Abstract
Besides controlling epithelial-to-mesenchymal transition (EMT) and cell invasion, the Snail1 transcriptional factor also provides cells with cancer stem cell features. Since telomere maintenance is essential for stemness, we have examined the control of telomere integrity by Snail1. Fluorescence in situ hybridization (FISH) analysis indicates that Snail1-depleted mouse mesenchymal stem cells (MSC) have both a dramatic increase of telomere alterations and shorter telomeres. Remarkably, Snail1-deficient MSC present higher levels of both telomerase activity and the long non-coding RNA called telomeric repeat-containing RNA (TERRA), an RNA that controls telomere integrity. Accordingly, Snail1 expression downregulates expression of the telomerase gene (TERT) as well as of TERRA 2q, 11q and 18q. TERRA and TERT are transiently downregulated during TGFβ-induced EMT in NMuMG cells, correlating with Snail1 expression. Global transcriptome analysis indicates that ectopic expression of TERRA affects the transcription of some genes induced during EMT, such as fibronectin, whereas that of TERT does not modify those genes. We propose that Snail1 repression of TERRA is required not only for telomere maintenance but also for the expression of a subset of mesenchymal genes.
INTRODUCTION
Telomeres are essential for genome integrity and protect chromosome ends from being recognized as DNA double-strand breaks (1). Telomere repeats are lost by attrition during DNA replication due to the end-replication problem, and critically short telomeres elicit a DNA damage–associated cell cycle arrest and replicative senescence (2–3). Mammalian telomeres consist of TTAGGG DNA tandem repeats and a six-subunit complex (TRF1, TRF2, TIN2, TPP1, POT1 and Rap1), referred to as shelterin, that regulates telomere length maintenance and protects the ends from being recognized as damaged DNA (4–5). Telomere structure resembles constitutive heterochromatin as it also contains the histone mark H3K9me3 and HP1 proteins, which control chromatin architecture and are also involved in telomere length maintenance and function (6–8). Alike heterochromatin, telomeres are transcribed into telomeric repeat-containing RNA (TERRA) molecules (9–10) that remain partly associated with telomeres and play crucial functions (11). Recent work has suggested that TERRA RNAs act as a scaffold, recruiting different factors to chromosome ends and controlling telomeric function in distinct ways. TERRA promotes H3K9 trimethylation to control heterochromatin formation (12–13), enhances recruitment of chromatin modifiers to damaged telomeres (14–15), contributes to telomere length regulation through the modulation of exonuclease 1 and telomerase activities and participates in capping chromosome ends (16–18). Recently, TERRA has been found in extracellular fractions, suggesting that it acts as a mechanism of communication between telomeres and innate immune signals in tissue and tumor microenvironments (19). TERRA is heterogeneous in size and is transcribed by RNA polymerase II (RNA pol II) (20) from CpG island-containing subtelomeric promoters, located on average 1 kb upstream of TTAGGG repeats (10). In mouse, the bulk of TERRA transcripts seems to be generated from a single subtelomere and associates in trans with the remaining telomeres (15).
Telomeres control tumor growth by limiting the number of cell divisions, even in the presence of oncogenic mutations. Short telomeres elicit permanent DNA damage responses that trigger cellular senescence, which is a powerful anticancer barrier that suppresses unlimited proliferation in many cells that lack telomerases (21). Therefore, stabilization of telomere length is a key requirement for transformed cells to achieve infinite proliferation potential during oncogenesis, and it is obtained by activation of the telomerase reverse transcriptase or the recombination-based ALT (alternative lengthening of telomeres) mechanism (22). Telomerase is a ribonucleoprotein with a catalytic subunit product of the TERT gene and a RNA template (TERC) required for reverse transcription (23). Although TERT expression can also be controlled through differential splicing (24,25), transcriptional repression of TERT limits telomerase activity in somatic cells (26). The increased expression of telomerase in many cancer cells is often due to mutations in the TERT promoter, indicating a necessary up-regulation of TERT during tumor progression (27–29). Finally, TERT transcription is also activated in human cells with short telomeres due to the loss of repressive chromatin loops (30). The presence of these short telomeres has also been associated to increased chr5p TERRA expression (30).
Epithelial-to-mesenchymal transition (EMT) plays an important role in the progression of primary epithelial tumors (31). During EMT, epithelial cells lose their epithelial characteristics, including adherens junctions and apical-basal polarity, and acquire a mesenchymal phenotype and the ability to migrate and invade (32,33). The Snail1 transcription factor plays a key role in initiating the EMT process (34). Snail1 is rapidly induced by cytokines or by stress conditions that trigger EMT, and it binds and represses the expression of E-cadherin and other epithelial genes (34,35). Moreover, acting through the co-repressor LOXL2, Snail1 controls transcription of major satellite transcription and heterochromatin during EMT (36). Snail1 also participates in the activation of mesenchymal genes, such as fibronectin (37,38). Likely related to this, Snail1 is also required for fibroblast activation; the absence of this factor prevents induction of markers of active fibroblasts by TGF-β or other factors (39). Moreover, Snail1 expression in cancer-associated fibroblasts is also required for their role stimulating epithelial tumor cell invasion (39).
Besides inducing migratory and invasive properties, EMT provides cells with other features, such as a higher resistance to apoptosis and immunosuppression (31), and even generates cells with properties of stem cells (40,41). Indeed, transfection of Snail1 or Twist to immortalized human mammary cells confers them with the capability to form mammospheres, a property associated to the acquisition of stem cell characteristics (40). Since stemness requires the capability to maintain telomere length, we have now examined the action of Snail1 on these structures. Here, we report the identification of a new role for Snail1 transcription factor regulating TERT and TERRA transcription and telomere integrity. Our results reveal that Snail1 repression of TERRA is required for expression of a subset of mesenchymal genes as well as for EMT completion.
MATERIALS AND METHODS
Cell lines, transfection and infection
Cells were maintained in Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 10% fetal bovine serum (Invitrogen) at 37°C in 5% CO2. NMuMG cells were also supplemented with insulin (10 mg/ml). TGFβ (5 ng/ml) was used to induce EMT in NMuMG cells. MSCs were obtained from a conditional knockout mouse (Snail1Flox/Flox) (42,43), and Snail1 deletion was induced by transfection with pcDNA3-Cre or (as control) pcDNA3-GFP. RWP-1 cells transfected with empty plasmid or with pcDNA3-Snail1 (44), murine embryonic fibroblasts (MEFs) or cancer associated fibroblasts (CAFs) depleted of the Snail1 gene (36,39), and HCT75 cells (45) were generated as previously described. NMuMG cells were infected with retroviruses using pLU-Flag-VP16-TRF1ΔN, pLU-Flag VP16 or pLU-Flag-TRF1ΔN vectors and selected with puromycin (1 μg/ml) to overexpress TERRA (19). To overexpress murine Tert in NMuMG cells, retroviral viruses were generated using Plat-E Retroviral Packaging Cell Line. Plat-E were transfected (day 0) using Lipofectamine 2000 reagent (Invitrogen) either with pBabe-mTert or pBabe-empty vectors. The transfection medium was replaced with fresh medium after 24 h (day 1), and the cell-conditioned medium at days 2 and 3 was filtered and used to infect NMuMG cells with 8 mg/ml polybrene. Infected NMuMG cells were then incubated with fresh medium for a further 24 h and then selected with puromycin (1 μg/ml). Downregulation of Snail1 in NMuMG cells was performed by transfecting control or Snail1-specific siRNA as previously described (36).
PNA-FISH of prometaphase spreads
For prometaphase spread analysis, MSC were incubated with colcemide (0.5 mg/ml) for 90 min, collected by trypsinization, swollen in hypotonic buffer for 10 min at 37°C and then fixed and processed as described previously (46). Chromosomes were hybridized with 0.5 μg/ml of a Cy3-conjugated (CCCTAA)3 PNA telomere repeat probe (Applied Biosystems) and stained with DAPI (0.2 μg/ml). Images were acquired using a fluorescent microscope (Olympus BX61) and processed and merged using ImageJ software. For Q-FISH analysis, images were processed using TFL-Telo software (kindly provided by P. Lansdorp, Terry Fox Laboratory, Vancouver, Canada). Analyses were performed as a blind study as described previously (47).
Immunofluorescence/PNA-FISH analysis
(IF)-FISH staining was performed as previously described (48). Briefly, cells were blocked in 1% bovine serum albumin in PBS, followed by incubation with primary mouse α-γH2AX (1.5 mg/ml) (Upstate Millipore 05-636) and secondary FITC α-mouse (Jackson Laboratories) antibodies. Coverslips were washed with PBS, fixed in 2% paraformaldehyde in PBS for 5 min and then washed six times in PBS for 10 min. The samples were dehydrated (using a 70%, 95% and 100% ethanol series for 5 min), air dried and incubated in denaturing solution (70% formamide/2× SSC) for 10 min at 75°C. Chromosomes were hybridized with 0.5 μg/ml of a Cy3-conjugated (TTAGGG)3 PNA telomere repeat probe (Applied Biosystems) in hybridizing solution for 5 min at 85°C, followed by a 2-h incubation at room temperature. Preparations were then washed twice for 15 min each with 70% formamide in 10 mM Tris (pH 7.2), washed with PBS and stained with DAPI (0.2 μg/ml). Images were acquired using a fluorescent microscope (Axioplan 2, Carl Zeiss, Inc.) and processed using Openlab software (Perkin Elmer).
