Abstract
Chromosomal rearrangement involving the KMT2A gene is one of the most common genetic alteration in acute myeloid leukemia. A total of 135 different KMT2A rearrangements have been identified, where 94 translocation partner genes are now characterized at the molecular level. Of these 94 translocation partner genes, 35 translocation partner genes occur recurrently, but only 9 specific gene fusions account for more than 90% of cases. Translocation of KMT2A with SEPT5 gene at 22q11.2 is rare, with few reported cases in the literature. In this report, we are presenting a case of KMT2A-SEPT5 fusion in de novo acute myeloid leukemia with t(11;22)(q23;q11.2) with a review of the literature.
Keywords: Acute myeloid leukemia, KMT2A, SEPT5
Introduction
Acute myeloid leukemia (AML) is a heterogeneous disease, reflecting the complexities of myeloid cell differentiation. The recent 2016 WHO classification1 divides AML into four main categories: AML with recurrent genetic aberrations, AML with myelodysplasia-related features, therapy-related myeloid neoplasms, and AML not-otherwise-specified. There are minor refinements related to updates in gene names such as the change from MLL to KMT2A are also included in 2016 WHO classification.1 Many of the genetic rearrangements in the myeloid neoplasms disrupt genes that are required for normal myeloid differentiation and create new transcripts that are leukemogenic.2
Chromosomal rearrangement of the KMT2A gene at 11q23 has been reported in approximately 10% of acute leukemias.3 This rearrangement results in AMLs with predominantly monocytic or myelomonocytic phenotypes. KMT2A rearrangement is also associated with therapy-related myeloid neoplasm, specifically following topoisomerase II inhibitor.4 The MLL recombinome of acute leukemias in 20175 identified 11 novel translocation partner genes (TPGs). Thus, a total of 135 different KMT2A rearrangements have been identified, where 94 TPGs are now characterized at the molecular level. Of these 94 TPGs, 35 occur recurrently, but only 9 specific gene fusions account for more than 90% of cases.
Translocation of KMT2A with SEPT5 gene at 22q11.2 is extremely rare with few reported cases in the literature.6 We are presenting a case of KMT2A-SEPT5 fusion in AML with t(11;22)(q23;q11.2) and review of the literature.
Case report
A 43-year-old Hispanic female presented to the emergency room complaining of fatigue for 2 months with intermittent nose bleeding for 2 weeks. The patient had no family history of hematologic malignancies and no history of previous exposure to chemotherapeutic agents. At the time of presentation, she had pancytopenia with a hemoglobin of 7.2 g/dL, white blood cell count of 3.61 × 103/µL, platelet count of 21X103/µL, and the absolute neutrophil count of 260 cells/µL. The peripheral blood smear showed increased monocytes with rare immature and atypical monocytes.
Bone marrow biopsy was hypercellular for age (90% cellularity) with decreased trilineage hematopoiesis and involvement by 80% blast cells. The leukemic cells were predominantly formed of myeloblasts, monoblasts, immature, and abnormal monocytes. The monocyte precursors formed 38.4% of all nucleated cells. Leukemic cells showed folded nuclei (Figure 1). No Auer rods were noted. Some of the leukemic cells stained for non-specific esterase (alpha-naphthyl butyrate esterase cytochemical stain). Specific esterase (chloroacetate esterase) was positive in monocytes and other granulocytes. Bone marrow biopsy was consistent with the diagnosis of acute myelomonocytic leukemia (French-American-British (FAB) classification M4). Flow cytometry, cytogenetic analysis, and dual-color fluorescent in situ hybridization (FISH) assay were performed.
Figure 1.
