Abstract
Rhizobium sp. IRBG74 not only nodulates Sesbania cannabina but also can enhance rice growth; however, the underlying molecular mechanisms are not clear. Here, we show that Rhizobium sp. IRBG74 colonizes the roots of Arabidopsis thaliana, which leads to inhibition in the growth of main root but enhancement in the formation of lateral roots. The promotion of lateral root formation by Rhizobium sp. IRBG74 in the fls2-1 mutant, which is insensitive to flagellin, is similar to the wild-type plant, while the auxin response deficient mutant tir1-1 is significantly less sensitive to Rhizobium sp. IRBG74 than the wild type in terms of the inhibition of main root elongation and the promotion of lateral root formation. Further transcriptome analysis of Arabidopsis roots inoculated with Rhizobium sp. IRBG74 revealed differential expression of 50 and 211 genes at 24 and 48 h, respectively, and a majority of these genes are involved in auxin signaling. Consistent with the transcriptome analysis results, Rhizobium sp. IRBG74 treatment induces expression of the auxin responsive reporter DR5:GUS in roots. Our results suggest that in Arabidopsis Rhizobium sp. IRBG74 colonizes roots and promotes the lateral root formation likely through modulating auxin signaling. Our work provides insight into the molecular mechanisms of interactions between legume-nodulating rhizobia and non-legume plants.
Keywords: Arabidopsis, auxin signaling, Rhizobium, RNA-seq, root development
Introduction
Legume nodulating rhizobia are among the most effective plant growth-promoting rhizobacteria as these bacteria form nitrogen-fixing symbiotic associations with legumes which are highly efficient in supplying nitrogen to the host plants (Graham and Vance, 2003; Peoples et al., 2009; Gyaneshwar et al., 2011). The legume-rhizobia symbiosis involves a complex signal exchange between rhizobia and the legume host (Oldroyd and Downie, 2006; Stacey et al., 2006; Jones et al., 2007). Apart from forming nitrogen-fixing symbiosis with legumes, many rhizobial strains are known to form growth promoting associations with non-legumes, such as maize (Zea mays), wheat (Triticum aestivum), and rice (Oryza sativa) (Biswas et al., 2000a,b; Chaintreuil et al., 2000; Gutiérrez-Zamora and Martínez-Romero, 2001; Ladha and Reddy, 2003; Yanni and Dazzo, 2010; Mitra et al., 2016). It has been suggested that the ability of rhizobia to secrete plant growth-promoting hormones, especially indole acetic acid (IAA), might be important for plant growth promotion (Biswas et al., 2000b; Mishra et al., 2008).
To better understand the mechanisms underlying the interactions between legume nodulating rhizobia and non-legumes, we have chosen Rhizobium sp. IRBG74, which nodulates several species of Sesbania, as a model rhizobia (Cummings et al., 2009). In addition, Rhizobium sp. IRBG74 was shown to enhance the growth of wetland rice cultivars by the mechanisms other than nitrogen fixation (Biswas et al., 2000a,b). We recently showed that Rhizobium sp. IRBG74 colonizes rice tissues internally and that rhizobial lipopolysaccharide plays an important role in Rhizobium colonization (Mitra et al., 2016). In contrast to detailed knowledge of the mechanisms involved in rhizobial-legume interactions, there is significant lack of understanding about the molecular details of how bacterial endophytes recognize and enter their plant hosts as well as the mechanisms by which endophytes enhance plant growth.
Arabidopsis is a model species for plant genetics (Arabidopsis Genome Intiative, 2000; Zhao et al., 2001, 2002) and in recent years it has been utilized as a model system to elucidate plant microbiome and to determine mechanisms of plant interactions with beneficial microorganisms, especially the endophytes (Lebeis et al., 2015; Müller et al., 2016; Wintermans et al., 2016; Asari et al., 2017). Earlier studies showed that Arabidopsis can be colonized by Azorhizobium caulinodans ORS571, a rhizobial symbiont of aquatic legume Sesbania rostrata (Gough et al., 1997; Stone et al., 2001). A recent report has suggested that Mesorhizobium loti colonizes Arabidopsis roots and enhances plant growth likely through changes in plant auxin levels (Poitout et al., 2017). However, the mechanistic details of interactions between Arabidopsis and legume nodulating rhizobia has not yet been studied. The present study is focused on examining whether Rhizobium sp. IRBG74 can colonize Arabidopsis and the response of Arabidopsis to rhizobial inoculation at morphological and molecular levels.
Materials and methods
Bacterial strains, plant materials, and growth conditions
The Rhizobium sp. IRBG74 wild-type strain as well as strains marked with GUS (Rhizobium sp. IRBG74-GUS) and GFP (Rhizobium sp. IRBG74-GFP) (Mitra et al., 2016) reporters were maintained on LB agar plates. Arabidopsis thaliana Landsberg erecta (Ler) and Columbia (Col-0), fls2-1 (Gómez-Gómez and Boller, 2000) and tir1-1 (Gray et al., 2001) mutants (Col-0), as well as the DR5:GUS transgenic line (Ulmasov et al., 1997) were used for studying colonization by Rhizobium sp. IRBG74. Seedlings were grown on the Murashige and Skooge (MS) medium in square Petri dishes under a 16-h light/8-h dark photoperiod regime at 22°C and 50% humidity.
Colonization analysis of Arabidopsis roots inoculated by Rhizobium sp. IRBG74
The inoculation of Arabidopsis seedlings by Rhizobium sp. IRBG74 (wild type), Rhizobium sp. IRBG74-GUS and Rhizobium sp. IRBG74-GFP strains was carried out as described earlier (Mitra et al., 2016). Briefly, Arabidopsis seeds were surface-sterilized by soaking in 50% of household bleach for 10 min and then in 70% ethanol for further 10 min. Sterilized seeds were then planted on Petri dishes containing MS salt and stratified for 3 days at 4°C and then germinated at 22°C in a growth chamber. Roots of 4-day old seedlings were inoculated with LB grown bacterial strains (0.1 O.D.) or plain LB as control. Seedlings were then transferred to square Petri dishes with the MS medium and incubated in a growth chamber at 22°C. Three biological repeats were performed with 10 seedlings per treatment.
The colonization of Arabidopsis roots was examined at 2 and 7 days after inoculation using histochemical GUS staining (Liu et al., 2010; Huang et al., 2016d) and fluorescence microscopy (Huang et al., 2016b,c, 2017) as described in our previous studies. The seedlings were removed from the plates and washed three times with sterile water and then used for GUS staining and GFP fluorescence analyses. The images of GUS stained roots were photographed by an Olympus SZX7 dissection microscope equipped with the Olympus DP 70 digital camera (Olympus, Center Valley, PA, USA). For confocal microscopy analysis, root samples were observed with a Leica TCS SP2 laser scanning confocal microscope using a 63×/1.4 water immersion objective lens. A 488-nm laser was used to excite GFP and FM4-64. The emission was captured using PMTs set at 505–530, 500–550, and 644–719 nm, respectively.
Analysis of Arabidopsis root development after treatment with Rhizobium sp. IRBG74
As described above, Arabidopsis wild-type, fls2-1 and tir1-1 mutant seedling were inoculated with the LB grown Rhizobium sp. IRBG74 (wild type) strain and the plain LB, respectively. The effect of bacterial inoculation on root development was determined by measuring the length of primary root and the numbers of lateral roots 7 days after inoculation. Three biological repeats were performed with 10 seedlings per treatment. Data were analyzed for statistical significance by t-test.
