Abstract
Objective
In peanut, the DNA polymorphism is very low despite enormous phenotypic variations. This limits the use of genomics-assisted breeding to enhance peanut productivity. This study aimed to develop and validate new AhMITE1 and cleaved amplified polymorphic sequences (CAPS) markers.
Results
In total, 2957 new AhMITE1 markers were developed in addition to identifying 465 already reported markers from the whole genome re-sequencing data (WGRS) of 33 diverse genotypes of peanut. The B sub-genome (1620) showed more number of markers than the A sub-genome (1337). Distribution also varied among the chromosomes of both the sub-genomes. Further, 52.6% of the markers were from genic regions; where 31.0% were from intronic regions and 5.2% were from exonic regions. Of the 343 randomly selected markers, 82.2% showed amplification validation, with up to 35.5% polymorphism. From the SNPs on the A03, B01, B02 and B03 chromosomes, 11,730 snip-SNPs (potential CAPS sites) were identified, and 500 CAPS markers were developed from chromosome A03. Of these markers, 30.0% showed validation and high polymorphism. This study demonstrated the potential of the WGRS data to develop AhMITE1 and CAPS markers, which showed high level of validation and polymorphism. These marker resources will be useful for various genetic studies and mapping in peanut.
Electronic supplementary material
The online version of this article (10.1186/s13104-017-3121-8) contains supplementary material, which is available to authorized users.
Keywords: Peanut, Diverse genotypes, WGRS, AhMITE1 markers, CAPS markers, Validation and polymorphism
Introduction
High resolution trait mapping in peanut (Arachis hypogaea L. 2n = 4x = 40) demands a considerably large number of evenly distributed genome-wide markers to identify marker-trait associations. The fact that the genotypic polymorphism is very limited despite enormous phenotypic differences among peanut genotypes signifies the requirement for a large number of markers. Different types of markers have been developed and employed for diversity analysis, DNA finger-printing, trait mapping and genomics-assisted breeding (GAB). The prominent markers were RFLP, AFLP, DAF, SSR, DArT etc. These DNA markers and other protein markers showed low polymorphism among the peanut genotypes [1–3]. Though single nucleotide polymorphism (SNP) markers are abundant and highly polymorphic in different systems, they showed low polymorphism (14.0%) in cultivated peanut [4].
Transposons are widely distributed in genomes, and their polymorphic insertions allowed development of transposon-based markers [5, 6]. Both class I and class II transposon-based markers have been developed and used for various genetic analysis and mapping [7, 8]. In peanut, use of DNA transposon markers was proposed [9], and one such marker was developed to track the activity of A. hypogaea miniature inverted-repeat transposable element (AhMITE1) to associate its transposition with high-frequency origin of late leaf spot disease resistant mutants [10], and differentiation of two subspecies [11]. Subsequently, 1039 AhMITE1 markers were developed [12, 13], and used for mapping [14–16].
Use of diverse genotypes including the genetically unstable peanut mutants which show hyperactivity of AhMITE1 for marker discovery might detect a large number of AhMITE1 insertion polymorphic sites (AIPs), which could be employed to develop new markers. In the past, transposon markers were developed using transposon display [5, 17–21], transposon-enriched library [12, 13] and in silico analysis [22]. But, analysis of whole genome re-sequencing (WGRS) data from a large number of diverse genotypes is expected to capture all AIPs when the short reads are analyzed using the computational method polymorphic TEs and their movement detection (PTEMD) [23] for the de novo discovery of AIPs. This study reports the development of new AhMITE1 markers using diverse genotypes, and their validation using the parents of various mapping populations and backcross populations. SNPs discovered from the WGRS data were also used to develop cleaved amplified polymorphic sequences (CAPS) markers from selected chromosomes (A03, B01, B02 and B03), harboring quantitative trait loci (QTL) for the important agronomical and productivity traits [15, 24, 25].