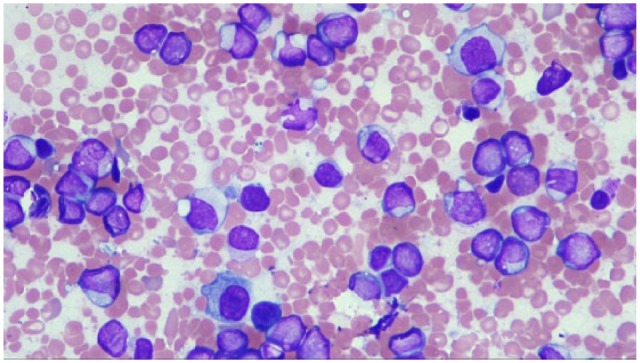
Bone marrow biopsy shows myeloblast population, some with folded nuclei (arrow).
Material and methods
Flow cytometry analysis
Flow cytometry was conducted on the suspended cells utilizing the Beckman Coulter FC500 instrument. Briefly, 100 mL of suspended cells were incubated with 20 mL of the antibody for 15 min, then lysed with fluorescence-activated cell sorting (FACS) solution for 10 min, and finally washed with a phosphate buffer saline (PBS) solution. The data were acquired on the Beckman Coulter FC500 Flow Cytometer and analyzed using the Beckman CXP software.
Cytogenetic analysis
A representative sample of the bone marrow was received for cytogenetic analysis. Standard culture and harvest procedures were performed. Briefly, the tissue was disaggregated and cultured in RPMI 1640 media supplemented with 20% fetal bovine serum for 24–48 h. The cells were exposed to colcemid for 45 min (0.02 μg/mL), were treated with hypotonic solution (0.075M KCl for 20 min), and fixed with methanol and glacial acetic acid (3:1). Metaphase cells were banded with Leishman stain.
Dual-color FISH assay
In an attempt to identify the putative genes involved in t(11;22)(q23;q11.2) translocation, KMT2A dual color, break-apart probe (Cytocell, Tarrytown, NY, USA) and SEPT5 aqua color probe (Empire Genomics, Buffalo, NY, USA) were used. FISH probe set was designed in such a way that the two probes flank both sides of the MLL breakpoint region without overlap. The MLL (KMT2A) product consists of an 87kb probe, labeled in red, covering a region telomeric to the MLL (KMT2A) gene including the marker D11S3207 and a green probe covering a 168kb region centromeric to the MLL (KMT2A) gene spanning the CD3G and UBE4A genes. The SEPT5 FISH probe was labeled in aqua, looking for 22q11.21 rearrangements. These two probes were commercially procured and mixed in equal proportions, and this probe mix was used to confirm the MLL/SEPT5 rearrangement.
FISH analysis was performed on both interphase and metaphase cells. FISH procedures were carried out on fixed bone marrow cells. Briefly, the slides was prepared and incubated in 2 SSC for 10 min at 37°C, followed by an alcohol series of dehydration. Co-denaturation was carried out at 75°C for 5 min, followed by overnight hybridization at 37°C using the HYBrite instrument (Vysis). Post-hybridization, the slides were washed with 2× SSC/0/3% NP-40 working solution at 70°C for 2 min and counterstained with 4′,6-diamidino-2-phenylindole (DAPI II; Vysis). Evaluation of the FISH signals was performed using the Olympus BX51 fluorescent microscope under 100× magnification. In total, 400 interphase nuclei and 10 metaphase cells were evaluated. Images were acquired by use of the CytoVision Image Analysis System (Applied Imaging, Santa Clara, CA, USA). The FISH results were interpreted in the context of routine G-banded karyotypes, and metaphases were used to verify the interphase FISH pattern as well as to confirm the chromosomal position of the rearrangement.7
The 2016 edition of the International System for Human Cytogenomic Nomenclature (ISCN 2016)8 was used for the nomenclature that is used to describe any genomic rearrangement identified by karyotyping to FISH techniques.