Transcriptome analysis of Arabidopsis root development in response to treatment with Rhizobium sp. IRBG74
To examine how the Arabidopsis root development responds to the Rhizobium sp. IRBG74 treatment at the molecular level, we performed RNA-seq experiments as described in our previous studies (Huang et al., 2016d). One hundred seedlings were inoculated with the LB grown Rhizobium sp. IRBG74 (wild type) strain and the plain LB (control). Roots were collected at 24 and 48 h after inoculation and then were used for RNA extraction by RNeasy Plant Mini Kit (Qiagen). On-column DNase digestion was carried out using the RNase-free DNase (Qiagen). RNA amounts were determined by the Qubit 3.0 Fluorimeter (Fisher scientific) and 500 ng of total RNA was used for preparing RNA-seq library. RNA-seq libraries were constructed using the Illumina's TruSeq Stranded Total RNA with Ribo-Zero Plant kit. Three biological repeats were performed; therefore, totally 12 libraries were sequenced (single-end sequencing, 1 × 100 bp) in the Biotechnology Center of University of Wisconsin-Madison on a HiSeq2500 (Illumina) using a TruSeq SBS sequencing kit version 3 (Illumina) and processed with Casava 1.8.2. The average reads for each library are 27.2 M.
RNA-seq data evaluation and pre-processing were performed by the CyVerse Discovery Environment (Goff et al., 2011; Merchant et al., 2016). The quality of RNA-seq reads was evaluated by FastQC 0.10.1. Adapter sequences were removed by Scythe-adapter-trimming. Low quality sequences were trimmed and filtered by Sickle-quality-based-trimming. The read-quality of the cleaned reads was re-evaluated by FastQC 0.10.1. RNA-Seq reads were aligned to the Arabidopsis genome using TopHat2-SE. Cufflinks2 was used to assemble transcripts from the RNA-seq data. Cuffmerge2 was used to merge all Cufflinks transcripts into a single transcriptome annotation file. Cuffdiff2.2.1a was used to compare differentially expressed genes. Genes with p-values < 0.05 and log2 fold changes more than 1 or less than−1 were selected as differentially expressed genes and were further analyzed. GO annotation search and functional categorization were performed from TAIR (https://www.arabidopsis.org).
Genes were clustered using the Database for Annotation, Visualization and Integrated Discovery (DAVID) (http://david.abcc.ncifcrf.gov/) (Huang et al., 2007, 2009, 2016d). The files of output DAVID chart records were used to generate enrichment map (Merico et al., 2011). Only gene-sets passing conservative significance thresholds (p-value < 0.001, False Discovery Rate (FDR) < 2%, the overlap coefficient = 0.5) were selected to present in the Enrichment Map, which results in 106 significantly enriched gene-sets. Heat-maps were generated by the Enrichment Map software. The complete datasets in this study are available in the NCBI GEO database under accession numbers of GSM2863566, GSM2863567, GSM2863568, GSM2863569, GSM2863570, GSM2863571, GSM2863572, GSM2863573, GSM2863574, GSM2863575, GSM2863576, and GSM2863577.
Characterization of auxin response in Arabidopsis roots inoculated with Rhizobium sp. IRBG74
Seedlings of the DR5:GUS transgenic line were inoculated with the LB grown Rhizobium sp. IRBG74 (wild type) strain and the plain LB (control). As described in our previous studies (Liu et al., 2010; Huang et al., 2016d), changes in GUS expression were determined using histochemical GUS staining and quantification of GUS activity using a Synergy HT multi-mode microplate reader at 360 nm (excitation) and 460 nm (emission). The concentration of protein was measured using the Bradford method and GUS activity was calculated as nM 4-MU/min/mg protein.
Results and discussion
Rhizobium sp. IRBG74 can colonize Arabidopsis roots
To test whether Rhizobium sp. IRBG74 can colonize the Arabidopsis root, we inoculated Arabidopsis (Col-0 and Ler) roots with strains of Rhizobium sp. IRBG74-GUS and IRBG74-GFP which were utilized to study colonization of Sesbania cannabina and rice before (Mitra et al., 2016). From 30 Col-0 seedlings (3 repeats, each repeat contains 10 seedlings) treated by Rhizobium sp. IRBG74-GUS, GUS signals were observed throughout the main roots of all seedlings by 2 Days After Inoculation (DAI) (Figure 1A and Supplementary Figure 1) and on both the main root and lateral roots of all seedlings at 7 DAI (Figure 1B). A similar result was obtained from Rhizobium sp. IRBG74-GUS treated Ler seedlings (Figure 1C,D). Confocal microscopy further demonstrated that although no GFP signal was observed in the root tips of 30 examined seedlings (3 repeats, each repeat contains 10 seedlings) treated by Rhizobium sp. IRBG74-GFP at 2 DAI (Figure 1E), the GFP expression was detected in root hairs (the surface of main root) at 7 DAI (Figure 1F), which agrees with the results from the GUS staining (Figures 1A,C). The fluorescent bacteria were found mainly on the root surface. These results suggest that Rhizobium sp. IRBG74 can colonize Arabidopsis roots epiphytically under the studied conditions.
Figure 1.

Rhizobium sp. IRBG74 can colonize Arabidopsis Root. (A,B) Columbia (Col-0) 4-day seedlings without (left) and with (right) Rhizobium sp. IRBG74-GUS treatment for 2 (A) and 7 (B) days. (C,D) Landsberg erecta (Ler) 4-day seedlings without (right) and with (right) Rhizobium sp. IRBG74-GUS treatment for 2 (C) and 7 (D) days. (E) No GFP signal was found in the root tip of Col-0 4-day seedlings treated with Rhizobium sp. IRBG74-GFP for 2 days. (F) GFP signal was detected in root hairs of Col-0 4-day seedlings treated with Rhizobium sp. IRBG74-GFP for 2 days.
Rhizobium sp. IRBG74 promotes lateral root formation in Arabidopsis
Recent studies have suggested that plant beneficial bacteria enhance the formation of lateral roots in host plants, which in turn allow more surface area for bacteria to colonize (Gyaneshwar et al., 2002; James et al., 2002). As shown in Figures 1A–D, Rhizobium sp. IRBG74 promoted the lateral root formation but inhibited the main root elongation. To further study this effect, the length of the main root and the number of lateral roots were quantified. Inoculation of 30 (3 repeats, each repeat contains 10 seedlings) Arabidopsis seedlings with Rhizobium sp. IRBG74 resulted in significant reduction in growth of the main root and significant enhancement in the number of latera roots at 7 DAI in both Col-0 (Figures 2A–D) and Ler ecotypes (Figures 2E–H). These results are consistent with earlier reports showing the enhancement of Arabidopsis root growth by Baccillus amyloliquefaciens SQR9 (Chen et al., 2017) and the increase in rice root growth by Rhizobium sp. IRBG74 (Mitra et al., 2016). Additionally, earlier studies have shown that the natural openings created as a result of emergence of lateral roots from the main root could allow point of entry for plant beneficial bacteria through “crack entry,” especially in non-legumes (Gyaneshwar et al., 2002; James et al., 2002). Rhizobium sp. IRBG74 is known to colonize its plant-hosts through “crack entry” (Mitra et al., 2016). Our results suggests that Rhizobium sp. IRBG74 likely induces the formation of lateral roots which in turn might provide additional openings for the bacteria to invade the internal tissues.
Figure 2.