Telomeric repeat amplification protocol (TRAP) assay
Quantitative measurement of telomerase activity was performed using the TRAPeze RT Telomerase detection kit (Millipore) according to the manufacturer's instructions. Briefly, the cells were lysed on ice for 30 min in CHAPS buffer and then centrifuged at 12 000 g for 20 min at 4°C. The supernatant was collected, and the protein concentrations were determined by BCA protein assay. Each PCR reaction used 200 ng of total protein extract. PCR products were separated in a 12.5% non-denaturing polyacrylamide gel. Images were quantified with ImageJ software.
Chromatin immunoprecipitation (ChIP)
ChiP experiments were performed as described previously (36). Briefly, cells were crosslinked in 1% formaldehyde for 10 min at 37°C. Crosslinking was stopped by adding glycine to a final concentration of 0.125 M for 2 min at room temperature. Cell monolayers were scraped in cold soft lysis buffer (50 mM Tris, pH 8, 10 mM EDTA, 0.1% NP-40 and 10% glycerol) and incubated 20 min on ice. Nuclei pellets were lysed with SDS lysis buffer (1% SDS, 10 mM EDTA and 50 mM Tris, pH 8). Extract was sonicated to generate 200 to 500 bp DNA fragments. Protein (1 mg) was diluted 1:10 with dilution buffer (0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris pH 8 and 167 mM NaCl) and precleared by incubating with 30 μl of protein A or protein G magnetic beads (Millipore) and 5 μg of irrelevant immunoglobulin G at 4°C for 3 h. Samples were immunoprecipitated with the primary antibody (mouse α-Snail1 (36), α-p53 (Santa Cruz, SC-6243) or α-myc (ATCC 9E10); an irrelevant G immunoglobulin was used as control. After incubating on a rotating wheel at 4°C overnight, the antibody-chromatin complexes were rescued by incubating with 20 μl pre-blocked protein G magnetic beads at 4°C for 3 h. The beads were then washed with the following buffers (four times each): low-salt washing buffer (20 mM Tris–HCl pH 8, 5 mM NaCl, 2 mM EDTA 0.1% SDS, 1% Triton X-100), high-salt washing buffer (20 mM Tris–HCl pH 8, 150 mM NaCl, 2 mM EDTA 0.1% SDS, 1% Triton X-100) and LiCl buffer (10 mM Tris–HCl pH 8, 1 mM EDTA, 250 mM LiCl, 15% NP-40, 1% sodium deoxycholate). Chromatin was eluted from beads by adding 100 μl elution buffer (1% SDS, 0.1 M Na2CO3) and shaking at 37°C for 1 h. The eluted samples were de-crosslinked by adding NaCl at 200 mM final concentration and incubated at 65°C overnight. The day after, the proteins complexes were treated with proteinase K solution (10 μl 0.5 M EDTA, 20 μl 1 M Tris, pH 6.5 and 40 μg proteinase K) and incubated 1 h at 55°C. DNA was purified with the Qiagen PCR Purification kit, eluted in water and analyzed by RT-qPCR.
RNA extraction and analysis
RNA extraction was performed using TRIzol (Invitrogen), and samples were treated with DNase Turbo (Ambion) to eliminate DNA contamination. One μg of total RNA was retro-transcribed with Transcriptor First Strand cDNA Synthesis Kit (Roche) at 65°C for 10 min using random hexamers as primers. When amplifying human TERRAs, retrotranscription was carried out using a C-strand telomeric oligonucleotide consisting of five CCCTAA repeats. A control retrotranscription reaction without enzyme was performed when required. Real-time PCR was performed with SYBR Green I Master Reagent using a LightCycler 480 Real-Time PCR machine (Roche) with the following conditions: 40 cycles amplification, 94°C for 30 s, 60°C for 15 s, 72°C for 10 s. Oligonucleotides sequences are indicated in Supplementary Table S1. Relative quantification for the studied genes was calculated using Light Cycler 480 Software 1.5.0 by the ΔCt method, using Pumilio or Gapdh as controls.
TERRA RNA was also determined by dot blot analysis. Five μg RNA were resuspended in 1 mM EDTA (pH 8.0) to a final volume of 50 μl, mixed with denaturing solution (30 μl 20 × SSC and 20 μl 37% formaldehyde), incubated at 65°C for 30 min and then immediately cooled down on ice. The RNA was spotted on a Hybond-N membrane (Amersham) using a dot blot apparatus. The membrane was then UV cross-linked at 125 mJ in UV Stratalinker 2400 (Stratagene) and pre-hybridized in Church buffer (0.5 N Na-phosphate, pH 7.2, 7% SDS, 1 mM EDTA, 1% BSA) at 55°C for 2 h. After addition of the 32P-labeled (TAACCC)4 probe, the sample was incubated overnight at 55°C. The blot was washed twice in wash buffer 1 (0.2 N Na-phosphate, 2% SDS, and 1 mM EDTA at room temperature) and once in wash buffer 2 (0.1 N Na-phosphate, 2% SDS and 1 mM EDTA) at 50°C. Radioactive signals were collected with a phosphorimaging screen, and the signal was measured using a Typhoon 9410 Imager (GE Healthcare). A 18s rRNA probe (5′-CCATCCAATCGGTAGTAGCG-3′) was 5′ end-labeled with 32P using T4 Polynucleotide Kinase (New England Biolabs); this was used as an internal control.
RNA-seq analysis
RNA was isolated with Gene Elute Mammal Total RNA Miniprep Kit (Sigma), and its integrity was analyzed by electropherogram (Agilent Technologies). mRNA libraries were prepared with the rRNA depletion protocol and sequenced on an Illumina HiSeq2500 with a read length of 50 bases. Quality check was performed using tool FastQC. Reads were aligned with STAR (Spliced Transcripts Alignment to a Reference) to release 86 of the Mus musculus ENSEMBL version of the genome (GRMm38/mm10 assembly). HTSeq-count tool from HTSeq package was used to count the number of reads mapping each gene. DESeq2 method was used for analyzing differential gene expression of count data.
Additional methods are provided in the Supplemental Information.
RESULTS
Snail1 is essential for telomere maintenance in mouse mesenchymal stem cells
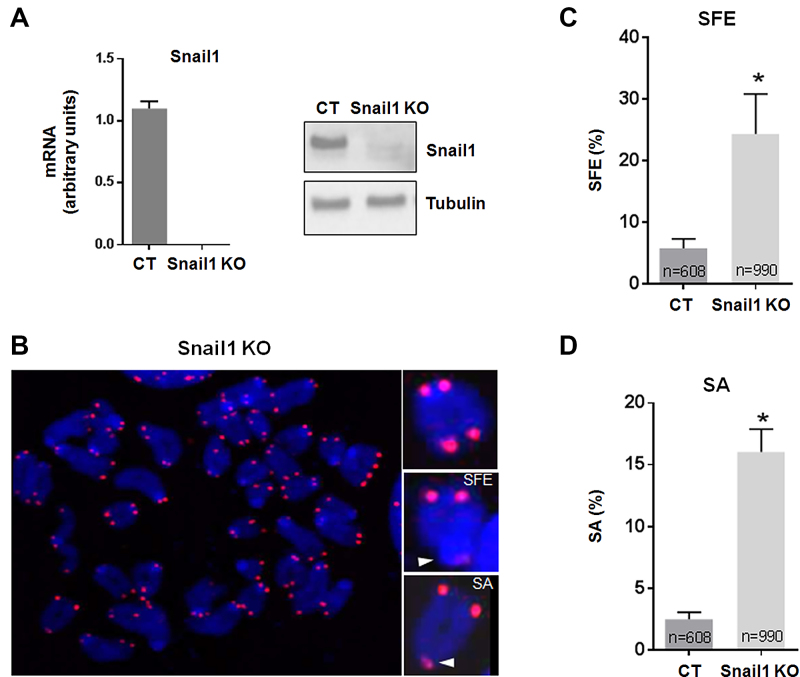
We initially studied the contribution of Snail1 to telomeric integrity using mouse MSC derived from the bone marrow of Snail1-conditional mice (Snail1Flox/Flox) (43). Snail1 was efficiently ablated by Cre-mediated recombination to generate the Snail1 knockout (KO) subline, as shown by analysis of mRNA levels and by western blot (Figure 1A). These cells grew slightly slower than control MSC, with no evidence of apoptosis (43); consequently, they could be maintained for at least 150 population doublings (PDs). Telomere integrity of MSC metaphase spreads was analyzed by FISH at 146 PD using a specific PNA probe for telomeric TTAGGG repeat (Figure 1B). We observed that Snail1 KO MSC presented chromatids lacking telomeric signal (signal-free ends [SFE]) as well as chromatids with fused sister telomeres (sister association [SA]) in 24% and 16% of the analyzed chromosomes, respectively. In contrast, only a small fraction of control cells (Snail1 CT) displayed these telomeric alterations (2.4% for SFE, and 5.8% for SA) (Figure 1C and D). The averages for SFE and SA were also significantly higher in Snail 1 KO mouse embryonic fibroblasts (MEFs) as compared to that of the controls (Supplementary Figure S1A).
