. The membrane was rinsed twice with washing buffer for 15 min then equilibrated for 3 min in detection buffer (0.1 M Tris–HCl, 0.1 M NaCl, pH to 9.5 with NaOH), then placed in 1 ml CDP-Star reagent (Roche Cat#: 12041677001) for 5 min at room temperature, dried and exposed using BioBlot BXR film (Laboratory Product Supply). Films were scanned on a HP Scanjet 5590 (Hewlett-Packard) and images quantified using ImageJ using densitometry (NIH).
Transgenic mice
Mice were studied in an Association for Assessment and Accreditation of Laboratory Animal Care (AAALAC)-accredited facility using procedures approved by the institutional animal welfare committee. Human skeletal actin-long repeat (HSALR, ∼220 CTG repeats in the 3′ UTR) and human skeletal actin-short repeat (HSASR, 5 CTG repeats) transgenic mice were previously described (3). Human skeletal actin transgenic mice with extra-long CTG repeats (extra-long repeat, HSAXLR) were generated by targeted integration of the ACTA1 transgene at the ROSA26 locus. The transgene design is similar to HSALR transgenic mice, except that the repeat tract is twice as long (CTG-440 versus CTG-220) and the expression of CUGexp RNA is conditional. As initially derived, transcription of the expanded repeat is blocked by an upstream transcription termination cassette (non-recombined mice, HSANR). HSANR mice were bred to E2A-Cre transgenic mice to obtain progeny with excision of the transcription terminator cassette and expression of (CUG)440 RNA (HSAXLR mice) (Supplemental Figure S1). LC15 transgenic mice harbor a construct in which expression of Gaussia luciferase is driven by the CMV enhancer and chicken beta-actin promoter. The luciferase cDNA is fused to the human DMPK 3′ UTR containing an expanded CTG repeat. The integration is multi-copy with repeat lengths ranging from ∼220–400 CTG repeats (Supplemental Figure S1).
Cell line expressing GGGGCC repeats
Mouse N2a neuroblastoma cells were stably transfected with construct LC19 (no repeats) or LC21 (160 GGGGCC repeats) using PhiC31 integrase-mediated insertion of mini-circle DNA (Supplemental Figure S2). Cells were cultured at 37°C and 5% CO2 in DMEM, 10% fetal bovine serum, and 1% penicillin/streptomycin (Gibco).
Antisense oligonucleotide (ASO) knockdown of CUG-repeat RNA in transgenic mice
2′-O-(2-Methoxyethyl) phosphorothioate ASOs 190401 and 445569 were previously described (18,30). These ASOs were a gift from Dr F. Bennett at Ionis Pharmaceuticals, Carlsbad, CA, USA. HSALR and HSAXLR mice were treated with ASO 190401 as previously described (18) using subcutaneous injection of 25 mg/kg twice weekly for 4 weeks. The ASO targets sequence in the coding region of the HSALR and HSAXLR transgenes. LC15 transgenic mice were treated with ASO 445569 75 mg/kg twice weekly for 6 weeks. This ASO targets sequence in the 3′ UTR of the LC15 transgene. Mice were euthanized for tissue harvest two days following the last injection.
Quantitative RT-PCR (qRT-PCR)
Transgene mRNA was quantified using Taqman gene expression mastermix (ThermoFisher) and the primer/probe set for HSALR, HSAXLR, HSANR and HSASR mice: hACTA1-Ex1F, hACTA1-Ex2R and hACTA1-probe, as seen in Table 1. Primer/probe sets for LC15 were hDMPK 3′UTR-F, hDMPK 3′UTR-R and hDMPK 3′UTR-probe (Table 1). Relative quantitation was performed using the 2−ΔΔCt method. Gtf2b (ThermoFisher Mm00663250_m1) was used for normalization of hACTA1, and Gapdh (ThermoFisher Mm99999915_g1) was used for normalization of hDMPK 3′ UTR.
RT-PCR splicing analysis
RTn was performed using 1 μg total cellular RNA, Superscript II (ThermoFisher), and oligo dT15 plus random hexamer primers, per the manufacturer's protocol. PCR was then performed using 5′ FAM-labeled primers flanking mSerca1 exon 22 and mClcn1 exon 7A in quadriceps cDNA (18,31), or mTmem63b exon 5 and mCacna1s exon 29 in heart cDNA (32) (Table 1). The PCR products were separated by agarose gel electrophoresis and detected with a laser fluorimager (Typhoon, GE Healthcare). The products were quantified using ImageQuant software (GE Healthcare).
Statistical Analysis
Comparison of knockdown and multiple displacement amplification efficiency was performed using Student′s t-test. Alternative splicing was analyzed by one-way analysis of variance (ANOVA), followed by multiple comparisons using Student's t-test with Bonferroni correction. All plots show average ± the standard error of the mean (SEM).
RESULTS
RTn of ER-RNA
Slot blot analysis of ER-RNA has the advantage that detection is relatively independent of repeat size or flanking sequence. The disadvantage, however, is that hybridization specificity for probes having tandem repeats is limited, giving rise to signal from non-expanded repeats or repeat-like sequences. In an effort to improve specificity we decided to convert ER-RNA to cDNA. However, this first required that we identify an enzyme capable of RTn on RNA having high GC content and strong propensity to form secondary or higher order structures, such as long hairpins or G quadruplexes (33,34). We focused on TGIRT-III, a thermostable mobile group II intron RT isolated from Geobacillus stearothermophilus, because of its high processivity and good activity on structured templates, such as tRNA (26,35–37). For initial experiments we examined a panel of enzymatically synthesized ER-RNAs, including r(CUG)100, r(CCUG)140, r(GGGGCC)40 and r(GGCCCC)40 (Figure 1), using RTn primers complementary to each of the respective repeat tracts [denoted (repeat sequence)# of repeats + remainder nts, e.g. (CAG)6+1 for r(CUG)100, (CAGG)4+2 for r(CCUG)140, (GGCCCC)3+3 for r(GGGGCC)40, and (GGGGCC)3+3 for r(CCCCGG)40, shown in Table 1]. Note that expanded CUG- and CCUG-repeats are expressed in DM1 and DM2, respectively, and that GGGGCC- and CCCCGG-repeats are both expressed in chromosome 9-associated amyotrophic lateral sclerosis and frontotemporal dementia (6,38). The molar ratios of primer:template were set at 5–10:1, depending on the transcript, to partially tile the templates with repeat primers (Figure 2A). Although our initial testing was performed using individual ER-RNAs in isolation, we used conditions designed to minimize off-target cDNA synthesis when later applied to total cellular RNA. For example, we omitted dNTPs not needed for the respective repeat sequence. r(CUG)100 and r(CCUG)140 were reverse transcribed with dCTP, dATP, dGTP, but not dTTP, whereas r(GGGGCC)40 and r(GGCCCC)40 were reverse transcribed with dCTP and dGTP. Also, we included dideoxy-UTP (ddUTP, 0.05 mM) to terminate off-target extension and block template switching.