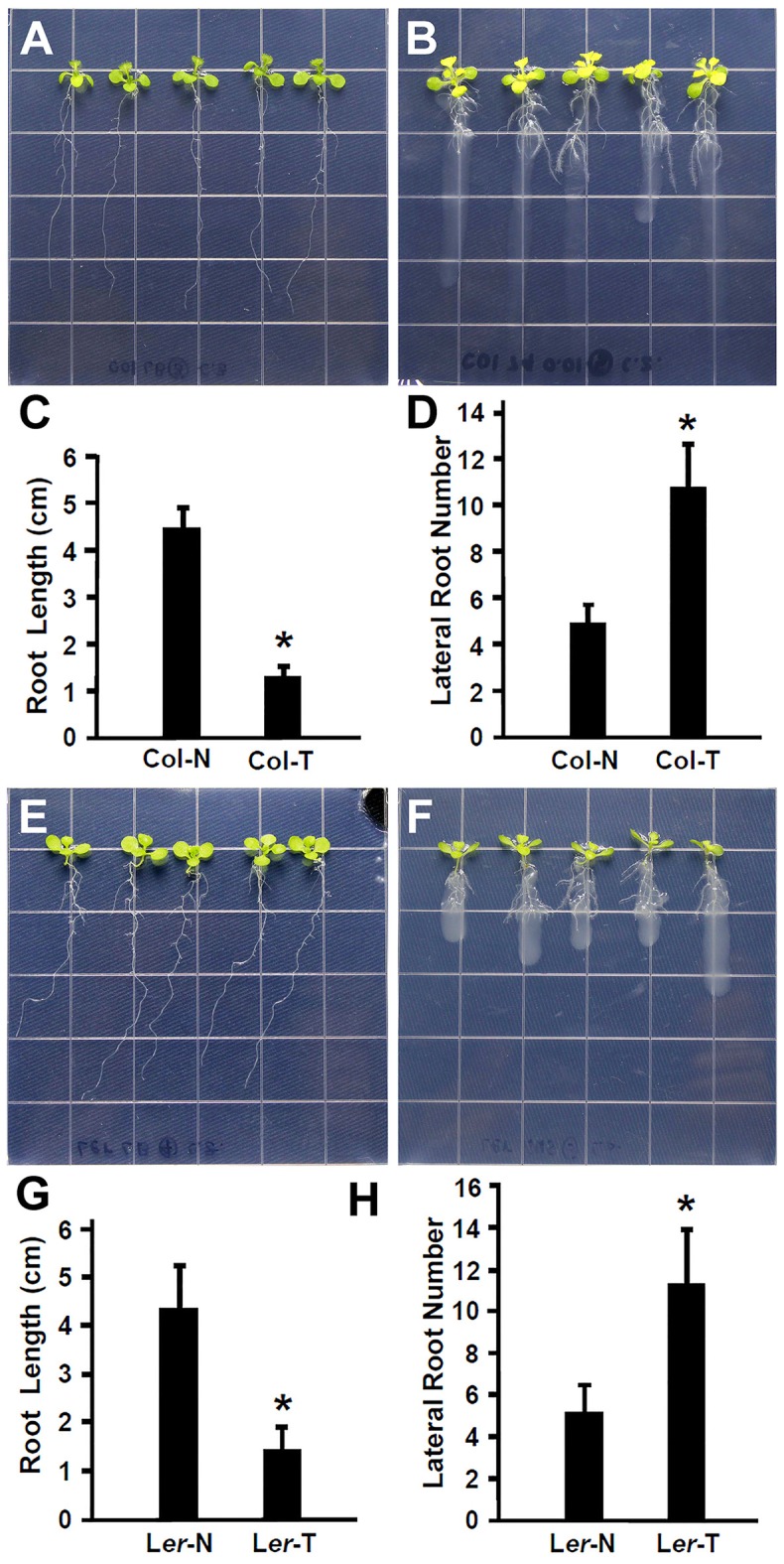
Rhizobium sp. IRBG74 promotes lateral root formation but inhibits the primary root growth in Arabidopsis. (A,B) Columbia (Col-0) 4-day seedlings without (A) and with (B) Rhizobium sp. IRBG74 treatment for 7 days. (C) Primary root length of Col-0 4-day seedlings without (Col-N) and with (Col-T) Rhizobium sp. IRBG74 treatment for 7 days. (D) Lateral root numbers of Col-0 4-day seedlings without (Col-N) and with (Col-T) Rhizobium sp. IRBG74 treatment for 7 days. (E,F) Landsberg erecta (Ler) 4-day seedlings without (E) and with (F) Rhizobium sp. IRBG74 treatment. (G) Primary root length of Ler 4-day seedlings without (Ler-N) and with (Ler-T) Rhizobium sp. IRBG74 treatment for 7 days. (H) Lateral root numbers of Ler 4-day seedlings without (Ler-N) and with (Ler-T) Rhizobium sp. IRBG74 treatment for 7 days. *Indicates the difference is significant (P < 0.01).
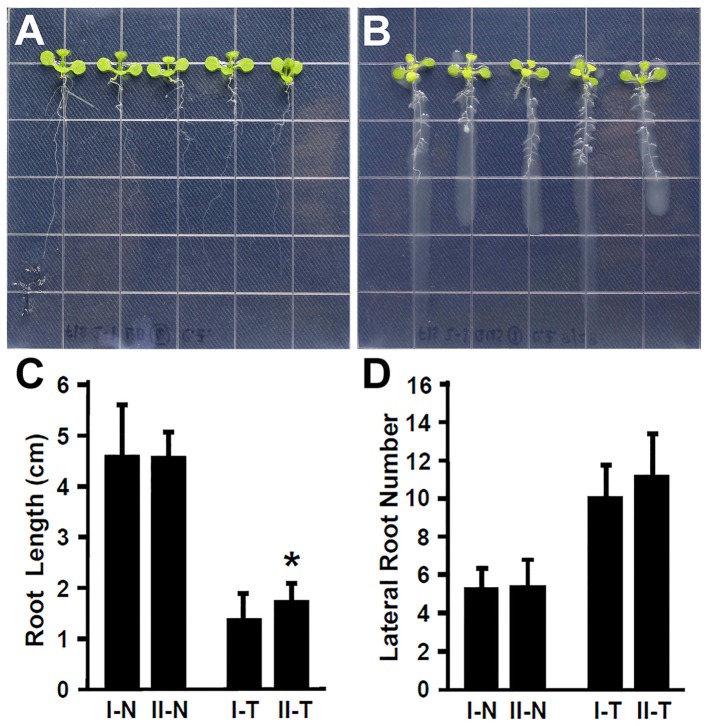
The enhancement of lateral root formation is not altered in fls2-1 seedlings treated by Rhizobium sp. IRBG74
Plant recognize the beneficial and pathogenic microorganisms through the Microbe-Associated Molecular Patterns (MAMPs) (Dangl and Jones, 2001). Bacterial flagellin is one of the MAMPs that triggers plant defense response upon its recognition by the FLAGELLIN SENSITIVE 2 (FLS2) receptor (Gómez-Gómez and Boller, 2000; Zipfel et al., 2004). The FLS2 receptor in Arabidopsis recognizes bacterial flagella and induces the plant defense response (Zipfel et al., 2004; Schwessinger and Ronald, 2012). It has been suggested that the lack of flagella might enhance the endophytic colonization (Capdevila et al., 2004; Iniguez et al., 2005). In contrast, rice colonization by growth promoting Azoarcus BH72 requires bacterial flagella (Buschart et al., 2012). To determine whether the flagellin recognition by Arabidopsis plays a role in colonization by Rhizobium sp. IRBG74, 30 (3 repeats, each repeat contains 10 seedlings) fls2-1 mutant seedlings (Gómez-Gómez and Boller, 2000) were inoculated and changes in root development were determined. Similar to the wild type, Rhizobium sp. IRBG74 inhibits the elongation of main roots of fls2-1 mutant seedlings; however the inhibition efficacy is lower (Figures 3A–C). In addition, Rhizobium sp. IRBG74 showed a similar effect in terms of enhancing lateral root formation in wild-type and fls2-1 seedlings (Figures 3A,B,D). Our results suggest that flagellin recognition is unlikely involved in the ability of Rhizobium sp. IRBG74 to colonize Arabidopsis roots.