Figure 1.
Snail1 loss causes telomere dysfunctions. (A) Expression of Snail1 at late passages (PD 146) of control MSC (CT) or MSC depleted of the Snail1 gene (Snail1 KO) as shown by RT-qPCR (left) and western blot (right). (B) A metaphase spread from Snail1 KO MSC showing telomere alterations (SFE, signal-free ends; SA, sister associations). Telomeres were stained with a Cy3-labeled PNA telomere probe (red). (C and D) Frequency of telomere alterations observed in control (CT) and Snail1 KO MSC. N indicates the number of chromosomes analyzed. The mean ± SEM of three independent experiments is shown. *P < 0.05.
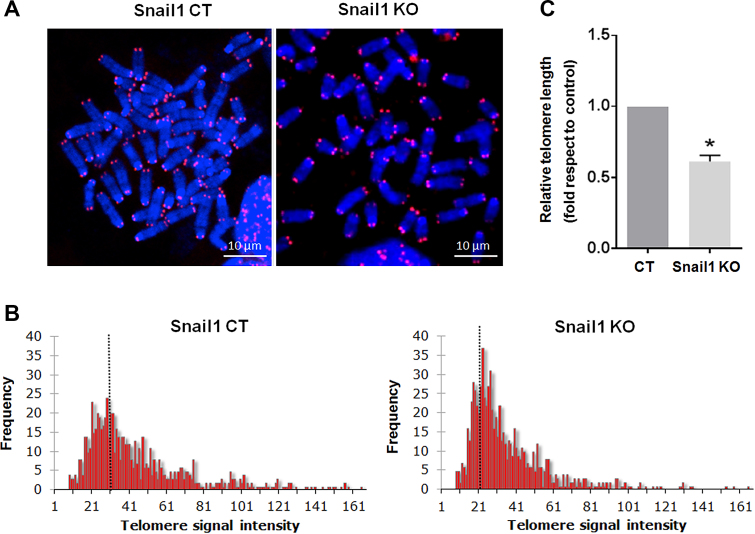
To determine if the telomere alterations observed in the Snail1 KO cells affect telomere extension, we analyzed the telomere length in these cells. MSC at metaphase were analyzed by quantitative fluorescence in situ hybridization (Q-FISH) at late passages after Snail1 depletion. Analysis of fluorescence intensities (Figure 2A–C) revealed that Snail1 KO MSC had shorter telomeres than control cells, indicating a positive role of Snail1 on telomere length. These results were also reproduced in MEFs, in which absence of Snail1 led to decreased telomere length (Supplementary Figure S1B).
Figure 2.
Snail1 regulates telomere length in MSC. (A) Representative metaphase spreads at late passages (PD 146) of Snail1 control MSC (CT) or Snail1 KO MSC. Telomeres were stained with a Cy3-labeled PNA telomere probe. (B) Q-FISH analysis showing the distribution of telomere fluorescent intensity in metaphase chromosomes from MSC. Data were accumulated using 40 metaphases for each histogram. Telomere intensity is expressed with arbitrary units; the average is indicated for each histogram. (C) Quantification of (B) from three independent Q-FISH analyses. *P < 0.05.
To verify these observations, telomere length was measured in the human HCT75 cell line at different passages with the telomeric terminal restriction fragment (TRF) assay. HCT75 is a clonal derivative of the human fibrosarcoma HT1080 cell line, which maintains its telomeres at a constant length. We generated stable cell lines expressing Snail1-HA, either wild-type (Snail1-HA) or with Pro2 mutated to Ala (Snail1 P2A-HA), which binds DNA but cannot repress transcription (34). Cell lines were analyzed at early PD (PD2–4) and late PD (PD64–68) stages. Immunoblot analysis confirmed that the cells stably expressed Snail1 (Supplementary Figure S2A). TRF assays indicated that Snail1 expression promoted an increase in telomere length when cells were passaged (Supplementary Figure S2B and C). This lengthening was not observed in the control cells or in cells over-expressing the transcriptionally-inactive form of Snail1P2A. Thus, Snail1 transcriptional repressive activity is required for its effect on telomere elongation.
Snail1 regulates Tert expression in MSC
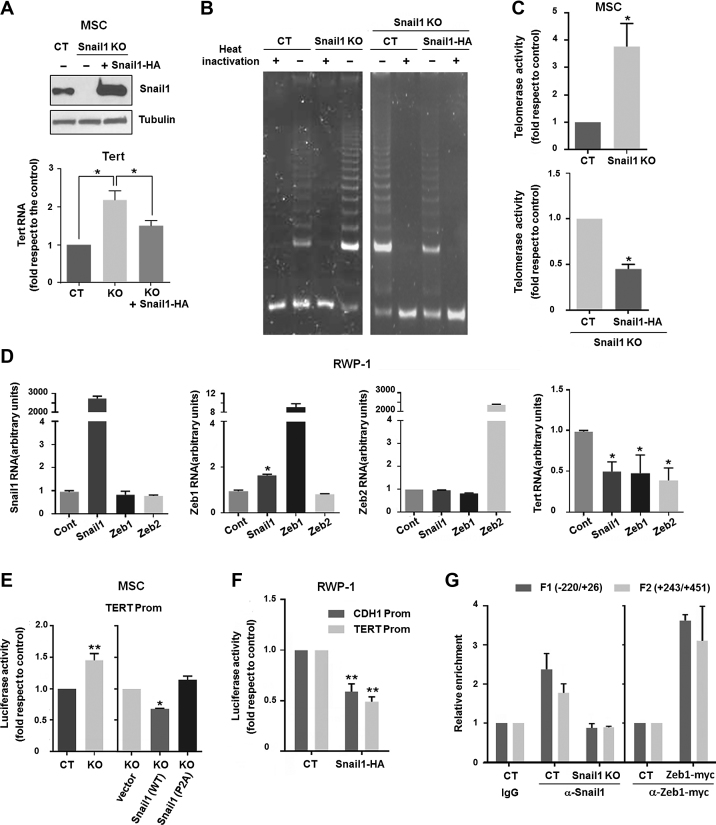
As telomere maintenance is greatly dependent on Tert expression, we next characterized the role of Snail1 in regulating the expression of this enzyme in mouse MSC. Although Snail1 KO MSC exhibited shorter telomeres (see Figure 2B and C), these cells displayed a 2-fold increase in the levels of Tert RNA with respect to the control (Figure 3A). In contrast, the levels of the telomerase RNA component, Terc, were not significantly different in Snail1 KO cells versus the control (Supplementary Figure S3). When Snail1 expression was rescued in the KO cells by stable transfection, the levels of Tert RNA were downregulated (Figure 3A). Up-regulation of Tert in MSC Snail1 KO was further confirmed by quantification of telomerase activity in cell extracts using the TRAP assay (Figure 3B and C). Also in this case, transfection of Snail1 repressed the telomerase activity (Figure 3B and C). Transcription of TERT was also controlled by Snail1 in the human cell line RWP-1, in which Snail1 overexpression down-regulated TERT RNA levels (Figure 3D). Snail1 up-regulates Zeb transcriptional repressors and cooperates with them in E-cadherin gene (CDH1) repression (49). Indeed, ZEB1 RNA was upregulated by ectopic Snail1 expression in RWP-1 cells (Figure 3D) and both Zeb1 and Zeb2 potently downregulated TERT levels when over-expressed in these cells (Figure 3D).
Figure 3.
Snail1 regulates Tert RNA expression and telomerase activity. (A) Western blot for Snail1 (upper panel) and RT-qPCR for Tert RNA (lower panel) in control MSC (CT), Snail1 KO and Snail1 KO after Snail1 rescue. *P < 0.05. (B) Representative image of a TRAP assay in MSC. PCR products were separated by 12.5% non-denaturing polyacrylamide gel electrophoresis. Activity was determined in control MSC, Snail1 KO and Snail1 KO after Snail1-HA transfection. (C) Quantification of three independent TRAP assays. *P < 0.05. (D) RT-qPCR analysis of Snail1 (1st left panel), Zeb1 (2nd left panel), Zeb2 (3rd left panel) or Tert RNA (right panel) in RWP-1 cells transfected with Snail1, Zeb1 or Zeb2 cDNA in eukaryotic expression plasmids. *P < 0.05. (E, F) Luciferase assays showing the activity of the Tert promoter (–599 to +53 promoter fragment cloned in pGL3) in MSC (E) or RWP-1 (F) cells. Cells were transfected with control, wild-type Snail1-HA or the Snail1 P2A mutant, as indicated. The CDH1 promoter activity was also assayed in (F). Data represent the mean ± SEM of three independent experiments. **P < 0.01; *P < 0.05. (G) ChIPs were performed using α-Snail1 antibody in CT and Snail1 KO MSC (left), or with α-myc antibody in CT cells or cells overexpressing ectopic myc-ZEB1 (right). Different fragments (F1 and F2) of the Tert promoter were amplified by RT-qPCR. An irrelevant IgG was used as a negative control in the same regions, whose values are set as 1 (left); the control corresponding to the value obtained with α-myc antibody in non-transfected cells is shown in the right panel.
