Abstract
RCC2 (also known as TD60) is a highly conserved protein involved in prognosis in colorectal cancer. However, its relationship with tumor development is less understood. Here we demonstrate a signaling pathway defining regulation of RCC2 and its functions in tumor progression. We report that p53 is a transcriptional regulator of RCC2 that acts through its binding to a palindromic motif in the RCC2 promoter. RCC2 physically interacts and deactivates a small GTPase Rac1 that is known to be involved in metastasis. We solved a high-resolution crystal structure of RCC2 and revealed one RCC1-like domain with a unique β-hairpin that is requisite for RCC2 interaction with Rac1. p53 or RCC2 deficiency leads to activation of Rac1 and deterioration of extracellular matrix sensing (haptotaxis) of surface-bound gradients. Ectopic expression of RCC2 restores directional migration in p53-null cells. Our results demonstrate that p53 and RCC2 signaling is important for regulation of cell migration and suppression of metastasis. We propose that the p53/RCC2/Rac1 axis is a potential target for cancer therapy.
Introduction
Cell migration is fundamental to development and maintenance of organisms during embryonic development and wound healing. However, defects in cell migration such as loss of directional migration, and especially such defects occurring during tumor formation, contribute to metastasis that leads to therapy failure and ultimately results in cancer death. Tumor cell migration and directionality are under the spatiotemporal control of complex signaling pathways and cytoskeletal networks.
The roles of tumor suppressor p53 in DNA damage, cell cycle and apoptosis have been extensively investigated.1 However, the function and mechanism of p53 in regulation of cell migration and metastasis is incompletely understood. We recently demonstrated that p53 is essential for suppression of metastasis in a mouse model.2 It has been reported that p53 regulates E-cadherin expression and restricts the progress of epithelial–mesenchymal transition.3, 4 Several targets of p53 in cell motility control have also been identified.5, 6, 7 However, deficiencies in motility control are not sufficient to drive migration. Failure to recognize and respond to directional cues such as extracellular matrix (ECM) leads to aberrant cell migration. The relationship of p53 with matrix sensing is currently not well understood.
RCC2 has previously been identified as a component of the chromosome passenger complex that plays a critical and fundamental role in ensuring that the mitotic process proceeds uniformly and accurately.8, 9, 10 The role of RCC2 in cancer has increasingly come under scrutiny in recent years. Several genome-scale sequencing projects have revealed the existence of RCC2 mutations in cancer, and most of these are found in colorectal cancers with a relatively high rate of incidence.11, 12, 13 There are reports that suggest abnormal RCC2 status is essential to the development of colorectal cancer (CRC), and that reduction of RCC2 expression is associated with poor outcome in patients with microsatellite stable (MSS) tumors.14, 15 These studies indicate there may be a link between RCC2 and colon cancer that exists outside of the area of mitotic control. However, the relation of RCC2 to cancer development and the function of this molecule have until now received little attention.
Rac1 is a member of the Rho family of small GTPases that plays a fundamental role in a wide variety of cellular processes. Together with its effectors, Rac1 participates in cytoskeleton remodeling, directional migration control and cell transformation.16, 17, 18 The activity of Rac1 is synergistically controlled by a group of proteins, and a small upregulation in total Rac1 activity can make cells switch from directionally persistent cell migration to random migration,19 facilitating the wandering and invasion of cancer cells. It is reported that p53 loss may lead to overactivation of Rac1, but this mechanism is incompletely understood.3, 20
In this study we found that RCC2 is transcriptionally activated by p53 through binding to a palindromic motif in the promoter of RCC2. RCC2 deficiency results in changes in cell morphology and Rac1 activation that causes deterioration in matrix sensing and directionality and increases cell migration. RCC2 overexpression in p53-null cells, or Rac1 inhibition in RCC2-null cells, restores directional migration and suppresses cell metastasis. We also solved the crystal structure of RCC2 at high resolution and found that RCC2 contains one RCC1-like domain with a β-hairpin that is important for interaction with Rac1. Our observations thus define a p53/RCC2/Rac1 signaling pathway and reveal its importance for suppression of tumor metastasis.
Results
Identification of RCC2 as a novel target of p53 in cell migration control
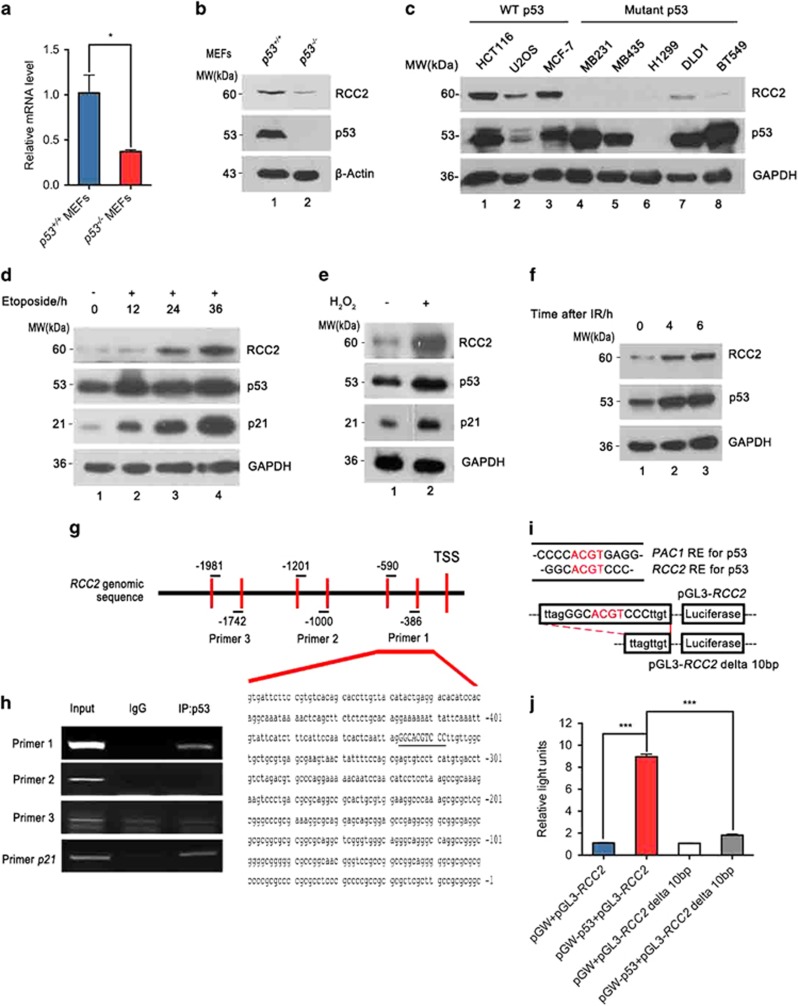
We have recently demonstrated that the tumor suppressor p53 mainly acts at later stages of tumor progression, and is essential for suppression of metastasis in a mouse model.2 Through microarray analysis using mouse embryonic fibroblasts (MEFs) derived from p53+/+ and p53−/− mice, we found expression of the gene RCC2 is altered under p53 depletion. RCC2 was previously identified as a potential target in malfunctioning DNA mismatch repair-deficient CRC.15 Weak RCC2 protein expression is significantly associated with poor prognosis in MSS-type CRC patients14 and RCC2 was found in the α5β1–fibronectin signaling network that reveals its role in cell migration and metastasis.21 This raised the possibility that p53 suppresses metastases by regulation of RCC2 expression. Quantitative PCR analysis showed that RCC2 mRNA levels are significantly reduced when p53 is deficient (Figure 1a). This result was confirmed using p53+/+HCT116 and p53−/− HCT116 cells (Supplementary Figure S1a). We also found that RCC2 protein levels are decreased when p53 is lost (Figure 1b and Supplementary Figure S1b). Evaluating expression of RCC2 in several cell lines carrying wild-type or mutant p53 consistently showed that RCC2 protein is dramatically reduced when p53 is dysfunctional (Figure 1c, lanes 4–8 versus lanes 1–3). RCC2 expression is upregulated in a time-dependent manner under genotoxic stress (Figure 1d) or other cellular stress, including oxidative stress and gamma irradiation that are classic means of p53 activation (Figures 1e and f and Supplementary Figure S1c). Moreover, in EB-1 cells where ZnCl2 can conditionally induce expression of transgenic p53 that is under the control of the metallothionein promoter,22 RCC2 upregulation is positively correlated with p53 induction (Supplementary Figure S1d). These results indicate that RCC2 expression is induced by p53.