Figure 1.
ER-RNAs produced by in vitro transcription. Bioanalyzer traces (A) and denaturing polyacrylamide gel electrophoresis (B) of ER-RNAs produced by T7 polymerase. The expected ER-RNAs lengths are: r(CUG)100, 300 nt; r(CCUG)140, 560 nt; r(GGGGCC)40 and r(CCCCGG)40; 240 nt. FU, fluorescence units. Ladder is ss100 DNA (Coli Genetic Stock Center, Yale). An empty lane between ladder and samples was removed for presentation.
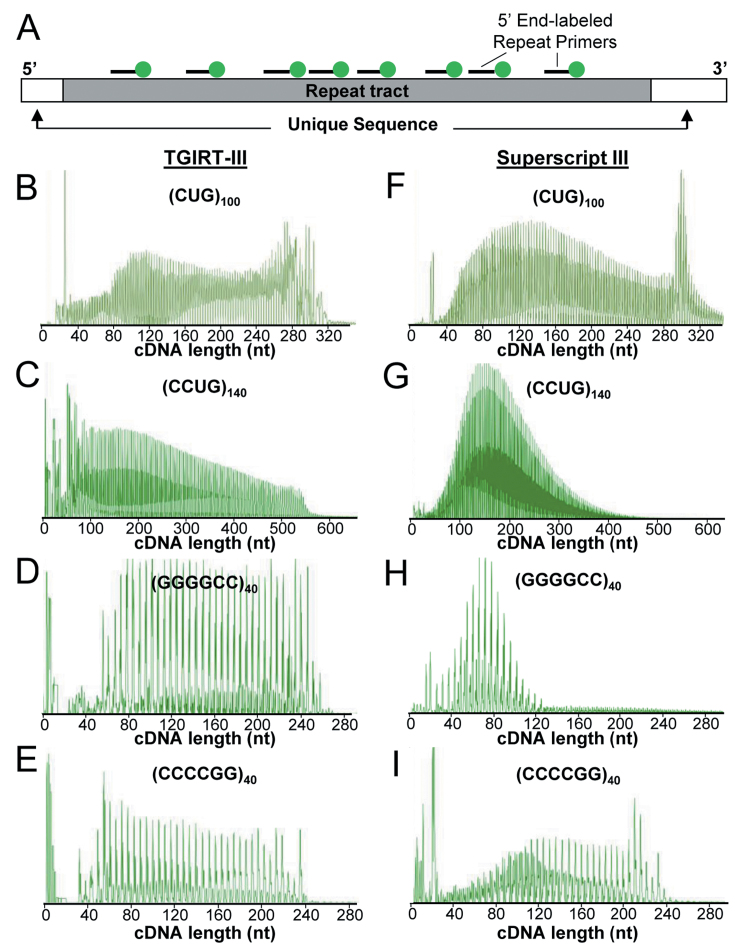
Figure 2.
Reverse transcription of enzymatically synthesized ER-RNAs. (A) Diagram of ER-RNA template, multiply primed with 5′ HEX end-labeled oligonucleotides. Representative chromatograms of HEX-labeled cDNA generated by TGIRT-III (B–E) or Superscript-III (F–I). Each trace represents analysis of a single cDNA synthesis.
Initial experiments were performed using HEX-labeled primers for analysis of extension products at high resolution by capillary electrophoresis. TGIRT-III transcribed all four ER-RNA templates, generating a series of cDNAs. As expected, the spacing of peaks on the electropherogram corresponded to the length of the respective repeat motif, 3 nt for r(CUG), 4 nt for r(CCUG), and 6 nt for r(GGGGCC) and r(CCCCGG) (Figure 2B–E). By comparison, Superscript-III (SS-III) also generated repeat cDNAs (Figure 2F–I), but the products from r(CCUG)140 and r(GGGGCC)40 were skewed towards shorter lengths (Figure 2G–H).
To further examine cDNA elongation and strand displacement we used a HEX-labeled primer that anneals 3′ of the repeat tract (Figure 3A). cDNA extension across the repeat tract was performed in the presence or absence of unlabeled repeat primers. In the absence of repeat primers, SS-III produced a full-length cDNA from r(CUG)100, but also generated a broad range of incomplete extension products (Figure 3D, upper). However, the production of full-length cDNA was inhibited when CAG-repeat primers were present (Figure 3D, lower). Notably, SS-III produced neither full-length nor incomplete extension products on the r(GGGGCC)40 template, regardless of whether CCCCGG-repeat primers were present (Figure 3E, three independent experiments).
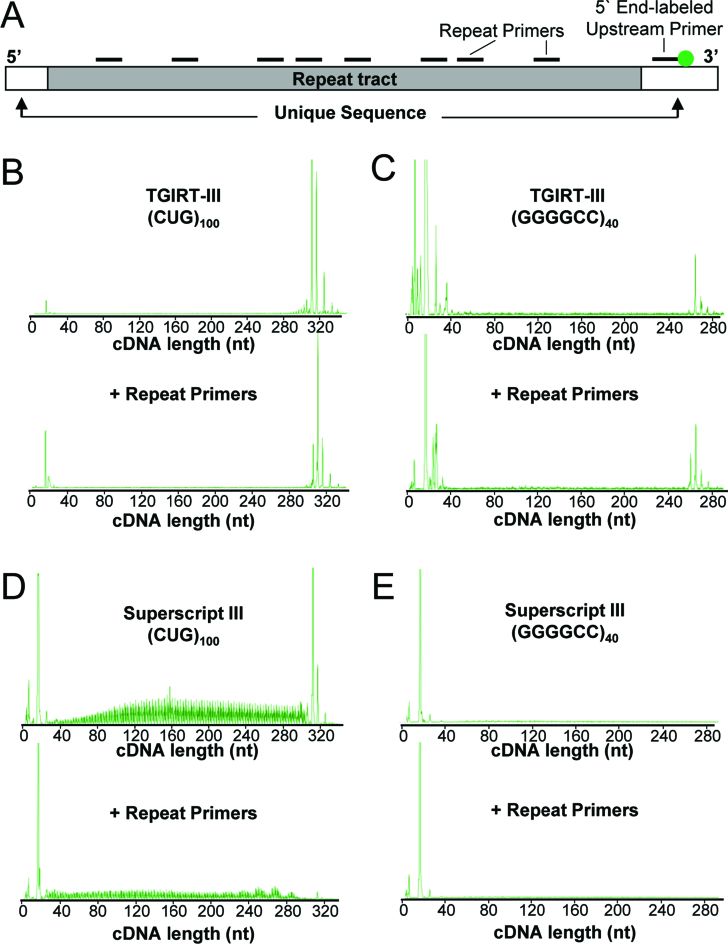
Figure 3.
