Abstract
Diversity-generating retroelements (DGRs) are novel genetic elements that use reverse transcription to generate vast numbers of sequence variants in specific target genes. Here, we present a detailed comparative bioinformatic analysis that depicts the landscape of DGR sequences in nature as represented by data in GenBank. Over 350 unique DGRs are identified, which together form a curated reference set of putatively functional DGRs. We classify target genes, variable repeats and DGR cassette architectures, and identify two new accessory genes. The great variability of target genes implies roles of DGRs in many undiscovered biological processes. There is much evidence for horizontal transfers of DGRs, and we identify lineages of DGRs that appear to have specialized properties. Because GenBank contains data from only 10% of described species, the compilation may not be wholly representative of DGRs present in nature. Indeed, many DGR subtypes are present only once in the set and DGRs of the candidate phylum radiation bacteria, and Diapherotrites, Parvarchaeota, Aenigmarchaeota, Nanoarchaeota, Nanohaloarchaea archaea, are exceptionally diverse in sequence, with little information available about functions of their target genes. Nonetheless, this study provides a detailed framework for classifying and studying DGRs as they are uncovered and studied in the future.
INTRODUCTION
Diversity-generating retroelements (DGRs) are a remarkable class of domesticated retroelements that have evolved useful functions to benefit their hosts. Having presumably originated from a retroelement that lost mobility functions, they use their reverse transcription activity to introduce a vast number of sequence variants into defined sites of specific target protein genes. The enormous number of protein variants allows the host to adapt rapidly to changing environmental conditions (1,2).
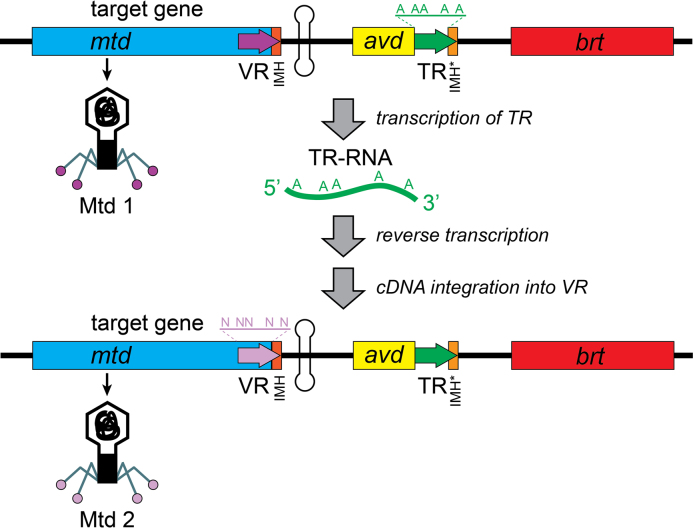
The prototypical DGR was discovered in a bacteriophage, BPP-1, which infects mammalian respiratory pathogens of the Bordetella genus (Figure 1) (3). The Bordetella cell surface is highly dynamic due to programmed changes in gene expression during its infectious cycle (4). During phage infection, BPP-1 adheres to Bordetella through its protein Mtd, which is positioned at the tips of the phage tail fibers (5–7). When the cellular surface changes, the BPP-1 phage is capable of switching its tropism through the action of its DGR on the mtd gene. The process, called mutagenic retrohoming, introduces multiple mutations into the 3′ portion of the phage’s mtd gene, at positions corresponding to the protein surface that binds to the bacterial receptors.
Figure 1.
Structure and mechanism of the Bordetella phage DGR. The prototypic Bordetella phage DGR contains a target gene (mtd) with a variable repeat (VR), an accessory gene (avd), a template repeat (TR) and a reverse transcriptase gene (brt). During mutagenic retrohoming, a transcript of the TR is reverse transcribed, and the cDNA is integrated into the VR sequence of the target gene. During this process, A’s in the template are subject to mutagenesis by the incorporation of random nucleotides opposite the A in the template. This results in a new VR sequence that codes for a diversified phage tail protein. Additional sequence elements important for mutagenesis are the initation of mutagenic homing (IMH) sequence at the end of the VR, and a nonidentical repeat IMH* at the end of TR. A GC-rich inverted repeat is found downstream of IMH.
The Bordetella phage DGR consists of four adjacent genes in the phage genome: a target gene (mtd) that contains a variable repeat (VR) at its 3′ end, a reverse transcriptase gene (brt), a template repeat (TR) and an accessory variability determinant (avd) (Figure 1) (1,3,8). During mutagenic retrohoming, a transcript of TR is reverse transcribed and the cDNA sequence is transferred to the VR locus. All reverse transcribed A’s in the TR are subject to an A-to-N mutagenic mechanism, which alters the sequence of the VR and the corresponding C-terminal ∼50 amino acid (aa) of the Mtd protein.
Because the TR sequence contains 23 A’s that can direct VR mutagenesis, mutagenic retrohoming can produce 1014 (423) VR DNA sequences, which correspond to 1013 aa sequences in the Mtd protein (3). Crystal structures of the ∼300 aa Mtd protein show that the C-terminal ∼125 aa comprise a C-type lectin fold, of which the final ∼45 aa correspond to the VR sequence (6). The residues subject to diversification are within the solvent exposed region of the C-type lectin fold, whereas the several constant aa encoded by the VR sequence are internalized in the structure, helping to form a structural scaffold from which the variable residues are displayed. A co-crystal structure between Mtd and the bacterial pertactin receptor confirms that the diversified residues make a network of direct interactions with the bacterial ligand (7).
Two additional sequence elements within the DGR cassette are important for mutagenic homing. The IMH (initiation of mutagenic homing) is an essential 34 bp sequence at the end of the VR whose first 14 bp are GC-rich. IMH marks the 3′ boundary of A-to-N mutagenesis in the VR (9). Downstream of IMH is a GC-rich inverted repeat that, while not essential, is required for efficient mutagenic homing (10). This repeat has been proposed to form a cruciform structure in vivo (10).
The TR sequence, which shares ∼90% sequence identity with VR, contains an analogous sequence called IMH*. IMH* differs in sequence somewhat from IMH and is not followed by an inverted repeat (10), thereby distinguishing the TR donor sequence from the recipient target DNA sequence.
Whereas the 3′ boundary for A-to-N mutagenesis is demarcated by the IMH sequence, the 5′ boundary appears to be less precise and is determined by the degree of 5′ sequence homology between TR and VR sequences (1,8,10). This is consistent with a reverse transcription mechanism in which cDNA synthesis begins at a fixed location at a 3′ position of the TR and proceeds upstream without a fixed stop point.
The accessory gene avd encodes an essential 128 aa protein that has a barrel structure and forms a homopentamer (11). Avd is highly basic and binds to both DNA and RNA in vitro, but without detectable sequence specificity. Avd also binds the reverse transcriptase (RT), and association between these two proteins is required for mutagenic retrohoming (11).
While the Bordetella phage DGR has been most extensively studied, two other DGRs have been characterized, and both operate on bacterial genes rather than phage genes. In Legionella pneumophila, a DGR is present within an integrative and conjugative element (12), whose target gene ldtA encodes a lipoprotein predicted to have a C-type lectin fold. The LdtA protein is exported to the outer leaflet of the outer membrane of the cell with its C-terminal VR region exposed. Based on the adenosine residues in the TR sequence, the DGR can theoretically produce 1026 DNA sequence variants, corresponding to 1019 protein variants. The precise roles of the LdtA protein and the biological significance of its diversification by the DGR are not defined.
The DGR of Treponema denticola can theoretically produce 1020 aa variants in its target–protein TvpA. An X-ray crystal structure shows that despite TvpA’s low sequence identity to Mtd, it has a C-type lectin fold with the variable aa exposed (13). TvpA belongs to the family of formylglycine-generating enzymes (FGE’s), which form a subclass of the C-type lectin fold and whose active sites contain the FGE–sulfatase motif (14,15). Despite the FGE motif, TvpA does not have formylglycine-generating activity and its C-type lectin domain is thought to have a binding function instead, with the diversified VR region mediating interactions with the biofilm/host cell surfaces in the oral cavity where T. denticola resides (13). In addition to tvpA, the DGR is thought to diversify seven related target genes scattered across the genome (13). Interestingly, there is substantial variation in DGR numbers and sequences among Treponema strains (16).