Figure 1.
RCC2 is a transcriptional target of p53. (a) mRNA expression of RCC2 in p53−/−mouse embryonic fibroblasts (MEFs) compared with p53+/+ MEFs. Data are presented as mean±s.e.m. and were analyzed with the unpaired t-test (n=3). *P<0.05. (b) Protein expression of RCC2 in p53+/+MEFs and p53−/−MEFs. (c) Cell lines are grouped according to p53 status (wild-type (WT) or mutant). The protein levels of RCC2 and p53 in these cell lines are shown by western blot. (d) p53 and RCC2 expression under treatment with etoposide for indicated times. Expression of p21 represents p53 activity. (e) Expression of RCC2 is induced by oxidative stress. p21 is used as a marker for p53 activation. (f) HCT116 cells were irradiated with 12 Gy and recovered for the indicating time. Expression of RCC2 and p53 were evaluated with specific antibodies. (g) Locations of three pairs of primers used to amplify the RCC2 genomic sequence. Numbers indicate primer location upstream of the transcriptional start site (TSS). (h) Genomic DNA fragments precipitated from HCT116 cells by p53 were used as templates to amplify indicated RCC2 genomic sequences with specific RCC2 primers. The DNA sequence covered by the first primer is shown and the palindromic motif is underlined. Anti-p53 antibody was used to immunoprecipitate p53. Nonspecific IgG was used as a negative control and p21 primer was used as a positive control. (i) Comparison of PAC1 and RCC2 sequences, and pGL3-RCC2 and pGL3-RCC2 delta 10 bp plasmid sequences. (j) p53−/− HCT116 cells were transfected with indicated combinations of pGW-p53 and various RCC2 promoter reporter plasmid constructs using pGW and pGL3 as negative controls. Luciferase activity was measured. All data are presented as mean±s.e.m. and were analyzed with the unpaired t-test (n=3). ***P<0.001. See also Supplementary Figure S1.
In order to identify the binding site of p53, three pairs of primers were designed to target different RCC2 promoter sequences over the region −1 to −2000 (Figure 1g) and we found a sequence near the RCC2 transcriptional start site that can be specifically immunoprecipitated by p53 (Figure 1h, left panel). Analysis of this promoter sequence did not reveal a consensus p53 binding site, but there was a 10 bp palindromic sequence (underlined in Figure 1h, right panel) ∼360 bp upstream from the transcriptional start site that is similar to the nonconsensus p53 binding site that has been characterized in known p53 targets such as PAC1 (Figure 1i) and DUSP1.23, 24, 25 To determine whether this palindromic sequence is recognized by p53, we constructed a RCC2 reporter vector (pGL3-RCC2) containing this motif (Figure 1j) and transfected it into p53−/−HCT116 cells together with empty or wild-type p53-expressing pGW vectors, and evaluated luciferase activity. As shown in Figure 1j, activity of the luciferase reporter was markedly induced by wild-type p53 as compared with the controls, and there was no response to p53 when this motif was deleted (designated pGL3-RCC2 delta 10bp, see Figure 1i; Figure 1j). This strongly suggests that this palindromic sequence is essential for positive regulation of RCC2 transcription by p53. Moreover, we also tested binding of p53 to this palindromic motif using the electrophoretic mobility shift assay. As shown in Supplementary Figure S1e, p53 protein generated a shift band (lane 2). When excess unlabeled competitive probes (100 ×) were added, the shift band could not be detected (lanes 3 and 4). However, the excess mutant probes (100 ×) were not able to compete with the labeled probes because of lack of the palindromic sequence (lanes 5 and 6). This shows that p53 can bind to this palindromic sequence and indicates that RCC2 is a novel p53 transcriptional target.
