Abstract
Pancreatic cancer is among the deadliest malignancies; however, the genetic events that lead to pancreatic carcinogenesis in adults remain unclear. In vivo models in which these genetic alterations occur in adult animals may more accurately reflect the features of human cancer. In this study, we demonstrate that inactivation of Cdkn2b (p15ink4b) is necessary for induction of pancreatic cancer by oncogenic KRASG12D expression and inactivation of Tp53 and Cdkn2a in adult mouse pancreatic ductal cells (P60 or older). KRASG12D overexpression in these cells activated transforming growth factor-β signaling and expression of CDKN2B, which, along with CDKN2A, led to cellular senescence and protected cells from KRAS-mediated transformation via inhibition of retinoblastoma phosphorylation. These results show a critical role of CDKN2B inactivation in pancreatic carcinogenesis, and provide a useful adult animal model by genetic engineering via lentiviral delivery.
Introduction
Only 6% of pancreatic cancer patients survive longer than 5 years after initial diagnosis.1, 2 Kirsten rat sarcoma (KRAS), tumor protein (TP)53 and cyclin-dependent kinase inhibitor (CDKN)2A are the most frequently mutated genes in human pancreatic cancer.3, 4, 5 Expression of oncogenic KRAS during development induces postnatal pancreatic intraepithelial neoplasia (PanIN), which occasionally progresses to pancreatic cancer with long latency. Inactivation of Tp53, Cdkn2a and/or mothers against decapentaplegic homolog (Smad)4 tumor-suppressor genes has been shown to promote cancer progression and metastasis.6, 7, 8
Most pancreatic cancer models have been generated by overexpressing oncogenic KRAS and inactivating Tp53, Cdkn2a, and other tumor suppressors such as retinoblastoma (Rb).9, 10, 11, 12, 13, 14 The most widely used pancreatic cancer models are KC model developed in LSL-KrasG12D/Pdx1-Cre mice15 and KPC model developed in LSL-KrasG12D/LSL-tp53R172H;Pdx1-Cre.8 Expression of KRAS and Tp53 mutants were initiated since early embryonic stage in these models. Recently some models were reported to be induced in adulthood before P60 (postnatal day 60).10 However, it was reported that the capacity of KRAS to induce PanIN in mouse pancreatic acinar cells decreases with age and is completely abolished after P60.10 Moreover, KRASG12D expression in mature acinar cells did not induce obvious lesions even in combination with Tp53 or Cdkn2a deficiency.10 Adult rat acinar cells also showed resistance to KRAS-induced transformation,16 whereas pancreatic ductal cells were >100 times more resistant to transformation.13 Given that most human malignancies including pancreatic cancer result from accumulation of mutations during adulthood, in vivo models induced by genetic engineering in adult animals may more accurately reflect the features of human cancer.
To this end, in this study we established a pancreatic cancer model in mouse elder than P60 by lentiviral delivery to express oncogenic KRASG12D and short hairpin (sh)RNAs targeting tumor-suppressor genes frequently mutated in human pancreatic cancer. We found that pancreatic cancer was induced in adult mice by the combination of KRASG12D overexpression and loss of Tp53 and Cdkn2a only if Cdkn2b was concomitantly inactivated. Moreover, CDKN2B expression was induced by oncogenic KRASG12D via transforming growth factor (TGF)-β signaling. Finally, inactivation of both Cdkn2b and Cdkn2a was necessary for Rb phosphorylation and to encompass oncogene-induced cellular senescence.
Results
Induction of pancreatic cancer in adult mice
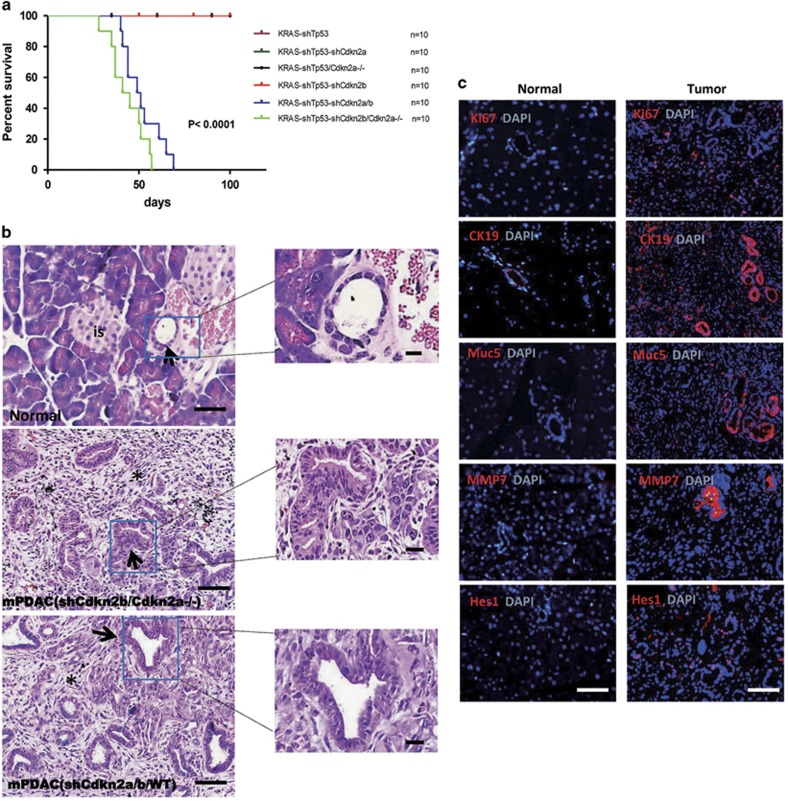
A search of The Cancer Genome Atlas (TCGA) revealed that alterations in KRAS, TP53 and CDKN2A are the most frequent genetic aberrations in human pancreatic cancer (86%, 70% and 52% of cases, respectively) (Supplementary Figure 1). Similar data are reported in other cancer genome databases.17, 18 We therefore decided to test if pancreatic cancer can be induced by dysregulating KRAS, TP53 and CDKN2A genes via lentiviral infection in 9-week-old mice, which were previously reported to be resistant to pancreatic cancer induction.10 The lentiviral vectors used in this study are illustrated in Supplementary Figure 2a. Lentiviral delivery of KRASG12D along with shRNA targeting Tp53 (KRAS-shTp53) was used as negative control because KRASG12D plus p53 knockout were reported to fail to induce cancer in adult. However, lentiviral delivery of KRASG12D along with shRNAs targeting Tp53 and Cdkn2a (KRAS-shTp53-shCdkn2a) into adult pancreas failed to induce pancreatic cancer (Figure 1a), although the Tp53-specific shRNA cassette has been used to induce tumors in previous studies19, 20 and the knockdown efficiency of Cdkn2a-specific shRNA targeting both p16Ink4a and p19Arf sequences was confirmed in mouse embryonic fibroblasts (Supplementary Figure 2b). To exclude the possibility that Cdkn2a knockdown efficiency was insufficient to induce tumor formation, we injected KRAS-shTp53 lentivirus into the pancreas of adult Cdkn2a−/− mice and still failed to develop tumors (Figure 1a).
Figure 1.