DGRs inhabit a wide range of organisms including proteobacteria, firmicutes, cyanobacteria and archaea (1,2,17–21). Two computer programs have been created to identify DGRs in genomic sequences. The program DiGReF uses a Position-Specific Iterative Basic Local Alignment Search Tool (PSI-BLAST) search to identify RTs and then screens for TR-VR repeats (19,20). The program DGRscan uses additional search criteria such as homology-based searches to known DGR genes and can locate incomplete DGR cassettes within small sequence contigs (16,22). More recently, an analysis of candidate phylum radiation (CPR) and Diapherotrites, Parvarchaeota, Aenigmarchaeota, Nanoarchaeota, Nanohaloarchaea (DPANN) metagenomic data identified a large number of highly diverse DGRs (23). Based on these reports it is clear that DGRs in nature have gene organizations differing from the DGRs of the Bordetella phage, Legionella and Treponema. Differences include multiple target genes (2,19,22), target genes with bacterial Ig domains instead of C-type lectin domains (16,17), variant gene orders within the DGR cassette (2,19,22) and an alternative accessory gene in some DGRs that consists of an HRDC (helicase and RNaseD C-terminal) domain (1,2,16).
In this work, we aim to produce a more complete and defined picture of the landscape of DGRs in nature. Our analysis begins with a bioinformatic approach similar to those used previously, but we then systematically examine all components and known features of DGRs in order to identify both conserved and unique features. In doing so, we have cataloged a wide spectrum of DGR types in detail and produced a reference set of presumably functional DGRs that should be a useful resource as more DGRs are identified and studied.
RESULTS AND DISCUSSION
Collection of sequence data and identification of DGR components
DGR sequences were collected from GenBank using a combination of automated and manual steps (‘Materials and Methods’ section). Repeated cycles of comparisons among DGRs were required to thoroughly identify known genes and elements and identify new ones. Only DGRs most likely to be functional were retained, while over half of potential DGRs were discarded because of premature stops, truncations, poor TR-VR matches or incomplete sequence data due to short contigs. While the high level of inactivated DGRs is perhaps not surprising for a component of migrant DNAs and phages, it suggests the use of caution when inferring functionality of DGRs found in genomic sequences.
In all, 372 non-redundant, putatively functional DGRs were compiled (Supplementary Table S1). An approximately equal number were ‘duplicates’, having >95% identity to the ‘unique’ set based on aa identity of the RT (not shown). A subset of 246 DGRs is referred to here as the ‘core set’ while 126 DGRs are called the ‘CPR’ set (Supplementary Table S1). The CPR set includes DGRs mainly from a groundwater metagenomic study of CPR bacteria and DPANN archaea (21,23–25). CPR bacteria are only distantly related to characterized bacteria, and not surprisingly, DGRs of the CPR set are not closely related to other DGRs, or to one other. A separate study, which includes sequences not present in GenBank, focuses on these highly diverse DGRs and presents more detailed information about the unusual elements (23).
Distribution of DGRs across species
As previously reported (1,2,19–21), DGRs are present widely among prokaryotes, and are not restricted to a narrow set of host organisms or living conditions (Supplementary Table S1). Of the core set of DGRs, ∼80% are in the phyla Firmicutes, Proteobacteria, Cyanobacteria and Bacteroidetes, which are among the most sequenced microbial groups. Smaller numbers of DGRs in the set are found in less sequenced phyla such as Actinobacteria, Chlorobi, Deinococcus and Spirochaetes.
It should be noted that the compilation probably does not saturate the DGR diversity present in nature. The data set appears to be biased toward the human microbiome and groundwater metagenomes, and many DGR subtypes are found only once in the compilation. GenBank contains sequence from only 10% of described species (https://www.ncbi.nlm.nih.gov/taxonomy); furthermore, the relatively recent discovery of bacterial ‘black matter’ (including CPR bacteria and DPANN archaea) raises the possibility that additional groups of microbes have eluded detection, and may contain novel types of DGRs.
Genomic locations of DGRs on chromosomes, plasmids and phages
Considering only the fraction of GenBank entries that specify the DNA source, it can be concluded that DGRs are most often found on chromosomes (69%; 60 of 87, including prophages), with lower numbers on free phages (24%; 21 of 87) or plasmids (7%; 6 of 87). These figures exclude data denoted as metagenomic and whole genome sequencing (WGS). If one considers metagenomics and WGS data to be chromosomal, then the proportion of chromosomal DGRs rises to 90% (336 of 372).
For chromosomal DGRs, it is highly challenging to judge whether or not they are phage-associated (i.e. within a prophage or inactivated remnant of a prophage). This is due to the lack of universal indicator genes for phages, the lack of conservation among phage genes and the large number of inactive phage remnants within genomes. Even when a phage is identified, the exact boundaries are often unclear, making it difficult to know if a DGR is within or merely adjacent to a prophage sequence.
To address this issue, we searched for homologs to known phage structural genes (e.g. tape measure protein, base plate protein) within the 20 kb segments containing the DGRs (‘Materials and Methods’ section). Using a conservative criterion, we considered DGRs to be clearly phage-associated only when homologs of phage structural genes are found on both sides of the DGR (E-value cutoff of e-20). By this criterion, only 9 of the 246 core DGRs are within phages; however, if a more relaxed criterion is used in which phage homologs are on only one side of the DGR, then 102 of 246 DGRs are within phages. An independent source of evidence comes from the fact that the target genes of 11 DGRs are homologs of the Bordetella phage tail protein Mtd, and hence those DGRs can be concluded to be phage-associated (below). By combining the three criteria (GenBank annotation, e-20 phage homologs, Mtd homologs), the number of DGRs that are phage-associated is conservatively estimated at 34 of 246 (14%; homologs on both sides) and less conservatively at 111 of 246 (45%; homolog on one side). There is essentially no information about whether CPR DGRs are associated with phages. One CPR DGR is annotated by GenBank as being within a free phage, but no conclusion can be made about the remainder, because few CPR genes show sequence similarities to any known genes, preventing the identification of phage homologs in flanking sequences.
In the end, we conclude that a large majority of DGRs are chromosomally encoded (∼70–90%), with >10% of the core set being phage-associated (either free phages, prophages or inactive remnants), and with a few DGRs on plasmids (2%; 6 DGRs). Of completed genome sequences that contain DGRs, most have only one DGR, although ∼10% contain multiple DGRs, with the most being five, for Syntrophobotulus glycolicus DSM 8271 (all are nearly identical copies and may be in prophages).
Components of DGRs
VR sequences
Five major classes of VR sequences
The VR sequences of target genes were divided into classes based on sequence alignments (Figure 2). Three VR classes correspond to C-type lectin folds (here denoted CLec1, CLec2 and CLec3), while two classes correspond to Ig fold proteins (Ig1 and Ig2). CLec1 is the most abundant class (25%; 94 of 372), and includes the prototypic Bordetella phage DGR. Six VRs of the CPR set correspond to a novel C-type lectin sub-type whose crystal structure has been determined (26) but they are left ungrouped as CLec proteins because of limited alignment with each other (Supplementary Figure S1 and Table S1). In addition, six clusters of alignable VRs were found among the CPR DGRs, and are denoted unknown VRs 1–6 (UVRs 1–6), because it cannot be concluded whether they are within a C-type lectin domain, an Ig domain or another domain. The remaining 45 VRs (all in the CPR set) were left ungrouped. WebLogo depictions of the major VR classes are in Figure 2, while UVR1–6 and CLec depictions are in Supplementary Figure S1; all VR aa alignments are available in Supplementary Data 1.
Figure 2.