Reduction of RCC2 facilitates cell migration and tumor metastasis
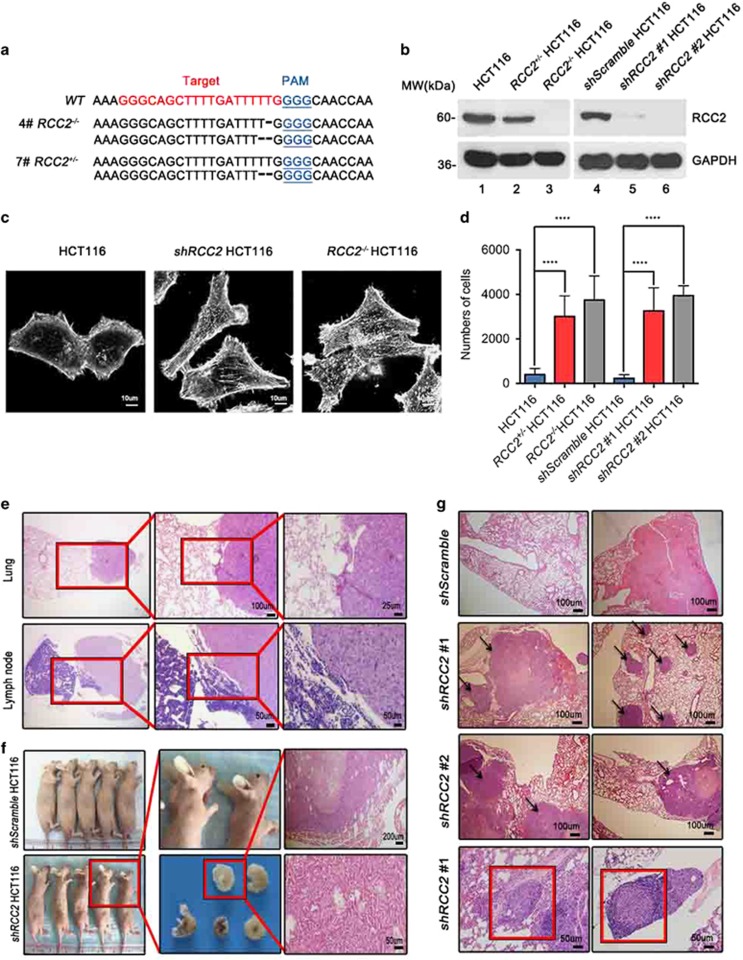
Protein distribution shows that RCC2 is predominantly expressed in the large intestine, spleen and lung tissues (Supplementary Figure S2a), reflecting the potential importance of RCC2 in CRC development. In order to study the function of RCC2, we generated somatic RCC2 heterozygous or homozygous knockout in a colon cancer cell line (RCC2+/−HCT116, RCC2−/−HCT116) using the CRISPR-Cas9 approach (Figure 2a). We generated stable shRCC2 HCT116 (shRCC2#1, shRCC2#2 HCT116) and control shScramble cell lines by introducing shRCC2 expression vectors or a control shScramble vector into HCT116 cells, and confirmed that RCC2 protein expression was significantly reduced as expected (Figure 2b, lanes 2 and 3 versus lane 1, lanes 5 and 6 versus lane 4). We observed that RCC2 deficiency results in prominent elongated morphologic changes in shRCC2 and RCC2−/−HCT116 cells as compared with control HCT116 cells (Figure 2c and Supplementary Figure S2b). These results argue that RCC2 plays a role in regulation of cell motility. To address this hypothesis, we tested the capacity of shScramble and shRCC2 HCT116 cells for migration with the wound-healing assay. shRCC2 HCT116 cells showed a much stronger tendency to migrate than shScramble cells (Supplementary Figures S2c and d). Loss of RCC2 increased cell migration as quantified using the transwell assay, consistent with findings in the wound-healing assay (Figure 2d). At the same time, these cell lines showed no significant differences in cell proliferation (Supplementary Figure S2e). A xenograft nude mouse model was employed to test the potential of RCC2-deficient cells for tumor metastasis in vivo. shScramble and shRCC2 HCT116 cells were injected into the subcutaneous tissues of nude mice, and distant metastases in the lungs and abdominal lymph nodes were observed more frequently in nude mice injected with shRCC2 cells (Figure 2e). All mice injected with shRCC2 cells died within 3 months, but only one mouse in the shScramble group died (Supplementary Figure S2f). To further confirm metastases are caused by RCC2 depletion, we injected nude mice intravenously with shScramble and shRCC2 cells. In these groups, 40% of the mice injected with shRCC2 cells produced metastatic tumors in the subcutaneous tissues of the chest or on the back (Figure 2f), as confirmed by hematoxylin and eosin (H&E) staining. We found that intravenous injection of RCC2-depleted cells enhanced tumor metastasis to lungs and lymph nodes, and was associated with more frequent tumor formation in nude mice compared with the group injected with shScramble cells where no metastatic tumor was found (Figure 2g). These results demonstrate that RCC2 is a powerful suppressor of tumor cell metastasis in vivo.
Figure 2.
Reduction of RCC2 facilitates cell migration and tumor metastasis. (a) Generation of CRISPR-mediated RCC2 knockout cell lines. Target site and PAM motif are illustrated with red and blue. Mutations of number 4 and 7 clones are shown with specific nucleotide deletions. (b) Elimination of RCC2 protein expression in shRCC2 #1, shRCC2 #2, RCC2+/− and RCC2−/−cells. (c) Morphology of HCT116, shRCC2 and RCC2−/− HCT116 cells shown with phalloidin staining. Cells were scanned with confocal microscopy. (d) Average numbers of migrated HCT116, RCC2+/−, RCC2−/−, shScramble, shRCC2 #1 and shRCC2 #2 HCT116 cells were calculated using the transwell assay. All data are presented as mean±s.e.m. and were analyzed by the unpaired t-test (n=4). ****P<0.0001. (e) Hematoxylin and eosin (H&E)-stained tissues from nude mice inoculated with shScramble and shRCC2 cells. Metastases found in lung tissues and lymph nodes are shown at various magnifications. Red boxes indicate the margins of normal tissue (left) and tumors (right). (f) Of the nude mice intravenously injected with shRCC2 cells, 40% produced tumors of the back or neck compared with the control group injected with shScramble cells. Each group contained five individual nude mice. (g) H&E staining shows shRCC2 cell metastases to lung tissue with formation of multiple tumor colonies in nude mice, but shScramble cells caused only necrosis. Red boxes show cells invading a blood vessel (left) and a lymph node (right), and arrows indicate metastatic tumor. See also Supplementary Figure S2.