Figure 3.
The enhancement of lateral root formation is not altered in fls2-1 seedlings treated by Rhizobium sp. IRBG74. (A,B) fls2-1 mutant 4-day seedlings without (A) and with (B) Rhizobium sp. IRBG74 treatment for 7 days. (C) Primary root length of Col-0 and fls2-1 4-day seedlings without and with Rhizobium sp. IRBG74 treatment for 7 days. (D) Lateral root numbers of Col-0 and fls2-1 4-day seedlings without and with Rhizobium sp. IRBG74 treatment for 7 days. I-N, Col-0 without Rhizobium sp. IRBG74 treatment; II-N, fls2-1 without Rhizobium sp. IRBG74 treatment; I-T, Col-0 with Rhizobium sp. IRBG74 treatment; and II-T, fls2-1 with Rhizobium sp. IRBG74 treatment. *Indicates the difference is significant (P < 0.01).
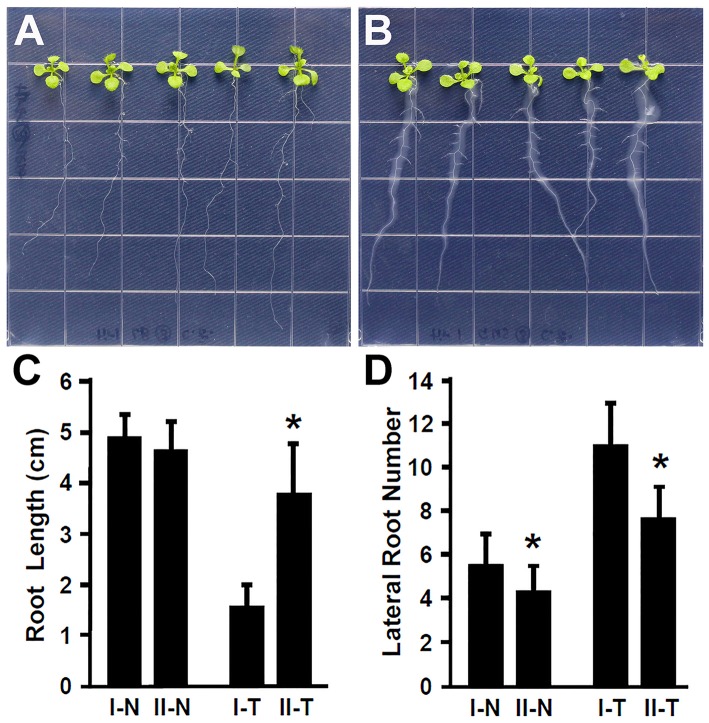
The tir1-1 mutant is insensitive to the treatment with Rhizobium sp. IRBG74
Plant hormone auxin plays an important role in lateral root development (Fukaki et al., 2007; Gutierrez et al., 2009; Huang et al., 2016a). Our results showed that inoculation with Rhizobium sp. IRBG74 promoted the lateral root formation, suggesting that auxin is likely involved in the response of Arabidopsis to Rhizobium sp. IRBG74. To further explore this idea, the Arabidopsis tir1-1 mutant which is defective in auxin perception (Gray et al., 2001; Dharmasiri et al., 2005; Kepinski and Leyser, 2005; Tan et al., 2007) was inoculated with Rhizobium sp. IRBG74 and its effect on root development was examined. In contrast to wild-type seedlings that showed a strong inhibition of main root elongation (Figure 2), the tir1-1 mutant seedlings (30 total, 3 repeats) exhibited a significantly less reduction in main root growth (Figures 4A–C). The tir1-1 mutant seedlings produced a less number of lateral roots than the wild type without Rhizobium sp. IRBG74 treatment; however, with the Rhizobium sp. IRBG74 treatment, the number of lateral roots in wild-type seedlings increased 98.19%, but the number of lateral roots in tir1-1 seedlings increased 76.94% (Figures 4A,B,D). These results indicate that the tir1-1 mutant is less sensitive than the wild type regarding the inhibition of main root elongation and promotion of lateral root formation. Auxin synthesized by plant beneficial bacteria is postulated to be a major mechanism by which these bacteria promote plant growth (Bloemberg and Lugtenberg, 2001; Lugtenberg and Kamilova, 2009). Our results suggest that Rhizobium sp. IRBG74 affects the main root growth and lateral root formation possibly by changing auxin signaling.
Figure 4.
The tir1-1 mutant is insensitive to Rhizobium sp. IRBG74 treatment. (A,B) tir1-1 mutant 4-day seedlings without (A) and with (B) Rhizobium sp. IRBG74 treatment for 7 days. (C) Primary root length of Col-0 and tir1-1 4-day seedlings without and with Rhizobium sp. IRBG74 treatment for 7 days. (D) Lateral root numbers of Col-0 and tir1-1 4-day seedlings without and with Rhizobium sp. IRBG74 treatment for 7 days. I-N, Col-0 without Rhizobium sp. IRBG74 treatment; II-N, tir1-1 without Rhizobium sp. IRBG74 treatment; I-T, Col-0 with Rhizobium sp. IRBG74 treatment; and II-T, tir1-1 with Rhizobium sp. IRBG74 treatment. *Indicates the difference is significant (P < 0.01).
Rhizobium sp. IRBG74 leads to differential expression of genes involved in auxin signaling, cell wall and cell membrane integrity, and transport during root development
To examine the molecular mechanisms by which Rhizobium sp. IRBG74 affects root development, we performed RNA-seq analysis using Arabidopsis seedling treated by Rhizobium sp. IRBG74. Roots of 4-day seedlings that were un-inoculated and inoculated with Rhizobium sp. IRBG74 for 24 and 48 h were sampled. Each sample contained 100 roots. Three biological repeats were performed; therefore, totally 12 libraries were constructed and sequenced. Analysis of transcriptome changes showed that 50 and 211 genes were differentially expressed at 24 and 48 h (Supplementary Tables 1, 2), respectively. Examination of these genes using Gene Ontology (GO) cellular component analysis demonstrated that proteins encoded by these genes may function in the extracellular region (13.884%), nucleus (10.896%), the plasma membrane (10.193%), and cell wall (6.854%, Supplementary Figure 2). In addition, GO molecular function analysis showed that the coding proteins have transferase (10.385%), hydrolase (8.462%), transporter (7.885%), protein binding (7.5%), kinase (5.769%), and transcription factor (5%) activities (Supplementary Figure 3). Moreover, GO biological process analysis further found that these genes are possibly involved in responses to biotic and abiotic stimulus/stress (19.423%), development (7.308%), transport (6.346%), signal transduction (5.577%), and gene transcription (4.712%, Supplementary Figure 4).