Major classes of VR sequences. Five major classes of VR sequences are shown in WebLogo format, and were generated from the VR alignments in Supplementary Data 1. Under each profile, the regions corresponding to aa variability are indicated by a black bar. Additional minor classes are shown in Supplementary Figure S1.
Interestingly, the initial attempts to form groups from the VR sequences failed to recognize that CLec2 and CLec3 VR’s belong to C-type lectin motifs, due to lack of alignability with the VR of the Bordetella phage DGR, or to any other protein motifs. Similarly, Ig1 and Ig2 VR sequences were not clearly identifiable as parts of Ig folds. Domain identities were eventually assigned after the target genes’ protein sequences were queried using the Phyre2 web server (27), which predicts protein folds ab initio based on sensitive homology comparisons with known three-dimensional protein structures. The predicted structural similarities among the CLec or Ig domains of the target proteins provide a satisfying commonality despite the highly diverse sequences of target genes and VR sequences. However, C-type lectin and Ig folds are unrelated structurally, and so DGRs must have acquired target genes on at least two occasions.
A feature of each VR class is that both the start and end of the VR sequence has 3–7 aa that are constant within the class and are not generally subject to A-to-N mutagenesis, whereas the internal sequence is variable and corresponds to the region of A-to-N mutagenesis (Figure 2). The central region still possesses a few conserved aa positions, which probably correspond to the protein’s structural scaffold that presents the diversified surface residues (6). The 3′ constant aa are presumably encoded by the IMH.
The position of VRs in the target genes
The great majority of VR sequences are located at the C-termini of their target genes, as seen for DGRs of the Bordetella phage, Legionella and Treponema. There are exceptions, however, with 9 (of 34) Ig1 DGRs, 9 (of 10) Ig2 DGRs and one CLec2 DGR having VR sequences internal to the target protein genes. In addition, one CLec1 target gene has three VRs, of which two are internal, and all three appear to be subject to mutagenesis (Supplementary Table S1). Three DGRs have a VR that is diversified at its N-terminus (Supplementary Table S1). Mechanistically, the internal and N-terminal VR sites are notable because the IMH for each VR must overlap with and be constrained by the proteins’ coding sequences.
Multiple target genes
Approximately 15% of DGRs have more than one target gene within the cassette, the most common being two targets (50 of 372, 13%). The greatest number of targets are found for the DGR in Stenotrophomonas sp., which has eight target genes arranged in tandem. However, the DNA sequence assembly might contain errors because there are only five unique VR sequences (Supplementary Table S1).
Non-adjacent target genes
Because the Treponema DGR has multiple remote targets (13), we screened genomes and sequence contigs for matches to TRs that might be non-adjacent targets. Non-adjacent target sites were assigned when there were only A-to-N differences with the TR sequence (allowing minor exceptions), and if the targets were not part of another DGR.
In the end, 45 DGRs (of 372) were assigned as having 73 putative target genes at non-adjacent target sites (Supplementary Table S1). Distances ranged from a few kbs to millions of bps away from the RT gene. (It should be noted that it is fairly arbitrary to assign a gene as non-adjacent when the gene is <10 kb from a DGR; there is a continuum of distances with no obvious cutoff.) Ten DGRs had 29 target genes that were >100 kb away. The greatest number of non-adjacent targets is seven, for the Treponema DGR.
It is expected that remote target genes (>100 kb away) will not be found for phage-associated DGRs, because remote target genes would not be inherited along with the phage upon phage exit from the cell. Indeed, all but one of the DGRs with remote target genes have no evidence for being phage-associated.
Considering the DGRs that have evidence for phage-association, there are many examples of non-adjacent target genes and in nearly all cases the targets are located within 50 kb of the RT gene (Supplementary Table S1), suggesting that the targets are within the same prophage, with one target adjacent to other DGR genes and one not. Interestingly, two putatively phage-associated DGRs have no adjacent target genes and only one non-adjacent target located 20 kb away.
Together, these examples demonstrate that DGRs are capable of acting on target genes that are not directly adjacent to other DGR genes, including at sites distant in the genome. Indeed it has been shown experimentally that the avd, TR and brt genes can be expressed in trans on a plasmid, which is consistent with the ability of DGRs to act on remote genomic sites (9).
Target gene domain compositions
Target genes vary dramatically in size, from 130 to 8091 bp (44–2697 aa) (full sequences in Supplementary Data 2). The smallest target proteins consist of Ig or C-type lectin folds alone, while larger proteins contain many domains appended to the Ig or C-type lectin domain. Domain compositions were systematically examined for motifs using the CDD of NCBI and the Pfam databases (‘Materials and Methods’ section). A total of 39 variant compositions were distinguished that include 27 protein motifs defined by the CDD and Pfam databases (Figure 3; Supplementary Tables S1 and 3). Approximately a quarter of target genes have no defined motifs outside the diversified domains, but have sizeable ‘extensions’ that presumably hold functions corresponding to undefined motifs (Figure 3). Given the diversity in target genes, the domain compositions were grouped into a hierarchy of categories, divided on the first level by the VR-containing motif.
Figure 3.
Protein domain structures of target genes. Schematics are shown for 39 domain variations of target genes, with one example of each (drawn to scale). Domain compositions are grouped by similarity; they are referred to as categories ‘a’ to ‘h’ in the text, and are displayed as (A-H) in the figure. Parentheses indicate the number of DGRs for each variation. The domains present are named according to the abbreviations used by CDD; a listing of the names of domains and their descriptions is in Supplementary Table S3. Asterisks over domains indicate the positions of diversification by mutagenic retrohoming. The abbreviation ‘(ext)’ indicates an extension of >250 aa with no identified motif. Codes for the domains are shown to the right, and correspond to abbreviations used in Supplementary Table S1.
Category ‘a’. Approximately 3% of target genes (12 of 372) have VR sequences within a putative Mtd domain, which is exemplified by the Mtd tail protein of the Bordetella BPP-1 phage (14,15) (Figure 3A). All of these proteins have CLec1 VR sequences, and are similar in size to the Bordetella Mtd, except for one that has an N-terminal extension containing a motif for a phage tail-collar fiber protein (Figure 3A). All but one of these are flanked by identifiable phage gene homologs, indicating that their DGRs act to diversify phage tail proteins, similar to the Bordetella phage DGR.
Category ‘b’. The largest category of target genes has CLec1 VR sequences at their C-termini, with the C-type lectin domain being a portion of a larger FGE–sulfatase domain. These account for 19% (71 of 372) of the DGRs, and are exemplified by the Treponema DGR protein TvpA (13). Over half of these target proteins have no additional appended motifs (52 of 71), although some have extensions of unknown function on the N-terminus (16 of 71). One target gene has three FGE–sulfatase domains in tandem, all of which appear to become diversified during mutagenic retrohoming.
The FGE–sulfatase-containing target genes show a remarkable variety of motifs appended to the C-type lectin domain, with 13 distinguishable domain compositions (Figure 3B). Most are represented by only one or a few examples, consistent with the idea that GenBank sequence data do not saturate the types of DGRs found in nature. In most cases, the functions of the target proteins are not obvious even when motifs are identified. For example, two protein domain compositions have protease-related domains (b4 and b5); one has a kinase domain (b12); a set of nine DGRs has different combinations of domains for Toll-like receptor, nucleoside-triphosphatase (NTPase), metallophosphatase and a dimerization domain (b7, b8, b9, b10 and b11). One unusual DGR has a pair of target genes, the first encoding 400 aa and containing IF2_N and FGE–sulfatase domains and the second located several kbs away and encoding 1247 aa with WD40 and FGE–sulfatase domains (Figure 3B and Supplementary Table S1), which suggests a coordinated biochemical process involving the two proteins. A third remote target in the same genome is short and consists of only an FGE–sulfatase domain.