RCC2 inactivates Rac1 and mediates p53 function to suppress Rac1 activation
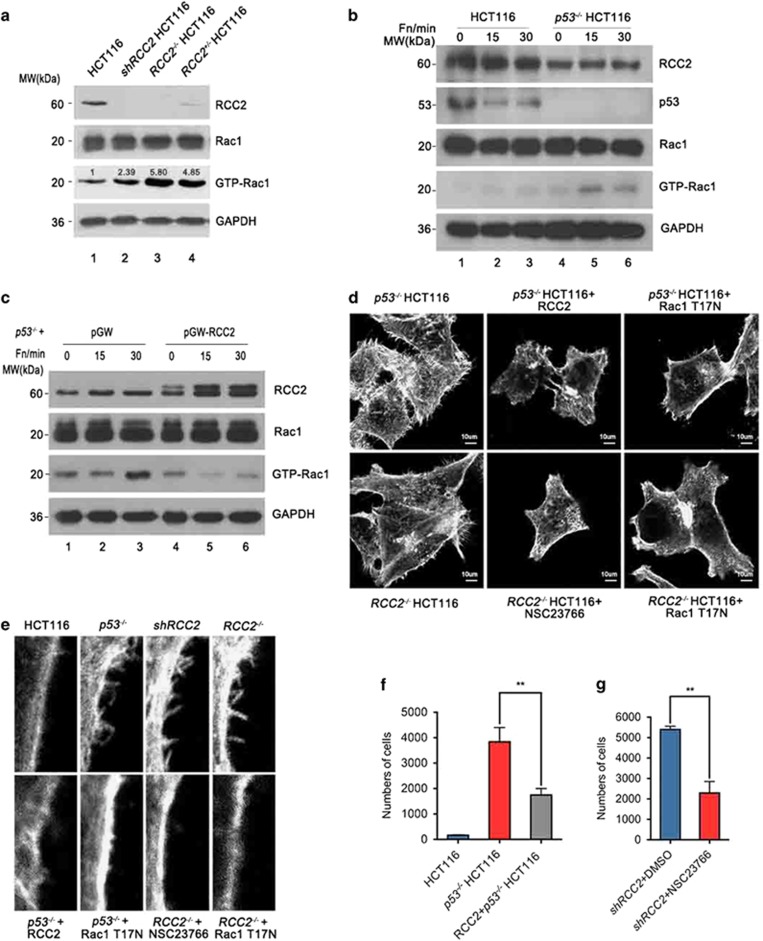
In order to find potential interaction proteins that account for RCC2 deficiency-induced cell migration, we conducted a flag-RCC2 pull-down assay and evaluated the proteins with mass spectrum analysis. We found that the small GTPase Rac1 that is closely associated with cell migration was pulled down by RCC2 (Supplementary Figure S3a), and this interaction was confirmed with immunoprecipitation assays (Supplementary Figure S3b). The activation or inactivation of Rac1 is coordinately controlled when Rac1 binds to its partners. Indeed, we found that endogenous GTP-Rac1 accumulates when RCC2 is knocked down or knocked out (Figure 3a, lanes 2–4 versus lane 1). Previous reports have shown how Rac1 activation promotes cell migration. In conjunction with this, there are controversial mechanisms that putatively account for p53 deficiency-induced Rac1 activation.20, 26 We were therefore led to consider that RCC2 is a mediator of p53 that inhibits cell migration through Rac1 inhibition. Our results showed that RCC2 or p53 deficiency triggers Rac1 activation (Figures 3a and b), and RCC2 overexpression in p53−/− cells is sufficient to suppress Rac1 activation (Figure 3c, lanes 5–6 versus lanes 2–3). This further supports our hypothesis. Moreover, we found that loss of RCC2 or p53 promotes filopodia formation, and artificial expression of RCC2 in p53−/− cells reduces filopodial protrusions on the cell surface (Figures 3d and e). An identical result is seen when the dominant negative mutant Rac1T17N is overexpressed in p53−/− and RCC2−/− cells, or by using the Rac1 inhibitor NSC23766 with RCC2−/− cells (Figures 3d and e). In addition, p53 deficiency enhanced cell migration (Figure 3f) and RCC2 overexpression in p53−/−cells inhibited cell migration as compared with the control group (Figure 3f). Increases in migration of RCC2-deficient cells can be inhibited by using NSC23766 (Figure 3g). These results show that RCC2 functions downstream of p53 to inactivate Rac1 and suppress cell migration.
Figure 3.
RCC2 inactivates Rac1 and mediates p53 function to suppress Rac1 activation. (a) Activation of Rac1 in RCC2 knockdown and knockout cells. Cells were lysed and equal amount of proteins were incubated with GST-PAK1 protein to pull down GTP-Rac1. Expression of GTP-Rac1 was evaluated with western blot using an anti-Rac1 antibody. Numbers represent the fold change of GTP-Rac1 compared with HCT116 cells. (b) HCT116 and p53−/− HCT116 cells were stimulated with fibronectin for specific lengths of time and evaluated for Rac1 activation. (c) RCC2 was overexpressed in p53−/− cells, and cells are stimulated with fibronectin for specific lengths of time and evaluated for Rac1 activation. (d) Changes in morphology in p53−/−and RCC2−/− cells. RCC2 and the Rac1 T17N mutant were transfected into specific cells. NSC23766 was used to inhibit Rac1 activation. (e) Magnified view of filopodia of p53−/− and RCC2−/− cells with RCC2 overexpression or Rac1 inactivation. (f) Capacity for migration of p53−/− cells and RCC2 overexpressing p53−/− cells. Data are presented as mean±s.e.m. and were analyzed with the unpaired t-test (n=3). **P<0.01. (g) Transwell assay showed the consequences of Rac1 inactivation by NSC23766 in shRCC2 cells. Data are presented as mean±s.e.m. and were analyzed with the unpaired t-test (n=2). **P<0.01. See also Supplementary Figure S3.
RCC2 protein folds as a seven-bladed propeller structure with a unique β-hairpin
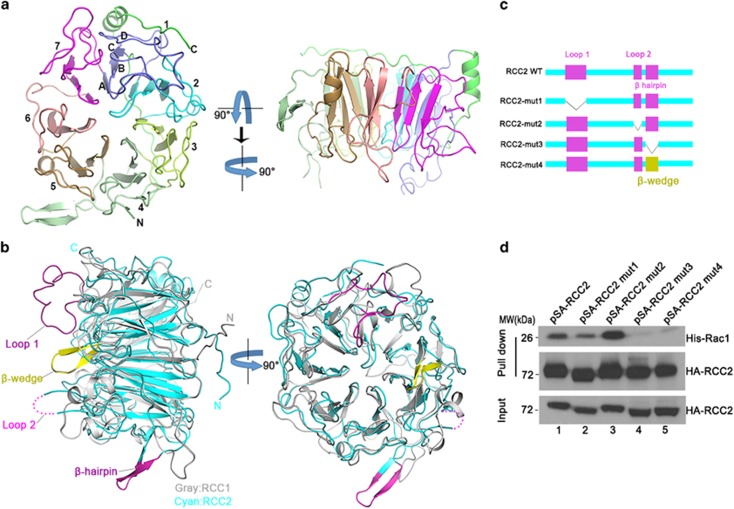
To better understand the mechanism by which RCC2 inactivates Rac1, we conducted structural biology studies. We first analyzed the structure of RCC2 by solving the crystal structure of a truncated RCC2 protein lacking the N-terminal residues 1–88 that are poorly conserved (Supplementary Figure S4a), and predicted to be flexible (Protein Data Bank (PDB) ID: 5GWN). This structure was determined by molecular replacement, and refined at 1.31 Å resolution (Table 1). Crystallized RCC2 adopts a seven-bladed propeller-like structure similar to that observed in its family member RCC1 (residues 21–421; PDB ID: 1A12)27 (Figure 4a). This structure comprises 1 α-helix, 8 310-helices and 31 β-strands (Supplementary Figure S4b). Each blade contains 46–76 residues and is formed by 4 β-strands designated βA, βB, βC and βD from the central shaft to the periphery of the propeller. These β-strands form an antiparallel β-sheet (Figure 4a). Residues 295–300 are not visible in the density map, possibly because of their high flexibility. Compared with RCC1, the connecting loops show different conformations, and the positions of the β-sheets differ slightly (with a root-mean-square deviation of 1.694 Å). In addition, the RCC1 β-hairpin extension required for its guanine exchange factor activity that is referred as a β-wedge is not found in RCC2. Instead, an extra switchback loop (loop 1) and an extra β-hairpin extension are found in blade 1 and between blades 4–5 of RCC2 respectively (Figure 4b and Supplementary Figure S4b). Complexes of RCC1-Ran28 and RCC1-nucleosome29 suggest extra structural elements are essential for protein–protein interaction. As RCC2 is similar to RCC1 and Rac1 is similar to Ran, we attempted to superimpose RCC2 and Rac1 onto the experimentally determined RCC1-Ran complex. However, we found that RCC2-Rac1 cannot interact in the same manner as RCC1-Ran, because the switchback loop (loop 1) resides on the top of the seven blade β-propeller and obstructs binding of Rac1 (Supplementary Figure S5). We therefore generated RCC2 deletion mutants to determine which parts of the protein structure account for interaction with Rac1 (Figure 4c). The extra switchback loop (loop1, 114–134, designated mut1), the disordered region (loop2, 295–300, designated mut2) and the β-hairpin (318–325, designated mut3) were individually deleted. We also swapped the β-hairpin with the β-wedge of RCC1 (designated mut4). These mutants were overexpressed in cells, and S-tag pull-down assays were performed. We found that the interaction between RCC2 and Rac1 is abolished only when the β-hairpin is deleted (Figure 4d, lanes 4–5 versus lanes 1–3). It therefore seemed likely that the β-hairpin is important for RCC2 binding of Rac1.