Regulation of Tert transcription by Snail1 was also verified by luciferase assays using a –599/+53 fragment of the mouse Tert promoter cloned in pGL3 plasmid. MSC Snail1 KO exhibited an increased activity of the Tert promoter with respect to the control; ectopic transfection of Snail1 in these cells repressed this activity (Figure 3E). As expected, the Snail1 P2A repression-deficient mutant did not affect the Tert promoter. A similar decrease of Tert promoter activity was also observed in RWP-1 cells after Snail1 expression, using as control the CDH1 promoter, a well-known Snail1-repressed gene (34) (Figure 3F). E-boxes corresponding to putative Snail1 binding elements were identified in the Tert promoter at –253 and –123 with respect to the transcription start site, as well as more 3′ downstream, at +301, +324 and +426. ChIP assays confirmed that Snail1 binds to –220/+26 and +243/+451 sequences in control MSC, while no binding was observed when an irrelevant IgG was used as a negative control or in MSC Snail1 KO (Figure 3G). Zeb1-inhibition of Tert expression (see above) was also associated with binding to the Tert promoter (Figure 3G).
Finally, we interrogated the RNA public databases from different tumors for the expression of TERT and its inhibitors. TERT RNA expression inversely correlated with that of SNAIL1 in pancreatic adenocarcinoma or liver hepatocellular carcinoma, with ZEB1 in pancreatic, prostatic and colorectal adenocarcinomas, and in serus ovarian cancer and head and neck ovarian carcinomas, and with ZEB2 in hepatocellular and prostate carcinomas and in serus ovarian cancer (Supplementary Figure S4). Taken together, these data demonstrate that Snail1 transcriptionally represses Tert RNA, either directly or by inducing the expression of Zeb proteins.
Snail1 inhibits telomeric transcription
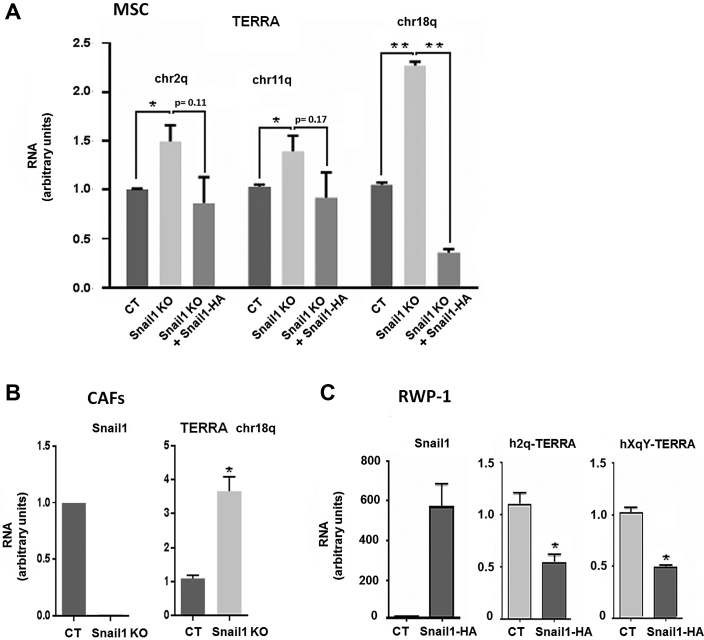
Although we observed that cells deficient for Snail1 had shorter telomeres, the presence of Snail1 did not increase but rather repressed telomerase transcription (see above). These results suggested that Snail1 regulates telomere maintenance by controlling mechanisms other than telomerase expression. Therefore, we next investigated a possible role of Snail1 in the regulation of telomere transcription. Telomeres are transcribed into telomeric repeat-containing RNAs (TERRA), large non-coding RNAs that form an integral part of telomeric heterochromatin. TERRA upregulation causes telomere shortening (16,50). We analyzed TERRA transcription by quantitative RT-qPCR, using sets of primers mapped to the subtelomeric region of different mouse chromosomes (chr2q, chr11q, chr18q). We observed a significant increase in the TERRA transcripts in the Snail1 KO MSC as compared to control cells, with chr18q TERRA presenting the highest Snail1-depending expression (Figure 4A). Accordingly, TERRA transcripts were downregulated after Snail1 ectopic expression in Snail1 KO MSC (Figure 4A). Similar results were obtained when we analyzed TERRA levels in CAFs obtained from murine tumors (39). Depleting Snail1 from these CAFs increased TERRA levels (Figure 4B), confirming a repressive role of Snail1 in telomere transcription. Moreover, an analysis in human cells showed that overexpression of Snail1 in the human RWP1 cell line downregulated TERRA h2q and hXqY (Figure 4C).
Figure 4.
Telomeric transcription is repressed by Snail1. (A) RT-qPCR showing the changes in expression of TERRA transcripts from different chromosomes (chr2q, chr11q or chr18q) in control MSC, Snail1KO MSC or Snail1KO+Snail1-HA MSC. Data represent the mean ± SEM of three independent experiments. *P < 0.05; **P < 0.01. (B, C) Levels of Snail1 and chr18q TERRA (B) or h2q and hXqY TERRA (C) were assessed by RT-qPCR in CAFs control (CT) and Snail1 KO (B) or in RWP-1 cells stably expressing Snail1-HA (C). **P < 0.01; *P < 0.05; P values higher than 0.05 are indicated.
TERRA downregulation during EMT is required for upregulation of a subset of mesenchymal genes
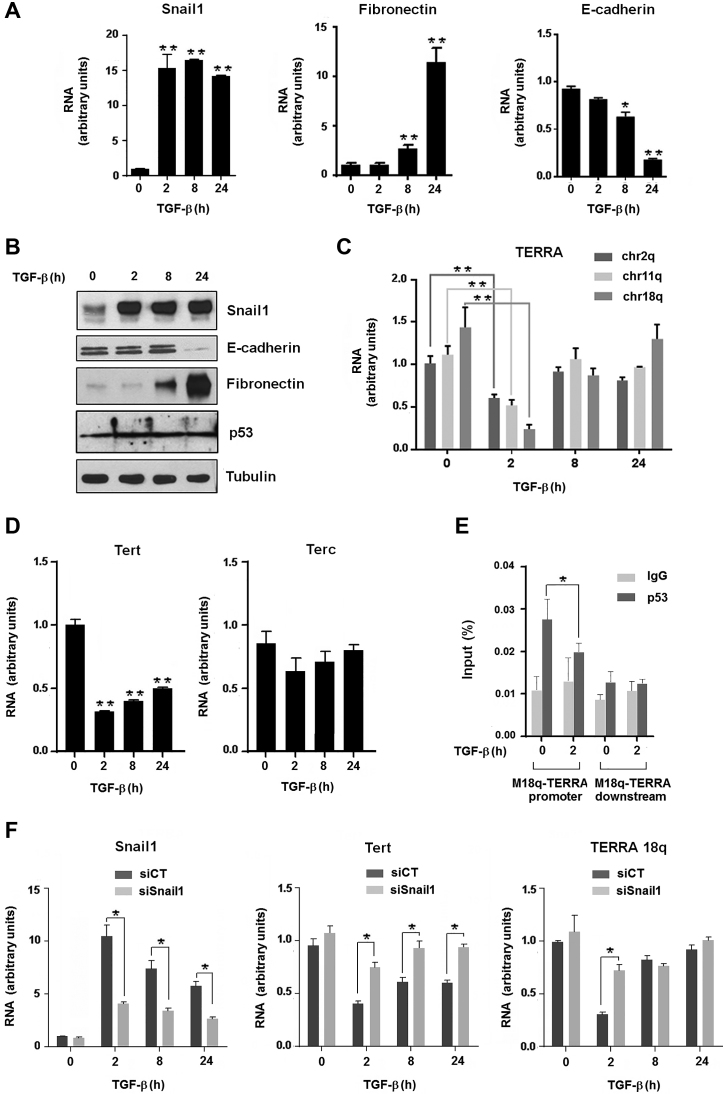
As Snail1 plays a key role in EMT, we next analyzed the levels of TERRA in the murine mammary gland NMuMG epithelial cell line treated with TGFβ, a widely used in vitro model of EMT. Addition of this cytokine induced Snail1 upregulation that preceded the expression of mesenchymal genes, such as fibronectin, and the silencing of E-cadherin (Cdh1) (Figure 5A and B). TERRA transcription was significantly decreased after 2 h of TGFβ exposure (Figure 5C), concomitantly with Snail1 upregulation. TERRA downregulation was transient, and TERRA levels after 24 h were equivalent to untreated control cells. In line with its regulation by Snail1, Tert RNA also decreased during TGFβ-induced EMT, reaching a minimal level after 2 h, whereas the Terc transcript did not change significantly (Figure 5D). TERRA expression is controlled by p53 (51), a protein whose function is antagonized by Snail1 (52). Since p53 activates TERRA transcription by direct binding to subtelomeric sequences (51), we determined whether p53 binding to the chr18q subtelomeric promoter was modulated during EMT. While p53 levels were not significantly altered during EMT (Figure 5B), its binding to the chr18q promoter was downregulated 2 h after TGFβ addition (Figure 5E), concomitant with Snail1 upregulation (Figure 5B).