In categorizing the protein motifs, most matches gave E-values of moderate significance (1e-5 to 1e-30), which is sufficient to indicate a structural motif but not to discern a biological function. However, ∼7 of the FGE–sulfatase motif matches gave E-values of high significance to GldJ and GldK motifs within the family of FGE–sulfatase proteins (as low as e-70) (Supplementary Table S1). Proteins with GldJ and GldK motifs have been characterized as being lipoproteins involved in surface gliding motility in the Bacteroidetes phylum (14). The DGRs with the GldJ and GldK matches are mostly found in Cyanobacteria rather than Bacteroidetes, but the highly significant scores suggest that the cyanobacterial DGRs may be involved in gliding motility or a related phenotype.
Category ‘c’. Three CLec1 target proteins do not give matches to domain motifs such as Mtd or FGE–sulfatase, nor did the Phyre2 web server predict a C-type lectin domain. These proteins are still considered to be C-type lectin proteins because their VR sequences align with other CLec1 VR’s. Since their target protein domains cannot be assigned to the Mtd or FGE–sulfatase categories, they are provisionally considered to be a separate category.
Category ‘d’. A fourth class corresponds to VR sequences that are detected by NCBI as overlapping DUF1566, DUF823 and/or Fib_succ_major motifs. This group accounts for 14% (51) of the DGRs and includes all CLec2 VRs. It is exemplified by the Legionella DGR. Matches to DUF1566, DUF823 and Fib_succ_major motifs are combined into one category because the matches were weak and mixed, and because all of the VR sequences align well with each other, suggesting a common protein structure (Figure 2 and Supplementary Data 1).
There is little functional information available for the motifs DUF1566, DUF823 and Fib_succ_major, except that Fib_succ_major proteins often have a lipoprotein signal and conserved cysteine residues that suggest extracellular disulfide bonds (14). The LdtA target protein of the Legionella DGR is consistent with the above features because it is a surface lipoprotein anchored to the outer leaflet of the outer membrane, with its diversified C-terminal sequence exposed (12).
About half of category ‘d’ target proteins are short and consist of the DUF1566 domain with no other motifs, while the other half have a variety of extensions and appended domains, including motifs for cellular adhesion or surface proteins such as the Bacteroides fragilis domain (BF2867_like_N), bacterial Ig-like domains 2 and 3 (Big_2 and Big_3) and major fimbrial subunit protein (P_gingi_FimA). One novel target has six bacterial Ig domains upsteam of the VR-containing C-type lectin domain, although none of the Ig domains are subject to A-to-N mutagenesis.
Category ‘e’. Interestingly, CDD and Pfam databases did not definitively detect the Ig motifs in the target genes. Most Ig1 and Ig2 target proteins were predicted to have no motifs, while three target genes had very weak matches to the eukaryotic FN3 domain (fibronectin type 3 domain), which belongs to the Ig superfamily. As described above, the Ig domain motifs were ultimately assigned because of Phyre2 structural predictions, which identified 3 (of 36) Ig1 and 5 (of 9) Ig2 VR sequences as being part of Ig domains. Given the relatively modest support for the motif, confirmation of the Ig fold awaits a detailed structural characterization.
Category ‘f’. Category ‘f’ corresponds to VRs of the CLec3 category. A set of 13 category f1 DGRs is notable because the target proteins are very long (∼2000 aa) and align well across their entire lengths, indicating closely related functions, while their matches to several protein motifs are weak and mixed. The primary motifs identified are CotH (spore coat protein H) and Laminin_G_3 (Concanavalin A-like lectin/glucanases superfamily). Further PSI-BLAST searches gave a few matches to genes annotated as phage head-tail adaptor proteins (not shown), which suggests a function of the DGR within phages. Consistent with this conclusion, seven of the thirteen category f1 DGRs had an identifiable homolog of a phage structural gene in the 10 kbs segments flanking each side of the RT gene (Supplementary Table S1).
Categories ‘g’ and ‘h'. As previously mentioned, only a small proportion of genes in CPR organisms show sequence similarities to characterized genes or known protein motifs. Consequently, there is little information about the functions of the target genes in the CPR subset. Only four motifs were found among 10 of the 126 CPR DGRs, and these were put into two categories, with the remaining left uncategorized (Supplementary Table S1).
Category ‘g’ consists of target genes that have a protein motif (either P-loop_NTPase, PcfJ-like protein or nucleotidyl transferase superfamily) upstream of the VR sequence. The P-loop NTPase subset (category g1) resembles categories b7 and b10 (Figure 3); however, they are not grouped together because the VR cannot be concluded to be in an FGE–sulfatase or C-type lectin domain. Category ‘h’ consists of a very unusual target protein consisting of a eukaryotic/archaeal type S3 ribosomal protein domain, with diversification occurring upstream at the N-terminus (Figure 3).
In an additional effort to identify functions for target genes in the CPR set, PSI-BLAST searches were done for target genes with UVRs 1–6. As a group, target genes of UVR1 showed matches to a domain of unknown function, DUF1127, while target genes of UVR4 showed matches to the Midasin domain. The Midasin domain is an AAA–Adenosinetriphosphatase (ATPase) motif related to the P-loop NTPase motif (which belongs to the AAA_16 family found in category b) (Figure 3 and Supplementary Table S3). Indeed, four of the UVR4-containing genes had been identified as P-loop_NTPase motifs by CCD and Pfam. Genes with UVRs 2, 3, 5, 6 showed no matches with any proteins having assigned motifs or functions. Despite the protein motifs identified for UVR1 and UVR4 target proteins, we leave them as unclassified VRs because it is not clear whether they contain a C-terminal C-type lectin fold, an Ig fold or another fold.
In summary, there is remarkable diversity in the protein domain structures of the target genes of DGRs, which suggests a large number of potential biological functions. A common theme is that the most commonly appended domains are related to functions in protein–protein binding, ligand binding or surface display.
TR and VR sequences
TR sequences
TR sequences are usually located directly adjacent to the RT genes, and most often upstream, with the portion corresponding to A-to-N mutagenesis ranging from ∼30–100 bp. On both sides of the mutagenic region are GC-rich sequences, ∼5–30 bp long on the 5′ side and 20–40 bp on the 3′ side, with both sequences repeated in the TR and VR. The two GC-rich sequences are generally not subject to sequence diversification, and they correspond to the ‘constant’ aa sequences that flank the sequences subject to mutagenesis (Figure 2). A sampling of TR-VR alignments is in Figure 4 and TR-VR alignments for all DGRs are in Supplementary Data 3, which allows detailed inspection of the mutagenesis properties of individual DGRs.
Figure 4.
Example TR and VR sequence alignments. Alignments are shown for the TR and VR DNA sequences and the aa sequence of the VR. The TR is not translated in vivo, but the aa corresponding to its unmutated sequence are shown for comparison with the VR sequences. The capitalized DNA sequences correspond to positions alignable between the TR and VR, while the lower-case DNA sequences are not alignable. Red DNA residues are TR-VR sequence differences consistent with A-to-N mutagenesis; green residues denote other differences compared to the TR. Yellow shading denotes aa in the VR that result from A-to-N mutations, while green shading shows other aa differences. The IMH sequences in VRs are indicated with purple shading, IMH* by blue shading and the inverted repeat by orange shading. (A) The Bordetella phage DGR. (B) A DGR with three target genes, each being diversified through mutagenic retrohoming. (C) An example of an indel in a VR sequence that is opposite AAC in the TR. (D) An extreme example of length difference between TR and VR. (E) An example of non-A-to-N substitutions and a frame shift.
As previously observed, the A residues in the TR most often correspond to the first and second codon positions, which maximizes aa recoding (3,6) and this is seen across all of the DGR subtypes. The trinucleotide AAC is exceptionally common, often occurring in tandem repeats (Figure 4); the AAC template sequence can produce 15 different aa after mutagenic retrohoming. Based on TR-VR alignments (Supplementary Data 3), the range of theoretical aa diversity generated for various DGRs varies widely from 105 (LCDE01000016.1_1809_2849) to 1030 (NZ_AHHG01000042.1_1256_2469).