Results
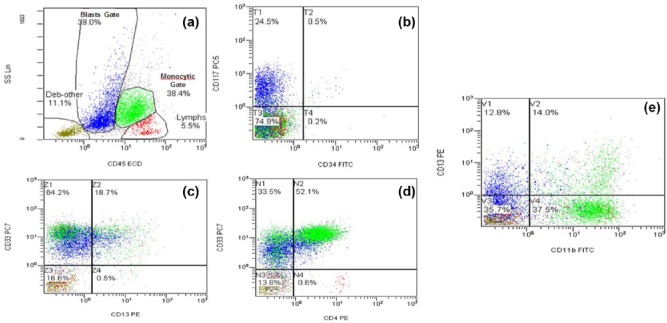
Concurrent flow cytometry analysis was performed on the entire population of cells. Aberrant population of monocytes forming 38.4% of cells expressed CD4, CD11b, CD13 (partial), CD14 (partial), CD15, CD33, and HLA-DR, CD64, CD36, and MPO (partial). On the lymphoid gate that represented 11.0% of total cells, B-lymphocyte population showed a polyclonal phenotype expressing CD19 and CD20. The ratio of Kappa to Lambda was normal at 1.45. The T-lymphocyte population had a normal mature phenotype expressing CD2, CD3, CD5, and CD4: CD8 ratio of 0.69. (Figure 2(a)). Both populations were negative for CD34. The cells in the monocyte gate (38.4% of total cells) expressed CD4, CD11b, CD13 (partial), CD14 (partial), CD15, CD33 and HLA-DR. (Figure 2).
Figure 2.
Flow cytometry analysis show 2 prominent populations in blast and monocyte gates (a). These 2 populations are negative for CD34 (b) and positive for CD33 (c). Blast population is positive for CD117 (b), and weakly for CD4 (d), and the monocytic population is positive for CD4 (d) and CD11b (e).
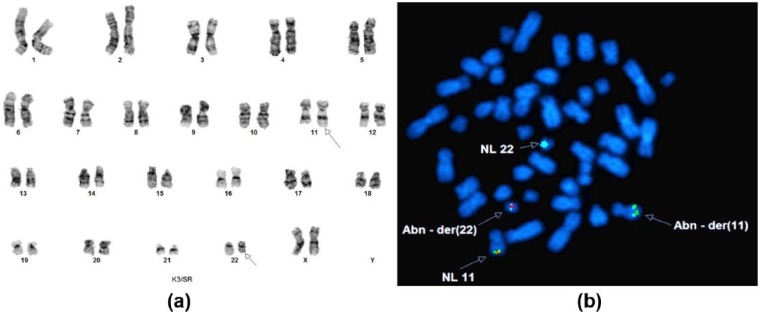
Chromosomal analysis showed an abnormal female karyotype with a balanced translocation between 11q23 and 22q11.2 in all 20 metaphase cells; 46, XX, t(11;22)(q23;q11.2) (Figure 3(a)). Molecular analyses were negative for NPM1, CEBPA, FLT-3 ITD, and FLT-3 TKD mutations. FISH on the metaphase cells of the patient using KMT2A break-apart probe (red and green) and SEPT5 probe (aqua) showed that the 3′KMT2A segment was translocated to the derivative chromosome 22, and the breakpoint on chromosome 22 was located near the SEPT5 gene at 22q11.2. The rearrangement of KMT2A with SEPT5 resulting in der(11) and der(22) chromosomes is shown in (Figure 3(b)). The patient was given induction chemotherapy using anthracycline and cytarabine; patient’s condition deteriorated rapidly, and she was transferred to the ICU. The patient developed ventricular tachycardia, then pulseless electrical activity, and unfortunately, she succumbed to the disease in 2 months.
Figure 3.
(a) Balanced translocation between the long arm of chromosome 11 and the long arm of chromosomes 22, with breakpoints at 11q23 and 22q11.2. (b) Metaphase FISH analysis confirmed that the 3′ end of KMT2A (red) has translocated to chromosome 22 and bind to SEPT5 (aqua), and part of SEPT5 (aqua) has fused to the 5′ end region (green) of KMT2A on the derivative 11.