Figure 5.
Telomeric transcription is downregulated during EMT.(A–D) Expression levels of Snail1, fibronectin, E-cadherin and p53 during TGFβ-induced EMT in NMuMG was assessed by RT-qPCR (A) and western blot (B). Levels of TERRA transcripts (from chr2q, chr11q and chr18q) (C), Tert and Terc (D) were determined by RT-qPCR during TGFβ-induced EMT in NMuMG cells. Data represent the mean ± SEM of three independent experiments. **P < 0.01; *P < 0.05. (E) NMuMG cells exposed to TGFβ were analyzed by ChIP with a p53-specific antibody. ChIP was quantified by qPCR with primers that anneal to the promoter of the murine chr18q-TERRA; a region downstream the TERRA transcript with no described promoter activity was used as a negative control. Data represent the mean ± SEM of three independent experiments. *P < 0.05. (F) TERRA and Tert levels in Snail1-deficient NMuMG cells. Snail1 was downregulated in NMuMG cells by transfection of a small interfering RNA (siSnail1) as indicated in Methods. A non-relevant siRNA was used as control (siCT). Levels of chr18q TERRA, Tert and Snail1 transcripts were determined by RT-qPCR during TGFβ-induced EMT in NMuMG cells. *P < 0.05.
The relevance of Snail1 expression for the TERRA transient inhibition during EMT was also demonstrated by RNA interference experiments. Downregulation of Snail1 in NMuMG cells by transfecting a specific siRNA prevented the TERRA decrease caused by TGFβ after 2 h of TGFβ exposure (Figure 5F). Tert RNA was also significantly less inhibited by TGFβ in Snail1-deficient cells than in controls (Figure 5F).
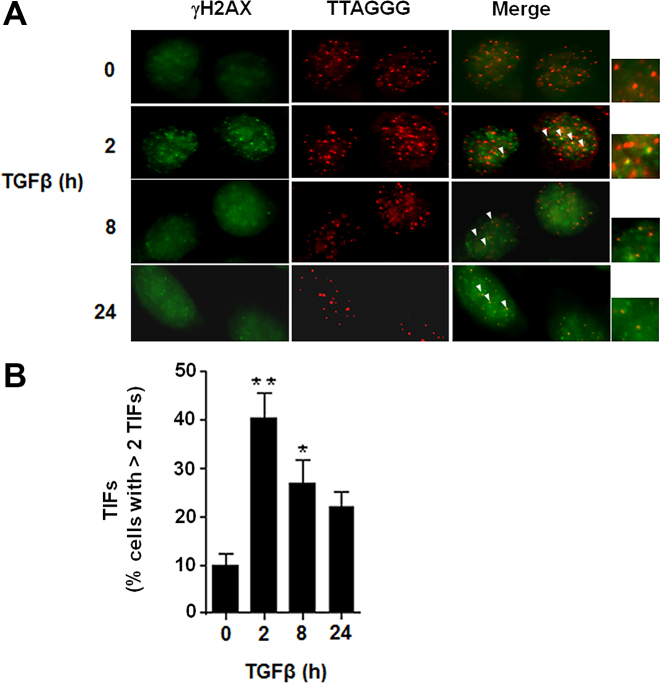
Reduction of chr18q TERRA increases the number of telomere dysfunction–induced foci (TIFs) (15). Since chr18q TERRA is downregulated during EMT, we determined the presence of TIFs in NMuMG cells during this transition by analyzing the presence of the γH2AX DNA damage marker at the telomeres (53). After 2 h of TGFβ exposure, and in parallel with Snail1 induction and chr18q TERRA downregulation, we observed a significant increase in the number of cells with two or more TIFs (Figure 6A and B). Following an inverse pattern over time as compared to TERRA, the number of TIF-positive cells was downregulated at later time points (Figure 6B).
Figure 6.
Downregulation of TERRA during EMT correlates with telomere deprotection. (A) NMuMG cells were treated with TGFβ (for 0, 2, 8, or 24 h) to induce EMT. Representative images of NMuMG stained with α-γH2AX (green) and processed for FISH with a Cy3-conjugated (TTAGGG)3 probe (red) are shown. Co-localizing events (arrowheads) indicate telomere dysfunction-induced foci (TIFs). (B) Quantification of the frequency of cells with > 2 TIFs/nuclei. Between 250 and 300 cells were analyzed in each time point. The value corresponds to the average of three experiments. **P < 0.01; *P < 0.05.
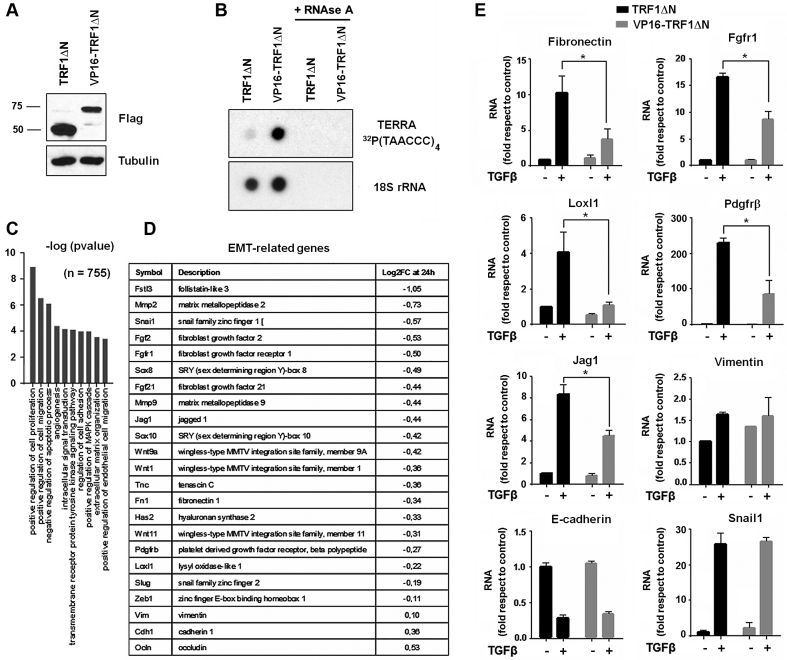
To determine whether TERRA downregulation is functionally important for EMT, we engineered NMuMG cells to produce elevated TERRA levels. We used the strategy developed by Lieberman and co-workers who transduced a mutant form of the telomere-binding factor TRF1 (TRF1ΔN) alone or fused to the VP16 transcriptional activation domain (VP16-TRF1ΔN) (19). Stable expression of these proteins was confirmed by western blot (Figure 7A), and upregulation of TERRA was validated by RNA dot blot (Figure 7B). Although TERRA upregulation did not modify morphology, migration or invasion of NMuMG cells after TGFβ-mediated EMT (Supplementary Figure S5), the transcription of EMT-related genes (such as that of fibronectin) was affected (Figure 7E). We compared the global transcriptomes of NMuMG control cells (TRF1ΔN) to those of cells that overexpressed TERRA (VP16-TRF1ΔN) following a 24-h treatment with TGFβ. RNA-seq analysis revealed that 1,508 genes were differentially regulated by TGFβ in cells after overexpressing TERRA with respect to control cells (Supplementary Table S2). Most of these genes are mesenchymal genes that were less stimulated by TGFβ when TERRA was present, and that are associated with pathways such as cell proliferation, cell migration, apoptosis, angiogenesis, cellular movement and extracellular matrix organization (Figure 7C). We also determined the relative expression of several genes classically associated with EMT (Figure 7D). Changes in the expression of several of these genes were validated by RT-qPCR (Figure 7E and Supplementary Figure S6). Fibronectin, Jag1, Loxl1, Fgfr1 and Pdgfrβ RNAs exhibited a lower upregulation by TGFβ in TERRA-expressing cells than in control cells. Expression of these genes was not affected by transfection of VP16 alone (Supplementary Figure S7). Regulation of other EMT-relevant genes, such as E-cadherin, Snail1 or Vimentin, was not significantly altered by TERRA over-expression (Figure 7E). NMuMG cells were also stably transduced with murine Tert cDNA in an expression plasmid, and the expression of TERRA-sensitive genes was also determined after TGFβ-induced EMT. None of the analyzed genes was expressed differently in cells with Tert as compared to control cells (Supplementary Figure S8).
Figure 7.