Oncogenic KRASG12D expression together with shRNA-mediated inactivation of TP53, CDKN2A and CDKN2B induced pancreatic cancer. (a) Survival curve. Nine-week-old male mice with C57B6/L background were injected with lentivirus on day 0. KRAS-shTP53 group was used as control. Only the mice in the KRAS-shTP53-shCDKNB/CDKN2A−/− and KRAS-shTP53-shCDKN2A/B group developed tumors. (b) Hematoxylin and eosin (H&E) staining of paraffin sections of normal and cancerous pancreatic tissues. The arrows in the image of the normal tissue indicates a normal duct and ‘is’ denotes pancreatic islet. The arrows in cancer tissue indicate the glandular structures and the asterisk indicate fibrous stroma. Scale bars: 40 μm. (c) Mouse pancreatic cancer express established molecular markers of human pancreatic cancer. The proportion of Ki67-positive cells in pancreatic cancer tissues indicates a high proliferation index. Scale bars: 40 μm.
CDKN2B encodes p15INK4B, which activates the Rb pathway and is co-deleted with CDKN2A in many cancers, including pancreatic cancer (Supplementary Figure 1). We therefore investigated the role of the Cdkn2b gene in pancreatic cancer. We verified the knockdown efficiency of shRNA targeting Cdkn2b (Supplementary Figure 2b). KRAS-shTp53-shCdkn2b lentivirus failed to induce pancreatic cancer in adult wild-type mice; however, simultaneous inactivation of Cdkn2a and Cdkn2b, either by injecting the KRAS-shTp53-shCdkn2b lentivirus into the pancreas of adult Cdkn2a−/− mice or the KRAS-shTp53-shCdkn2a-shCdkn2b lentivirus into Cdkn2a wild-type mice, resulted in pancreatic cancer development at 4 weeks post-injection (Figure 1a).
Pancreatic tumors induced by lentivirus KRAS-shTp53-shCdkn2b in Cdkn2a−/− mice or by KRAS-shTp53-shCdkn2a/b in wild-type mice showed similar morphology, ranging in size from 0.5 to 1.5 cm, with yellowish white cut surfaces and few signs of hemorrhage or necrosis. The tumors were frequently observed to obstruct the common bile duct and/or the main pancreatic duct, resulting in gall bladder dilatation, blockage of bile overflow and additional defects in digestive organs (Supplementary Figure 3).
Histological examination of the pancreatic tumors from both KRAS-shTp53-shCdkn2b in Cdkn2a−/− mice and KRAS-shTp53-shCdkn2a/b in wild-type mice revealed signs of moderately differentiated ductal adenocarcinoma with intensive stromal reaction (Figure 1b and Supplementary Figure 4), representative to human pancreatic ductal adenocarcinoma. Moreover, the tumors expressed other well-established markers of human pancreatic cancer, including cytokeratin (CK)19, Mucin (Muc) 5, matrix metalloproteinase-7 (MMP-7), and Hairy and enhancer of split 1(Hes1) (Figure 1c). The tumor cells were highly proliferative, as evidenced by Ki67 and proliferating cell nuclear antigen staining (Figure 1c and Supplementary Figure 5a). In contrast to pancreatic cancer induced by germline transmission of KRASG12D, the proliferation rate of distal pancreatic tissue in our model was comparable to that of normal pancreatic cells (Supplementary Figure 5b), which is similar to the situation in human cancers where tumor cells are surrounded by normal tissue.
Pancreatic cancers originate from pancreatic ductal cells
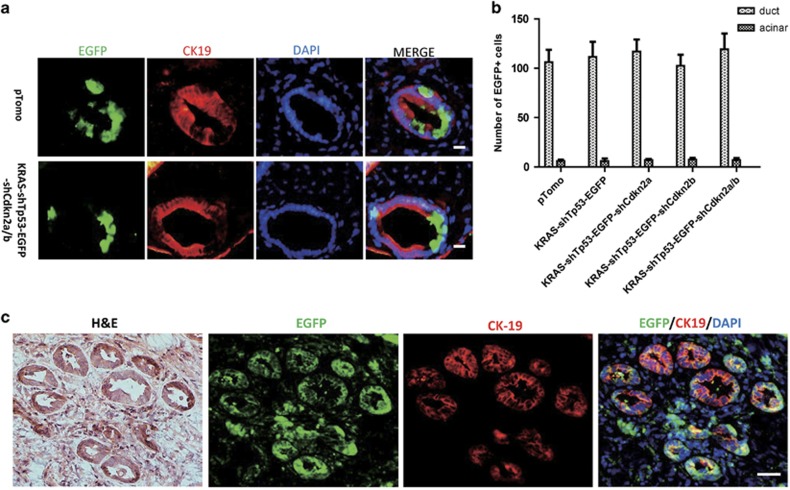
To identify the cell-of-origin of pancreatic cancer in our model, enhanced green fluorescent protein (EGFP)-expressing lentivirus was injected into the pancreas of adult mice. Infected (EGFP-positive) cells were detected 7 days after injection. In addition to the lentivirus shown in Supplementary Figure 2a, we also injected the empty pTomo vector expressing EGFP to exclude the possibility that the introduced genes induced a change in cell type. Although pancreatic cells are not sensitive to lentiviral infection,21 we found that mostly ductal cells were infected as demonstrated by CK-19 expression, whereas only a few acinar cells were EGFP positive (Figures 2a and b and Supplementary Figure 6). The small number of infected cells underscored the fact that most human cancers originate from a single or few cells transformed by accumulated mutations. No positive cells were observed in pancreatic islets. Strikingly, both EGFP-positive and -negative cells were often detected in a single duct, implying that infected cells were ductal rather than transformed acinar cells. To confirm that the ducts in tumor tissue were derived from lentivirus-transformed cells, we injected GFP-expressing oncogenic lentivirus (KRAS-shTp53-EGFP-shCdkn2a/b) to induce pancreatic cancer and detected EGFP and CK-19 expression by immunofluorescence. CK-19-positive ductal cells in tumor tissue also expressed EGFP, indicating the identity of tumor cells (Figure 2c).
Figure 2.
Mouse pancreatic cancers originate from pancreatic ductal cells. (a) Double staining of ductal cell marker CK-19 and EGFP. Scale bars: 20 μm. (b) The counted results of ductal cells and acinar cells positive for EGFP staining in a. (c) Double staining for EGFP and CK-19 on the pancreatic cancer tissue induced by oncogenic lentivirus-expressing EGFP. Scale bars: 40 μm.
To confirm that genetic alterations in ductal cells gave rise to pancreatic tumors, we injected lentiviral particle, in which KRASG12D was expressed under the control of the CK-19 promoter,22 into the pancreas of adult mice. As expected, pancreatic tumors appeared after 7 weeks, with histological features that were similar to those of ductal adenocarcinoma induced by cytomegalovirus promoter-driven KRASG12D expression (Supplementary Figure 7).