Reverse transcription across ER-RNA tracts using TGIRT-III or SS-III. (A) Diagram of ER-RNA showing HEX-labeled primer annealed 3′ of repeat tract and unlabeled primers tiled across the expanded repeat. (B) Representative chromatogram of cDNA generated by TGIRT-III from r(CUG)100 in the presence or absence of intervening repeat primers. (C) Representative chromatogram of cDNA generated by TGIRT-III from r(GGGGCC)40, as in (B). (D and E) Representative chromatograms from r(CUG)100 and r(GGGGCC)40 templates using Superscript-III, as in (B) and (C).
In contrast to SS-III, TGIRT-III produced full-length cDNAs on both r(CUG)100 and r(GGGGCC)40, without noticeable levels of incomplete extension, irrespective of whether intervening repeat primers were present (Figure 3B and C). Similarly, when cDNAs were analyzed on denaturing polyacrylamide gels, TGIRT-III was found to generate full-length cDNAs from the r(CUG)100 and r(GGGGCC)40 templates, even when repeat primers were pre-annealed (Supplemental Figure S3). Taken together, these results indicate that TGIRT-III is capable of primer extension and strand displacement on high-GC repeat templates, consistent with previous observations that TGIRTs have high strand displacement activity, as evidenced by their ability to reverse transcribe efficiently through a highly structured group II intron RNA (26) and give full-length, end-to-end reads of tRNAs, which is not possible for retroviral RTs (37,39).
Next, we examined cDNA synthesis using RNA/DNA hybrid primers, designed for post-synthesis removal by ribonucleases. In pilot experiments we determined that TGIRT-III and SS-III were both capable of extending hybrid primers having as few as 4 nt of DNA at the 3′ end (data not shown), therefore this design was used in subsequent experiments. We examined RT extension from the hybrid (CAG)6+1 primer on r(CUG)100. For this experiment the primer was unlabeled but the ddUTP terminator carried FAM, for exclusive detection of cDNAs that extended beyond the (CUG)100 tract to terminate in the flanking sequence. TGIRT-III and SS-III both showed primer extension from hybrid primers, but once again SS-III was skewed toward shorter extension products whereas TGIRT-III was not (Supplemental Figure S4).
Expanded repeat-cDNA (ER-cDNA) slot blots
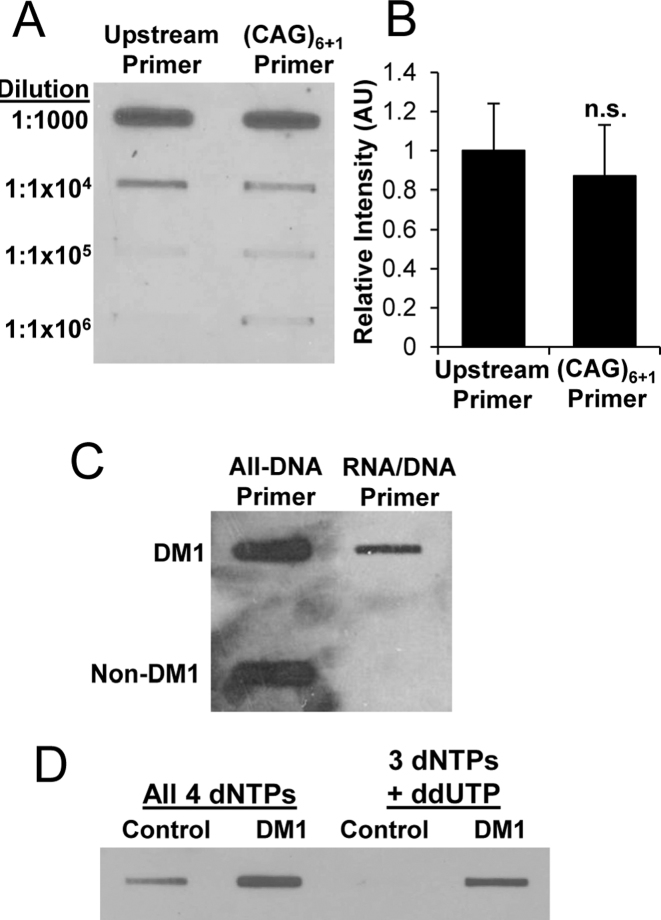
We next considered the possibility that action of TGIRT-III on ER-RNAs may produce multiple displacement amplification. For example, the r(CUG)100 template, if tiled to saturation with (CAG)6+1 primers and then fully extended by RTn, could generate up to 7.5-fold greater cDNA than ER-RNA input. To test for multiple displacement amplification we used cDNA slot blots to monitor d(CAG)n yield when r(CUG)100 was primed with (CAG)6+1, as compared to priming from a single upstream primer. The slot blots were hybridized with digoxigenin-labeled (CTG)7 probe. To maximize (CAG)6+1 tiling the molar ratio of primer to template was increased to 25:1. Notably, the cDNA yield from (CAG)6+1 priming was no greater than priming from a single site (P = 0.74, Figure 4A and B), which indicated that multiple displacement amplification did not occur on the multiply primed ER-RNA template (see below).
Figure 4.
cDNA slot blots of ER-RNA. (A) Dilution series of cDNA obtained when r(CUG)100 template was singly primed from upstream site versus multiply primed within the repeat tract by (CAG)6+1 (RNA/DNA hybrid primers). (B) Quantification of cDNA slot blot in (A), (n = 3 for each condition). (C) cDNA slot blot analysis of total cellular RNA (2 μg) from DM1 versus non-DM1 cardiac tissue using all-DNA versus RNA/DNA hybrid primers. Both reactions in (C) used 3 dNTPs plus ddUTP. (D) cDNA slot blot analysis of total cellular RNA (2 μg) from healthy control or DM1 tibialis anterior muscle biopsy samples using all 4 dNTPs versus 3 dNTPs (dCTP, dATP, dGTP plus ddUTP chain terminator) in the RT reaction. The reactions in (D) used RNA/DNA hybrid primers.