Discussion
To our knowledge to this date, nine cases of AML with t(11;22)(q23;q11.2) translocation resulting in KMT2A-SEPT5 fusion transcripts have been reported (Table 1); three of the cases were infants, four were in adults, and two cases were unknown (article in Chinese).9 All cases including ours were diagnosed as AML. The overall incidence of AML with monocytic differentiation in KMT2A-SEPTIN cases is 54%, higher with the KMT2A-SEPT9 and lower with the KMT2A-SEPT6 translocations.10 In the AMLs with KMT2A-SEPT5, less than half of the reported cases including the current case demonstrate monocytic or myelomonocytic phenotype (Table 1). Tatsumi et al. detected the expression of SEPT5 in 13 AML and 14 acute lymphoblastic leukemia (ALL) cell lines and found that its expression is significantly higher in AML cell lines than in ALL. This might indicate the importance of KMT2A-SEPT5 fusion in the leukemogenesis of AML with t(11;22)(q23;q11.2).
Table 1.
All the reported cases of AML with KMT2A-SEPT5 fusion.
| Reference | Age/sex | Diagnosis (FAB) | Karyotype | KMT2A break-apart probe FISH | SEPT5 FISH | Prognosis |
|---|---|---|---|---|---|---|
| Current case | 43/F | AML-M4 | t(11;22)(q23;q11.2) | Positive | KMT2A-SEPT5 | OS: 2 months |
| Gao et al.6 | 32/M | AML-M2 | t(11;22)(q23;q11.2) | Positive | ND | OS: 7 months |
| Megonigal et al.11 | 11.5 months/F | AML-M2 | t(11;22)(q23;q11.2) | Positive | ND | NR |
| Megonigal et al.11 | 13 months/F | AML-M1 | t(11;22)(q23;q11.2) | Positive | ND | NR |
| Launay et al.12 | 23 months/F | AML-M5 | t(11;22)(q23;q11) | Positive | MLL-SEPT5 | Survived >2 years after allograft |
| Tatsumi et al.13 | 39/M | AML-M2 | t(11;22)(q23;q11) | Positive | SEPT5 Split Signals | OS: 12 months |
| Wang et al.9 | Not available | AML-M2 | t(11;22)(q23;q11.2) | Not available | Not available | CR after induction but died during the consolidation therapy |
| Wang et al.9 | Not available | AML-M5 | t(11;22)(q23;q11.2) | Not available | Not available | CR by induction chemo |
| Wang et al.14 | 21/M | AML-M1 | t(11;22)(q23;q11.2) | Positive | ND | CR by induction chemo and BM transplant. In remission to date. |
| Wang et al.14 | 22/F | AML-M4 | t(11;22)(q23;q11.2) | NR | ND | NR |
AML: acute myeloid leukemia; CR: complete remission; FISH: fluorescent in situ hybridization; ND: not done; NR: not recorded; OS: over all survival.
The overall survival of the AML patients with KMT2A-SEPT5 varied according to the therapy protocol. Cases who received allogenic bone marrow graft showed the longest survival. One case survived 2 years12 and the other is in remission till the date of publication in 2016.12 All cases listed in Table 1 are de novo, and none were therapy-induced leukemia.
KMT2A translocation can be divided into four distinct categories: fusion with nuclear proteins, fusion with cytoplasmic proteins, fusion to histone acetyltransferases, and fusion to septin family members.10 Septins belong to GTP-binding proteins that are associated with cell membrane and cytoskeleton.15 There are at least 13 septin genes throughout the human genome that are expressed in almost all types of tissue; however, expression of some is higher in certain tissues.15 They have a role in cytokinesis, apoptosis, vesicle trafficking, cell motility, and possibly cell DNA damage checkpoint.16 Alterations in septin expression have been reported in many malignancies including colorectal,17 breast,18 ovarian,19 urogenital, and brain tumors,20 as well as non-malignant conditions such as Alzheimer21 and Parkinson diseases.22
Among the septin family genes, rearrangement of SEPT2, SEPT5, SEPT6, SEPT9, and SEPT11 with KMT2A have been reported; the majority of them in patients with myeloid neoplasm.10 The breakpoints in all the septin genes happen between exon 2 and exon 4, creating a fusion gene with KMT2A that contains almost the entire open reading frame of the septin genes.10 Interestingly, the majority of the translocation involving SETP6 happens in pediatric AML23 and the ones with SEPT2 in therapy-related myeloid neoplasms.24 There have been two reports of KMT2A-SEPT11, one in a patient with chronic neutrophilic leukemia25 and the other in ALL.26
Santos et al.16 have shown that expression of septin genes such as SEPT2 and SEPT6 are down-regulated in KMT2A-SEPT2- and KMT2A-SEPT6-related myeloid neoplasms.16 Moreover, they demonstrated that down-regulation of septin genes, including septin 5, is not limited to the myeloid neoplasms with KMT2A-SEPTIN and can be detected in other neoplasms including AML with PML-RARA.16 However, the significance of this observation is still unclear and requires additional studies.