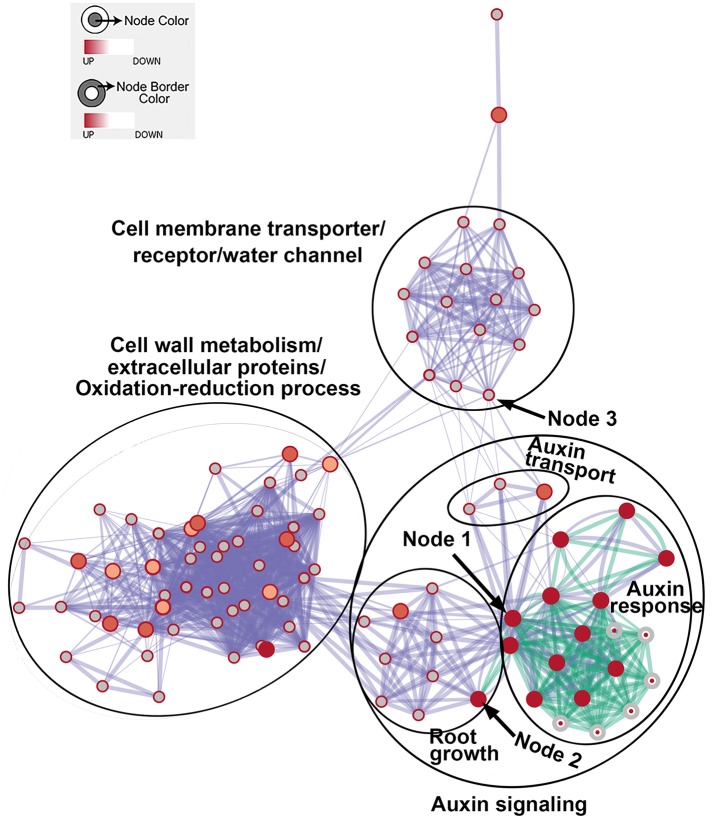
We then generated enriched GO gene-sets using the Database for Annotation, Visualization and Integrated Discovery (DAVID) (Huang et al., 2007, 2009, 2016d; Merico et al., 2011). The enrichment map contains 106 significantly enriched gene-sets (Figure 5). Enriched gene-sets from the 24-h treatment (indicated by the node center) were connected by green lines, while gene-sets from the 48-h treatment (indicated by the node border) are linked by light blue lines.
Figure 5.
The enrichment map showing differentially expressed gene-sets in roots treated by Rhizobium sp. IRBG74 for 24 and 48 h. The map displays enriched gene-sets in untreated and treated roots at 24 and 48 h. The node center represents gene-sets at 24 h, whereas the node border represents gene-sets at 48 h. Gene-sets at 24 h are connected by green lines, while gene-sets at 48 h are linked by light blue lines. The node size represents the gene-set size and line thickness indicates the degree of overlap between two gene-sets. Clusters of gene-sets were manually circled and labeled to demonstrate biological and molecular functions. Node 1 represents auxin response genes, node 2 contains genes with functions in root growth, and node 3 represents cell membrane transporter/receptor/water channel genes.
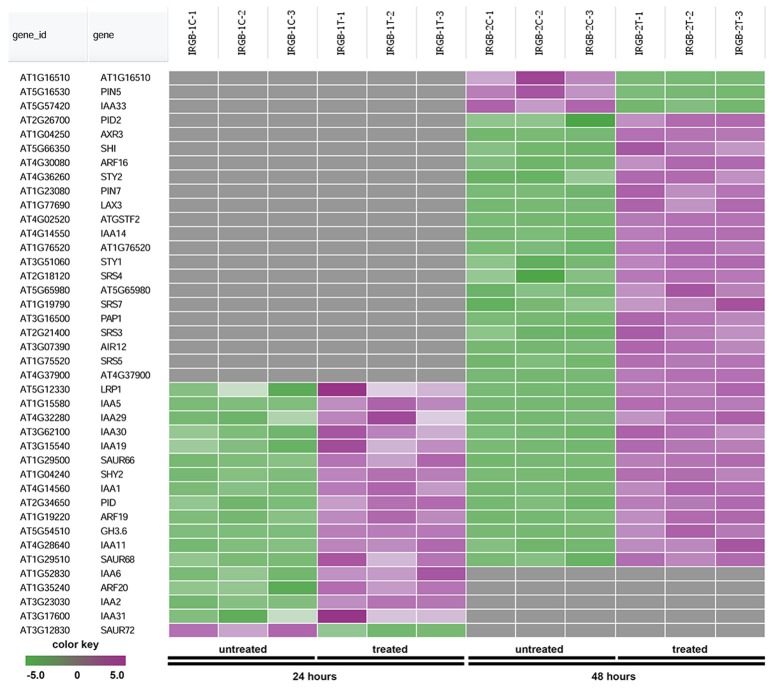
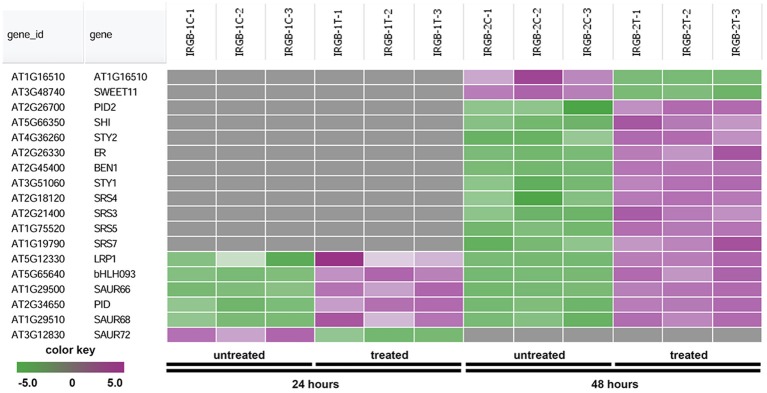
In the enrichment map, three distinct groups represent auxin signaling, cell wall metabolism/extracellular protein/ oxidation-reduction process, and cell membrane reporter/transporter/water channel (Figure 5). The mainly overlapped gene-sets between genes expressed at 24 and 48 h are clustered into the subgroup of auxin signaling. The heatmap generated from the node 1 showed 40 auxin response genes, such as IAA (IAA1, IAA2, IAA3/SHY2, IAA5, IAA6, IAA11, IAA19, IAA29, IAA30, and IAA31), SAUR (SAUR66 and SAUR68), ARF (ARF19 and ARF20), PIN (PIN5 and PIN7), PID, and GH3.6 genes (Figure 6 and Supplementary Table 3). More auxin signaling genes were expressed at 48 h, including auxin transport genes.
Figure 6.
Heatmap showing expression changes of representative genes related to auxin signaling. The auxin signaling subgroup in the node 1 gene-set showing in Figure 5 contains a total of 40 genes. Green color: down-regulated; Purple color: up-regulated; and Gray color: no significant expression change.
The heatmap generated from the node 2 found SHORT INTERNODES (SHI), SHI-RELATED SEQUENCE (SRS/STY), PINOID (PID), and SMALL AUXIN UPREGULATED RNA (SAUR) (Figure 7 and Supplementary Table 4). Genes in the node 2 function in root growth (Figure 7).
Figure 7.
Heatmap showing expression changes of representative genes regulating root growth. The root growth subgroup in the node 2 gene-set showing in Figure 5 consists of a total of 18 genes. Green color, down-regulated; Purple color, up-regulated; and Gray color, no significant expression change.
The node 3 represents the cell membrane transporter/receptor/water channel group which connects to the auxin transport subgroup (Figure 5). The node 3 contains genes encoding receptor kinases (PERK4, BRL3, FLS2, and ER), auxin transporters (PIN5, PIN7, and LAX3), ion transporters/water channels (ATFRO5, PIP1D, RD28, PIP2F, and PIP1A), proton transporters (PPI2 and HA3), sucrose transporters (SWEET11) (Figure 8 and Supplementary Table 5). Those proteins, which are involved in signal transduction, cell wall metabolism and oxidation-reduction process, mostly function in the plasma membrane, cell wall or the extracellular region. In this group, genes were mainly induced at 48 h. Our RNA-seq results suggest that expression of genes mainly involved in auxin signaling is altered in response to colonization by Rhizobium sp. IRBG74.