Main text
Methods
A total of 33 genotypes were employed for AhMITE1 marker development. Details on the genotypes [10, 26–31] used for WGRS are given in Additional file 1: Table S1. WGRS reads were generated using Illumina HiSeq 2000 for six genotypes and obtained from public DNA sequence databases for the remaining 27 genotypes (DRA004503–DRA004506 and SRA459965) (https://www.ncbi.nlm.nih.gov/pubmed/27902796 and https://www.ncbi.nlm.nih.gov/pubmed/27993622) (Additional file 1: Table S1), and used to detect AIPs using PTEMD [23] without replication since the results were subjected for confirmation (validation) using wet-lab experiment through PCR, and detection in multiple genotypes to support the results. The sequences flanking the AIPs were retrieved, and primers were designed using default parameters of BatchPrimer3 [32]. AhMITE1 markers were validated by checking the amplicons from DER, VL 1, 110 and 110(S) for the expected size. The seeds of these genotypes were collected from the Department of Genetics and Plant Breeding, University of Agricultural Sciences, Dharwad, India. DNA was isolated from the young leaves of the plants following the modified cetyl trimethyl ammonium bromide (CTAB) method [33]. The cells were lysed with CTAB buffer, and the debris were removed by centrifugation. Proteins were removed from the extract by phenol:chloroform extraction and the RNA was removed by RNase treatment. The DNA was washed with ethanol and finally dissolved in Tris–EDTA (TE) buffer. Polymerase chain reaction (PCR) for the AhMITE1 markers was carried out in a reaction volume of 10 µl with the standard ingredients and PCR profile (Additional file 2: Table S2a and S2b) [12, 15] using eppendorf Mastercycler® pro. The PCR products were resolved on 2% agarose gel. The markers amplifying the expected product, depending on the presence or absence of AhMITE1 at those marker loci, were considered to be validated. The validated markers were checked for polymorphic information content (PIC) using PowerMarker V3.25 [34]. For this, additional ten genotypes constituting the recombinant inbred line (RIL) populations [35] and backcross populations [36, 37] were employed. The seeds of these genotypes were collected from the Department of Genetics and Plant Breeding, University of Agricultural Sciences, Dharwad, India.
Single nucleotide polymorphism identification from the WGRS data was performed as described earlier [38]. A 1001 bp sequence was obtained for each SNP (500 bp on left and right), and they were analyzed for snip-SNPs (SNP sites which modify restriction enzyme recognition sites) using CLC Sequence Viewer 7 (CLC Bio: http://www.clcbio.com) for 25 restriction enzymes. Primers were designed for those sequences which contained snip-SNP using GeneTool Lite [39]. Preference was given to those primers which could amplify 200–900 bp amplicons, and generate restriction fragments of at least 100 bp.
Cleaved amplified polymorphic sequences markers were validated by PCR amplification and restriction digestion with the respective restriction enzyme (Additional file 2: Tables S2c–S2e and Additional file 7: Table S6). A reaction volume of 12 µl containing Emerald Amp® GT PCR Master Mix (Catalog No. RR310A, Clontech), 5 pmol of each primer and 50 ng of genomic DNA was amplified, and used for restriction digestion. The restriction fragments were separated on 2% agarose gel, and checked for the products of expected size (Additional file 7: Table S6). Those markers producing the PCR and restriction fragments of expected size were considered to be validated. PIC was calculated for the CAPS markers as described for AhMITE1 markers.
Results and discussion
Large copy number [40, 41], enormous genome-wide insertion variation [12] and association with genes to alter the function makes AhMITE1 a target for marker development [42]. A total of 3546 AIPs were identified from a total reads of 9.9 billion across 33 genotypes, of which 3081 were new and 465 were already reported [12, 13]. This high success rate of marker discovery could be attributed to the diverse genotypes and the software (PTEMD) used in this study. Primers could be designed for 2957 AIPs to amplify 100–405 bp amplicons depicting the presence or absence of AhMITE1 at each marker locus. Genotype-specific alleles were observed for all the genotypes at varying number of markers (Additional file 3: Table S3). At least two genotypes showed the same type of allele at 1342 marker loci.