Ectopic TERRA expression alters the EMT gene profile. (A) Western blot of NMuMG cells transduced with Flag-TRF1ΔN alone or fused with the transcriptional activator VP16. (B) Radioactive RNA dot blot for TERRA from NMuMG cells transduced with TRF1ΔN or VP16-TRF1ΔN. 18S served as a loading control. Samples treated with RNase A were used as negative controls. (C) Gene ontology analysis of differentially expressed genes in VP16-TRF1ΔN NMuMG cells as compared to control TRF1ΔN NMuMG cells after TGFβ treatment (24 h). The global transcriptome was analyzed by RNA-seq. (D) Short list of relevant EMT-related genes and their expression in VP16-TRF1ΔN-NMuMG cells as compared to control NMuMG at 24 h after TGFβ treatment. Log2 fold-changes are indicated. A more extended list of transcripts is available in Supplementary Table S2. (E) RT-qPCR was used to determine the expression of the selected EMT-related genes at 24 h after TGFβ treatment (except for Snail1 at 2 h). Data represent the mean ± SEM of three independent experiments. *P < 0.05.
DISCUSSION
In primary epithelial cells, chromosomes with short telomeres are more frequently involved in missegregation events than chromosomes with normal telomere lengths (54). Shortened telomeres have been linked to chromosome instability in the setting of age-related diseases associated with genetic disruption and carcinogenesis (55). Critically short telomeres expose chromosomal ends, engage the DNA damage response and precipitate end-to-end fusions and recombination events (SFE and SA). We show here that Snail1 depletion leads to telomere abnormalities in mesenchymal cells, in line with the described action of Snail1 inducing stemness properties (40). In contrast to our expectations, however, we observed that Snail1 represses rather than activates TERT expression; this effect has also been reported by others (56). Indeed, not only Snail1 but also other transcriptional factors involved in EMT, such as Zeb1, also inhibit Tert. Accordingly, Tert is downregulated during EMT. The precise functional outcome of this decrease is still unknown; it is possible that it is related to the non-canonical actions described for telomerase in controlling NFkB transcriptional activity (57).
Snail1 also represses the expression of TERRA transcripts. We suggest that the altered telomeres observed after Snail1 depletion are a consequence of the excessive levels of TERRA in these cells. Although the function of TERRA is not fully understood, the emerging view is that it acts as a molecular scaffold for proteins required for proper telomere function. Several findings indicate that telomere transcription controls telomere structure and function (13). Cells from the human pathology ICF (immunodeficiency, centromeric region instability, facial anomalies syndrome) have hypomethylated subtelomeric regions, abnormally elevated TERRA levels, shortened telomeres and associated telomere aberrations (such as SFE), but unaltered telomerase levels (58). In fact, it has been previously suggested that upregulated TERRA transcription leads to several forms of telomeric aberrations (including SFEs and SA), heterochromatin reorganization and telomere shortening (9,59). Some authors suggest that TERRA acts as direct inhibitor of telomerase activity (60); accordingly, TERRA ectopic expression decreases telomerase activity (61). It is also possible that TERRA-induced telomere shortening is independent of telomerase, since it has been reported to also act on exonuclease 1 (16).
How telomere transcription is controlled is still largely unknown, but transcription regulators of telomeres include the CTCF and ATRX chromatin remodelers, as well as the Rad21 cohesin subunit (62–64). Recently, the nuclear respiratory factor 1 (NRF1) has also been identified as a new activator of human telomere transcription, revealing a new link between telomeres and metabolism (65). A recent report has shown that 15q TERRA transcription is activated by Rb by its direct binding to this promoter (66). A similar activation of TERRA expression has been described for p53 related to its ability to prevent TIFs (51). Interestingly, p53 and Snail1 are mutually antagonistic, and Snail1 prevents p53 from functioning in distinct systems (44,52,67–69). Although this interference is general, the molecular mechanism might be cell-specific, since it is associated with p53 degradation in some systems (52), an effect that we however have not observed. We have not detected Snail1 binding to the TERRA subtelomeric promoter, suggesting that Snail1’s action on p53 is likely due to p53 inactivation, perhaps through covalent modification of p53 itself or the repression of an essential p53 cofactor. In any case, our results suggest that TERRA is another locus inversely regulated by Snail1 and p53.
The role of TERRA was investigated during EMT in the widely-used model of NMuMG cells treated with TGFβ. Telomere transcription is transiently downregulated at 2 h after exposure to this cytokine, correlating with the upregulation of Snail1 and prior to the expression of most mesenchymal genes (Figure 5). Simultaneously, TGFβ also promotes an increase in the number of cells displaying TIFs, suggesting that telomeres are deprotected but are still long (Figure 6). TIFs are a consequence of activation of DNA-damage response at chromosome ends, originated by TERRA downregulation (15,53). Although TERRA chr18q seems to be the main transcript responsible for this regulation (15), other telomeric transcripts, such as TERRA 2q and 11q, might also contribute to deprotection, since they are also transiently downregulated during EMT, similar to TERRA chr18q. An interesting question remaining to be investigated consists in determining the relevance of these TIFs in EMT. It is possible that this transient response is required to activate mesenchymal genes; alternatively, it might correspond to a transient lesion caused by telomere deprotection, necessary for the chromosome reorganization taking place during EMT.
Our results confirm and extend previous observations indicating that Snail1 regulates pericentromeric heterochromatin transcription (36). Snail1 repression of pericentromeric major satellite RNA also takes place early during EMT and is required for the completion of the process. Compared with major satellite, upregulation of ectopic TERRA had a reduced impact on EMT, with no changes visible for cell migration or invasion with respect to control cells. It is likely that the global contribution of TERRA is lower than that of major satellite in the cellular organization of heterochromatin; therefore, it would be expected that it would also have a lesser role in the proposed chromosome repositioning that takes place during this transition (36). Accordingly, we did not detect alterations in essential transcriptional factors, such as Zeb1 and Zeb2, in contrast to what we observed with overexpressed major satellite (36). In any case, global transcriptome analysis indicated that ectopic TERRA expression prevented the transcription of a subset of EMT-related genes. Several questions now remain to be addressed, such as what are the functions of these genes, and whether their lower levels of expression prevent some other actions of mesenchymal cells, such as communication with tumoral cells or other roles from the stroma.
As a final point, our results reveal a novel connection between EMT and stemness. Several transcriptional factors involved in EMT, such as Twist and Zeb1, also inhibit cell senescence (70). We now show that the Snail1 transcription factor, the key regulator of EMT, not only affects cell invasion, resistance to apoptosis and metabolic reprogramming (31,34,44,71–73) but also is essential for telomere maintenance and integrity. This multiple function in tumorigenesis might open new strategic therapeutic opportunities to develop Snail1-inhibitory drugs for fighting tumor cells.
Supplementary Material
ACKNOWLEDGEMENTS
We appreciate Raúl Peña's excellent technical assistance. We thank Agnel Sfeir, Antonio Postigo, Gabriel Gil, Alfonso Gutiérrez-Adán and Paul Lieberman for kindly providing γH2AX antibody, pcDNA3-Zeb1-Myc, anti myc antibody, 5k-mTERT-EGFP and VP16-TRF1ΔN vectors, respectively. We also thank the members of the Smith and García de Herreros laboratories for comments on the manuscript and helpful discussions. R.M. was supported by a MINECO 2013 postdoctoral fellowship and S.C., by a RYC-2013–12598 grant. The current address for S.P. and A.G.-G. is Vall d’Hebron Institute of Oncology, Barcelona, Spain, and for A.M.-R., the Institute for Research in Biomedicine, Barcelona, Spain.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Ministerio de Economía y Competitividad (MINECO) [SAF2013-40922-R1 to A.G.H.]; (MINECO) [RYC-2013–12598 to S.C]. Agencia Estatal de Investigación (AEI) and Fondo Europeo de Desarrollo Regional-FEDER [SAF2016-76461-R to A.G.H.]; Generalitat de Catalunya [2014 SGR 32]; Instituto de Salud Carlos III [PIE15/00008]. Funding for open access charge: MINECO RYC-2013–12598.
Conflict of interest statement. None declared.