In conclusion, we report here a new case of de novo AML with KMT2A-SEPT5 fusion. Up to date, there are only reported nine cases with the same fusion. The prognosis of the cases varied where our case survived for 2 months since the first induction chemotherapy, while others survived for >2 years after allogenic transplantation. Further studies are required to explore the role of septins in leukemogenic. And, the effect of KMT2A-SEPTIN fusion transcripts on the prognosis and response to treatment.
Footnotes
Declaration of conflicting interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Ethical approval: Texas Tech University Health Science Center does not require ethical approval for reporting individual cases or case series.
Funding: The author(s) received no financial support for the research, authorship, and/or publication of this article.
Informed consent: Informed consent for patient information to be published in this article was obtained.
References
- 1. Arber DA, Orazi A, Hasserjian R, et al. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood 2016; 127(20): 2391–2405. [DOI] [PubMed] [Google Scholar]
- 2. Licht JD, Sternberg DW. The molecular pathology of acute myeloid leukemia. Hematology Am Soc Hematol Educ Program 2005: 137–142. [DOI] [PubMed] [Google Scholar]
- 3. Marschalek R. Systematic classification of mixed-lineage leukemia fusion partners predicts additional cancer pathways. Ann Lab Med 2016; 36(2): 85–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Biondi A, Cimino G, Pieters R, et al. Biological and therapeutic aspects of infant leukemia. Blood 2000; 96(1): 24–33. [PubMed] [Google Scholar]
- 5. Meyer C, Burmeister T, Groger D, et al. The MLL recombinome of acute leukemias in 2017. Leukemia. Epub ahead of print 13 July 2017. DOI: 10.1038/leu.2017.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gao W, Wang T, Wu Y, et al. Mixed lineage leukemia-septin 5 fusion transcript in de novo adult acute myeloid leukemia with t(11;22)(q23;q11.2): a case report. Oncol Lett 2014; 7(6): 1930–1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cytocell. Instructions for use: MLL breakapart probe kit insert. DS083/CE v007.00/2014-03-19. [Google Scholar]
- 8. ISCN 2016: an international System for Human Cytogenomic Nomenclature. Reprint of Cytogenetic and Genome Research, Karger, 2016; 149(1–2). [Google Scholar]
- 9. Wang T, Gao W, Liu HX, et al. Acute myeloid leukemia with t(11;22) (q23;q11.2): two cases report and literature review. Zhonghua Xue Ye Xue Za Zhi 2013; 34(12): 1028–1031. [DOI] [PubMed] [Google Scholar]
- 10. Cerveira N, Bizarro S, Teixeira MR. MLL-SEPTIN gene fusions in hematological malignancies. Biol Chem 2011; 392(8–9): 713–724. [DOI] [PubMed] [Google Scholar]
- 11. Megonigal MD, Rappaport EF, Jones DH, et al. t(11;22)(q23;q11.2) In acute myeloid leukemia of infant twins fuses MLL with hCDCrel, a cell division cycle gene in the genomic region of deletion in DiGeorge and velocardiofacial syndromes. Proc Natl Acad Sci USA 1998; 95(11): 6413–6418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Launay E, Henry C, Meyer C, et al. MLL-SEPT5 fusion transcript in infant acute myeloid leukemia with t(11;22)(q23;q11). Leuk Lymphoma 2014; 55(3): 662–667. [DOI] [PubMed] [Google Scholar]