B sub-genome (1620) had marginally more number of AhMITE1 markers than the A sub-genome (1337). An unequal distribution of markers was observed across the chromosomes of both the sub-genomes (Table 1). A general correlation was observed between the number of markers and the length of the chromosome [40]. In the A sub-genome, the number of markers varied from 84 (A02 chromosome) to 210 (A03 chromosome); while it ranged from 124 (B02 chromosome) to 269 (B03 chromosome) in the B sub-genome. The recent efforts on sequencing of the diploid progenitors of peanut, Arachis duranensis (A genome) and Arachis ipaensis (B genome) showed that transposable elements occupy larger space (68.5%) in the B genome than in the A genome (61.7%), and DNA transposons make about 10% of both A and B genome [40]. Unequal distribution of DNA transposons was also observed in rice [43], Brassica [44] and foxtail millet [45].
Table 1.
Chromosome-wise distribution of AhMITE1 markers in peanut
| Chromosome | Total | Inter-genic | Exon | Intron | UTR | Upstream | Downstream |
|---|---|---|---|---|---|---|---|
| A01 | 140 | 82 | 5 | 20 | 5 | 20 | 8 |
| A02 | 84 | 42 | 2 | 12 | 4 | 17 | 7 |
| A03 | 210 | 100 | 5 | 30 | 16 | 41 | 18 |
| A04 | 116 | 55 | 3 | 20 | 9 | 21 | 8 |
| A05 | 190 | 80 | 5 | 33 | 13 | 40 | 19 |
| A06 | 138 | 73 | 1 | 18 | 11 | 21 | 14 |
| A07 | 94 | 38 | 5 | 18 | 7 | 19 | 7 |
| A08 | 122 | 61 | 0 | 22 | 7 | 20 | 12 |
| A09 | 117 | 59 | 6 | 28 | 2 | 14 | 8 |
| A10 | 126 | 61 | 4 | 16 | 6 | 29 | 10 |
| Sub-total | 1337 | 651 | 36 | 217 | 80 | 242 | 111 |
| B01 | 135 | 71 | 3 | 15 | 8 | 27 | 11 |
| B02 | 124 | 56 | 4 | 16 | 5 | 32 | 11 |
| B03 | 269 | 116 | 7 | 44 | 26 | 50 | 26 |
| B04 | 140 | 67 | 3 | 26 | 7 | 27 | 10 |
| B05 | 196 | 94 | 5 | 29 | 18 | 39 | 11 |
| B06 | 159 | 76 | 4 | 24 | 14 | 30 | 11 |
| B07 | 153 | 74 | 7 | 32 | 6 | 19 | 15 |
| B08 | 154 | 74 | 0 | 22 | 4 | 37 | 17 |
| B09 | 162 | 68 | 9 | 33 | 6 | 31 | 15 |
| B10 | 128 | 55 | 3 | 24 | 6 | 28 | 12 |
| Sub-total | 1620 | 751 | 45 | 265 | 100 | 320 | 139 |
| Total | 2957 | 1402 | 81 | 482 | 180 | 562 | 250 |
Analyzing the genomic location of these 2957 markers revealed that 1555 were genic and 1402 were intergenic. A maximum of 562 (36.1%) marker loci had AhMITE1 insertion at upstream regions (within 1 kb) followed by 482 (31.0%) in intronic, 250 in downstream regions (within 1 kb), 180 (11.6%) in UTRs and 81 (5.1%) in exonic regions. Insertion of MITE in the genic region as well as intergenic region is known to affect the gene expression [46]. Thus, the AhMITE1 markers developed in this study could have functional role as well. A sample of 343 markers (Additional file 4: Table S4) was employed for validation, and as high as 282 markers produced the amplicons of expected size (Fig. 1) in all the four genotypes (DER, VL 1, 110 and 110(S), indicating 82.2% marker validation.
Fig. 1.
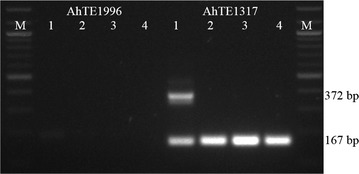
Validation of selected AhMITE1 markers in peanut. M 100 bp ladder, 1 DER, 2 VL 1, 3 110 and 4 110(S)
The validated markers (282) were tested for their PIC using additional ten genotypes of cultivated peanut. The PIC ranged from zero to 0.375 with an average of 0.155. In total, 221 and 61 markers were classified as low (≤ 0.25) and moderate (0.26–0.50) for PIC. A maximum of 35.5% polymorphism was observed between VL 1 and 110 (Table 2, Additional file 5: Figure S1), followed by 26.2% (TMV 2 and TMV 2-NLM), 23.1% (TMV 2 and ICGV 86699), 22.3% (TMV 2 and ICGV 99005; TG 26 and GPBD 4) and 16.7% (TAG 24 and GPBD 4). High rate of TE marker validation was also reported from foxtail millet (Setaria italica) [45] and Caenorhabditis elegans [47].