Application of cDNA slot blot to tissue samples
Next, we tested whether TGIRT-III is capable of reverse transcribing CUGexp RNA when present as a minor species in a complex mixture. First we examined total cellular RNA from postmortem DM1 cardiac muscle, a tissue showing conspicuous nuclear foci of CUGexp RNA (40). We primed the cDNA synthesis using all-DNA or RNA/DNA hybrid (CAG)6+1 primers, followed by ribonuclease A and H (RNase A/H) digest to remove RNA inputs and hybrid primers. When RTn was primed with the all-DNA primer, the cDNA slot blot showed strong hybridization signals from DM1 and non-DM1 samples alike, which likely reflects primer extension from non-expanded repeats, primer carry through on slot blot membranes, or both. In contrast, hybrid primers showed signal from DM1 samples but not from non-DM1 samples (Figure 4C), consistent with d(CAG)n synthesis from expanded but not from non-expanded repeats. Notably, the background signal from non-DM1 samples was also high when cDNA synthesis was performed using all four dNTPs, as compared to three dNTPs plus ddUTP (Figure 4D), indicating that hybrid primers and constrained dNTP mix were both contributing to the disease specificity of the cDNA slot blot.
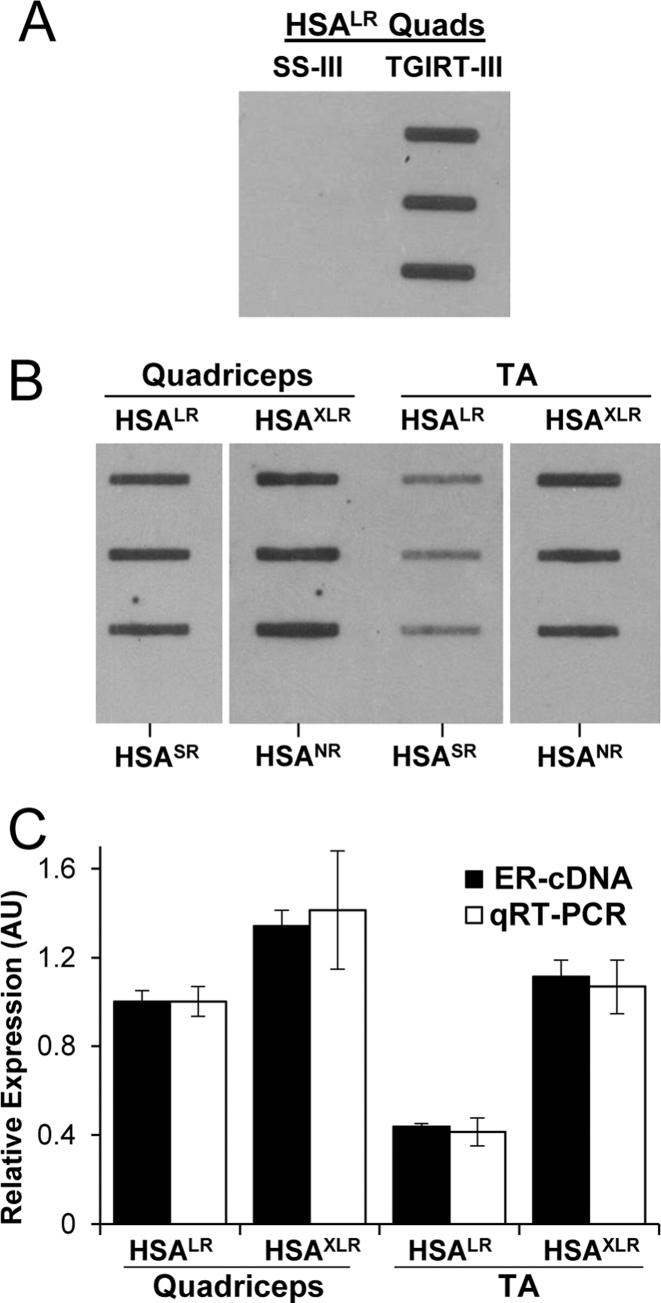
To further assess specificity and response characteristics of cDNA slot blots we used RNA from human skeletal actin-long repeat (HSALR) or HSA extra-long repeat (HSAXLR) transgenic mice. These mice carry (CTG)220 or (CTG)440 in the 3′ untranslated region of transgene mRNA, at levels several fold higher than human DM1 (41). In contrast to human DM, these transgenic lines do not exhibit major somatic expansion of the repeat tracts in muscle tissue. Also, there is no corresponding wild-type allele, and the ER-RNA fragments do not persist as decay intermediates (18). Altogether, these circumstances allow direct comparison of cDNA slot blots with qRT-PCR assays of flanking sequences. The cDNA slot blots of HSALR muscle showed robust signal when cDNA synthesis was performed using TGIRT-III, but no signal using SS-III (Figure 5A), therefore TGIRT-III was used for subsequent experiments. The cDNA slot blots also showed positive results from HSAXLR mice that express (CUG)440, but not from HSA-short repeat (HSASR) mice expressing a transgene with a non-expanded (CUG)5 repeat, or from non-recombined HSAXLR mice (HSANR) that harbor a (CTG)440 tract without expressing it (Figure 5B and supplemental Figure S1). Consistent with previous studies using qRT-PCR (18), the CUGexp expression by cDNA slot blot was higher in quadriceps than tibialis anterior muscle (Figure 5C). We used qRT-PCR data to calculate an inferred CUGexp level in HSALR and HSAXLR lines by multiplying the repeat length by normalized HSA expression levels. The results of cDNA slot blots showed good agreement with inferred CUGexp level in comparisons of HSALR versus HSAXLR mice or quadriceps versus tibialis anterior muscle (Figure 5C). Taken together, these results support the feasibility of using cDNA slot blots for CUGexp quantification and again suggest that longer repeats do not generate disproportionately greater ER-cDNA through multiple displacement amplification.
Figure 5.
ER-RNA expression in transgenic mouse models of DM1. (A) cDNA slot blot of total cellular RNA from HSALR quadriceps (Quads, 2 μg, three different mice) using Superscript-III (SS-III) or TGIRT-III. (B) cDNA slot blot of 2 μg total cellular RNA from quadriceps or tibialis anterior (TA) muscles from HSALR and HSAXLR mice. Bottom-most wells in each column are from HSASR or HSANR mice, which do not express ER-RNA. (C) Relative amount of ER-RNA by cDNA slot blot in HSALR and HSAXLR quadriceps and TA muscle (black bars, mean signal in HSALR quadriceps set to 1), as compared to inferred ER-RNA level based on qRT-PCR (white bars, calculated by multiplying expression level x repeat length, with the mean product in HSALR quadriceps set to 1). Results are based on n = 3 for HSALR and HSAXLR mice. HSA transgene expression by qRT-PCR was normalized to general transcription factor 2b (Gtf2b).