Mutagenic patterns inferred from TR and VR sequences
The majority of DGRs (65%; 243 of 372) exhibit exclusively the canonical pattern of A-to-N substitutions between TR and VR sequences (Supplementary Table S1). This confirms the general property of A-to-N mutagenesis, but it also leaves numerous examples of apparent noncanonical mutagenesis. Noncanonical mutagenesis can be categorized as either substitutions between the TR and VR that are not A-to-N differences, or differences in length between the TR and VR (indels). Non-A-to-N substitutions are the most common, being found in 23% of DGRs in at least one target gene (86 of 372 DGRs). Twenty-four DGRs have more than one non-A-to-N difference, with the highest number in a single target gene being 11 (Supplementary Table S1). Mechanistically, the non-A-to-N differences might be rationalized as being due to either atypical RT incorporation during reverse transcription, or mutations arising during cDNA integration and resolution. It is also possible that some might be mutations that arise independently of mutagenic retrohoming.
Approximately 7% (25 of 372) of DGRs have TR and VR sequences differing in length (Figure 4C and D). The indels can be either multiples of 3 bp, which do not affect the reading frame, or other lengths that cause a frame shift. Indels of multiples of 3 bp are more common (14 of 25 DGRs) while the other 11 examples have frame shifts that cause even greater levels of aa mutagenesis (Figure 4D and E; Supplementary Table S1 and Data 3). Interestingly the indels are almost always adjacent to AAC sequences in the TR (74% of the indels), suggesting template slippage during reverse transcription of the AAC repeats. It is plausible that an RT might go out of register when polymerizing through AAC repeats, because two out of three incorporations might be mismatches.
Also notable is the fact that when DGRs have more than one target gene, the mutagenesis pattern is often different for the targets. For example, in the DGR ANKO01000117.1_3531_4562, one target has seven non-A-to-N differences and one indel, while the other has only A-to-N differences. This implies that rather than being errors, noncanonical mutagenesis may be an additional mutagenic capability of DGRs to generate more sequence diversity, at least for some DGRs.
IMH, IMH* and GC-rich inverted repeats
Putative IMH and IMH* sequences are detected readily for nearly all DGRs. They are marked by imperfect TR-VR repeat sequences located after the region of A-to-N differences, and they usually begin with a GC-rich segment, as in the Boredetella DGR (Figure 4). In some cases the predicted IMH and IMH* sequences are identical, and recognition of IMH would have to extend downstream to distinguish it from the IMH* sequence. The fact that IMH and IMH* sequences can be predicted nearly universally predicts that the same IMH-dependent mechanism is used across DGRs.
GC-rich inverted repeats (of high quality confidence; see ‘Materials and Methods’ section) located downstream of IMH were found for a minority of DGRs (34%; 127 of 372). This suggests that the repeat is not a universal feature of DGRs, even though it is essential for efficient mutagenic retrohoming for the Bordetella and Legionella DGRs (9,12) (Supplementary Table S1).
Because inverted repeats are a characteristic of intrinsic terminator motifs, we considered the possibility that the repeats might have a transcriptional terminator function. When the sequences downstream of IMH were screened for terminator motifs, looking either for a tract of T’s downstream of the inverted repeats or using the web server ARNold (http://rna.igmors.u-psud.fr/toolbox/arnold/), only a minority of DGRs had evidence for terminators following IMH. This suggests that some DGRs have a terminator after the target genes, but most DGRs do not.
Boundaries of mutagenesis
From the TR-VR alignment data (Supplementary Data 3) it can be predicted for all DGRs that the 3′ boundary of A-to-N mutagenesis occurs directly upstream of the putative IMH sequences, consistent with reverse transcription initiating in the corresponding region of the TR. The 5′ boundary of mutagenesis, on the other hand, is not at a fixed position across DGRs. When there are multiple target genes within a single DGR, they sometimes have different boundaries. This agrees with experimental data for the Bordetella phage DGR, which indicates that the 5′ boundary is governed by sequence homology between the TR and VR (1). The expanded data set here suggests that the property is shared across DGRs.
RT genes
RTs of DGRs comprise a distinct subclass of reverse transcriptases related to those of group II introns, retrons and non-LTR elements (8,21,28). The DGR RTs range in size from ∼300–500 aa, and contain RT motifs 1–7, which correspond to the palm and finger domains of other polymerases (Supplementary Data 4). DGR RT’s contain motif 2a, located between motifs 2 and 3, which is found among group II introns, non-LTR retroelements and retrons, but not among other RTs such as retroviral or telomerase RTs (29). DGR RTs do not contain motif 0, which is upstream of motif 1 in group II introns and non-LTR elements (30,31), but they do share alignment with the thumb domain of group II intron RTs. Because thumb domain motifs do not generally align across RT types, this supports a close relationship between DGRs and group II introns as was predicted previously (8).
Approximately 20% of DGR RTs have an extension of up to ∼150 aa downstream of the thumb domain. Searches for protein motifs in the C-terminal extensions (as well as N-terminal sequences upstream of domain 1) did not reveal any RNase H or nuclease domains like those present in some other RT types. However, three related RTs showed weak sequence identity in their C-terminal extensions to a MutS_I motif (Supplementary Table S1 and Data 4). The MutS_I motif is the DNA-mismatch-binding domain of the mismatch repair protein MutS. The MutS gene was previously shown not to be required for mutagenic retrohoming of the Bordetella phage DGR (9). Still, the mismatch repair system might be involved in some way for other DGRs, especially because a newly identified accessory protein family is related to MutS (below).
Accessory genes: four classes
The majority of DGRs have homologs to avd (275 of 372; 74%), with one DGR having two avd genes (Supplementary Table S1). The avd genes are very poorly conserved but of similar length (Supplementary Data 5). Consistent with a role in nucleic acid binding (11), the proteins are basic with the average of calculated pI’s being 9.5 ± 0.7. We considered the possibility that DGR cassettes lacking avd may rely on an avd gene encoded elsewhere in the genome. A profile-based search for avd homologs failed to find such genes, suggesting that there is a mechanism of Avd-independent mutagenic homing in some organisms that differs from the mechanism of the Bordetella phage DGR.
Four percent (14 of 372) of DGRs in the collection have HRDC genes, with one DGR having two HRDC genes. About half of the HRDC-containing DGRs contain avd genes as well. HRDC proteins are predicted to bind nucleic acids (14,15), but unlike Avd proteins they are not basic, as the average of their calculated pI is 6.9 ± 1.3.
Two new accessory genes were identified, called MSL and CH1 (MutS-like and conserved hypothetical gene 1), which are found in 16 and 14 DGRs, respectively (Supplementary Table S1 and Data 5). MSL proteins are small (112–283 aa) with neutral charge (pIs of 7.2 ± 1.7), and one is present in the Legionella DGR. Three MSL proteins are annotated by GenBank as MutS homologs, while 11 of 16 were given strong to moderate evidence of being homologs to MutS by the Phyre2 server (not shown). DGRs that contain MSL genes also contain avd genes; however, none also contain HRDC genes (Supplementary Table S1).
CH1 proteins are small (160–169 aa) and acidic, with average calculated pI’s of 4.7 ± 0.2 and no identifiable motifs. Supporting evidence that CH1 is a functional DGR component comes from the fact that they are conserved as flanking genes among a set of 12 duplicate copies of the DGR ABQC02000022.1_61713_62543 (Supplementary Figure S3). CH1 genes are found only in DGRs that do not contain avd, HRDC or MSL genes.
The significance of other potential accessory genes is tenuous. Four potential accessory genes (denoted PAG1–4) were identified that are shared among only 3 or 4 DGRs (Supplementary Table S1 and Data 5). However, being shared among such a small number is weak evidence for DGR function because, for example, such a gene might be a phage gene conserved among related phages. Supplementary Table S3 lists the genes sandwiched between DGR genes, as well as those between DGRs and non-adjacent target genes that are located within 10 kb of the DGR’s RT. Most genes are hypothetical and the few genes with identifications do not provide obvious insights. One interesting observation, however, is that five CPR DGRs contain ribosomal protein genes between DGR genes, which evokes similarities with the CPR DGR with a ribosomal protein-related gene as the target.