Figure 8.
Heatmap showing expression changes of representative genes involved in cell membrane activities. The cell membrane transporter/receptor/water channel subgroup in the node 3 gene-set showing in Figure 5 has a total of 28 genes. Green color, down-regulated; Purple color, up-regulated; and Gray color, no significant expression change.
Expression of DR5:GUS is induced by Rhizobium sp. IRBG74 treatment
The root growth phenotype and transcriptome analysis suggest that Rhizobium sp. IRBG74 inoculation leads to altered auxin response in Arabidopsis roots. To further determine the changes in auxin response, Arabidopsis seedlings carrying the auxin response reporter DR5:GUS (Ulmasov et al., 1997; Liu et al., 2010) were inoculated with Rhizobium sp. IRBG74 and GUS activities were then assayed. After DR5:GUS seedlings (30 total, 3 repeats) growing for 24 (Figure 9A), 48 (Figure 9B), 72 (Figure 9C), and 120 (Figure 9D) hours, GUS signals were mainly observed in tips of main and lateral roots without Rhizobium sp. IRBG74 treatment; however, enhanced GUS signals were found along main and lateral roots when inoculated with Rhizobium sp. IRBG74. Quantitative analysis further showed that GUS activities were significantly increased in roots of DR5:GUS seedlings treated by Rhizobium sp. IRBG74 for 24, 48, and 72 h (Figure 9E). GUS expression changes were not observed in leaves after Rhizobium sp. IRBG74 treatments. Our results support that Rhizobium sp. IRBG74 enhances auxin response in Arabidopsis roots.
Figure 9.

Rhizobium sp. IRBG74 affects the DR5:GUS expression. (A–D) GUS staining showing DR5:GUS 4-day seedlings without (left) and with (right) Rhizobium sp. IRBG74 treatment for 24 (A), 48 (B), 72 (C), and 120 (D) h. (E) GUS activity [nM 4-methylumbelliferone (4-MU)/min/mg] quantification of DR5:GUS 4-day seedlings without and with Rhizobium sp. IRBG74 treatment for 24, 48, and 72 h. 24N: 24 h non-treated, 24T, 24 h treated; 48N, 48 h non-treated; 48T, 48 h treated; 72N, 72 h non-treated, and 72T, 72 h treated. *Indicates the difference is significant (P < 0.01).
In conclusion, our study shows that the legume nodulating rhizobia Rhizobium sp. IRBG74 can colonize roots of the non-legume model plant Arabidopsis. Rhizobium sp. IRBG74 enhances lateral root formation but inhibits growth of the main root. Analyses using the auxin perception deficient mutant, auxin response reporter, and RNA-seq suggest that Rhizobium sp. IRBG74 affects the main root growth and lateral root formation by altering auxin signaling. Earlier studies have shown that Rhizobium sp. IRBG74 synthesizes IAA when supplemented with tryptophan (Biswas et al., 2000a) and its genome (Crook et al., 2013) contains genes that are putatively involved in IAA synthesis. The requirement of supplemented tryptophan for IAA synthesis is a common feature of many plant beneficial bacteria (Idris et al., 2007), including the legume-nodulating rhizobia (Camerini et al., 2008). The biosynthesis of IAA by these bacteria is affected by root exudates (Bais et al., 2006). It has recently been shown that inoculation of cucumber with Bacillus amyloliquefaciens results in enhanced secretion of tryptophan by cucumber roots which increases the IAA production by B. amyloliquefaciens and thus enhanced plant growth (Liu et al., 2016). In our experiments, the lateral root formation of seedlings which were grown in the basal MS salt media without tryptophan was promoted by Rhizobium sp. IRBG74, suggesting that, similar to the cucumber B. amyloliquefaciens system, Rhizobium sp. IRBG74 likely utilizes tryptophan secreted from Arabidopsis roots to synthesize IAA. Further studies are underway to determine whether the observed affects are due to IAA synthesis by Rhizobium sp. IRBG74 or due to alteration in plant synthesized IAA upon inoculation with Rhizobium sp. IRBG74.
Author contributions
CZ, DZ, and GP conceived and designed the experiments. DZ and GP supervised the experiments. CZ and JH performed most of the experiments. All authors analyzed data. All authors contributed to writing the article.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank T. J. Guilfoyle for the DR5:GUS seed, the ABRC for the fls2-1 and tir1-1 seeds, and T. Schuck and P. Engevold for plant care.
Footnotes
Funding. This work was supported by the Research Growth Initiative (RGI) at the University of Wisconsin-Milwaukee to GP and DZ. Research in the Zhao Laboratory was supported by the National Science Foundation (IOS-0721192 and IOS-1322796), RGI, the Shaw Scientist Award from the Greater Milwaukee Foundation, the Bradley Catalyst Award from the UWM Research Foundation, and the CAS/SAFEA International Partnership Program for Creative Research Teams. Research in the Prasad Laboratory was supported by the grant from National Science Foundation (IOS-1256879).
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2017.02556/full#supplementary-material
References
- Arabidopsis Genome Intiative (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408, 796–815. 10.1038/35048692 [DOI] [PubMed] [Google Scholar]
- Asari S., Tarkowská D., Rolcík J., Novák O., Palmero D. V., Bejai S., et al. (2017). Analysis of plant growth-promoting properties of Bacillus amyloliquefaciens UCMB5113 using Arabidopsis thaliana as host plant. Planta 245, 15–30. 10.1007/s00425-016-2580-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bais H. P., Weir T. L., Perry L. G., Gilroy S., Vivanco J. M. (2006). The role of root exudates in rhizosphere interactions with plants and other organisms. Annu. Rev. Plant Biol. 57, 233–266. 10.1146/annurev.arplant.57.032905.105159 [DOI] [PubMed] [Google Scholar]
- Biswas J. C., Ladha J. K., Dazzo F. B. (2000a). Rhizobial inoculation influences seedling vigor and yield of rice. Agronomy J 92, 880–886. 10.2134/agronj2000.925880x [DOI] [Google Scholar]
- Biswas J. C., Ladha J. K., Dazzo F. B. (2000b). Rhizobia inoculation improves nutrient uptake and growth of lowland rice. Soil Sci. Soc. Am. J. 64, 1644–1650. 10.2136/sssaj2000.6451644x [DOI] [Google Scholar]
- Bloemberg G. V., Lugtenberg B. J. (2001). Molecular basis of plant growth promotion and biocontrol by rhizobacteria. Curr. Opin. Plant Biol. 4, 343–350. 10.1016/S1369-5266(00)00183-7 [DOI] [PubMed] [Google Scholar]
- Buschart A., Sachs S., Chen X., Herglotz J., Krause A., Reinhold-Hurek B. (2012). Flagella mediate endophytic competence rather than act as MAMPS in rice-Azoarcus sp. strain BH72 interactions. Mol. Plant Microb. Interact. 25, 191–199. 10.1094/MPMI-05-11-0138 [DOI] [PubMed] [Google Scholar]