Table 1. Data collection and structure refinement statistics.
| PDB | 5GWN:RCC2 |
|---|---|
| Data collection | |
| Wavelength, Å | 0.97918 |
| Space group | C121 |
| Cell dimensions | |
| a, b, c (Å) | 94.45, 84.06, 60.67 |
| α, β, γ (°) | 90, 101.57, 90 |
| Resolution (Å) | 50–1.31 (1.36–1.31)a |
| Rpim (%) | 1.9 (39.6) |
| I/δI | 37.8 (2.0) |
| Completeness (%) | 98.1 (96.6) |
| Redundancy | 12.8(8.8) |
| Refinement | |
| Resolution (Å) | 40.68–1.31 |
| No. of reflections | 109 282 |
| Rwork/Rfree | 16.8/18.9 |
| No. of atoms | |
| Protein | 3298 |
| Ligand/ion | 15 |
| Water | 557 |
| B-factors | |
| Protein | 19.4 |
| Ligand/ion | 39.3 |
| Water | 29.6 |
| RMSD | |
| Bond lengths (Å) | 0.006 |
| Bond angles (°) | 1.122 |
| Ramachandran plot | |
| Favored, % | 98.6 |
| Allowed, % | 1.4 |
| Disallowed, % | 0 |
Abbreviations: PDB, Protein Data Bank; RMSD, root-mean-square deviation.
Values in parentheses are for highest-resolution shell. One crystal was used for the data set.
Figure 4.
Overall structure of RCC2 and the structural elements accounting for interaction with Rac1. (a) Ribbon diagram of the RCC2 propeller structure viewed along or perpendicular to the central shaft. The blades are numbered (1–7) along the sequence. (b) Superimposition of human RCC2 (cyan) and human RCC1 (gray) structures viewed perpendicular to or along the central shaft. The extra structural elements in RCC2 (loop 1, loop 2 and β-hairpin) relative to RCC1 are shown in magenta. The extra β-wedge required for RCC1 guanine exchange factor activity is shown in yellow. (c) RCC2 deletion mutants used in this study. (d) S-tag-HA-tag RCC2 mutants were expressed in vivo and lysates were incubated with purified His-Rac1 protein. Rac1 or RCC2 was then evaluated with anti-Rac1 or anti-HA antibody respectively. See also Supplementary Figure S4.
RCC2 deficiency leads to defective responses to absolute or gradient concentration changes of fibronectin
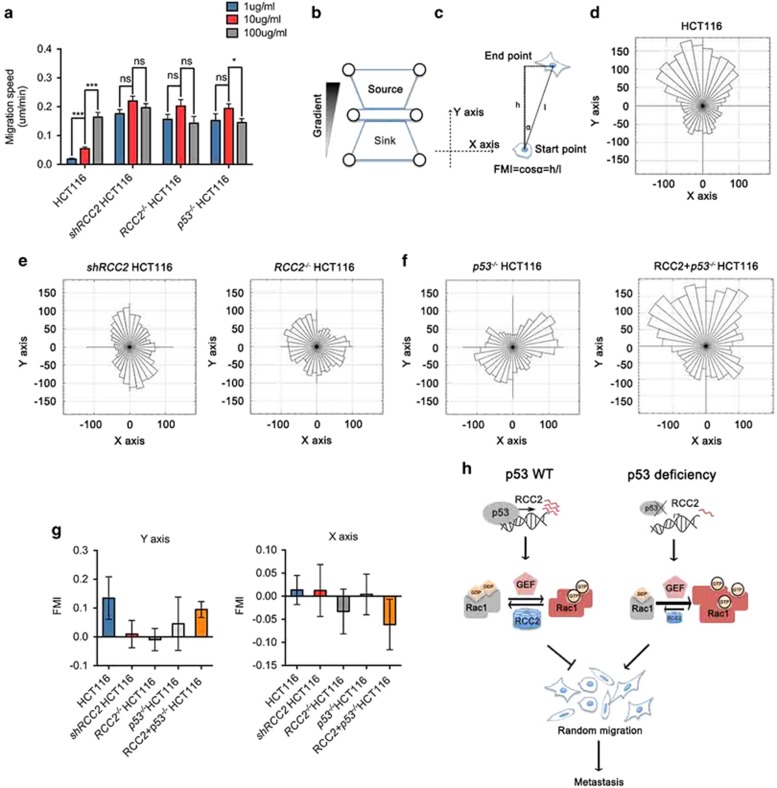
The activity of Rac1 modulates cell migration.16, 19 Morphologic changes such as increased membrane ruffles observed in RCC2-deficient cells are a strong indication of altered cell motility. Changes in motility occur when cells are plated at different concentrations of ECM, and optimal motility is displayed at intermediate ECM concentrations.16, 30 Single-cell tracking was performed to follow individual cells plated on fibronectin-coated culture dishes over time, and the velocity of these cells was calculated. Upon stimulation by different concentrations of fibronectin, RCC2- or p53-deficient cells migrated at a higher speed than HCT116 control cells. The migration speed of RCC2- or p53-deficient cells was not enhanced as fibronectin concentration increased, but HCT116 cells exhibited increased migration speed (Figure 5a). Based on these observations, we postulated that cells with intact RCC2 or p53 recognize substrate surface ECM content, but RCC2-null or p53-null cells are refractory to this directional cue. Inability to respond to changes in ECM concentration suggests that RCC2- or p53-deficient cells migrate in a random manner. To further evaluate this, we used a previously modified microfluidic system16 to establish a fibronectin gradient on the surface of the cell culture chamber between a fibronectin source and a phosphate buffer saline sink, and plated cells onto it (Figure 5b). Single-cell tracking was carried out to measure speed and direction of cell migration. We transposed the tracks so that the fibronectin gradient started along the positive y axis, and computed the start and end points of individual cells to calculate the forward migration index16 (Figure 5c). A forward migration index with s.d. (shown by error bars) that includes the value ‘0’ represents absence of directional migration. We found RCC2−/− and shRCC2 cells show no directional response to concentration gradients of fibronectin on the y axis, despite the significant response of HCT116 wild-type cells plated in the same chamber to the fibronectin source on the y axis (Figures 5d–f). Identical results were observed with p53−/− cells, but expression of RCC2 in p53−/−cells promoted directional migration (Figures 5f and g). These results confirm that RCC2 is essential for the cellular response to changes in either absolute or gradient concentrations of fibronectin, and that RCC2 acts downstream of p53 to mediate directional migration. Cellular RCC2 deficiency may therefore result in random cell migration, and thus promote cell migration to sites favorable for metastasis.