Responsivity of ER-cDNA slot blot to therapeutic intervention
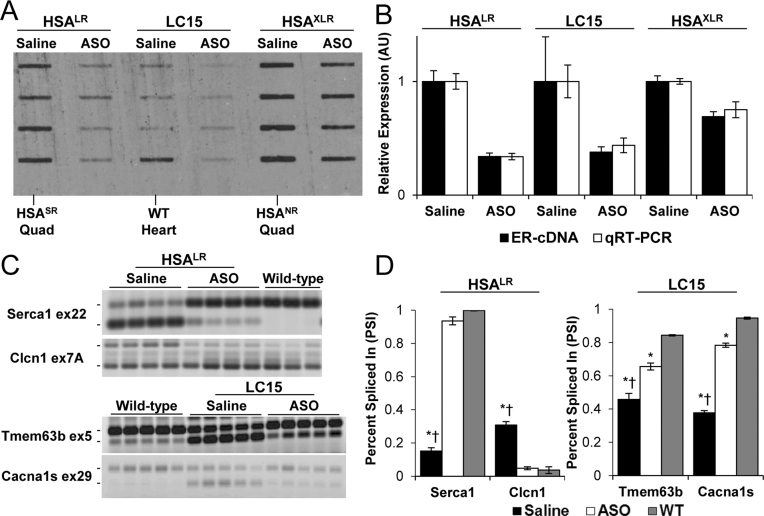
Next, we used cDNA slot blots to examine CUGexp knockdown in transgenic mice. We administered RNase H-active ASOs by subcutaneous injection in three independent lines of CUGexp-expressing mice. In each case, the targeting sequences for the ASO were located 5′ to the CUGexp tract. HSALR transgenic mice were injected with ASO 190401 at 25 mg/kg twice weekly for four weeks as previously described (18). HSAXLR mice were treated with the same regimen. In both of these lines the CUGexp RNA is expressed exclusively in skeletal muscle. In contrast, LC15 transgenic mice express CUGexp RNA predominantly in cardiac muscle. They were treated with ASO 445569 (30) by subcutaneous injection of 75 mg/kg twice weekly for six weeks. We compared results of cDNA slot blots with qRT-PCR to assess transgene silencing in quadriceps muscle (HSALR and HSAXLR) or heart (LC15). Across all three lines the knockdown efficiency was similar by both assays (Figure 6, n = 4 ASO-treated and four saline-treated mice for each line). We also tested whether the CUGexp reductions observed on cDNA slot blots were accompanied by improvements of alternative splicing. In HSALR mice the 66 ± 3% reduction of CUGexp RNA in skeletal muscle was associated with near-complete normalization of alternative splicing for Serca1 exon 22 and Clcn1 exon 7a (Figure 6C and D), and in LC15 mice the 31 ± 4% reduction of CUGexp RNA in cardiac muscle was associated with partial correction of splicing regulation for Tmem63b exon 5 and Cacna1s exon 29. These results indicate that cDNA slots are capable of detecting ER-RNA reductions that are therapeutically relevant.
Figure 6.
Responsivity of cDNA slot blot and alternative splicing to treatment with antisense oligonucleotides (ASOs) in transgenic mice. (A) cDNA slot blot of 2 μg total RNA from quadriceps muscle (HSALR and HSAXLR mice) or left ventricle (LC15 mice) treated with subcutaneous injections of saline (n = 4) or ASO (n = 4 in each group). Negative control samples from HSASR quadriceps, HSANR quadriceps, or wild-type (WT) heart are indicated. (B) Relative ER-RNA quantification by cDNA slot blot (black bars) versus qRT-PCR (white bars). Saline-treated groups set to 1 for both assays. (C) RT-PCR analysis of alternative splicing in saline- versus ASO-treated HSALR quadriceps (Serca1 and Clcn1) and LC15 heart tissue (Tmem63b and Cacna1s), as compared to wild-type controls. The mean ± S.D. values of ‘percent spliced in’ for saline versus ASO versus wild-type groups were Serca1 exon 22: 15 ± 2%, 94 ± 2%, 100 ± 0%; Clcn1 exon 7A: 31 ± 2%, 5 ± 1%, 4 ± 2%); Tmem63b exon 5: 46 ± 4%, 66 ± 2%, 84 ± 0%; Cacna1s exon 29: 38 ± 1%, 78 ± 1%, 95 ± 1%). (D) Quantification of (C). ANOVA of Serca1 exon 22 (P = 2.2 × 10−9), Clcn1 exon 7A (P = 4.4 × 10−6), Tmem63b exon 5 (P = 3.1 × 10−9), and Cacna1s (P = 3.7 × 10−9). *Student's t-test versus WT P < 0.0005, † = Student's t-test versus ASO P < 0.00005.
ER-cDNA slot blots of DM1 skeletal muscle
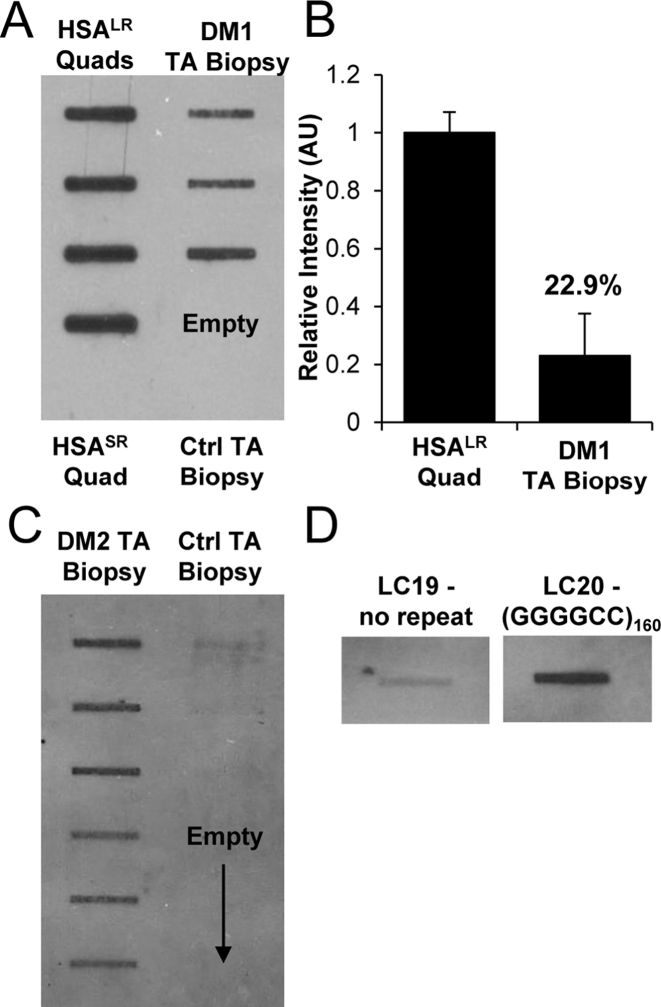
Next, we tested whether CUGexp expression can be detected in small tissue samples, such as those obtained by needle biopsies in clinical studies. For these experiments we focused on tibialis anterior (TA), a muscle that is preferentially affected by DM1. Once again, TA samples from DM1 patients but not healthy controls showed positive results on cDNA slot blots. Notably, the CUGexp levels were variable among patients, consistent with the known clinical heterogeneity of DM1 (Figure 7A). Comparison to transgenic mice indicated that levels of CUGexp RNA in DM1 biopsy samples were 4- to 5-fold lower than in HSALR mice (Figure 7B).