- Camerini S., Senatore B., Lonardo E., Imperlini E., Bianco C., Moschetti G., et al. (2008). Introduction of a novel pathway for IAA biosynthesis to rhizobia alters vetch root nodule development. Arch. Microbiol. 190, 67–77. 10.1007/s00203-008-0365-7 [DOI] [PubMed] [Google Scholar]
- Capdevila S., Martínez-Granero F. M., Sánchez-Contreras M., Rivilla R., Martín M. (2004). Analysis of Pseudomonas fluorescens F113 genes implicated in flagellar filament synthesis and their role in competitive root colonization. Microbiology 150, 3889–3897. 10.1099/mic.0.27362-0 [DOI] [PubMed] [Google Scholar]
- Chaintreuil C., Giraud E., Prin Y., Lorquin J., Bâ A., Gillis M., et al. (2000). Photosynthetic bradyrhizobia are natural endophytes of the African wild rice Oryza breviligulata. Appl. Environ. Microbiol.ogy 66, 5437–5447. 10.1128/AEM.66.12.5437-5447.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Liu Y., Wu G., Zhang N., Shen Q., Zhang R. (2017). Beneficial rhizobacterium Bacillus amyloliquefaciens SQR9 induces plant salt tolerance through spermidine production. Mol. Plant Microb. Interact. 30, 423–432. 10.1094/MPMI-02-17-0027-R [DOI] [PubMed] [Google Scholar]
- Crook M. B., Mitra S., Ané J. M., Sadowsky M. J., Gyaneshwar P. (2013). Complete genome sequence of the Sesbania Symbiont and rice growth-promoting endophyte Rhizobium sp. Strain IRBG74. Genome Announ. 1:e00934-13 10.1128/genomeA.00934-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummings S. P., Gyaneshwar P., Vinuesa P., Farruggia F. T., Andrews M., Humphry D., et al. (2009). Nodulation of Sesbania species by Rhizobium (Agrobacterium) strain IRBG74 and other rhizobia. Environ. Microbiol. 11, 2510–2525. 10.1111/j.1462-2920.2009.01975.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dangl J. L., Jones J. D. (2001). Plant pathogens and integrated defence responses to infection. Nature 411, 826–833. 10.1038/35081161 [DOI] [PubMed] [Google Scholar]
- Dharmasiri N., Dharmasiri S., Estelle M. (2005). The F-box protein TIR1 is an auxin receptor. Nature 435, 441–445. 10.1038/nature03543 [DOI] [PubMed] [Google Scholar]
- Fukaki H., Okushima Y., Tasaka M. (2007). Auxin-mediated lateral root formation in higher plants. Int. Rev. Cytol. 256, 111–137. 10.1016/S0074-7696(07)56004-3 [DOI] [PubMed] [Google Scholar]
- Goff S. A., Vaughn M., McKay S., Lyons E., Stapleton A. E., Gessler D., et al. (2011). The iPlant collaborative: cyberinfrastructure for plant biology. Front. Plant Sci. 2:34 10.3389/fpls.2011.00034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-Gómez L., Boller T. (2000). FLS2: an LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol. Cell 5, 1003–1011. 10.1016/S1097-2765(00)80265-8 [DOI] [PubMed] [Google Scholar]
- Gough C., Galera C., Vasse J., Webster G., Cocking E. C., Dénarié J. (1997). Specific flavonoids promote intercellular root colonization of Arabidopsis thaliana by Azorhizobium caulinodans ORS571. Mol. Plant Microb. Interact. 10, 560–570. 10.1094/MPMI.1997.10.5.560 [DOI] [PubMed] [Google Scholar]
- Graham P. H., Vance C. P. (2003). Legumes: importance and constraints to greater use. Plant Physiol. 131, 872–877. 10.1104/pp.017004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray W. M., Kepinski S., Rouse D., Leyser O., Estelle M. (2001). Auxin regulates SCF(TIR1)-dependent degradation of AUX/IAA proteins. Nature 414, 271–276. 10.1038/35104500 [DOI] [PubMed] [Google Scholar]
- Gutierrez L., Bussell J. D., Pacurar D. I., Schwambach J., Pacurar M., Bellini C. (2009). Phenotypic plasticity of adventitious rooting in Arabidopsis is controlled by complex regulation of AUXIN RESPONSE FACTOR transcripts and microRNA abundance. Plant Cell 21, 3119–3132. 10.1105/tpc.108.064758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutiérrez-Zamora M. L., Martínez-Romero E. (2001). Natural endophytic association between Rhizobium etli and maize (Zea mays L.). J. Biotechnol. 91, 117–126. 10.1016/S0168-1656(01)00332-7 [DOI] [PubMed] [Google Scholar]
- Gyaneshwar P., Hirsch A. M., Moulin L., Chen W. M., Elliott G. N., Bontemps C., et al. (2011). Legume-nodulating betaproteobacteria: diversity, host range, and future prospects. Mol. Plant Microb. Interact. 24, 1276–1288. 10.1094/MPMI-06-11-0172 [DOI] [PubMed] [Google Scholar]
- Gyaneshwar P., James E. K., Reddy P. M., Ladha J. K. (2002). Herbaspirillum colonization increases growth and nitrogen accumulation in aluminium-tolerant rice varieties. N. Phytol. 154, 131–145. 10.1046/j.1469-8137.2002.00371.x [DOI] [Google Scholar]
- Huang D. W., Sherman B. T., Lempicki R. A. (2009). Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57. 10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
- Huang D. W., Sherman B. T., Tan Q., Kir J., Liu D., Bryant D., et al. (2007). DAVID Bioinformatics Resources: expanded annotation database and novel algorithms to better extract biology from large gene lists. Nucleic Acids Res. 35, W169–W175. 10.1093/nar/gkm415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J., Li Z., Zhao D. (2016a). Deregulation of the OsmiR160 target gene OsARF18 causes growth and developmental defects with an alteration of auxin signaling in rice. Sci. Rep. 6:29938 10.1038/srep29938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J., Li Z., Biener G., Xiong E., Malik S., Eaton N., et al. (2017). Carbonic anhydrases function in anther cell differentiation downstream of the receptor-like kinase EMS1. Plant Cell 29, 1335–1356. 10.1105/tpc.16.00484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J., Smith A. R., Zhang T., Zhao D. (2016b). Creating completely both male and female sterile plants by specifically ablating microspore and megaspore mother cells. Front. Plant Sci. 7:30 10.3389/fpls.2016.00030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J., Wijeratne A. J., Tang C., Zhang T., Fenelon R. E., Owen H. A., et al. (2016d). Ectopic expression of TAPETUM DETERMINANT1 affects ovule development in Arabidopsis. J. Exp. Bot. 67, 1311–1326. 10.1093/jxb/erv523 [DOI] [PubMed] [Google Scholar]
- Huang J., Zhang T., Linstroth L., Tillman Z., Otegui M. S., Owen H. A., et al. (2016c). Control of anther cell differentiation by the small protein igand TPD1 and its receptor EMS1 in Arabidopsis. PLoS Genet. 12:e1006147 10.1371/journal.pgen.1006147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Idris E. E., Iglesias D. J., Talon M., Borriss R. (2007). Tryptophan-dependent production of indole-3-acetic acid (IAA) affects level of plant growth promotion by Bacillus amyloliquefaciens FZB42. Mol. Plant Microb. Interact. 20, 619–626. 10.1094/MPMI-20-6-0619 [DOI] [PubMed] [Google Scholar]
- Iniguez A. L., Dong Y., Carter H. D., Ahmer B. M., Stone J. M., Triplett E. W. (2005). Regulation of enteric endophytic bacterial colonization by plant defenses. Mol. Plant Microb. Interact. 18, 169–178. 10.1094/MPMI-18-0169 [DOI] [PubMed] [Google Scholar]