Gene expression profile in the pancreatic cancer model is similar to that of human pancreatic cancer
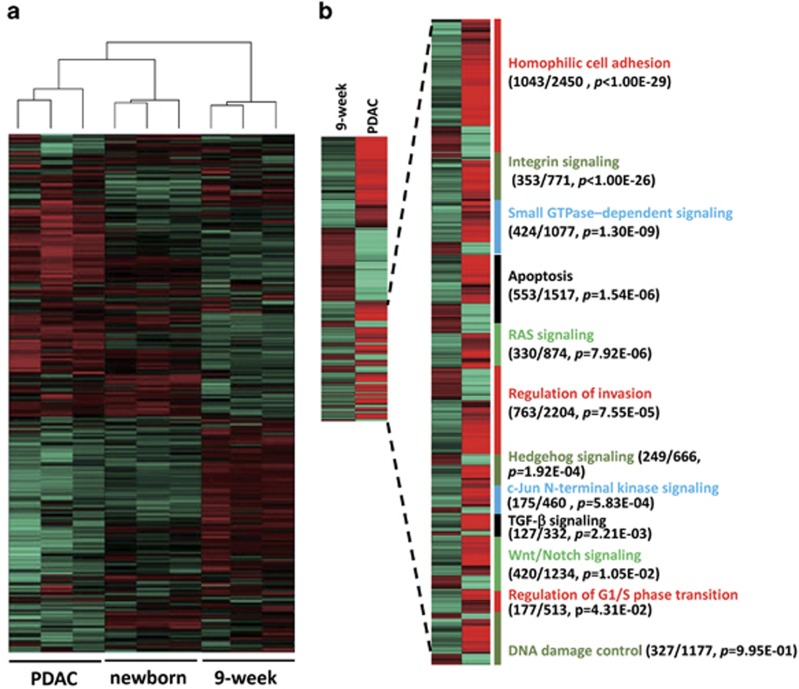
We investigated the gene expression profile of pancreatic tumors induced by KRAS-shTp53-shCdkn2a/b injection in wild-type mice as compared with pancreas transcriptomes of neonatal and 9-week-old mice. The global expression profile in pancreatic cancer was correlated with that in neonatal mice (P<0.01) (Figure 3a). Specifically, genes that were differentially expressed between cancerous and normal pancreatic tissue from 9-week-old mice enriched in Ras, P53 and Rb, as well as other pathways that contribute to human pancreatic cancer development including Homophilic cell adhesion, integrin signaling, small GTPase-dependent signaling, apoptosis, regulation of invasion and Hedgehog signaling, and others4 (Figure 3b and Supplementary Table 2).
Figure 3.
Transcriptome analysis of pancreatic cancer samples. (a) The global gene expression pattern of pancreatic cancer tissues is similar to that of newborn pancreatic tissue and different from that of adult pancreatic tissue. (b) Genes differentially expressed in adult pancreas and cancer. The 12 core pathways associated with pancreatic cancer was shown with the number of differentially expressed genes, the total number of genes in the indicated pathway and enrichment P-value in parentheses.
Cdkn2b inactivation leads to Rb phosphorylation
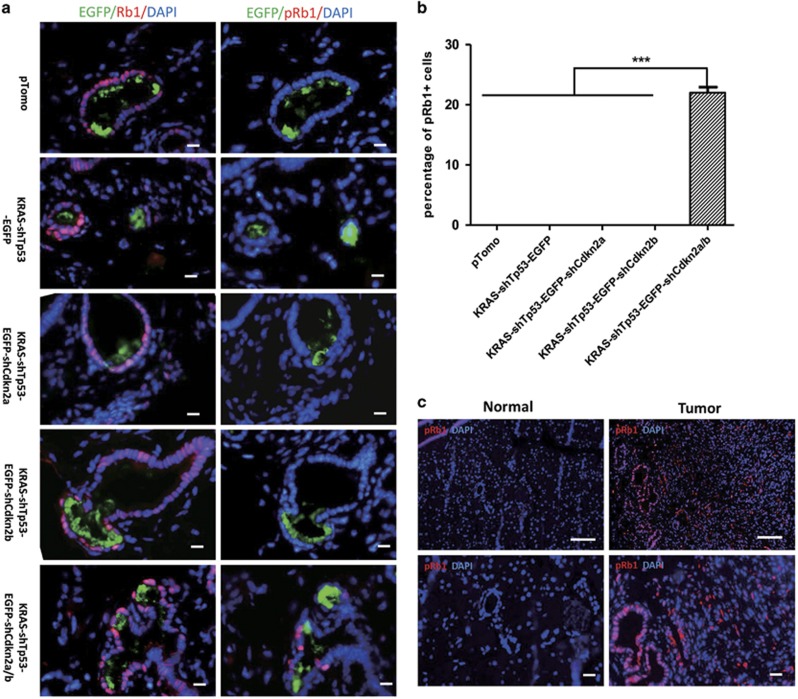
To investigate the role of Cdkn2b inactivation in pancreatic cancer induction, we injected mice with EGFP-labeled lentivirus and examined the total level of Rb and Rb phosphorylation at Ser780, which is inhibited by both p16ink4a and p15ink4b via abrogation of CDK4 and CDK6 activities. The total level of Rb was not significantly different among these groups, such as KRAS-shTp53, KRAS-shTp53-shCdkn2a, KRAS-shTp53-shCdkn2b and KRAS-shTp53-shCdkn2a/b (Figure 4a). Rb phosphorylation at Ser780 (phospho-Rb (Ser780)) was rarely detected in cells infected by lentivirus KRAS-shTp53, KRAS-shTp53-shCdkn2a or KRAS-shTp53-shCdkn2b (Figures 4a and b). However, it was detected in a fraction of ductal cells infected by lentivirus KRAS-shTp53-shCdkn2a/b in which both Cdkn2a and Cdkn2b were inactivated (Figures 4a and b). Furthermore, phospho-Rb (Ser780) was highly expressed in pancreatic cancer but detected rarely in acinar and ductal cells in the normal pancreas (Figure 4c).
Figure 4.
Cdkn2b inactivation is indispensable for Rb phosphorylation of ductal cells. (a) Immunohistochemistry staining for total Rb and Rb phosphorylation at S780 was conducted on fresh adjacent sections from pancreases infected with the pTomo empty vector, KRAS-shp53-EGFP, KRAS-shp53-EGFP-shCdkn2a, KRAS-shp53-EGFP-shCdkn2b or KRAS-shP53-EGFP-shCdkn2a/b lentiviral particles. Scale bars: 20 μm. (b) The ratio of Rb phosphorylation-positive cell to EGFP-positive cells in a. (c) Immunohistochemistry staining for Rb1 phosphorylation in mouse pancreatic cancer. Scale bars: 40 μm. The quantification value is presented as the mean±s.e.m. ***P<0.001.
Cdkn2b inactivation is necessary to overcome cellular senescence induced by oncogenic KRAS
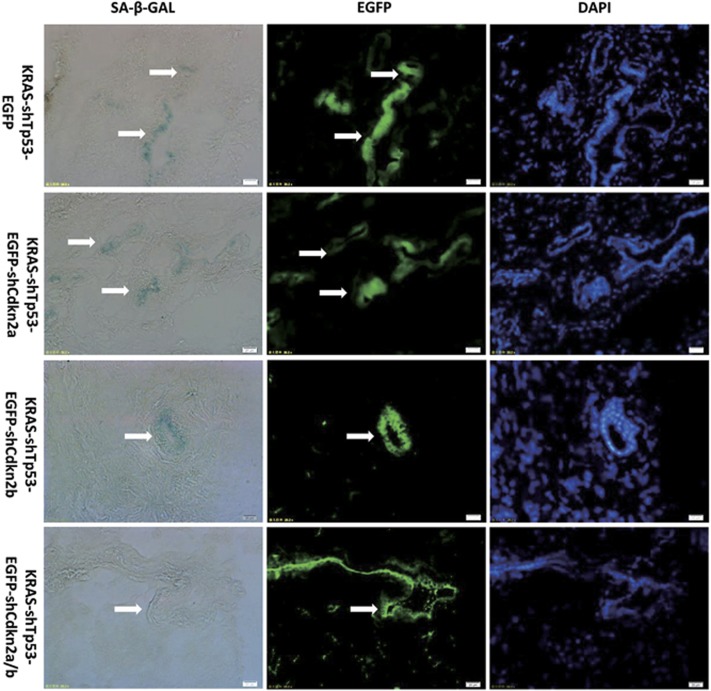
Cellular senescence induced by oncogenic KRAS in adult pancreas has been shown to block pancreatic cancer progression.10 To investigate whether Cdkn2b inactivation is indispensable to overcome cellular senescence induced by oncogenic KRAS, we injected mice with EGFP-labeled lentivirus. Senescence-associated β-galactosidase and EGFP staining demonstrated obvious senescence-associated β-galactosidase activity in the cells transduced by KRAS-shTp53, KRAS-shTp53-shCdkn2a, KRAS-shTp53-shCdkn2b, but not in the cells transduced by KRAS-shTp53-shCdkn2a/b where Cdkn2b and Cdkn2a were both inactivated (Figure 5). These results suggested that Cdkn2b inactivation is indispensable to overcome cellular senescence induced by oncogenic KRAS.