Figure 7.
cDNA slot blot of DM1 and DM2 muscle biopsy samples and (GGGGCC)160-expressing cells. (A) cDNA slot blot of total cellular RNA (2 μg) from tibialis anterior (TA) biopsy samples of DM1 patients (n = 3) or healthy controls, as compared to quadriceps muscle from HSALR transgenic mice. The bottom-most wells in each column are HSASR or healthy controls. (B) Relative quantification of (A), with the mean value from HSALR mice set to 1. (C) cDNA slot blot of 2 μg total cellular RNA from TA biopsy samples from DM2 patients (n = 6) or healthy controls (n = 3, upper-most wells). Reverse transcription was primed with (CAGG)4+2 RNA/DNA hybrid primer, as shown in table 1. (D) Representative ER-cDNA slot blot of 2 μg total RNA from pLC19 (no repeat) versus pLC21 [expressing (GGGGCC)160] cell lysates. Blots were run simultaneously, with an intervening slot removed for presentation.
ER-cDNA slot blot detection of intronic or all-GC repeats
Next, we applied cDNA slot blots to intronic repeats, using muscle biopsy samples from DM2 patients, and to all-GC repeats, using cells that integrated an r(GGGGCC)160 expression construct. Hybrid primers specific to each repeat sequence were used to generate cDNA, and slot blots were hybridized with the corresponding repeat probes (Table 1). We observed strong signals on cDNA slot blots from intronic CCUG-repeats in DM2 patients (Figure 7C), and from r(GGGGCC)160-expressing cells (Figure 7D), but not from muscle samples of healthy controls or cells expressing a no-repeat construct.
DISCUSSION
RNA dominant effects may potentially occur at any point in the metabolic cycle of ER-RNA, from nascent transcript to decay intermediate, creating a need for analysis methods that are free of assumptions about repeat length or continuity with flanking sequences. Fluorescence in situ hybridization (FISH) and RNA slot blots arguably come closest to meeting these objectives, but have trade-offs regarding specificity and precision. For example, FISH probes have photo bleaching, potential for auto-quenching when fluorophores are crowded in foci (42,43), and variable access to nuclear targets in tissue sections, whereas RNA slot blots have limited specificity due to off-target hybridization of repeat probes (25).
Our results show that conversion of ER-RNA to cDNA using TGIRT-III can reduce non-specific background on slot blots. The specificity of the assay stems partly from antisense-repeat primers that are used for priming the cDNA synthesis and sense-repeat probes that are used for hybridizing slot blots. Beyond these measures, several additional conditions were used to limit off-target RTn. These included restricting the pool of dNTPs, including an off-target chain terminator (ddUTP), and using RNA/DNA chimeric primers. Taken together, these conditions can terminate cDNA synthesis from off-target sequences, eliminate cDNA products from short repeats that are spanned by repeat primers, and truncate cDNA products generated from repeats of intermediate length. For example, even if the size limit of CAG-repeats that are retained on the slot blot and detected by repeat probes is as low as 10 nts, then r(CUG)n tracts ≥ 25 nt would be required to generate signal, assuming that the cagcagcagcagcagCAGC (rna/DNA) primers are fully annealed on repeat RNA. Global reference data indicate that 194 human loci have ≥25 nts of CTG repeats, but only 16 of these express CUG-repeats in exons (44,45). To estimate the expected signal from these loci in aggregate, we multiplied the expression level (RNAseq data from 430 muscle samples in the GTEx Project) by the length of the repeat tract (major allele in the 1,000 Genomes Project). Notably, the predicted aggregate signal across non-expanded repeats was 60-fold lower than DMPK having 2000 CTG repeats (Supplemental Table S1), consistent with our slot blot findings in DM1 versus healthy muscle samples.
The studies in DM1 mouse models indicate that results of cDNA slots blots correlate well with conventional qRT-PCR, when compared across (1) muscles having different levels of CUGexp expression; (2) mice treated with ASOs versus saline; and (3) transgenic lines having CTG expansions of different length. Furthermore, a therapeutic intervention causing full or partial correction of CUGexp-induced splicing defects in mice was readily detected on cDNA slot blots. While these observations are useful for validation purposes, the more useful application is for human samples having bi-allelic expression, repeat expansions that are much larger and more heterogeneous, and especially for intronic repeats, where alternative methods are lacking.
While our results suggest that cDNA slot blots may be useful in these situations, the current study has limitations and poses several unresolved questions. First, we observed differences between SS-III and TGIRT-III for reverse transcription of ER-RNAs, consistent with evidence that ER-RNAs form stable hairpins or G-quadruplexes (33,34). Our results fit with the observations that TGIRT-III is active on highly structured templates (26,35,37,39,46), and show that TGIRT-III was more effective for reverse transcribing expanded repeats than SS-III. However, it remains possible that other RTs may share this activity. Furthermore, given that both enzymes exhibited RTn for r(CUG)100 templates in isolation, it remains unclear why TGIRT-III but not SS-III generated cDNA slot blot signals from CUGexp-expressing mice. Second, our results indicate that multiple displacement amplification does not appreciably occur during ER-cDNA synthesis by TGIRT-III. Presumably this indicates that the overall efficiency of primer annealing, RT initiation, cDNA elongation, and strand displacement was insufficient for multiple displacement amplification, but the underlying biochemical explanations remain unclear. Finally, we observed variable levels of CUGexp accumulation in TA muscle among DM1 patients. It is unknown, however, how these differences relate to the variability of repeat length, DMPK transcription, or CUGexp turnover among patients. Prospective studies in large cohorts are needed to determine whether the levels of ER-RNA are determinative for disease severity in DM1 and DM2.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Kirti Bhatt at the University of Rochester for technical assistance and C. Frank Bennett at Ionis Pharmaceuticals for providing antisense oligonucleotides.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR online.
FUNDING
National Institute of Health [GM37949 to A.M.L., NS094393 and NS048843 to C.A.T.]; Welch Foundation [F-1607 to A.M.L.]. Funding for open access charge: Departmental funds.