Table 2.
Percent polymorphism exhibited by AhMITE1 markers among the parents of RIL and backcross populations in peanut
| Parents | No. of polymorphic markersa | Polymorphism (%) |
|---|---|---|
| VL 1 × 110 | 100 | 35.5 |
| TAG 24 × GPBD 4 | 47 | 16.7 |
| TG 26 × GPBD 4 | 63 | 22.3 |
| GPBD 4 × JL 24 | 36 | 12.8 |
| GPBD 4 × TMV 2 | 41 | 14.5 |
| JL 24 × IL 1 | 41 | 14.5 |
| JL 24 × IL 2 | 42 | 14.9 |
| TMV 2 × ICGV 86699 | 65 | 23.1 |
| TMV 2 × ICGV 99005 | 63 | 22.3 |
| TMV 2 × IL 1 | 44 | 15.6 |
| TMV 2 × IL 2 | 45 | 16.0 |
| TMV 2 × TMV 2-NLM | 74 | 26.2 |
| Mean | 19.5 |
aout of 282 markers screened
In total, 5,36,072 SNPs were identified when the WGRS data from four peanut genotypes [DER, VL 1, 110 and 110(s)] were compared to the reference genomes [40]. Considering the mapped QTL for resistance to bacterial wilt, late leaf spot and rust, and other important productivity traits [15, 24, 25], the SNPs on A03, B01, B02 and B03 were selected for identifying the snip-SNPs. Screening of 64,416 SNPs from these four chromosomes identified 11,730 (potential CAPS sites) for 25 restriction enzymes (Additional file 6: Table S5). No significant differences were found between the chromosomes for the snip-SNPs. Further, 500 snip-SNPs on A03 chromosome were used to develop CAPS markers. Currently, only two CAPS markers are available in peanut [48]. They were developed to detect specific mutations in AhFAD2A and AhFAD2B leading to high oleic acid content [49].
Of the 500 CAPS markers identified, 30 were checked for PCR amplification and restriction digestion (Additional file 7: Table S6). Twenty markers showed PCR amplification, and 10 amplicons showed restriction digestion; of which nine showed restriction fragments of expected size (Additional file 8: Figure S2), and one (CAPS0100) failed to show the fragments of expected size with the enzyme BstKTI, indicating 30.0% validation for CAPS markers. The CAPS sites for AluI and BamHI showed maximum validation (100%) followed by AseI with 57.1% validation. On the other hand, CAPS for AclI, BglII and BstKTI showed no validation.
Polymorphic information content was calculated for the nine markers showing the expected PCR product and restriction products. PIC ranged from 0.195 (CAPS0072) to 0.305 (CAPS0002, CAPS0043, CAPS0047, CAPS0050, CAPS0053, CAPS0054, CAPS0058 and CAPS0059) with a mean PIC of 0.293 (Additional file 7: Table S6). Parents of the RILs and backcross populations like TAG 24 versus GPBD 4, JL 24 versus GPBD 4, TMV 2 versus GPBD 4, TMV 2 versus ICGV 86699, TMV 2 versus ICGV 99005, TMV 2 versus IL 1 and TMV 2 versus IL 2 showed the same level of polymorphism (88.9%), whereas VL 1 versus 110 failed to show any polymorphism for these markers.
The number of CAPS markers was more as compared to AhMITE1 markers in peanut genome. However, different families of class I and class II elements can be considered to develop more AhMITE1 markers. Handling of AhMITE1 markers in the laboratory is easy and requires fewer resources. The transposition of the element from a “donor” site can be validated by sequencing the empty-site-related PCR, and searching for footprints (duplicated regions). Thus, AhMITE1 markers can give an indication not only of the genetic divergence that was caused by AhMITE1 transposition, but also of the history of transposition in each species [22]. Further, AhMITE1 markers offer a DNA tag (AhMITE1) for gene discovery and cloning [20].