Figure 5.
Cdkn2b inactivation is necessary to make cell encompass senescence induced by oncogenic KRAS. Obvious senescence-associated β-galactosidase (SA-β-GAL) activity was detected in cells infected by KRAS-shTp53, KRAS-shTp53-shCdkn2a, KRAS-shTp53-shCdkn2b, but that was not detected in infected pancreatic cells by KRAS-shTp53-shCdkn2a/b where Cdkn2b and Cdkn2a were both inactivated. Representative images are shown for SA-β-GAL (blue), EGFP and 4,6-diamidino-2-phenylindole (DAPI) staining of DNA. Scale bars: 20 μm.
CDKN2B expression is induced by oncogenic KRAS via TGF-β signaling
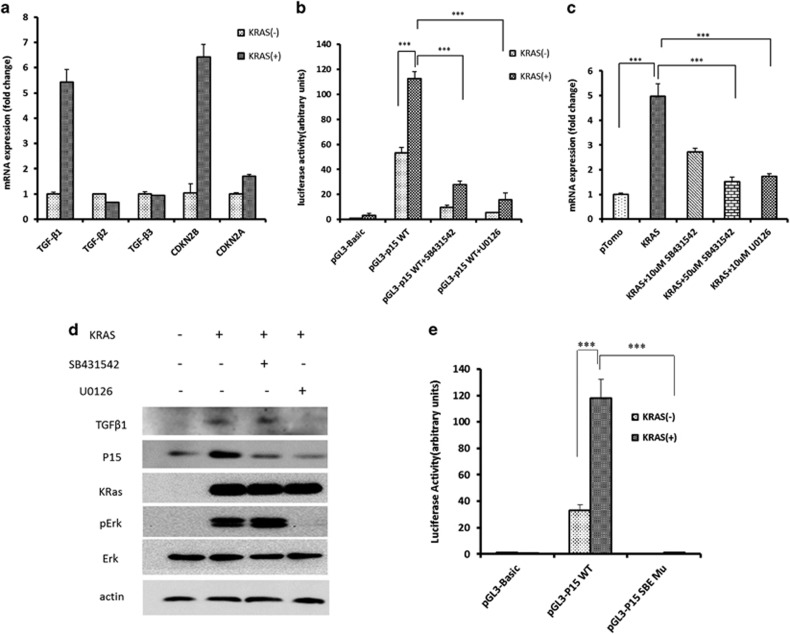
Oncogenic BRAF was reported to increase CDKN2B expression in melanocytes via activation of TGF-β signaling.23 We investigated whether CDKN2B expression is induced upon oncogenic KRAS expression in HPDE6-C7 human pancreatic ductal cells. KRASG12D overexpression upregulates CDKN2B and TGF-β1 expression at both mRNA level and protein level (Figures 6a and d) and CDKN2B promoter activity as shown by reporter assay (Figure 6b). This effect was abolished in the presence of the inhibitor U0126 of mitogen-activated protein kinase kinase (MEK) that mediates KRAS signaling or TGF-β inhibitor SB431542 (Figures 6c and d). The Smad-binding elements in the Cdkn2b promoter, as well as TGF-β signaling were required for increased CDKN2B expression, as determined by a reporter assay (Figures 6c and e). These results showed that oncogenic KRAS expression could upregulate CDKN2B gene transcription via TGF-β signaling.
Figure 6.
KRAS signaling induces CDKN2B expression via TGFβ pathway. (a) Expression of oncogenic KRAS induced the expression of TGFβ1 and CDKN2B in HPDE6-C7 cell line quantified by real-time PCR. (b) KRAS increased the CDKN2B promoter activity, and the effect was inhibited by MEK inhibitor U0126. (c) The expression of CDKN2B induced by KRAS was abolished by TGFβ receptor kinase inhibitor SB431542. (d) Western blot analysis of TGFβ1, CDKN2B, p-ERK and ERK. (e) P15 promoter with mutated Smad-binding element (SBE) site lost promoter activity as shown by luciferase reporter assay. The value is presented as the mean±s.e.m. **P<0.01, ***P<0.001.
TGF-β1 expression induced by oncogenic KRAS was mediated by early growth response 1 (EGR1)
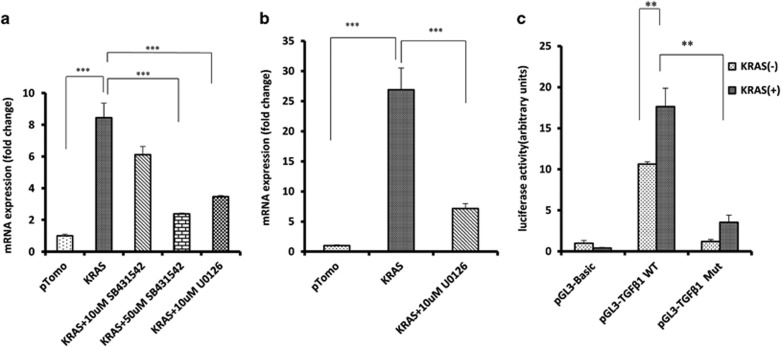
To understand how KRAS induces TGF-β1 expression, U0126 inhibitor of MEK was used to block KRAS signaling and found that the induction of TGF-β1 expression was abolished at both protein and mRNA level (Figures 6d and 7a). EGR1 is reported to be regulated by RAS signaling and in turn regulates TGF-β1 expression.24, 25, 26 Here we found that EGR1 was significantly upregulated in HPDE6-C7 cells overexpressing oncogenic KRAS (Figure 7b), and the upregulation was abolished in the presence of U0126. Mutation of the EGR1-binding site in TGF-β1 promoter inhibited the reporter activity induced by KRAS overexpression (Figure 7c). These results suggested that TGF-β1 induction by KRAS was mediated by EGR1.
Figure 7.
EGR1 mediates the expression of TGF-β1 induced by oncogenic KRAS. (a) Oncogenic KRAS upregulated TGF-β1 expression and this was inhibited by MEK inhibitor U0126. (b) KRAS-induced EGR1 expression. MEK inhibitor U0126 suppressed the induction. (c) Induction of TGFb1 by oncogenic KRAS relies on EGR1-binding site in promoter as shown by luciferase reporter assay. The value is presented as the mean±s.e.m. ** P<0.01, ***P<0.001.