Architectural variations of the DGR cassettes
Architectures of DGR genes were grouped hierarchically based first on the order of the TR, RT and accessory genes (if present), and second, on the number, order and orientation of the target genes. Five major families of architectures were established (A, B, C, D and E) (Figure 5; Supplementary Figure S3 and Table S1), of which Architectures A and B account for 88% of the DGRs. Each major family has subgroupings, and there is a total of 48 architectural variations. Many architectures have only one or two examples, again suggesting that the DGR sequence coverage in GenBank is underrepresented for some varieties of DGRs present in nature.
Figure 5.
Major architectures of genes in DGR cassettes. Architecture A1 has a core organization of TR-RT with different positions of the target gene (only one is shown). Architectures B1-B3 have core organizations of avd-TR-RT, avd-RT-TR and TR-(avd)-RT, respectively, with different arrangements of target genes. DGRs of Architectures C-E contain HRDC, MSL and CH1 accessory genes, respectively. Only one example is shown for Architectures A, C and E (A1, C1, E1); however, additional examples are in Supplementary Figure 3. Genes in parentheses indicate the reverse strand orientation of the gene. Numbers in parentheses indicatethe count of examples in the data set (Supplementary Table S1).
The simplest architecture, Architecture A, accounts for 29% (75 of 372) of DGRs and is distinguished by the lack of accessory genes (Figure 5). The core components for this class are TR-RT (for A1) or RT-TR (for A2), and there are further variations in the position and number of target genes, as well as spacing between components (Supplementary Figure S3). Architecture B has the core components of avd, TR and RT, with no additional accessory genes. It is the most common architecture, found for 68% (253 of 372) of DGRs and is exemplified by the Bordetella and Legionella DGRs. Subtypes of Architecture B (e.g. B1, B2 and B3) vary in the order and orientation of the avd and TR sequences as well as the number and order of target genes. Interestingly, the B3 architecture accounts for 90% of the CPR set and is nearly absent from the core set.
Architecture C is the family of DGRs containing HRDC genes, and is represented by the Treponema DGR. Among the 14 HRDC-containing DGRs, there are 11 architectural variations, which probably reflects poor representation of HRDC-containing DGRs in the dataset, rather than a propensity for architectural rearrangements. Architecture D is the family of DGRs that contain MSL genes, while Architecture E contains CH1 genes.
For most DGRs, genes in the cassette are all encoded on the same strand, allowing the potential for transcription in a single unit. However, some DGRs encode components on different strands, requiring at least two transcription units (Supplementary Table S1). Even the TR and RT genes can be transcribed from different strands, indicating that they do not have to be transcribed together to form the RNP complex needed for mutagenic retrohoming.
Another notable observation is that across all architectures there are examples of DGRs with open reading frames nested between DGR components, in most cases separating the target gene(s) from the core components. The genes do not appear to be accessory genes, or involved in DGR function, because they are not conserved among DGRs. They are indicated by an ‘X’ in Supplementary Table S1 (e.g. A1-X) and further information about them is in Supplementary Table S2.
Phylogeny and evolution of DGRs
In order to divide DGRs into groupings that reflect evolutionary lineages, phylogenetic analysis was performed on their RT sequences. While, it cannot be assumed that RT phylogeny exactly represents evolution of the entire DGR units, nevertheless the RT is the only component alignable across all DGRs. Hence an RT tree provides a starting point for dividing DGRs into formal groupings with shared characteristics.
Phylogenetic analysis was done for 372 RTs using maximum likelihood analysis (see ‘Materials and Methods’ section). Although some clades had strong support, there was poor resolution for the tree as a whole, making it impossible to place all RTs into defined groups (Figure 6). In particular, there was very little resolution for the CPR RTs. The trees presented in Figure 6 and in Supplementary Data 6 are not collapsed for unsupported nodes in order to make it easier to distinguish all DGRs, and also because some unsupported nodes share notable features (below). Nodes having significant support (>75% bootstrap) are indicated by black dots in figures, and other nodes should be considered as having weak to no support. We have chosen four subsets of DGRs to name as lineages (Lineages 1, 2, 3, 3a and 4), because their members share characteristics that appear to distinguish them from other DGRs (below).
Figure 6.
Phylogenetic tree of RTs and major VR classes. An unrooted maximum likelihood tree of RTs is shown with the VR class of each DGR indicated by color, and black dots indicating nodes with >75% bootstrap support. Colored arrows indicate the position of the Bordetella (red), Legionella (green) and Treponema (blue) DGRs. The four lineages identified in the text are shaded in gray, while the tan shading indicates DGRs from the CPR set (Supplementary Table S1). The order of taxa in the tree is the same order as in Supplementary Table S1, clockwise starting from the left boundary of the CPR DGRs.
The RT tree appears to be a good approximation of DGR evolution
Overall, there is good correspondence between the RT tree and major DGR features, which supports the notion that DGR genes largely coevolve as a cassette. These features include the VR class, target protein domain structures (e.g. a1 and b2), the presence of accessory genes, and the architecture of the DGR cassette (e.g. A and B).
Target genes: VR classes and protein domain structures
Of all DGR features, the VR classes correlate most directly with RT phylogeny (Figure 6). Classes CLec2, CLec3, CLec, Ig1 and Ig2 all form clusters that are consistent with the RT tree. While it is perhaps surprising that Ig1 and Ig2 VRs do not cluster with each other, the two classes are not also separated by supported nodes in the RT tree, leaving open the possibility that Ig1 and Ig2 are a single lineage that descended from a DGR that acquired the Ig domain. The CLec1 class, which is the largest, is found among several clades rather than being monophyletic, which might reflect an ancestral status. There is little clustering of UVR classes, which is consistent with the lack of phylogenetic resolution of CPR RTs due to the great sequence diversity (Supplementary Data 6A).
VRs that are internal or N-terminal within target genes correspond to clades on the RT tree (Supplementary Data 6B). Ig2-containing DGRs (Lineage 2) have internal VRs, as do about a quarter of Ig1-containing DGRs. Six additional DGRs scattered across the tree have either internal or N-terminal VR locations in their target genes, suggesting independent evolution of the feature.
Protein domain structures of target genes likewise correspond with the RT tree. All high-level domain structures (e.g., ‘a’, ‘b’ and ‘c’) form clusters on the tree, either as single clades or several clades that are not separated by supported nodes (Supplementary Data 6C). Many domain subclasses (e.g. ‘a1’ and ‘a2’) are found in supported clades or unsupported clusters (not shown). That said, the sheer number of protein domain compositions (39 variants; Figure 3) indicates that the protein-domain structures evolve more rapidly than many other DGR features.
Accessory genes and DGR cassette architectures
Accessory genes form groupings that are mostly consistent with the RT tree. The avd gene is found in the majority of DGRs in the tree, perhaps because it was an ancestral accessory gene; however, Lineage 3 notably lacks avd genes, suggesting that these DGRs evolved a distinct variation of mutagenic retrohoming that does not require Avd (Supplementary Data 6D). In addition, the accessory genes HRDC, MSL and CH1 are mostly found within clades in the RT tree, although imperfectly in some cases (Supplementary Data 6E).
With regard to gene cassette architectures, there is considerable agreement between the RT tree and the high-level architectures (e.g. A, B and C) as well as variations of architectures (e.g. A1, B1 and B2) (Supplementary Data 6F and Table S1). For example, Architecture C is only found among a clade that contains the Treponema DGR, and Architecture E1 is only found among Lineage 3a DGRs. Taking into account the resolution of the tree, it appears that DGR cassette architectures as defined in Figure 5 and Supplementary Figure S3 are inherited fairly stably over time. Again, it should be noted that the number of cassette architectures of DGRs (48) indicates substantial plasticity in gene organizations, although much of it is due to target gene variations, which appear to change comparatively rapidly (below).