Figure 5.
Loss of RCC2 leads to defective haptotaxis and induces random cell migration. (a) Migration speed test of indicated cells under stimulation by different concentrations of fibronectin. Cells were seeded onto glass bottom dishes precoated with different concentrations of fibronectin. Cells were then cultured and imaged with live-cell confocal imaging for 24 h. Cell migration speeds (n>30) were calculated by using the manual tracking program in ImageJ software. All data are presented as mean±s.e.m. and analyzed by unpaired t-test. *P<0.05, ***P<0.001, NS, no significance. (b) Schematic diagram of the microfluidic system. (c) Schematic diagram of forward migration index (FMI) calculation. (d) Rose plots for HCT116 cells migrating in a fibronectin gradient. (e) Rose plots for shRCC2 and RCC2−/−HCT116 cells migrating in a fibronectin gradient. (f) Rose plots for p53−/− and RCC2+p53−/−HCT116 cells migrating in a fibronectin gradient. (g) FMIs of HCT116, shRCC2, RCC2−/−, p53−/− and RCC2+p53−/−HCT116 cells on the y and x axes are computed and calculated. Data are presented as mean±s.d. (h) Schematic representation depicting the effects of p53 and RCC2 on Rac1 activity control and cell metastasis.
Discussion
Metastasis is a complex process that involves multiple steps, leading to therapy failure and cancer death. p53 is known to control transcription of genes that are involved in metastasis pathways, including cell adhesion, motility, invasion, epithelial–mesenchymal transition, stemness, ECM interactions and anoikis.4 Haptotaxis is the ability to sense gradients of extracellular matrix and loss of haptotaxis is reported to promote metastasis.31 However, it has heretofore been unclear whether p53 participates in haptotaxis, and if so, what mechanism is involved. In this study we report that RCC2 is transcriptionally activated by p53, and p53 deficiency diminishes RCC2 expression that induces Rac1 activation and brings about deterioration of haptotaxis. Diminished haptotaxis in turn leads to random cell migration, and promotes tumor cell metastasis. In summary, we show p53 suppresses metastasis by inducing RCC2-mediated inactivation of Rac1 (Figure 5h).
It is well known that the consensus binding site for p53 consists of two internal symmetrical copies of the 10 bp motif 5'-PuPuPuC(A/T)(T/A)GPyPyPy-3' separated by 0–13 bp.32 There are a number of p53 target genes that contain this consensus motif in their regulatory regions,33 such as p21 (WAF1),34 Bax35 and PUMA.36 However, some p53-responsive genes do not contain this type of consensus site. In these cases, p53 uses palindromic binding sites to regulate its target genes, such as PAC1,23 DUSP1,25 and MKP2.24 Here we identify another 10 bp palindromic motif (GGCACGTCCC) in the RCC2 promoter as a new binding site for p53 regulation of RCC2. Our observations together with previous studies of this phenomenon strongly suggest that binding of palindromic motifs may be a ubiquitous mechanism by which p53 regulates its target genes.
A previous report demonstrated that RCC2 reduction is associated with poor outcome of CRC patients with MSS tumors.14 CRC has become one of the most common types of cancer, and is the third and fourth leading cause of cancer death worldwide in females and males respectively.37 MSS tumors and microsatellite instable (MSI) tumors are the two major types of CRC. Patients with MSS tumors usually have a significantly worse prognosis than patients with MSI tumors.14 Our findings regarding promotion of cancer metastasis by RCC2 deficiency provides a novel mechanistic explanation for poor prognosis of MSS tumors. These results suggest that RCC2/Rac1 signaling is a potential therapeutic target in MSS tumors.
It was recently proposed that RCC2 is a regulator of Rac1, but various studies presented inconsistent results regarding the mechanism of regulation.9, 21 RCC2 was thought to be a guanine exchange factor, as its family member RCC1 acts as a guanine exchange factor for Ran GTPase.9 However, reduction of RCC2 expression results in the activation of Rac1, indicating RCC2 inhibits Rac1 activation.21 Activation of Rac1 usually facilitates cancer progression and several spontaneously activating mutants have been found in melanoma.38 Structural studies of Rac1 mutants and their complexes are important for us to understand how it is regulated. Therefore, it is vital to determine the three-dimensional structure of the RCC2–Rac1 complex to understand the relationship more precisely. Determination of the structure of RCC2 is a first step toward this goal. In this study we solved the crystal structure of RCC2 and found that RCC2 interacts with Rac1 through a unique β-hairpin that may help us understand the function of RCC2.
A previous study showed that deletion of Arf and/or p53 in MEFs induces Rac1 activation in a phosphatidylinositol 3-kinase-dependent manner, promoting actin cytoskeleton reorganization and cell migration.26 However, this mechanism was shown to operate independently of phosphatidylinositol 3-kinase in p53-deficient B- and T-cell lines.20 This argues that the mechanism that accounts for p53 deficiency-induced Rac1 activation is still incompletely understood and raises the possibility that an alternative pathway exists. Here we have identified a new mechanism whereby p53 suppresses Rac1 activation through transcriptional upregulation of RCC2, thus strengthening the understanding of p53 and Rac1 regulation.
We show that p53 and RCC2 deficiency result in promotion of random cell migration when cells are placed on an ECM substratum, and loss of RCC2 facilitates tumor metastasis in vivo. We therefore postulate cell migration in RCC2-sufficient cells is limited by cues from the ECM. It is now accepted that when tumor proliferates in vivo, the preexisting collagen that surrounds cells is pushed outward, resulting in formation of a collagen ‘capsule’ that surrounds the tumor periphery. This high concentration ECM component forms a natural three-dimensional barrier that constrains tumor cell migration and invasion.39 This is particularly important for restriction of cell migration with normal haptotaxis. Loss of haptotaxis has been reported to induce metastasis associated with the LKB1 gene in which LKB1 loss in melanoma cells brings about defects in matrix sensing and haptotaxis that promote cell metastasis.31, 40 We thus speculate that loss of capacity for haptotaxis resulting from RCC2 loss confers a greater degree of random migration, and causes failure to respect natural tumor boundaries. Cells are then free to wander and invade sites that lack restriction. These results together reveal the mechanism that RCC2 reduction induces cell metastasis.