Discussion
In this study, we established a mouse model of pancreatic ductal adenocarcinoma by overexpressing KRASG12D and inactivating Tp53 and Rb via lentiviral infection. In addition to signaling pathways associated with these genes, others that are dysregulated in human pancreatic cancer were enriched in our animal model. Thus, our model recapitulates the molecular mechanisms of human pancreatic cancer.
Lentiviral infection provides a rapid and low-cost means of inducing pancreatic ductal adenocarcinoma in animal models. One drawback of expressing oncogenic KRAS via lentiviral infection is the variability in expression levels as compared with Lox-Stop-Lox-KRAS transgenic mice that express KRAS under the control of an endogenous promoter.27 However, mutant KRAS expression and activity were highly upregulated in human and mouse pancreatic cancer as compared with endogenous levels before tumor development.28 In fact, endogenous levels of mutant KRAS in differentiated acinar cells may induce PanIN without progression to pancreatic cancer, whereas overexpression can lead to tumorigenesis,29 possibly by inducing inflammation.10, 29
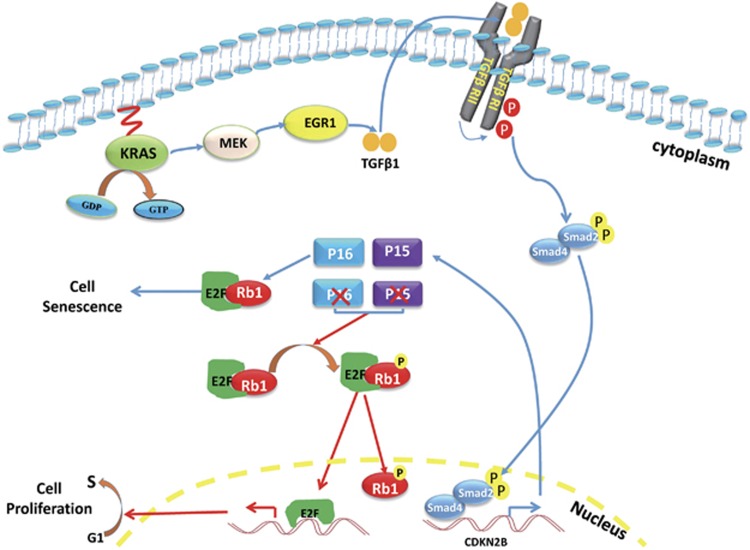
Simultaneous deletion of CDKN2B and CDKN2A is frequently observed in human cancer, including pancreatic cancer.30, 31 The CDKN2B gene encodes p15ink4b, which, like p16Ink4a, inhibits the Rb1 regulators CDK4 and CDK6. The role of the CDKN2B gene in tumor development has not been well studied because of the modest impact of genetic CDKN2B inactivation32 and the rarity of CDKN2A-independent CDKN2B mutations in human cancers. A previous study found that Cdkn2a/b−/− mice were more prone to spontaneous tumor development.33 In an animal model of lung cancer induced by mutant KRAS, Cdkn2b inactivation was found to promote tumorigenesis.34 Although these studies link Cdkn2b to cancer development, it was only recently that the definitive role of p15ink4b rather than p16Ink4a in the proliferation of primary melanocytes and development of melanoma was demonstrated.23 Here, we showed that pancreatic cancer was induced by Cdkn2b inactivation in adult mice only with simultaneous overexpression of oncogenic KRAS and Tp53 and Cdkn2a silencing. In addition, inactivation of both Cdkn2a and Cdkn2b was necessary for Rb phosphorylation and to encompass cellular senescence (Figure 8). Thus, CDKN2B is likely important in other cancer types, especially when co-deleted with CDKN2A.
Figure 8.
The KRAS/TGF-β/p15Ink4b pathway controls the cell fate. Although oncogenic KRAS activates the proliferative signals, it also stimulates the TGF-β expression through MEK-EGR1 cascade. TGF-β further activates Cdkn2b expression via Smads component of TGF-β pathway. The p15Ink4b encoded by Cdkn2b gene together with p16Ink4a encoded by Cdkfn2a inhibit the Rb phosphorylation and activate Rb to induce cellular senescence and prevent tumor formation. However, when both Cdkn2a and Cdkn2b are inactivated, Rb is phosphorylated by CDK4/6 and make the cells encompass cellular senescence, which allow tumor formation.
Oncogenic KRAS expression in pancreatic cells is known to induce senescence, which blocks cell cycle progression and inhibits cellular transformation and tumor formation. p53/p19ARF and/or p16INK4A are major mediators of KRAS-induced senescence. Bmi1 binds to the promoter of INK4A and represses its transcription. KRAS stimulates INK4A expression by phosphorylating Bmi1 via p38-MAPKAP kinase 3, resulting in the release of Bmi1 from the INK4A promoter.35, 36 KRAS also activates INK4A transcription by inducing H3K27 demethylation in the INK4A promoter via inhibition of EZH2 gene and upregulation of Jmjd3 gene.35, 36 INK4A inhibits CDK4/6 and subsequently induces the dephosphorylation of Rb, which is thought to be an essential component in the senescence pathway. In this study, we found KRAS also induces CDKN2B (INK4B) expression via MEK-ERK-EGR1-TGF-β signaling; concurrent inactivation of INK4B and INK4A induced Rb phosphorylation and consequently make cells encompass senescence.
Smad4 is a critical factor in TGF-β signaling and CDKN2B regulation that is mutated and inactivated in approximately 40% of human pancreatic cancer cases reported in the TCGA database. CDKN2B deletion is also frequently detected in glioblastoma and bladder, esophageal, and lung cancers. Whether CDKN2B expression can be induced by activated oncogenes via TGF-β signaling in these cancers is an open question.
Cellular context contributes to the induction of pancreatic cancer; various differentiated cell types in the pancreas can serve as the cell-of-origin for tumors, with acinar and ductal cells considered as the most likely sources.14, 37, 38 Acinar cells in adult mice older than P60 are resistant to transformation by KRAS even in combination with Tp53 or Cdkn2a inactivation. Whether genetic manipulations that induce the transformation of ductal cells in adults have similar effects on mature acinar cells, and the role of the Rb pathway regulator CDKN2B in the transformation of adult acinar cells are important questions for future studies.
In conclusion, we established a new animal model of pancreatic cancer by inducing genetic alterations in adult mice and demonstrated the critical role of Cdkn2b in pancreatic tumorigenesis. Our findings provide a basis for the development of improved therapeutic strategies for pancreatic cancer treatment.
Materials and methods
Animal models
All animal experimental were approved by the Animal Ethic Committee of Kunming Institute of Zoology. All animals were maintained under specific pathogen-free conditions. Cdkn2afl/fl mice were provided by Dr Ronald DePinho.39 Cdkn2a−/− mice were obtained by crossing Cdkn2afl/fl mice with germline Cre-expressing (MVH-Cre) mice.40 Mice were maintained in a 129SvJ/C57BL/6 background. Nine-week-old male mice were used in this study. Twenty Cdkn2a−/− were divided into two groups. One group of mice (n=10) were injected with KRAS-shTp53. Another group (n=10) were injected with KRAS-shTp53-shCdkn2b. Forty Cdkn2a+/+ mice were divided into four groups. The first group of mice (n=10) were injected with KRAS-shTp53. The second group (n=10) were injected with KRAS-shTp53-shCdkn2a. The third group (n=10) were injected with KRAS-shTp53-shCdkn2b. The last group (n=10) were injected with KRAS-shTp53-shCdkn2a/b.