DGR features that correspond less well with the RT tree and undergo more frequent reassortment and change
Location of DGRs on chromosomes, plasmids and phages
There is little relation between the RT tree and the location of DGRs on chromosomes, plasmids or free phage genomes, indicating that DGRs are readily transferred between those DNAs (Supplementary Data 6G). However, the presence of DGRs in prophages deserves further scrutiny. As described previously, it is difficult to identify prophage-associated DGRs (i.e. DGRs in a prophage or fragment of a prophage). We concluded conservatively that 14% of the DGRs are phage-associated, and less conservatively that 45% might be phage-associated (Supplementary Table S1). When plotted onto the RT tree, the conservatively assigned phage-associated DGRs are concentrated in clusters near the Bordetella phage DGR, while the less conservatively assigned are concentrated in Lineages 1, 2 and 3 (Supplementary Data 6H). In contrast, the DGRs with the least evidence for phage association are concentrated in Lineage 4 and neighboring DGRs, including the Treponema and Legionella DGRs. Overall, this pattern suggests that some lineages of DGRs are adapted to perform a phage function, and other lineages are adapted to serve a cellular function.
Target gene numbers and arrangements
Although the VR classes correspond closely with the RT tree, the number of target genes, their positions, and the protein domains appended to the VR domain appear to change relatively rapidly. For example, the number of target genes varies even among closely related DGRs (Supplementary Data 6I), indicating that this feature is not highly stable. That said, Lineages 1, 2, 3 and CPR DGRs tend to have a single target gene, while DGRs near Lineage 4 in the tree (i.e. probably not phage-associated) tend to have multiple genes.
Remote and non-adjacent target genes
Non-adjacent target genes are found across the tree, indicating that non-adjacent targets are utilized by diverse DGRs (Supplementary Data 6J). However, remote target genes (>100 kb away from the DGR RT) are only found among Lineage 4 DGRs and neighbors on the tree, again suggesting that remote targets may be used only for a subset of DGRs that are not phage associated. As noted above, this is logical because remote cellular targets would not be inherited along with phages.
TR-VR mutational patterns
Most mutational characteristics do not cluster in the tree, indicating that they are not specialized properties but are characteristics of many DGRs. For example, the number of predicted mutagenic A’s in the TR template ranges widely, as does the number of DNA and aa differences in TR-VR alignment pairs (Supplementary Data 6K–M). In general, there are not great differences across the tree in mutagenic potential, although the DGRs near the Legionella DGR in the tree have somewhat higher mutagenic potential which might be an adapted characteristic among a set of related DGRs.
Non-A-to-N substitutions also occur throughout the tree, but are less common among Lineages 1, 2, 3 and nearby DGRs, and are more frequent among Lineage 4 and nearby DGRs (Supplementary Data 6N). Consistent with this, Lineages 1, 2 and 3 and nearby DGRs usually have one or no non-A-to-N substitution, whereas the Lineage 4-related and CPR DGRs frequently have multiple non-A-to-N substitutions, suggesting less fidelity in reverse transcription (Supplementary Data 6N). Interestingly, the Lineage 4-related DGRs include nearly all of the examples of indels in multiples of 3 bp (not causing frame shifts) (Supplementary Data 6O), whereas indels causing frameshifts occur among Lineage 4-related DGRs as well as other DGRs across the tree (Supplementary Data 6P). Together, the data suggest differences in mutagenic potential among DGR groups, including differences in non-canonical mutagenesis properties, with some DGRs having greater adherence to the canonical A-to-N mutagenesis mechanism.
In summary, DGRs occur in lineages with features that appear to mostly correspond with the RT tree. The features that appear most constant over time are the VR class, accessory genes, cassette gene organization and to a lesser extent the target protein domain composition. The characteristics of DGRs that appear to change most rapidly are the location of DGRs on chromosomes, plasmids and phages, and the number and organization of target genes.
Horizontal and verical inheritance of DGRs
There is abundant evidence for lateral inheritance of DGRs, as might be expected of an element found in plasmids and phages. Evidence for horizontal transfer includes the sporadic occurrence of DGRs in strains of a species, atypical GC content relative to the host genome, the existence of nearly identical DGRs in different species, and discordance between RT phylogeny and species phylogeny.
Sporadic presence in strains is exemplified by the Legionella DGR, where a DGR is found in the sequenced genome of L. pneumophila strain Corby, but not in the sequenced strains Philadelphia, Paris and Lens (32). With regard to GC content, roughly 20% of DGRs are suggested to have moved horizontally relatively recently because the DGRs’ GC content deviates by ≥5% from the host GC content (Supplementary Figure S4). DGRs with similar GC content may also have been acquired horizontally, but may have been in the host long enough to take on the host’s GC content.
In a third type of example, NC_010113.1_49890_51157 from Vibrio sp. 0908 is 99% identical to a DGR in Alteromonas macleodii, while the 16S rRNAs from the two organisms share only 86% identity. In the fourth type of example, phylogenetic trees show that DGRs from different lineages are found in the same species or genus (Supplementary Table S1). When phylum data are plotted onto the RT tree it can be seen that DGRs of Firmicutes, Bacteroidetes and Proteobacteria are present in different lineages in the tree, reflecting independent introductions of DGRs into these bacterial phyla (Supplementary Data 6Q). Similar patterns are seen when Class, Order or Family data are plotted onto the RT tree (not shown).
Conversely, there is evidence for a level of vertical inheritance for DGRs, because some lineages of DGRs appear to be present mainly in the same species or related species, indicating mostly vertical inheritance, and/or horizontal inheritance only among related organisms. For example, Lineage 4 is only found in cyanobacteria, and Lineage 3A is only present in Bacteroides species and relatives. This pattern could be due to either dependence of these DGR on the genetic environment of the hosts, or the kinetics of horizontal transfer among different microbes.
CONCLUSION
DGRs are a unique class of genetic elements that evolved from selfish retroelements to become useful genetic elements within their phage, bacterial and archaeal hosts. While only the Bordetella and Legionella DGRs have been characterized experimentally in detail, there is a plethora of DGRs in nature that have remarkably varied structures and target genes, and have the potential to participate in many undiscovered biological functions. The range of biological functions performed by DGRs has only begun to be explored.
On a practical level, the action of DGRs provides a powerful system for use in biotechnology. Mutagenic retrohoming produces sequence variability exceeding that of any known biological system. While the mammalian humoral immune system can generate up to 1016 nucleotide sequence variations within the hypervariable region of its antibody scaffold (33,34), DGRs have the theoretical potential to generate 1030 protein variants. Defining the full natural scope of DGR variants and their mutagenic properties will allow maximal utilization of DGR mechanisms for protein engineering and biotechnological applications.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Santa Naorem for comments on the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Natural Sciences and Engineering Research Council of Canada (NSERC) [RGPIN/203717–2012, RGPIN/05871–2017 to S.Z.]; National Institutes of Health [R01 AI096838 to J.F.M., P.G.]; National Science Foundation [MCB1413158 to M.P.]; University of Missouri Startup Fund (to H.G). Funding for open access charge: NSERC Grant RGPIN/05871–2017 (to S.Z.).
Conflict of interest statement. J.F.M. is a cofounder, equity holder and chair of the scientific advisory board of AvidBiotics Inc., a biotherapeutics company in San Francisco.