REFERENCES
- 1. de Lange T. How telomeres solve the end-protection problem. Science. 2009; 326:948–952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Palm W., de Lange T.. How Shelterin Protects Mammalian Telomeres. Annu. Rev. Genet. 2008; 42:301–334. [DOI] [PubMed] [Google Scholar]
- 3. O’Sullivan R.J., Karlseder J.. Telomeres: protecting chromosomes against genome instability. Nat. Rev. Mol. Cell Biol. 2010; 11:171–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Van Steensel B., Smogorzewska A., De Lange T.. TRF2 protects human telomeres from end-to-end fusions. Cell. 1998; 92:401–413. [DOI] [PubMed] [Google Scholar]
- 5. Karlseder J., Broccoli D., Dai Y., Hardy S., de Lange T.. p53- and ATM-dependent apoptosis induced by telomeres lacking TRF2. Science. 1999; 283:1321–1325. [DOI] [PubMed] [Google Scholar]
- 6. Schoeftner S., Blasco M.A.. A ‘higher order’ of telomere regulation: telomere heterochromatin and telomeric RNAs. EMBO J. 2009; 28:2323–2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. García-Cao M., O’Sullivan R., Peters A.H.F.M., Jenuwein T., Blasco M.A.. Epigenetic regulation of telomere length in mammalian cells by the Suv39h1 and Suv39h2 histone methyltransferases. Nat. Genet. 2003; 36:94–99. [DOI] [PubMed] [Google Scholar]
- 8. Blasco M.A., Gonzalo S., García-Cao M., Fraga M.F., Schotta G., Peters A.H.F.M., Cotter S.E., Eguía R., Dean D.C., Esteller M. et al. . Role of the RB1 family in stabilizing histone methylation at constitutive heterochromatin. Nat. Cell Biol. 2005; 7:420–428. [DOI] [PubMed] [Google Scholar]
- 9. Azzalin C.M., Reichenbach P., Khoriauli L., Giulotto E., Lingner J.. Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science. 2007; 318:798–801. [DOI] [PubMed] [Google Scholar]
- 10. Nergadze S.G., Farnung B.O., Wischnewski H., Khoriauli L., Vitelli V., Chawla R., Giulotto E., Azzalin C.M.. CpG-island promoters drive transcription of human telomeres. RNA. 2009; 15:2186–2194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Rippe K., Luke B.. TERRA and the state of the telomere. Nat. Struct. Mol. Biol. 2015; 22:853–858. [DOI] [PubMed] [Google Scholar]
- 12. Arnoult N., Van Beneden A., Decottignies A.. Telomere length regulates TERRA levels through increased trimethylation of telomeric H3K9 and HP1α. Nat. Struct. Mol. Biol. 2012; 19:948–956. [DOI] [PubMed] [Google Scholar]
- 13. Deng Z., Norseen J., Wiedmer A., Riethman H., Lieberman P.M.. TERRA RNA binding to TRF2 facilitates heterochromatin formation and ORC recruitment at telomeres. Mol. Cell. 2009; 35:403–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Azzalin C.M., Lingner J.. Telomere functions grounding on TERRA firma. Trends Cell Biol. 2015; 25:29–36. [DOI] [PubMed] [Google Scholar]
- 15. López de Silanes I., Grana O., De Bonis M.L., Dominguez O., Pisano D.G., Blasco M.A.. Identification of TERRA locus unveils a telomere protection role through association to nearly all chromosomes. Nat. Commun. 2014; 5:4723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Pfeiffer V., Lingner J.. TERRA promotes telomere shortening through exonuclease 1-mediated resection of chromosome ends. PLoS Genet. 2012; 8:e1002747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Luke B., Panza A., Redon S., Iglesias N., Li Z., Lingner J.. The Rat1p 5′ to 3′ Exonuclease Degrades Telomeric Repeat-Containing RNA and Promotes Telomere Elongation in Saccharomyces cerevisiae. Mol. Cell. 2008; 32:465–477. [DOI] [PubMed] [Google Scholar]
- 18. Cusanelli E., Romero C., Chartrand P.. Telomeric noncoding RNA TERRA is induced by telomere shortening to nucleate telomerase molecules at short telomeres. Mol. Cell. 2013; 51:780–791. [DOI] [PubMed] [Google Scholar]
- 19. Wang Z., Deng Z., Dahmane N., Tsai K., Wang P., Williams D.R., Kossenkov A. V, Showe L.C., Zhang R., Huang Q. et al. . Telomeric repeat-containing RNA (TERRA) constitutes a nucleoprotein component of extracellular inflammatory exosomes. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:E6293–E6300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Blasco M., Schoeftner S.. Developmentally regulated transcription of mammalian telomeres by DNA-dependent RNA polymerase II. Nat. Cell Biol. 2008; 10:228–236. [DOI] [PubMed] [Google Scholar]
- 21. Hanahan D., Weinberg R.A.. The hallmarks of cancer: the next generation. Cell. 2011; 144:646–674. [DOI] [PubMed] [Google Scholar]
- 22. Shay J.W., Wright W.E.. Hallmarks of telomeres in ageing research. J. Pathol. 2007; 211:114–123. [DOI] [PubMed] [Google Scholar]
- 23. Blackburn E.H. Telomeres and telomerase: their mechanisms of action and the effects of altering their functions. FEBS Lett. 2005; 579:859–862. [DOI] [PubMed] [Google Scholar]
- 24. Yi X., White D.M., Aisner D.L., Baur J.A., Wright W.E., Shay J.W.. An alternate splicing variant of the human telomerase catalytic subunit inhibits telomerase activity. Neoplasia. 2000; 2:433–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wong M.S., Wright W.E., Shay J.W.. Alternative splicing regulation of telomerase: a new paradigm?. Trends Genet. 2014; 30:430–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Meyerson M., Counter C.M., Eaton E.N., Ellisen L.W., Steiner P., Caddle S.D., Ziaugra L., Beijersbergen R.L., Davidoff M.J., Qingyun L. et al. . hEST2, the putative human telomerase catalytic subunit gene, is up- regulated in tumor cells and during immortalization. Cell. 1997; 90:785–795. [DOI] [PubMed] [Google Scholar]
- 27. Huang F.W., Hodis E., Xu M.J., Kryukov G.V., Chin L., Garraway L.A., Berger M.F., Barretina J., Hodis E., Krauthammer M. et al. . Highly recurrent TERT promoter mutations in human melanoma. Science. 2013; 339:957–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Horn S., Figl A., Rachakonda P.S., Fischer C., Sucker A., Gast A., Kadel S., Moll I., Nagore E., Hemminki K. et al. . TERT promoter mutations in familial and sporadic melanoma. Science. 2013; 339:959–961. [DOI] [PubMed] [Google Scholar]
- 29. Eckel-Passow J.E., Lachance D.H., Molinaro A.M., Walsh K.M., Decker P.A., Sicotte H., Pekmezci M., Rice T., Kosel M.L., Smirnov I.V. et al. . Glioma groups based on 1p/19q, IDH, and TERT promoter mutations in tumors. N. Engl. J. Med. 2015; 372:2499–2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kim W., Ludlow A.T., Min J., Robin J.D., Stadler G., Mender I., Lai T.P., Zhang N., Wright W.E., Shay J.W.. Regulation of the human telomerase gene TERT by telomere position effect-over long distances (TPE-OLD): implications for aging and cancer. PLoS Biol. 2016; 14:e20000168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Thiery J.P., Acloque H., Huang R.Y.J., Nieto M.A.. Epithelial-Mesenchymal transitions in development and disease. Cell. 2009; 139:871–890. [DOI] [PubMed] [Google Scholar]
- 32. Yang J., Weinberg R.A.. Epithelial-mesenchymal transition: at the crossroads of development and tumor metastasis. Dev. Cell. 2008; 14:818–829. [DOI] [PubMed] [Google Scholar]
- 33. Nieto M.A., Huang R.Y., Jackson R.A., Thiery J.P.. EMT: 2016. Cell. 2012; 166:21–45. [DOI] [PubMed] [Google Scholar]
- 34. Batlle E., Sancho E., Francí C., Domínguez D., Monfar M., Baulida J., García de Herreros A.. The transcription factor Snail is a repressor of E-cadherin gene expression in epithelial tumour cells. Nat. Cell Biol. 2000; 2:84–89. [DOI] [PubMed] [Google Scholar]
- 35. Vincent T., Neve E.P., Johnson J.R., Kukalev A., Rojo F., Albanell J., Pietras K., Virtanen I., Philipson L., Leopold P.L. et al. . A SNAIL1-SMAD3/4 transcriptional repressor complex promotes TGF-beta mediated epithelial-mesenchymal transition. Nat. Cell Biol. 2009; 11:943–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Millanes-Romero A., Herranz N., Perrera V., Iturbide A., Loubat-Casanovas J., Gil J., Jenuwein T., García de Herreros A., Peiró S.. Regulation of heterochromatin transcription by Snail1/LOXL2 during epithelial-to-mesenchymal transition. Mol. Cell. 2013; 52:746–757. [DOI] [PubMed] [Google Scholar]
- 37. Stanisavljevic J., Porta-de-la-Riva M., Batlle R., García de Herreros A., Baulida J.. The p65 subunit of NF- B and PARP1 assist Snail1 in activating fibronectin transcription. J. Cell Sci. 2011; 124:4161–4171. [DOI] [PubMed] [Google Scholar]
- 38. Hsu D.S., Wang H.J., Tai S.K., Chou C.H., Hsieh C.H., Chiu P.H., Chen N.J., Yang M.H.. Acetylation of Snail modulates the cytokinome of cancer cells to enhance the recruitment of macrophages. Cancer Cell. 2014; 26:534–548. [DOI] [PubMed] [Google Scholar]
- 39. Alba-Castellón L., Olivera-Salguero R.R., Mestre-Farrera A., Peña R., Herrera M., Bonilla F.F., Casal J.I., Baulida J., Peña C., García de Herreros A.. Snail1-dependent activation of cancer-associated fibroblast controls epithelial tumor cell invasion and metastasis. Cancer Res. 2016; 76:6205–6217. [DOI] [PubMed] [Google Scholar]