Conflict of interest statement. Thermostable group II intron reverse transcriptase (TGIRT) enzymes and methods for their use are the subject of patents and patent applications that have been licensed by the University of Texas and East Tennessee State University to InGex, LLC. A.M.L. and the University of Texas are minority equity holders in InGex, LLC, and S.M., A.M.L. and the University of Texas receive royalty payments from the sale of TGIRT enzymes and kits and from the licensing of intellectual property. S.T.C., Z.T. and C.A.T. have no conflicts to report.
REFERENCES
- 1. Nelson D.L., Orr H.T., Warren S.T.. The unstable repeats–three evolving faces of neurological disease. Neuron. 2013; 77:825–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Pieretti M., Zhang F.P., Fu Y.H., Warren S.T., Oostra B.A., Caskey C.T., Nelson D.L.. Absence of expression of the FMR-1 gene in fragile X syndrome. Cell. 1991; 66:817–822. [DOI] [PubMed] [Google Scholar]
- 3. Mankodi A., Logigian E., Callahan L., McClain C., White R., Henderson D., Krym M., Thornton C.A.. Myotonic dystrophy in transgenic mice expressing an expanded CUG repeat. Science. 2000; 289:1769–1773. [DOI] [PubMed] [Google Scholar]
- 4. Liquori C.L., Ricker K., Moseley M.L., Jacobsen J.F., Kress W., Naylor S.L., Day J.W., Ranum L.P.. Myotonic dystrophy type 2 caused by a CCTG expansion in intron 1 of ZNF9. Science. 2001; 293:864–867. [DOI] [PubMed] [Google Scholar]
- 5. Xu Z., Poidevin M., Li X., Li Y., Shu L., Nelson D.L., Li H., Hales C.M., Gearing M., Wingo T.S. et al. . Expanded GGGGCC repeat RNA associated with amyotrophic lateral sclerosis and frontotemporal dementia causes neurodegeneration. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:7778–7783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mori K., Weng S.M., Arzberger T., May S., Rentzsch K., Kremmer E., Schmid B., Kretzschmar H.A., Cruts M., Van Broeckhoven C. et al. . The C9orf72 GGGGCC repeat is translated into aggregating dipeptide-repeat proteins in FTLD/ALS. Science. 2013; 339:1335–1338. [DOI] [PubMed] [Google Scholar]
- 7. Ash P.E., Bieniek K.F., Gendron T.F., Caulfield T., Lin W.L., Dejesus-Hernandez M., van Blitterswijk M.M., Jansen-West K., Paul J.W., Rademakers R. et al. . Unconventional translation of C9ORF72 GGGGCC expansion generates insoluble polypeptides specific to c9FTD/ALS. Neuron. 2013; 77:639–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Taneja K.L., McCurrach M., Schalling M., Housman D., Singer R.H.. Foci of trinucleotide repeat transcripts in nuclei of myotonic dystrophy cells and tissues. J. Cell Biol. 1995; 128:995–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. DiFiglia M., Sapp E., Chase K.O., Davies S.W., Bates G.P., Vonsattel J.P., Aronin N.. Aggregation of huntingtin in neuronal intranuclear inclusions and dystrophic neurites in brain. Science. 1997; 277:1990–1993. [DOI] [PubMed] [Google Scholar]
- 10. Zu T., Gibbens B., Doty N.S., Gomes-Pereira M., Huguet A., Stone M.D., Margolis J., Peterson M., Markowski T.W., Ingram M.A. et al. . Non-ATG-initiated translation directed by microsatellite expansions. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:260–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Freibaum B.D., Lu Y., Lopez-Gonzalez R., Kim N.C., Almeida S., Lee K.H., Badders N., Valentine M., Miller B.L., Wong P.C. et al. . GGGGCC repeat expansion in C9orf72 compromises nucleocytoplasmic transport. Nature. 2015; 525:129–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhang K., Donnelly C.J., Haeusler A.R., Grima J.C., Machamer J.B., Steinwald P., Daley E.L., Miller S.J., Cunningham K.M., Vidensky S. et al. . The C9orf72 repeat expansion disrupts nucleocytoplasmic transport. Nature. 2015; 525:56–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Miller J.W., Urbinati C.R., Teng-Umnuay P., Stenberg M.G., Byrne B.J., Thornton C.A., Swanson M.S.. Recruitment of human muscleblind proteins to (CUG)(n) expansions associated with myotonic dystrophy. EMBO J. 2000; 19:4439–4448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wang G.S., Kuyumcu-Martinez M.N., Sarma S., Mathur N., Wehrens X.H., Cooper T.A.. PKC inhibition ameliorates the cardiac phenotype in a mouse model of myotonic dystrophy type 1. J. Clin. Invest. 2009; 119:3797–3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Brockhoff M., Rion N., Chojnowska K., Wiktorowicz T., Eickhorst C., Erne B., Frank S., Angelini C., Furling D., Rüegg M.A. et al. . Targeting deregulated AMPK/mTORC1 pathways improves muscle function in myotonic dystrophy type I. J. Clin. Invest. 2017; 127:549–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jones K., Wei C., Iakova P., Bugiardini E., Schneider-Gold C., Meola G., Woodgett J., Killian J., Timchenko N.A., Timchenko L.T.. GSK3β mediates muscle pathology in myotonic dystrophy. J. Clin. Invest. 2012; 122:4461–4472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Thornton C.A., Wang E., Carrell E.M.. Myotonic dystrophy: approach to therapy. Curr. Opin. Genet. Dev. 2017; 44:135–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wheeler T.M., Leger A.J., Pandey S.K., MacLeod A.R., Nakamori M., Cheng S.H., Wentworth B.M., Bennett C.F., Thornton C.A.. Targeting nuclear RNA for in vivo correction of myotonic dystrophy. Nature. 2012; 488:111–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sareen D., O’Rourke J.G., Meera P., Muhammad A.K., Grant S., Simpkinson M., Bell S., Carmona S., Ornelas L., Sahabian A. et al. . Targeting RNA foci in iPSC-derived motor neurons from ALS patients with a C9ORF72 repeat expansion. Sci. Transl. Med. 2013; 5:208ra149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Harper S.Q., Staber P.D., He X., Eliason S.L., Martins I.H., Mao Q., Yang L., Kotin R.M., Paulson H.L., Davidson B.L.. RNA interference improves motor and neuropathological abnormalities in a Huntington's disease mouse model. Proc. Natl. Acad. Sci. U.S.A. 2005; 102:5820–5825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Thornton C.A., Johnson K., Moxley R.T.. Myotonic dystrophy patients have larger CTG expansions in skeletal muscle than in leukocytes. Ann. Neurol. 1994; 35:104–107. [DOI] [PubMed] [Google Scholar]