Cell culture
Cells were regularly tested for mycoplasma contaminations free. HPDE6-C7 human pancreatic duct epithelial cells were obtained from China Center for Type Culture Collection (Wuhan, Hubei Province, China). Cells were cultured in Roswell Park Memorial Institute-1640 medium containing 100 U/ml penicillin, 100 μg/ml streptomycin and 10% fetal bovine serum (Life Technologies, Carlsbad, CA, USA) at 37 °C and 5% CO2. HPDE6-C7 cell line was confirmed on the basis of its morphology and expression markers of pancreatic ductal cells, cytokeratins (CK8/18/19) and sox9.
Chemicals and reagents
TGFβ receptor kinase inhibitor SB431542 was purchased from Selleckchem (cat. no. S1067, Houston, CA, USA). The MEK inhibitor U0126 was purchased from LC Laboratories (cat. no. U-6770, Boston, MA, USA). All restriction enzymes were purchased from Thermo Fisher Scientific (Waltham, MA, USA). Polyethylenimine was purchased from Sigma (cat. no. 408727, St Louis, MO, USA). Tribromoethanol (Avertin) was purchased from Sigma (cat. no. T48402). TRIzol reagent was purchased from Sigma (cat. no. T9424). Primary antibodies were used for IHC: CK-19 (cat. no. TROMA-III; Developmental Studies Hybridoma Bank, Iowa City, IA, USA), Ki67 (cat. no. VP-K452; Vector Laboratories, Burlingame, CA, USA), proliferating cell nuclear antigen (cat. no. 610664; BD Biosciences, Franklin Lakes, NJ, USA), Muc5 (cat. no. NCL-MUC-5AC; Novocastra, Newcastle Upon Tyne, UK), Hes1 (cat. no. ARP32372_T100; Aviva Systems Biology, San Diego, CA, USA), and MMP-7 (Vanderbilt Antibody and Protein Resource, Nashville, TN, USA), Rb1 (cat. no. 10048-2-Ig; Proteintech, Chicago, IL, USA), Rb-phosphor S780 (cat. no. ab47763; Abcam, Cambridge, UK). Fluorophore-conjugated secondary antibodies were purchased from Life Technologies. Primary antibodies were used for WB: RASG12D (cat. no. 26036; NewEast Bioscience, Malvern, PA, USA), P15 (cat. no. 12877-1-AP; Proteintech), TGF-β1 (cat. no. ab64715; Abcam), p-ERK (cat. no. sc-7383; Santa Cruz, Santa Cruz, CA, USA), ERK (cat. no. sc-154; Santa Cruz).
Lentiviral vector construction
We constructed lentiviral vectors containing KRASG12D and shRNAs targeting Tp53, Cdkn2a and Cdkn2b using pTomo as the backbone. First, the red fluorescent protein-IRES-EGFP fragment in pTomo between the XbaI and SalI restriction sites was replaced with KRASG12D. An shRNA targeting Tp53 was inserted upstream of the cytomegalovirus promoter via the ClaI restriction site, and an shRNA targeting Cdkn2a or Cdkn2b was inserted downstream of KRASG12D between the AgeI and SalI restriction sites. KRASG12D was amplified using the following primers: XbaI-KRASG12D-forward and SalI-AgeI-KRASG12D-reverse. The shRNA targeting Tp53 was amplified using the following primers: ClaI-shTp53-forward and ClaI-shTp53-reverse. The shRNA targeting Cdkn2b was amplified using the following primers: Kpn2I-shCdkn2b-forward and SalI-shCdkn2b-reverse. The shRNA targeting Cdkn2a was amplified using the following primers: SalI-shCdkn2a-forward and SalI-shCdkn2a-reverse. The sequence of all primers and target sequences of shTp53, shCdkn2b and shCdkn2a were shown in Supplementary Table 3.
Preparation of lentiviral particles
The lentiviral DNA vector along with the packaging plasmids pCMVΔ8.9 and pMD2.G prepared at a ratio of 5:2.5:1 were introduced into HEK293T cells in logarithmic growth phase by polyethylenimine-mediated transfection.41 The culture medium was collected and centrifuged at 3000 g for 10 min; the supernatant was passed through a sterile 0.45-μm filter to remove loose cells and cellular debris, and centrifuged at 12500 g at 4 °C for 2.5 h. The supernatant was discarded, and the pellet containing viral particles was resuspended in phosphate-buffered saline. Aliquots of the virus were stored at −80 °C.
Lentiviral injection
Mice were anesthetized by intraperitoneal injection of tribromoethanol (Avertin) at a dose of 250 mg/kg.42 Oculentum was used to prevent drying of the eyes. Following hair removal and sterilization of the skin surface, a 1-cm incision was made at the midline of the abdomen, and 15 μl of lentivirus containing 0.1% Trypan blue was slowly injected into the head of the pancreas with a 30-gauge needle. The needle was held in place for an additional 10 s before removal to prevent leakage. The skin of the abdomen was sutured, and mice were maintained under a heating lamp until recovery from anesthesia.
RNA isolation and RNA sequencing
Total RNA was extracted from the pancreases of newborn and 9-week-old mice and adult mice with pancreatic cancer. Three biological replicates were obtained for each group. RNA sequencing libraries were constructed using the mRNA-Seq Prep kit (Illumina, San Diego, CA, USA), and sequencing was carried out on an Illumina HiSeq 2000 platform as paired-end reads with a length of 150 bp. Approximately 10 Gb of raw, high-quality sequence data were obtained for each sample (Supplementary Table 1).
Raw data and expression matrix files for this work have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus (accession no. GSE87388). We filtered out low-quality reads with Q-score values lower than 20 (99.99% accuracy) and reads shorter than 70 nucleotides using GSQC Toolkit v.2.3.3.43 Clean reads were mapped to the mouse genome (version GRCm38.p4 that includes 21,936 protein-coding and 3,495 lincRNA genes) using Tophat-2.1.0.44 Gene expression levels (fragments per kilobase of transcript per million fragments) were calculated using Cufflinks 2.2.1.44 The Pearson correlation coefficient was used for sample transcriptome clustering.45
Quantitative real-time–PCR
Total RNA was isolated with TRIzol reagent (Sigma-Aldrich, St Louis, MO, USA) according to the manufacturer’s protocol. Reverse transcription was carried out using the RevertAid First Strand cDNA Synthesis kit (Thermo Fisher Scientific). quantitative real-time–PCR was performed with the SYBR Select Master Mix kit (Life Technologies) and the following primers: TP53 RT-forward, TP53 RT-reverse, CDKN2A RT-forward, CDKN2A RT-reverse, CDKN2B RT-forward, CDKN2B RT-reverse, sequence of primers were shown in Supplementary Table 3.
Histology and immunohistochemistry
Pancreatic tissue specimens were dissected and immediately fixed in 4% formalin for 48 h, then dehydrated in a graded series of alcohol and embedded in paraffin. Specimens were sectioned at a thickness of 4 μm and stained with hematoxylin and eosin for histopathological analysis. For immunolabeling, sections were deparaffinized and rehydrated, and antigen retrieval was performed in citric acid solution (pH 6.0) for 5 min at 125 °C in an autoclave. Endogenous peroxidase activity was quenched by incubation in 3% hydrogen peroxide for 15 min, followed by blocking in 10% goat serum for 2 h at room temperature. The sections were incubated overnight at 4 °C with primary antibodies, followed by incubation at room temperature for 1 h with fluorescent secondary antibodies. Nuclei were stained with 4,6-diamidino-2-phenylindole. The slides were mounted with Aqua-Poly/Mount (cat. no. 18606; Polysciences, Warminster, PA, USA).