- James E. K., Gyaneshwar P., Mathan N., Barraquio W. L., Reddy P. M., Iannetta P. P., et al. (2002). Infection and colonization of rice seedlings by the plant growth-promoting bacterium Herbaspirillum seropedicae Z67. Mol. Plant Microb. Interact. 15, 894–906. 10.1094/MPMI.2002.15.9.894 [DOI] [PubMed] [Google Scholar]
- Jones K. M., Kobayashi H., Davies B. W., Taga M. E., Walker G. C. (2007). How rhizobial symbionts invade plants: the Sinorhizobium-Medicago model. Nat. Rev. Microbiol. 5, 619–633. 10.1038/nrmicro1705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kepinski S., Leyser O. (2005). The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature 435, 446–451. 10.1038/nature03542 [DOI] [PubMed] [Google Scholar]
- Ladha J. K., Reddy P. M. (2003). Nitrogen fixation in rice systems: state of knowledge and future prospects. Plant Soil 252, 151–167. 10.1023/A:1024175307238 [DOI] [Google Scholar]
- Lebeis S. L., Paredes S. H., Lundberg D. S., Breakfield N., Gehring J., McDonald M., et al. (2015). PLANT MICROBIOME. Salicylic acid modulates colonization of the root microbiome by specific bacterial taxa. Science 349, 860–864. 10.1126/science.aaa8764 [DOI] [PubMed] [Google Scholar]
- Liu X., Huang J., Wang Y., Khanna K., Xie Z., Owen H. A., et al. (2010). The role of floral organs in carpels, an Arabidopsis loss-of-function mutation in MicroRNA160a, in organogenesis and the mechanism regulating its expression. Plant J. 62, 416–428. 10.1111/j.1365-313X.2010.04164.x [DOI] [PubMed] [Google Scholar]
- Liu Y., Chen L., Zhang N., Li Z., Zhang G., Xu Y., et al. (2016). Plant-microbe communication enhances auxin biosynthesis by a root-associated bacterium, Bacillus amyloliquefaciens SQR9. Mol. Plant Microb. Interact. 29, 324–330. 10.1094/MPMI-10-15-0239-R [DOI] [PubMed] [Google Scholar]
- Lugtenberg B., Kamilova F. (2009). Plant-growth-promoting rhizobacteria. Annu. Rev. Microbiol. 63, 541–556. 10.1146/annurev.micro.62.081307.162918 [DOI] [PubMed] [Google Scholar]
- Merchant N., Lyons E., Goff S., Vaughn M., Ware D., Micklos D., et al. (2016). The iPlant collaborative: cyberinfrastructure for enabling data to discovery for the life sciences. PLoS Biol. 14:e1002342 10.1371/journal.pbio.1002342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merico D., Isserlin R., Bader G. D. (2011). Visualizing gene-set enrichment results using the Cytoscape plug-in enrichment map. Methods Mol. Biol. 781, 257–277. 10.1007/978-1-61779-276-2_12 [DOI] [PubMed] [Google Scholar]
- Mishra R. P. N., Singh R. K., Jaiswal H. K., Singh M. K., Yanni Y. G., Dazzo F. B. (2008). Rice-rhizobia association: evolution of an alternate niche of beneficial plant-bacteria association, in Plant-Bacteria Interactions: Strategies and Techniques to Promote Plant Growth, eds Ahmad I., Pichtel J., Hayat S. (Weinheim: Wiley-VCH Verlag; ), 165–194. [Google Scholar]
- Mitra S., Mukherjee A., Wiley-Kalil A., Das S., Owen H., Reddy P. M., et al. (2016). A rhamnose-deficient lipopolysaccharide mutant of Rhizobium sp. IRBG74 is defective in root colonization and beneficial interactions with its flooding-tolerant hosts Sesbania cannabina and wetland rice. J. Exp. Bot. 67, 5869–5884. 10.1093/jxb/erw354 [DOI] [PubMed] [Google Scholar]
- Müller D. B., Vogel C., Bai Y., Vorholt J. A. (2016). The plant microbiota: systems-level insights and perspectives. Annu. Rev. Genet. 50, 211–234. 10.1146/annurev-genet-120215-034952 [DOI] [PubMed] [Google Scholar]
- Oldroyd G. E., Downie J. A. (2006). Nuclear calcium changes at the core of symbiosis signalling. Curr. Opin. Plant Biol. 9, 351–357. 10.1016/j.pbi.2006.05.003 [DOI] [PubMed] [Google Scholar]
- Peoples M. B., Brockell J., Herridge D. F., Rochester I. J., Alves B. J. R., Urquiaga S., et al. (2009). The contributions of nitrogen-fixing crop nodules to the productivity of agricultural systems. Symbiosis 48, 1–17. 10.1007/BF03179980 [DOI] [Google Scholar]
- Poitout A., Martinière A., Kucharczyk B., Queruel N., Silva-Andia J., Mashkoor S., et al. (2017). Local signalling pathways regulate the Arabidopsis root developmental response to Mesorhizobium loti inoculation. J. Exp. Bot. 68, 1199–1211. 10.1093/jxb/erw502 [DOI] [PubMed] [Google Scholar]
- Schwessinger B., Ronald P. C. (2012). Plant innate immunity: perception of conserved microbial signatures. Annu. Rev. Plant Biol. 63, 451–482. 10.1146/annurev-arplant-042811-105518 [DOI] [PubMed] [Google Scholar]
- Stacey G., Libault M., Brechenmacher L., Wan J., May G. D. (2006). Genetics and functional genomics of legume nodulation. Curr. Opin. Plant Biol. 9, 110–121. 10.1016/j.pbi.2006.01.005 [DOI] [PubMed] [Google Scholar]
- Stone P. J., O'Callaghan K. J., Davey M. R., Cocking E. C. (2001). Azorhizobium caulinodans ORS571 colonizes the xylem of Arabidopsis thaliana. Mol. Plant Microb. Interact. 14, 93–97. 10.1094/MPMI.2001.14.1.93 [DOI] [PubMed] [Google Scholar]
- Tan X., Calderon-Villalobos L. I., Sharon M., Zheng C., Robinson C. V., Estelle M., et al. (2007). Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature 446, 640–645. 10.1038/nature05731 [DOI] [PubMed] [Google Scholar]
- Ulmasov T., Murfett J., Hagen G., Guilfoyle T. J. (1997). Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 9, 1963–1971. 10.1105/tpc.9.11.1963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wintermans P. C., Bakker P. A., Pieterse C. M. (2016). Natural genetic variation in Arabidopsis for responsiveness to plant growth-promoting rhizobacteria. Plant Mol. Biol. 90, 623–634. 10.1007/s11103-016-0442-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanni Y., Dazzo F. (2010). Enhancement of rice production using endophytic strains of Rhizobium leguminosarum bv. trifolii in extensive field inoculation trials within the Egypt Nile delta. Plant Soil 336, 129–142. 10.1007/s11104-010-0454-7 [DOI] [Google Scholar]
- Zhao D., Wang G. F., Speal B., Ma H. (2002). The EXCESS MICROSPOROCYTES1 gene encodes a putative leucine-rich repeat receptor protein kinase that controls somatic and reproductive cell fates in the Arabidopsis anther. Genes Dev. 16, 2021–2031. 10.1101/gad.997902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao D., Yu Q., Chen M., Ma H. (2001). The ASK1 gene regulates B function gene expression in cooperation with UFO and LEAFY in Arabidopsis. Development 128, 2735–2746. [DOI] [PubMed] [Google Scholar]
- Zipfel C., Robatzek S., Navarro L., Oakeley E. J., Jones J. D., Felix G., et al. (2004). Bacterial disease resistance in Arabidopsis through flagellin perception. Nature 428, 764–767. 10.1038/nature02485 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.