- 40. Mani S.A., Guo W., Liao M.J., Eaton E.N., Ayyanan A., Zhou A.Y., Brooks M., Reinhard F., Zhang C.C., Shipitsin M et al. . The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008; 133:704–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Morel A.P., Lievre M., Thomas C., Hinkal G., Ansieau S., Puisieux A.. Generation of breast cancer stem cells through epithelial-mesenchymal transition. PLoS One. 2008; 3:e2888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Murray S.A., Carver E.A., Gridley T.. Generation of a Snail1 (Snai1) conditional null allele. Genesis. 2006; 44:7–11. [DOI] [PubMed] [Google Scholar]
- 43. Batlle R., Alba-Castellón L., Loubat-Casanovas J., Armenteros E., Francí C., Stanisavljevic J., Barderas R., Martin-Caballero J., Bonilla F., Baulida J. et al. . Snail1 controls TGF-β responsiveness and differentiation of mesenchymal stem cells. Oncogene. 2013; 32:3381–3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Escrivà M., Peiró S., Herranz N., Villagrasa P., Dave N., Montserrat-Sentís B., Murray S., Francí C., Gridley T., Virtanen I. et al. . Repression of PTEN phosphatase by Snail1 transcriptional factor during gamma radiation-induced apoptosis. Mol. Cell. Biol. 2008; 28:1528–1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. van Steensel B., de Lange T.. Control of telomere length by the human telomeric protein TRF1. Nature. 1997; 385:740–743. [DOI] [PubMed] [Google Scholar]
- 46. Canudas S., Smith S.. Differential regulation of telomere and centromere cohesion by the Scc3 homologues SA1 and SA2, respectively, in human cells. J. Cell Biol. 2009; 187:165–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hao L.Y.Y., Armanios M., Strong M.A., Karim B., Feldser D.M., Huso D., Greider C.W.. Short telomeres, even in the presence of telomerase, limit tissue renewal capacity. Cell. 2005; 123:1121–1131. [DOI] [PubMed] [Google Scholar]
- 48. Canudas S., Houghtaling B.R., Bhanot M., Sasa G., Savage S.A., Bertuch A.A., Smith S.. A role for heterochromatin protein 1γ at human telomeres. Genes Dev. 2011; 25:1807–1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Guaita S., Puig I., Franci C., Garrido M., Dominguez D., Batlle E., Sancho E., Dedhar S., García de Herreros A., Baulida J.. Snail induction of epithelial to mesenchymal transition in tumor cells is accompanied by MUC1 repression and ZEB1 expression. J. Biol. Chem. 2002; 277:39209–39216. [DOI] [PubMed] [Google Scholar]
- 50. Maicher A., Kastner L., Dees M., Luke B.. Deregulated telomere transcription causes replication-dependent telomere shortening and promotes cellular senescence. Nucleic Acids Res. 2012; 40:6649–6659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Tutton S., Azzam G.A., Stong N., Vladimirova O., Wiedmer A., Monteith J.A., Beishline K., Wang Z., Deng Z., Riethman H. et al. . Subtelomeric p53 binding prevents accumulation of DNA damage at human telomeres. EMBO J. 2016; 35:193–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ni T., Li X.Y., Lu N., An T., Liu Z.P., Fu R., Weng-Cong L., Zhang L.W., Xu X.J., Rowe R.G. et al. . Snail1-depndent p53 repression regulates expansion and activity of tumor-initiating cells in breast cancer. Nat. Cell Biol. 2016; 18:1221–1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Takai H., Smogorzewska A., De Lange T.. DNA damage foci at dysfunctional telomeres. Curr. Biol. 2003; 13:1549–1556. [DOI] [PubMed] [Google Scholar]
- 54. Pampalona J., Soler D., Genescà A., Tusell L.. Whole chromosome loss is promoted by telomere dysfunction in primary cells. Genes Chromosom. Cancer. 2010; 49:368–378. [DOI] [PubMed] [Google Scholar]
- 55. Campisi J., Kim S., Lim C.S., Rubio M.. Cellular senescence, cancer and aging: the telomere connection. Exp. Gerontol. 2001; 36:1619–1637. [DOI] [PubMed] [Google Scholar]
- 56. Yoo Y.S., Park S., Gwak J., Ju B.G., Oh S.. Involvement of transcription repressor Snail in the regulation of human telomerase reverse transcriptase (hTERT) by transforming growth factor-β. Biochem. Biophys. Res. Commun. 2015; 465:131–136. [DOI] [PubMed] [Google Scholar]
- 57. Ghosh A., Saginc G., Leow S.C., Khattar E., Shin E.M., Yan T.D., Wong M., Zhang Z., Li G., Sung W.K. et al. . Telomerase directly regulates NF-κB-dependent transcription. Nat. Cell Biol. 2012; 14:1270–1281. [DOI] [PubMed] [Google Scholar]
- 58. Yehezkel S., Segev Y., Viegas-Péquignot E., Skorecki K., Selig S.. Hypomethylation of subtelomeric regions in ICF syndrome is associated with abnormally short telomeres and enhanced transcription from telomeric regions. Hum. Mol. Genet. 2008; 17:2776–2789. [DOI] [PubMed] [Google Scholar]
- 59. Sandell L.L., Gottschling D.E., Zakian V.A.. Transcription of a yeast telomere alleviates telomere position effect without affecting chromosome stability. Proc. Natl. Acad. Sci. U.S.A. 1994; 91:12061–12065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Redon S., Reichenbach P., Lingner J.. The non-coding RNA TERRA is a natural ligand and direct inhibitor of human telomerase. Nucleic Acids Res. 2010; 38:5797–5806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Kreilmeier T., Mejri D., Hauck M., Kleiter M., Holzmann K.. Telomere transcripts target telomerase in human cancer cells. Genes. 2016; 7:E46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Deng Z., Wang Z., Stong N., Plasschaert R., Moczan A., Chen H.-S.S., Hu S., Wikramasinghe P., Davuluri R. V, Bartolomei M.S. et al. . A role for CTCF and cohesin in subtelomere chromatin organization, TERRA transcription, and telomere end protection. EMBO J. 2012; 31:4165–4178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Episkopou H., Draskovic I., Van Beneden A., Tilman G., Mattiussi M., Gobin M., Arnoult N., Londoño-Vallejo A., Decottignies A.. Alternative Lengthening of Telomeres is characterized by reduced compaction of telomeric chromatin. Nucleic Acids Res. 2014; 42:4391–4405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Eid R., Demattei M.V., Episkopou H., Augé-Gouillou C., Decottignies A., Grandin N., Charbonneau M.. Genetic inactivation of ATRX leads to a decrease in the amount of telomeric cohesin and of telomere transcription in human glioma cells. Mol. Cell. Biol. 2015; 35:2818–2830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Diman A., Boros J., Poulain F., Rodriguez J., Purnelle M., Episkopou H., Bertrand L., Francaux M., Deldicque L., Decottignies A.. Nuclear respiratory factor 1 and endurance exercise promote human telomere transcription. Sci. Adv. 2016; 2:e1600031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Gonzalez-Vasconcellos I., Schneider R., Anastasov N., Alonso-Rodriguez S., Sanli-Bonazzi B., Fernández J.L., Atkinson M.J.. The Rb1 tumour suppressor gene modifies telomeric chromatin architecture by regulating TERRA expression. Sci. Rep. 2017; 7:42056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Kajita M., McClinic K.N., Wade P.A.. Aberrant expression of the transcription factors snail and slug alters the response to genotoxic stress. Mol. Cell. Biol. 2004; 24:7559–7566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Kurrey N.K., Jalgaonkar S.P., Joglekar A.V., Ghanate A.D., Chaskar P.D., Doiphode R.Y., Bapat S.A.. Snail and slug mediate radioresistance and chemoresistance by antagonizing p53-mediated apoptosis and acquiring a stem-like phenotype in ovarian cancer cells. Stem Cells. 2009; 27:2059–2068. [DOI] [PubMed] [Google Scholar]
- 69. Lee S.H., Lee S.J., Jung Y.S., Xu Y., Kang H.S., Ha N.C., Park B.J.. Blocking of p53-Snail binding, promoted by oncogenic K-Ras, recovers p53 expression and function. Neoplasia. 2009; 11:22–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Smit M.A., Peeper D.S.. Epithelial-mesenchymal transition and senescence: Two cancer-related processes are crossing paths. Aging. 2010; 2:735–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Baulida J., García de Herreros A.. Snail1-driven plasticity of epithelial and mesenchymal cells sustains cancer malignancy. Biochim. Biophys. Acta - Rev. Cancer. 2015; 1856:55–61. [DOI] [PubMed] [Google Scholar]
- 72. Vega S., Morales A. V., Ocaña O.H., Valdés F., Fabregat I., Nieto M.A.. Snail blocks the cell cycle and confers resistance to cell death. Genes Dev. 2004; 18:1131–1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Jiang L., Xiao L., Sugiura H., Huang X., Ali A., Kuro-o M., Deberardinis R.J., Boothman D.A.. Metabolic reprogramming during TGFβ1-induced epithelial-to-mesenchymal transition. Oncogene. 2015; 34:3908–3916. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.