Western blot
Cells were lysed in RIPA buffer and were centrifuged to remove the debris. Protein in supernatant was quantified with BCA method (Beyotime; cat. no. P0009, Shanghai, China). Protein was analyzed by sodium dodecyl sulfate–polyacrylamide gel electrophoresis followed by immunoblotting with primary antibodies. Specific signals were detected with horseradish peroxidase-conjugated secondary antibodies (Sigma) and chemiluminescent horseradish peroxidase substrate reagents (cat. no. WbKLS0500; Millipore, Billerica, MA, USA).
Senescence-associated β-galactosidase staining
Senescence-associated β-galactosidase staining was carried out according to the manufacturer’s instructions (cat. no. 9860; Cell Signaling Technology, Danvers, MA, USA). Dissected tissue specimens were flash frozen in liquid nitrogen and immediately mounted in optimum cutting temperature compound (Sakura Finetek INC, Torrance, CA, USA).46 Sections were cut at a thickness of 4 μm and incubated in 1 × fixation solution for 1 min at room temperature, then washed three times in phosphate-buffered saline followed by overnight incubation at 37 °C in staining solution. Blue-stained senescent cells were visualized by light microscopy.
Promoter analysis
The −463/+70 region of p15Ink4b promoter47 and −250/+1 region of the TGF-β148 promoter were separately cloned into the pGL3-Basic luciferase vector (Promega, Madison, WI, USA) at KpnI and XhoI sites to generate pGL3-p15 and pGL3-TGF-β1 wild-type luciferase reporter plasmids. The two Smad-binding elements in the −463/+70 region of p15Ink4b were mutated from GCCT and AGAC to GCTT and AAAC, respectively, to obtain the pGL3-p15SBE mutant vector,49 whereas the EGR1 binding site in the −250/+1 region of TGFβ1 was mutated from cgcccccgc to cgcTTTcgc48 to obtain the pGL3-TGF-β1 EGR1 mutant plasmid. The vectors were used to transfect HPDE6-C7 cells with Lipofectamine 2000 (Life Technologies). Luciferase activity was evaluated with a dual-luciferase reporter assay system (Promega).
Statistical analysis
Data were analyzed using GraphPad Prism software (GraphPad Inc., San Diego, CA, USA) and represented as the mean±s.e.m. Comparisons between two groups were analyzed by two tailed Student’s t-test for statistical significance. One-way analysis of variances was applied for multiple comparisons. All experiments were repeated at least three times. Statistical significance was provided for all cases and the variance among groups was not statistically different. The sample sizes were determined on the basis of previous experience in the laboratory and pre-specified effect sizes considered to be biological significant. No samples or animals were excluded from any analyses and all replicates were authentic biological replicates. Animals were randomly assigned to lentivirus injection. Blind analysis was not performed in this study. The levels of significance was set at P<0.01 (**) and P<0.001 (***).
Acknowledgments
We thank Dr Ronald Depinho, Dr Gerry Chu and Dr Haoqiang Ying for constructive opinions on this study. We thank Dr Antonio Lavarone for the useful discussion on the regulation of CDKN2B. We thank Dr Jumin Zhou for manuscript revision. We thank Yujie Xia for technical assistance with the histological analysis. This work was supported by by the ‘Strategic Priority Research Program’ of the Chinese Academy of Sciences (grant no. XDB13020400 to PS and XDA 01040403 to XZ), the National Natural Science Foundation of China (NSFC, 81472862) and the Top Talents Program of Yunnan Province, China (2012HA014) to XZ.
Author contributions
QT and X Zhou designed and performed most experiments. JH and SA performed the computational works. LY, HD, BS, DY and LL performed lentivirus preparation and animal experiments. BJ, CC and RL participated in some experimental design and manuscript revision. SP and X Zhao supervised the project and wrote the manuscript.
Footnotes
Supplementary Information accompanies this paper on the Oncogene website (http://www.nature.com/onc)
The authors declare no conflict of interest.
Supplementary Material
References
- Ying H, Dey P, Yao W, Kimmelman AC, Draetta GF, Maitra A et al. Genetics and biology of pancreatic ductal adenocarcinoma. Genes Dev 2016; 30: 355–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan DP, Hong TS, Bardeesy N. Pancreatic adenocarcinoma. N Engl J Med 2014; 371: 1039–1049. [DOI] [PubMed] [Google Scholar]
- Waddell N, Pajic M, Patch AM, Chang DK, Kassahn KS, Bailey P et al. Whole genomes redefine the mutational landscape of pancreatic cancer. Nature 2015; 518: 495–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones S, Zhang XS, Parsons DW, Lin JCH, Leary RJ, Angenendt P et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science 2008; 321: 1801–1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang LH, Tsutsumi S, Kawaguchi T, Nagasaki K, Tatsuno K, Yamamoto S et al. Whole-exome sequencing of human pancreatic cancers and characterization of genomic instability caused by MLH1 haploinsufficiency and complete deficiency. Genome Res 2012; 22: 208–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardeesy N, Aguirre AJ, Chu GC, Cheng KH, Lopez LV, Hezel AF et al. Both p16(Ink4a) and the p19(Arf)-p53 pathway constrain progression of pancreatic adenocarcinoma in the mouse. Proc Natl Acad Sci USA 2006; 103: 5947–5952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguirre AJ, Bardeesy N, Sinha M, Lopez L, Tuveson DA, Horner J et al. Activated Kras and Ink4a/Arf deficiency cooperate to produce metastatic pancreatic ductal adenocarcinoma. Genes Dev 2003; 17: 3112–3126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hingorani SR, Wang L, Multani AS, Combs C, Deramaudt TB, Hruban RH et al. Trp53R172H and KrasG12D cooperate to promote chromosomal instability and widely metastatic pancreatic ductal adenocarcinoma in mice. Cancer Cell 2005; 7: 469–483. [DOI] [PubMed] [Google Scholar]
- Bailey JM, Hendley AM, Lafaro KJ, Pruski MA, Jones NC, Alsina J et al. p53 mutations cooperate with oncogenic Kras to promote adenocarcinoma from pancreatic ductal cells. Oncogene 2016; 35: 4282–4288. [DOI] [PubMed] [Google Scholar]
- Guerra C, Collado M, Navas C, Schuhmacher AJ, Hernandez-Porras I, Canamero M et al. Pancreatitis-induced inflammation contributes to pancreatic cancer by inhibiting oncogene-induced senescence. Cancer Cell 2011; 19: 728–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris JPt, Cano DA, Sekine S, Wang SC, Hebrok M. Beta-catenin blocks Kras-dependent reprogramming of acini into pancreatic cancer precursor lesions in mice. J Clin Invest 2010; 120: 508–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habbe N, Shi G, Meguid RA, Fendrich V, Esni F, Chen H et al. Spontaneous induction of murine pancreatic intraepithelial neoplasia (mPanIN) by acinar cell targeting of oncogenic Kras in adult mice. Proc Natl Acad Sci USA 2008; 105: 18913–18918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopp JL, von Figura G, Mayes E, Liu FF, Dubois CL, Morris JPt et al. Identification of Sox9-dependent acinar-to-ductal reprogramming as the principal mechanism for initiation of pancreatic ductal adenocarcinoma. Cancer Cell 2012; 22: 737–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gidekel Friedlander SY, Chu GC, Snyder EL, Girnius N, Dibelius G, Crowley D et al. Context-dependent transformation of adult pancreatic cells by oncogenic K-Ras. Cancer Cell 2009; 16: 379–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hingorani SR, Petricoin EF, Maitra A, Rajapakse V, King C, Jacobetz MA et al. Preinvasive and invasive ductal pancreatic cancer and its early detection in the mouse. Cancer Cell 2003; 4: 437–450. [DOI] [PubMed] [Google Scholar]