In conclusion, our findings demonstrate that RCC2 regulated by p53 acts as inhibitor of Rac1 function, thus providing new insights into the molecular mechanism by which p53 regulates Rac1. We propose that p53, RCC2 and Rac1 function as a signaling axis for regulation of cell migration and suppression of metastasis. Our observations regarding p53 control of cell migration by inducing RCC2-mediated inactivation of Rac1 may offer new targets for prevention and treatment of cancer metastasis.
Materials and methods
More detailed information is described in Supplementary Materials and methods.
Migration speed test
shRCC2 HCT116, RCC2−/−HCT116, p53−/−HCT116 and control cells were separately labeled with cell tracker Red CMPTX or green CMFDA dye (Life Technologies, Carlsbad, CA, USA). Equal amounts of these cells together with HCT116 control cells were mixed and seeded onto 1, 10 or 100 μg/ml fibronectin-coated glass bottom culture dishes for 6 h. Time-lapse images were captured using an A1+ confocal laser microscope system (Nikon, Tokyo, Japan) at 10 min intervals for 24 h. Cell speed (n>30) was measured with ImageJ (NIH, Bethesda, MD, USA) using the Manual Tracking plug-in.
Xenograft mouse studies
Female Balb/c nude mice (4–6 weeks) were randomly grouped and intravenously injected with 2 × 106 shScramble and shRCC2 cells separately, or subcutaneously injected with 4 × 106 shScramble and shRCC2 HCT116 cells separately. Each group contained 5 mice and kept for 2 months. The mice were maintained in laminar airflow units under aseptic conditions, and the care and treatment of experimental animals was in accordance with institutional guidelines. Nude mice were killed, and lungs and lymph nodes were fixed and hematoxylin and eosin stained to evaluate for metastasis.
Statistical analysis
Data from three independent experiments were analyzed with the unpaired t-test, and error bars represent s.e.m., unless otherwise stated. The statistical significances between data sets was expressed as a P-value, and P<0.05 was considered statistically significant.
Protein purification
Different truncations of human RCC2 protein were cloned and screened for expression and purification in both bacteria and insect cells. The recombinant proteins were expressed using the pFastBac baculovirus system (Invitrogen, Carlsbad, CA, USA). Briefly, bacmid DNAs were generated in DH10Bac cells, and the resulting baculoviruses were generated and amplified in Sf9 insect cells (Invitrogen). Proteins were overexpressed in Sf9 insect cells (Invitrogen) grown in SIM SF medium (Sino Biological Inc., Beijing, China). The protein human RCC2 (89-522) was overexpressed with an N-terminal TEV (tobacco etch virus)-cleavable 6-His tag. The recombinant proteins were purified with nickel affinity chromatography. After removing the 6-His tag with TEV protease, proteins were further purified by gel filtration chromatography. Protein concentrations were determined with microspectrophotometry using the theoretical molar extinction coefficients at 280 nm, and protein purity was evaluated with Coomassie blue staining of SDS–PAGE gels.
Crystallization, diffraction data collection and structure determination
Crystals of RCC2 (89–522) were grown at 20 °C using the hanging-drop vapor diffusion method by mixing 1 μl of protein (12 mg/ml in 20 mM Tris-HCl, 0.15 mM NaCl, pH 8.0, 1% glycerol, 1 mM TCEP) with 1 μl of reservoir solution (0.1 M BIS-TRIS 5.5, 0.2 M (NH4)2SO4, 16% PEG3350), followed by microseeding after balancing for 1 h. Crystals appeared in 1 day and rapidly grew to full size. The 1.31 Å native data were collected at 100 K on beamline 19ID at the Advanced Photon Source (Argonne National Laboratory, Chicago, IL, USA).
The data sets were processed using the HKL3000 package (HKL Research Inc., Charlottesville, VA, USA). The structures were solved by molecular replacement with PHASER41 using PDB entries 1A12 as the search models for RCC2, and were manually refined with COOT and PHENIX. Data collection and refinement statistics are listed in Table 1. All the figures were prepared using PyMOL software (http://pymol.org/).
Protein data bank accession number
Coordinates and structural factors have been deposited in the Protein Data Bank under accession code PDB 5GWN.
Acknowledgments
We thank the staff of the Argonne National Laboratory (ID19) and the Shanghai SSRF Beamline (BL17U1 and BL19U1) for technical assistance. We thank X Chen, L Yuan, D Lu, G Wang, J Feng, C Zhang and S He for technical assistance and critical discussion. This work was supported by the following grants to YY including: National Natural Science Foundation of China (Key Grants 81430056, 31420103905, 81301801 and 81621063), National Key Research and Development Program of China (Grant 2016YFA0500302), Beijing Natural Science Foundation (Key Grant 7161007) and the Lam Chung Nin Foundation for Systems Biomedicine. C-HY was supported by the grant 31270769 from the National Natural Science Foundation of China.
Footnotes
Supplementary Information accompanies this paper on the Oncogene website (http://www.nature.com/onc)
The authors declare no conflict of interest.
Supplementary Material
References
- Kruse J-P, Gu W. Modes of p53 regulation. Cell 2009; 137: 609–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z, Huang C, He J, Lamb KL, Kang X, Gu T et al. PTEN C-terminal deletion causes genomic instability and tumor development. Cell Rep 2014; 6: 844–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller PA, Vousden KH, Norman JC. p53 and its mutants in tumor cell migration and invasion. J Cell Biol 2011; 192: 209–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell E, Piwnica-Worms D, Piwnica-Worms H. Contribution of p53 to metastasis. Cancer Discov 2014; 4: 405–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei CL, Wu Q, Vega VB, Chiu KP, Ng P, Zhang T et al. A global map of p53 transcription-factor binding sites in the human genome. Cell 2006; 124: 207–219. [DOI] [PubMed] [Google Scholar]
- Mashimo T, Watabe M, Hirota S, Hosobe S, Miura K, Tegtmeyer PJ et al. The expression of the KAI1 gene, a tumor metastasis suppressor, is directly activated by p53. Proc Natl Acad Sci USA 1998; 95: 11307–11311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins CP, Brown-Swigart L, Evan GI. Modeling the therapeutic efficacy of p53 restoration in tumors. Cell 2006; 127: 1323–1334. [DOI] [PubMed] [Google Scholar]
- Adams RR, Carmena M, Earnshaw WC. Chromosomal passengers and the (aurora) ABCs of mitosis. Trends Cell Biol 2001; 11: 49–54. [DOI] [PubMed] [Google Scholar]