- 13. Tatsumi K, Taki T, Taniwaki M, et al. The CDCREL1 gene fused to MLL in de novo acute myeloid leukemia with t(11;22)(q23;q11.2) and its frequent expression in myeloid leukemia cell lines. Genes Chromosomes Cancer 2001; 30(3): 230–235. [DOI] [PubMed] [Google Scholar]
- 14. Wang N, Wu X, Sheng G, et al. MLL-SEPT5 fusion transcript in two de novo acute myeloid leukemia patients With t(11;22)(q23;q11). Ann Lab Med 2016; 36(5): 501–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hall PA, Russell SE. The pathobiology of the septin gene family. J Pathol 2004; 204(4): 489–505. [DOI] [PubMed] [Google Scholar]
- 16. Santos J, Cerveira N, Bizarro S, et al. Expression pattern of the septin gene family in acute myeloid leukemias with and without MLL-SEPT fusion genes. Leuk Res 2010; 34(5): 615–621. [DOI] [PubMed] [Google Scholar]
- 17. Tanaka M, Kijima H, Itoh J, et al. Impaired expression of a human septin family gene Bradeion inhibits the growth and tumorigenesis of colorectal cancer in vitro and in vivo. Cancer Gene Ther 2002; 9(6): 483–488. [DOI] [PubMed] [Google Scholar]
- 18. Kalikin LM, Sims HL, Petty EM. Genomic and expression analyses of alternatively spliced transcripts of the MLL septin-like fusion gene (MSF) that map to a 17q25 region of loss in breast and ovarian tumors. Genomics 2000; 63(2): 165–172. [DOI] [PubMed] [Google Scholar]
- 19. Burrows JF, Chanduloy S, McIlhatton MA, et al. Altered expression of the septin gene, SEPT9, in ovarian neoplasia. J Pathol 2003; 201(4): 581–588. [DOI] [PubMed] [Google Scholar]
- 20. Scott M, Hyland PL, McGregor G, et al. Multimodality expression profiling shows SEPT9 to be overexpressed in a wide range of human tumours. Oncogene 2005; 24(29): 4688–4700. [DOI] [PubMed] [Google Scholar]
- 21. Kinoshita A, Kinoshita M, Akiyama H, et al. Identification of septins in neurofibrillary tangles in Alzheimer’s disease. Am J Pathol 1998; 153(5): 1551–1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ihara M, Tomimoto H, Kitayama H, et al. Association of the cytoskeletal GTP-binding protein Sept4/H5 with cytoplasmic inclusions found in Parkinson’s disease and other synucleinopathies. J Biol Chem 2003; 278(26): 24095–24102. [DOI] [PubMed] [Google Scholar]
- 23. De Braekeleer E, Meyer C, Douet-Guilbert N, et al. Complex and cryptic chromosomal rearrangements involving the MLL gene in acute leukemia: a study of 7 patients and review of the literature. Blood Cells Mol Dis 2010; 44(4): 268–274. [DOI] [PubMed] [Google Scholar]
- 24. Cerveira N, Correia C, Bizarro S, et al. SEPT2 is a new fusion partner of MLL in acute myeloid leukemia with t(2;11)(q37;q23). Oncogene 2006; 25(45): 6147–6152. [DOI] [PubMed] [Google Scholar]
- 25. Kojima K, Sakai I, Hasegawa A, et al. FLJ10849, a septin family gene, fuses MLL in a novel leukemia cell line CNLBC1 derived from chronic neutrophilic leukemia in transformation with t(4;11)(q21;q23). Leukemia 2004; 18(5): 998–1005. [DOI] [PubMed] [Google Scholar]
- 26. Stevens SJ, Meers LE, Albrechts JC, et al. A translocation in acute lymphoblastic leukemia that cytogenetically mimics the recurrent MLL-AFF1 translocation and fuses SEPT11 to MLL. Cancer Genet Cytogenet 2010; 201(1): 48–51. [DOI] [PubMed] [Google Scholar]