- Tanaka H, Fukamachi K, Futakuchi M, Alexander DB, Long N, Tamamushi S et al. Mature acinar cells are refractory to carcinoma development by targeted activation of Ras oncogene in adult rats. Cancer Sci 2010; 101: 341–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biankin AV, Waddell N, Kassahn KS, Gingras MC, Muthuswamy LB, Johns AL et al. Pancreatic cancer genomes reveal aberrations in axon guidance pathway genes. Nature 2012; 491: 399–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witkiewicz AK, McMillan EA, Balaji U, Baek G, Lin WC, Mansour J et al. Whole-exome sequencing of pancreatic cancer defines genetic diversity and therapeutic targets. Nat Commun 2015; 6: 6744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niola F, Zhao X, Singh D, Sullivan R, Castano A, Verrico A et al. Mesenchymal high-grade glioma is maintained by the ID-RAP1 axis. J Clin Invest 2013; 123: 405–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedmann-Morvinski D, Bushong EA, Ke E, Soda Y, Marumoto T, Singer O et al. Dedifferentiation of neurons and astrocytes by oncogenes can induce gliomas in mice. Science 2012; 338: 1080–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houbracken I, Baeyens L, Ravassard P, Heimberg H, Bouwens L. Gene delivery to pancreatic exocrine cells in vivo and in vitro. BMC Biotechnol 2012; 12: 74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brembeck FH, Moffett J, Wang TC, Rustgi AK. The keratin 19 promoter is potent for cell-specific targeting of genes in transgenic mice. Gastroenterology 2001; 120: 1720–1728. [DOI] [PubMed] [Google Scholar]
- McNeal AS, Liu KV, Nakhate V, Natale CA, Duperret EK, Capell BC et al. CDKN2B loss promotes progression from benign melanocytic nevus to melanoma. Cancer Discov 2015; 5: 1072–1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CT, Yao J, de Belle I, Huang RP, Adamson E, Mercola D. The transcription factor EGR-1 suppresses transformation of human fibrosarcoma HT1080 cells by coordinated induction of transforming growth factor-beta(1), fibronectin, and plasminogen activator inhibitor-1. J Biol Chem 1999; 274: 4400–4411. [DOI] [PubMed] [Google Scholar]
- Baron V, Adamson ED, Calogero A, Ragona G, Mercola D. The transcription factor Egr1 is a direct regulator of multiple tumor suppressors including TGF beta 1, PTEN, p53, and fibronectin. Cancer Gene Ther 2006; 13: 115–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Chetrit N, Tarcic G, Yarden Y. ERK-ERF-EGR1, a novel switch underlying acquisition of a motile phenotype. Cell Adhes Migr 2013; 7: 33–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson EL, Willis N, Mercer K, Bronson RT, Crowley D, Montoya R et al. Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Gene Dev 2001; 15: 3243–3248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji BA, Tsou L, Wang HM, Gaiser S, Chang DZ, Daniluk J et al. Ras activity levels control the development of pancreatic diseases. Gastroenterology 2009; 137: 1072–1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logsdon CD, Ji B. Ras activity in acinar cells links chronic pancreatitis and pancreatic cancer. Clin Gastroenterol Hepatol 2009; 7: S40–S43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beroukhim R, Mermel CH, Porter D, Wei G, Raychaudhuri S, Donovan J et al. The landscape of somatic copy-number alteration across human cancers. Nature 2010; 463: 899–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bignell GR, Greenman CD, Davies H, Butler AP, Edkins S, Andrews JM et al. Signatures of mutation and selection in the cancer genome. Nature 2010; 463: 893–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latres E, Malumbres M, Sotillo R, Martin J, Ortega S, Martin-Caballero J et al. Limited overlapping roles of P15(INK4b) and P18(INK4c) cell cycle inhibitors in proliferation and tumorigenesis. EMBO J 2000; 19: 3496–3506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krimpenfort P, Ijpenberg A, Song JY, van der Valk M, Nawijn M, Zevenhoven J et al. p15Ink4b is a critical tumour suppressor in the absence of p16Ink4a. Nature 2007; 448: 943–946. [DOI] [PubMed] [Google Scholar]
- Schuster K, Venkateswaran N, Rabellino A, Girard L, Pena-Llopis S, Scaglioni PP. Nullifying the CDKN2AB locus promotes mutant K-ras lung tumorigenesis. Mol Cancer Res 2014; 12: 912–923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiMauro T, David G. Ras-induced senescence and its physiological relevance in cancer. Curr Cancer Drug Tar 2010; 10: 869–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh SK, Ellenrieder V. Senescence in pancreatic carcinogenesis: from signalling to chromatin remodelling and epigenetics. Gut 2013; 62: 1364–1372. [DOI] [PubMed] [Google Scholar]
- Guerra C, Barbacid M. Genetically engineered mouse models of pancreatic adenocarcinoma. Mol Oncol 2013; 7: 232–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopinathan A, Morton JP, Jodrell DI, Sansom OJ. GEMMs as preclinical models for testing pancreatic cancer therapies. Dis Model Mech 2015; 8: 1185–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano M, Lee H, Chin L, Cordon-Cardo C, Beach D, DePinho RA. Role of the INK4a locus in tumor suppression and cell mortality. Cell 1996; 85: 27–37. [DOI] [PubMed] [Google Scholar]
- Gallardo T, Shirley L, John GB, Castrillon DH. Generation of a germ cell-specific mouse transgenic Cre line, Vasa-Cre. Genesis 2007; 45: 413–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y, Garson K, Li L, Vanderhyden BC. Optimization of lentiviral vector production using polyethylenimine-mediated transfection. Oncol Lett 2015; 9: 55–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohn DF, Wixson SK, White WJ, Benson GJ. Anesthesia and Analgesia in Laboratory Animals. Academic Press: San Diego, CA, USA, 1997. [Google Scholar]
- Patel RK, Jain M. NGS QC toolkit: a toolkit for quality control of next generation sequencing data. PLoS ONE 2012; 7: e30619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 2013; 14: R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 2010; 28: 511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimri GP, Lee X, Basile G, Acosta M, Scott G, Roskelley C et al. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc Natl Acad Sci USA 1995; 92: 9363–9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seoane J, Pouponnot C, Staller P, Schader M, Eilers M, Massague J. TGFbeta influences Myc, Miz-1 and Smad to control the CDK inhibitor p15INK4b. Nat Cell Biol 2001; 3: 400–408. [DOI] [PubMed] [Google Scholar]
- Nagarajan RP, Chen FF, Li W, Vig E, Harrington MA, Nakshatri H et al. Repression of transforming-growth-factor-beta-mediated transcription by nuclear factor kappa B. Biochem J 2000; 348: 591–596. [PMC free article] [PubMed] [Google Scholar]
- Feng XH, Lin X, Derynck R. Smad2, Smad3 and Smad4 cooperate with Sp1 to induce p15(Ink4B) transcription in response to TGF-beta. EMBO J 2000; 19: 5178–5193. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.