REFERENCES
- 1. Guo H., Arambula D., Ghosh P., Miller J.F.. Diversity-generating retroelements in phage and bacterial genomes. Microbiol. Spectr. 2014; 2:doi:10.1128/microbiolspec.MDNA3-0029-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Medhekar B., Miller J.F.. Diversity-generating retroelements. Curr. Opin. Microbiol. 2007; 10:388–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Liu M., Deora R., Doulatov S.R., Gingery M., Eiserling F.A., Preston A., Maskell D.J., Simons R.W., Cotter P.A., Parkhill J. et al. Reverse transcriptase-mediated tropism switching in Bordetella bacteriophage. Science. 2002; 295:2091–2094. [DOI] [PubMed] [Google Scholar]
- 4. Mattoo S., Foreman-Wykert A.K., Cotter P.A., Miller J.F.. Mechanisms of Bordetella pathogenesis. Front. Biosci. 2001; 6:E168–E186. [DOI] [PubMed] [Google Scholar]
- 5. Dai W., Hodes A., Hui W.H., Gingery M., Miller J.F., Zhou Z.H.. Three-dimensional structure of tropism-switching Bordetella bacteriophage. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:4347–4352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. McMahon S.A., Miller J.L., Lawton J.A., Kerkow D.E., Hodes A., Marti-Renom M.A., Doulatov S., Narayanan E., Sali A., Miller J.F. et al. The C-type lectin fold as an evolutionary solution for massive sequence variation. Nat. Struct. Mol. Biol. 2005; 12:886–892. [DOI] [PubMed] [Google Scholar]
- 7. Miller J.L., Le Coq J., Hodes A., Barbalat R., Miller J.F., Ghosh P.. Selective ligand recognition by a diversity-generating retroelement variable protein. PLoS Biol. 2008; 6:e131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Doulatov S., Hodes A., Dai L., Mandhana N., Liu M., Deora R., Simons R.W., Zimmerly S., Miller J.F.. Tropism switching in Bordetella bacteriophage defines a family of diversity-generating retroelements. Nature. 2004; 431:476–481. [DOI] [PubMed] [Google Scholar]
- 9. Guo H., Tse L.V., Barbalat R., Sivaamnuaiphorn S., Xu M., Doulatov S., Miller J.F.. Diversity-generating retroelement homing regenerates target sequences for repeated rounds of codon rewriting and protein diversification. Mol. Cell. 2008; 31:813–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Guo H., Tse L.V., Nieh A.W., Czornyj E., Williams S., Oukil S., Liu V.B., Miller J.F.. Target site recognition by a diversity-generating retroelement. PLoS Genet. 2011; 7:e1002414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Alayyoubi M., Guo H., Dey S., Golnazarian T., Brooks G.A., Rong A., Miller J.F., Ghosh P.. Structure of the essential diversity-generating retroelement protein bAvd and its functionally important interaction with reverse transcriptase. Structure. 2013; 21:266–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Arambula D., Wong W., Medhekar B.A., Guo H., Gingery M., Czornyj E., Liu M., Dey S., Ghosh P., Miller J.F.. Surface display of a massively variable lipoprotein by a Legionella diversity-generating retroelement. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:8212–8217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Le Coq J., Ghosh P.. Conservation of the C-type lectin fold for massive sequence variation in a Treponema diversity-generating retroelement. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:14649–14653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Marchler-Bauer A., Derbyshire M.K., Gonzales N.R., Lu S., Chitsaz F., Geer L.Y., Geer R.C., He J., Gwadz M., Hurwitz D.I. et al. CDD: NCBI's conserved domain database. Nucleic Acids Res. 2015; 43:D222–D226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Finn R.D., Bateman A., Clements J., Coggill P., Eberhardt R.Y., Eddy S.R., Heger A., Hetherington K., Holm L., Mistry J. et al. Pfam: the protein families database. Nucleic Acids Res. 2014; 42:D222–D230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Nimkulrat S., Lee H., Doak T.G., Ye Y.. Genomic and metagenomic analysis of diversity-generating retroelements associated with Treponema denticola. Front. Microbiol. 2016; 7:852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Minot S., Sinha R., Chen J., Li H., Keilbaugh S.A., Wu G.D., Lewis J.D., Bushman F.D.. The human gut virome: inter-individual variation and dynamic response to diet. Genome Res. 2011; 21:1616–1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Park J., Zhang Y., Buboltz A.M., Zhang X., Schuster S.C., Ahuja U., Liu M., Miller J.F., Sebaihia M., Bentley S.D. et al. Comparative genomics of the classical Bordetella subspecies: the evolution and exchange of virulence-associated diversity amongst closely related pathogens. BMC Genomics. 2012; 13:545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Schillinger T., Lisfi M., Chi J., Cullum J., Zingler N.. Analysis of a comprehensive dataset of diversity generating retroelements generated by the program DiGReF. BMC Genomics. 2012; 13:430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Schillinger T., Zingler N.. The low incidence of diversity-generating retroelements in sequenced genomes. Mob. Genet. Elements. 2012; 2:287–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Paul B.G., Bagby S.C., Czornyj E., Arambula D., Handa S., Sczyrba A., Ghosh P., Miller J.F., Valentine D.L.. Targeted diversity generation by intraterrestrial archaea and archaeal viruses. Nat. Commun. 2015; 6:6585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ye Y. Identification of diversity-generating retroelements in human microbiomes. Int. J. Mol. Sci. 2014; 15:14234–14246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Paul B.G., Burstein D., Castelle C.J., Handa S., Arambula D., Czornyj E., Thomas B.C., Ghosh P., Miller J.F., Banfield J.F. et al. Retroelement-guided protein diversification abounds in vast lineages of Bacteria and Archaea. Nat. Microbiol. 2017; 2:17045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Brown C.T., Hug L.A., Thomas B.C., Sharon I., Castelle C.J., Singh A., Wilkins M.J., Wrighton K.C., Williams K.H., Banfield J.F.. Unusual biology across a group comprising more than 15% of domain Bacteria. Nature. 2015; 523:208–211. [DOI] [PubMed] [Google Scholar]
- 25. Rinke C., Schwientek P., Sczyrba A., Ivanova N.N., Anderson I.J., Cheng J.F., Darling A., Malfatti S., Swan B.K., Gies E.A. et al. Insights into the phylogeny and coding potential of microbial dark matter. Nature. 2013; 499:431–437. [DOI] [PubMed] [Google Scholar]
- 26. Handa S., Paul B.G., Miller J.F., Valentine D.L., Ghosh P.. Conservation of the C-type lectin fold for accommodating massive sequence variation in archaeal diversity-generating retroelements. BMC Struct. Biol. 2016; 16:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kelley L.A., Mezulis S., Yates C.M., Wass M.N., Sternberg M.J.. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015; 10:845–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zimmerly S., Wu L.. An unexplored diversity of reverse transcriptases in bacteria. Microbiol. Spectr. 2015; 3:doi:10.1128/microbiolspec.MDNA3-0058-2014. [DOI] [PubMed] [Google Scholar]
- 29. Malik H.S., Burke W.D., Eickbush T.H.. The age and evolution of non-LTR retrotransposable elements. Mol. Biol. Evol. 1999; 16:793–805. [DOI] [PubMed] [Google Scholar]
- 30. Blocker F.J., Mohr G., Conlan L.H., Qi L., Belfort M., Lambowitz A.M.. Domain structure and three-dimensional model of a group II intron-encoded reverse transcriptase. RNA. 2005; 11:14–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Qu G., Kaushal P.S., Wang J., Shigematsu H., Piazza C.L., Agrawal R.K., Belfort M., Wang H.W.. Structure of a group II intron in complex with its reverse transcriptase. Nat. Struct. Mol. Biol. 2016; 23:549–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lautner M., Schunder E., Herrmann V., Heuner K.. Regulation, integrase-dependent excision, and horizontal transfer of genomic islands in Legionella pneumophila. J. Bacteriol. 2013; 195:1583–1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Alder M.N., Rogozin I.B., Iyer L.M., Glazko G.V., Cooper M.D., Pancer Z.. Diversity and function of adaptive immune receptors in a jawless vertebrate. Science. 2005; 310:1970–1973. [DOI] [PubMed] [Google Scholar]
- 34. Sidhu S.S., Li B., Chen Y., Fellouse F.A., Eigenbrot C., Fuh G.. Phage-displayed antibody libraries of synthetic heavy chain complementarity determining regions. J. Mol. Biol. 2004; 338:299–310. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.