- Mollinari C, Reynaud C, Martineau-Thuillier S, Monier S, Kieffer S, Garin J et al. The mammalian passenger protein TD-60 is an RCC1 family member with an essential role in prometaphase to metaphase progression. Dev Cell 2003; 5: 295–307. [DOI] [PubMed] [Google Scholar]
- Rosasco-Nitcher SE, Lan W, Khorasanizadeh S, Stukenberg PT. Centromeric Aurora-B activation requires TD-60, microtubules, and substrate priming phosphorylation. Science 2008; 319: 469–472. [DOI] [PubMed] [Google Scholar]
- Cancer Genome Atlas Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature 2012; 487: 330–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannakis M, Hodis E, Jasmine Mu X, Yamauchi M, Rosenbluh J, Cibulskis K et al. RNF43 is frequently mutated in colorectal and endometrial cancers. Nat Genet 2014; 46: 1264–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouradov D, Sloggett C, Jorissen RN, Love CG, Li S, Burgess AW et al. Colorectal cancer cell lines are representative models of the main molecular subtypes of primary cancer. Cancer Res 2014; 74: 3238–3247. [DOI] [PubMed] [Google Scholar]
- Bruun J, Kolberg M, Ahlquist TC, Royrvik E, Nome T, Leithe E et al. Regulator of chromosome condensation 2 identifies high-risk patients within both major phenotypes of colorectal cancer. Clin Cancer Res 2015; 21: 3759–3770. [DOI] [PubMed] [Google Scholar]
- Kim NG, Rhee H, Li LS, Kim H, Lee JS, Kim JH et al. Identification of MARCKS, FLJ11383 and TAF1B as putative novel target genes in colorectal carcinomas with microsatellite instability. Oncogene 2002; 21: 5081–5087. [DOI] [PubMed] [Google Scholar]
- Wu C, Asokan SB, Berginski ME, Haynes EM, Sharpless NE, Griffith JD et al. Arp2/3 is critical for lamellipodia and response to extracellular matrix cues but is dispensable for chemotaxis. Cell 2012; 148: 973–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall A. Rho GTPases and the actin cytoskeleton. Science 1998; 279: 509–514. [DOI] [PubMed] [Google Scholar]
- Vega FM, Ridley AJ. Rho GTPases in cancer cell biology. FEBS Lett 2008; 582: 2093–2101. [DOI] [PubMed] [Google Scholar]
- Pankov R, Endo Y, Even-Ram S, Araki M, Clark K, Cukierman E et al. A Rac switch regulates random versus directionally persistent cell migration. J Cell Biol 2005; 170: 793–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosco EE, Ni W, Wang L, Guo F, Johnson JF, Zheng Y. Rac1 targeting suppresses p53 deficiency-mediated lymphomagenesis. Blood 2010; 115: 3320–3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphries JD, Byron A, Bass MD, Craig SE, Pinney JW, Knight D et al. Proteomic analysis of integrin-associated complexes identifies RCC2 as a dual regulator of Rac1 and Arf6. Sci Signal 2009; 2: ra51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao R, Gish K, Murphy M, Yin Y, Notterman D, Hoffman WH et al. Analysis of p53-regulated gene expression patterns using oligonucleotide arrays. Genes Dev 2000; 14: 981–993. [PMC free article] [PubMed] [Google Scholar]
- Yin Y, Liu YX, Jin YJ, Hall EJ, Barrett JC. PAC1 phosphatase is a transcription target of p53 in signalling apoptosis and growth suppression. Nature 2003; 422: 527–531. [DOI] [PubMed] [Google Scholar]
- Shen WH, Wang J, Wu J, Zhurkin VB, Yin Y. Mitogen-activated protein kinase phosphatase 2: a novel transcription target of p53 in apoptosis. Cancer Res 2006; 66: 6033–6039. [DOI] [PubMed] [Google Scholar]
- Liu YX, Wang J, Guo J, Wu J, Lieberman HB, Yin Y. DUSP1 is controlled by p53 during the cellular response to oxidative stress. Mol Cancer Res 2008; 6: 624–633. [DOI] [PubMed] [Google Scholar]
- Guo F, Gao Y, Wang L, Zheng Y. p19Arf-p53 tumor suppressor pathway regulates cell motility by suppression of phosphoinositide 3-kinase and Rac1 GTPase activities. J Biol Chem 2003; 278: 14414–14419. [DOI] [PubMed] [Google Scholar]
- Renault L, Nassar N, Vetter I, Becker J, Klebe C, Roth M et al. The 1.7 A crystal structure of the regulator of chromosome condensation (RCC1) reveals a seven-bladed propeller. Nature 1998; 392: 97–101. [DOI] [PubMed] [Google Scholar]
- Renault L, Kuhlmann J, Henkel A, Wittinghofer A. Structural basis for guanine nucleotide exchange on Ran by the regulator of chromosome condensation (RCC1). Cell 2001; 105: 245–255. [DOI] [PubMed] [Google Scholar]
- Makde RD, England JR, Yennawar HP, Tan S. Structure of RCC1 chromatin factor bound to the nucleosome core particle. Nature 2010; 467: 562–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupton SL, Waterman-Storer CM. Spatiotemporal feedback between actomyosin and focal-adhesion systems optimizes rapid cell migration. Cell 2006; 125: 1361–1374. [DOI] [PubMed] [Google Scholar]
- Chan KT, Asokan SB, King SJ, Bo T, Dubose ES, Liu W et al. LKB1 loss in melanoma disrupts directional migration toward extracellular matrix cues. J Cell Biol 2014; 207: 299–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- el-Deiry WS, Kern SE, Pietenpol JA, Kinzler KW, Vogelstein B. Definition of a consensus binding site for p53. Nat Genet 1992; 1: 45–49. [DOI] [PubMed] [Google Scholar]
- Riley T, Sontag E, Chen P, Levine A. Transcriptional control of human p53-regulated genes. Nat Rev Mol Cell Biol 2008; 9: 402–412. [DOI] [PubMed] [Google Scholar]
- el-Deiry WS, Tokino T, Velculescu VE, Levy DB, Parsons R, Trent JM et al. WAF1, a potential mediator of p53 tumor suppression. Cell 1993; 75: 817–825. [DOI] [PubMed] [Google Scholar]
- Miyashita T, Reed JC. Tumor suppressor p53 is a direct transcriptional activator of the human bax gene. Cell 1995; 80: 293–299. [DOI] [PubMed] [Google Scholar]
- Nakano K, Vousden KH. PUMA, a novel proapoptotic gene, is induced by p53. Mol Cell 2001; 7: 683–694. [DOI] [PubMed] [Google Scholar]
- Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin 2015; 65: 87–108. [DOI] [PubMed] [Google Scholar]
- Davis MJ, Ha BH, Holman EC, Halaban R, Schlessinger J, Boggon TJ. RAC1P29S is a spontaneously activating cancer-associated GTPase. Proc Natl Acad Sci USA 2013; 110: 912–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hompland T, Erikson A, Lindgren M, Lindmo T, de Lange Davies C. Second-harmonic generation in collagen as a potential cancer diagnostic parameter. J Biomed Opt 2008; 13: 054050. [DOI] [PubMed] [Google Scholar]
- Liu W, Monahan KB, Pfefferle AD, Shimamura T, Sorrentino J, Chan KT et al. LKB1/STK11 inactivation leads to expansion of a prometastatic tumor subpopulation in melanoma. Cancer Cell 2012; 21: 751–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr 2007; 40: 658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.