- 22. Ashizawa T., Dubel J.R., Harati Y.. Somatic instability of CTG repeat in myotonic dystrophy. Neurology. 1993; 43:2674–2678. [DOI] [PubMed] [Google Scholar]
- 23. Margolis J.M., Schoser B.G., Moseley M.L., Day J.W., Ranum L.P.. DM2 intronic expansions: evidence for CCUG accumulation without flanking sequence or effects on ZNF9 mRNA processing or protein expression. Hum. Mol. Genet. 2006; 15:1808–1815. [DOI] [PubMed] [Google Scholar]
- 24. Kozlowski P., de Mezer M., Krzyzosiak W.J.. Trinucleotide repeats in human genome and exome. Nucleic Acids Res. 2010; 38:4027–4039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wheeler T.M., Sobczak K., Lueck J.D., Osborne R.J., Lin X., Dirksen R.T., Thornton C.A.. Reversal of RNA dominance by displacement of protein sequestered on triplet repeat RNA. Science. 2009; 325:336–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mohr S., Ghanem E., Smith W., Sheeter D., Qin Y., King O., Polioudakis D., Iyer V.R., Hunicke-Smith S., Swamy S. et al. . Thermostable group II intron reverse transcriptase fusion proteins and their use in cDNA synthesis and next-generation RNA sequencing. RNA. 2013; 19:958–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Osborne R.J., Thornton C.A.. Cell-free cloning of highly expanded CTG repeats by amplification of dimerized expanded repeats. Nucleic Acids Res. 2008; 36:e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rio D.C., Ares M., Hannon G.J., Nilsen T.W.. Purification of RNA using TRIzol (TRI reagent). Cold Spring Harb. Protoc. 2010; 2010:doi:10.1101/pdb.prot5439. [DOI] [PubMed] [Google Scholar]
- 29. Rio D.C., Ares M., Hannon G.J., Nilsen T.W.. Removal of DNA from RNA. Cold Spring Harb. Protoc. 2010; 2010:doi:10.1101/pdb.prot5443. [DOI] [PubMed] [Google Scholar]
- 30. Pandey S.K., Wheeler T.M., Justice S.L., Kim A., Younis H., Gattis D., Jauvin D., Puymirat J., Swayze E.E., Freier S.M. et al. . Identification and characterization of modified antisense oligonucleotides targeting DMPK in mice and nonhuman primates for the treatment of myotonic dystrophy type 1. J. Pharmacol. Exp. Ther. 2015; 355:329–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lin X., Miller J.W., Mankodi A., Kanadia R.N., Yuan Y., Moxley R.T., Swanson M.S., Thornton C.A.. Failure of MBNL1-dependent post-natal splicing transitions in myotonic dystrophy. Hum. Mol. Genet. 2006; 15:2087–2097. [DOI] [PubMed] [Google Scholar]
- 32. Lee K.Y., Li M., Manchanda M., Batra R., Charizanis K., Mohan A., Warren S.A., Chamberlain C.M., Finn D., Hong H. et al. . Compound loss of muscleblind-like function in myotonic dystrophy. EMBO Mol. Med. 2013; 5:1887–1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Reddy K., Zamiri B., Stanley S.Y., Macgregor R.B., Pearson C.E.. The disease-associated r(GGGGCC)n repeat from the C9orf72 gene forms tract length-dependent uni- and multimolecular RNA G-quadruplex structures. J. Biol. Chem. 2013; 288:9860–9866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Napierała M., Krzyzosiak W.J.. CUG repeats present in myotonin kinase RNA form metastable ‘slippery’ hairpins. J. Biol. Chem. 1997; 272:31079–31085. [DOI] [PubMed] [Google Scholar]
- 35. Zheng G., Qin Y., Clark W.C., Dai Q., Yi C., He C., Lambowitz A.M., Pan T.. Efficient and quantitative high-throughput tRNA sequencing. Nat. Methods. 2015; 12:835–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Shen P.S., Park J., Qin Y., Li X., Parsawar K., Larson M.H., Cox J., Cheng Y., Lambowitz A.M., Weissman J.S. et al. . Protein synthesis. Rqc2p and 60S ribosomal subunits mediate mRNA-independent elongation of nascent chains. Science. 2015; 347:75–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Qin Y., Yao J., Wu D.C., Nottingham R.M., Mohr S., Hunicke-Smith S., Lambowitz A.M.. High-throughput sequencing of human plasma RNA by using thermostable group II intron reverse transcriptases. RNA. 2016; 22:111–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zu T., Liu Y., Bañez-Coronel M., Reid T., Pletnikova O., Lewis J., Miller T.M., Harms M.B., Falchook A.E., Subramony S.H. et al. . RAN proteins and RNA foci from antisense transcripts in C9ORF72 ALS and frontotemporal dementia. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:E4968–4977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Katibah G.E., Qin Y., Sidote D.J., Yao J., Lambowitz A.M., Collins K.. Broad and adaptable RNA structure recognition by the human interferon-induced tetratricopeptide repeat protein IFIT5. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:12025–12030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Mankodi A., Lin X., Blaxall B.C., Swanson M.S., Thornton C.A.. Nuclear RNA foci in the heart in myotonic dystrophy. Circ. Res. 2005; 97:1152–1155. [DOI] [PubMed] [Google Scholar]
- 41. Gudde A.E., González-Barriga A., van den Broek W.J., Wieringa B., Wansink D.G.. A low absolute number of expanded transcripts is involved in myotonic dystrophy type 1 manifestation in muscle. Hum. Mol. Genet. 2016; 25:1648–1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Cox W.G., Beaudet M.P., Agnew J.Y., Ruth J.L.. Possible sources of dye-related signal correlation bias in two-color DNA microarray assays. Anal. Biochem. 2004; 331:243–254. [DOI] [PubMed] [Google Scholar]
- 43. Lichtman J.W., Conchello J.A.. Fluorescence microscopy. Nat. Methods. 2005; 2:910–919. [DOI] [PubMed] [Google Scholar]
- 44. Auton A., Brooks L.D., Durbin R.M., Garrison E.P., Kang H.M., Korbel J.O., Marchini J.L., McCarthy S., McVean G.A., Abecasis G.R. et al. . A global reference for human genetic variation. Nature. 2015; 526:68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Willems T., Gymrek M., Highnam G., Mittelman D., Erlich Y., Consortium G.P.. The landscape of human STR variation. Genome Res. 2014; 24:1894–1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Enyeart P.J., Mohr G., Ellington A.D., Lambowitz A.M.. Biotechnological applications of mobile group II introns and their reverse transcriptases: gene targeting, RNA-seq, and non-coding RNA analysis. Mob. DNA. 2014; 5:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.