Limitations
In this study, a large number of AhMITE1 and CAPS markers were developed in peanut, where marker polymorphism is a major limitation. In future, retrotransposons and other DNA transposons can also be considered for marker development. Similarly, snip-SNPs can be identified for the whole genome for the development of CAPS markers. The major limitation could be the fact that these markers are applicable only for peanut. Currently, the new AhMITE1 markers are being extensively used for trait mapping [15, 16] and backcross breeding [37, 50] to develop foliar disease resistant genotypes in our laboratory. It is necessary to test more number of CAPS markers to assess their true rate of polymorphism.
Additional files
Additional file 1: Table S1. Details on the genotypes used for marker discovery.
Additional file 2: Table S2a. PCR components for AhMITE1 marker assay. Table S2b. PCR temperature profile used for AhMITE1 markers. Table S2c. PCR components for CAPS marker assay. Table S2d. PCR temperature profile used for CAPS markers. Table S2e. Restriction digestion components for CAPS assay.
Additional file 3: Table S3. Genotype-specific AhMITE1 markers.
Additional file 4. Details on the newly developed AhMITE1 markers.
Additional file 5: Figure S1. Polymorphism survey for AhTE1131 among the parents of RIL and backcross populations of peanut. [M: 100 bp ladder, 1: DER, 2: VL 1, 3: 110, 4: 110(S), 5: TAG 24, 6: GPBD 4, 7: JL 24, 8: TMV 2, 9: ICGV 86699, 10: ICGV 99005, 11: IL 1 and 12: IL 2].
Additional file 6. Details on the potential CAPS sites for 25 restriction enzymes on A03, B01, B02 and B03 chromosomes of peanut.
Additional file 7. Details on the newly developed CAPS markers.
Additional file 8: Figure S2. Validation of selected CAPS markers in peanut. [M: 100 bp ladder, 1: DER, 2: VL 1, 3: 110 and 4: 110(S)].
Authors’ contributions
MG: Carried out bench work (DNA isolation, PCR, restriction digestion etc.). KS: Helped in developing the AhMITE1 and CAPS markers. RKV: Generated the WGRS data of GPBD 4 and TAG 24. MKP: Generated the WGRS data of GPBD 4 and TAG 24. RSB: Conceptualized the idea and prepared the manuscript. All authors read and approved the final manuscript.
Acknowledgements
We acknowledge the receipt of genotypes DER, VL 1, 110 and 110(S) from Dr. M. V. C. Gowda, Professor, Department of Genetics and Plant Breeding, University of Agricultural Sciences, Dharwad 580 005, India. We thank S. Nakayama (Kazusa DNA Research Institute) for technical assistance.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
The sequences have been submitted and the GenBank accession numbers DRA005803 (WGRS of DER and VL 1), DRA005805 [WGRS of 110 and 110(S)], and DRA006239 (WGRS of ICGV 86855 and VG 9514) have been obtained. The newly developed markers have been given under Additional file 1: Table S1, Additional file 2: Table S2, Additional file 3: Table S3, Additional file 4: Table S4, Additional file 6: Table S5, Additional file 7: Table S6. The genotypes used in this study are the released varieties or the breeding lines.
Consent to publish
Not applicable.
Ethics approval and consent to participate
This study involves peanut varieties that are publicly available and released to the farmers for cultivation. Other genotypes are the breeding lines developed and maintained at our university. Consent to participate: Not applicable.
Funding
Financial support received from the DST-JSPS Bilateral Program is gratefully acknowledged.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Abbreviations
- AhMITE1
Arachis hypogaea miniature inverted-repeat transposable element 1
- CAPS
cleaved amplified polymorphic sequences
- WGRS
whole genome re-sequencing
- SNP
single nucleotide polymorphism
- GAB
genomics-assisted breeding
- RFLP
restriction fragment length polymorphism
- AFLP
amplified fragment length polymorphism
- DAF
DNA amplification fingerprinting
- SSR
simple sequence repeat
- DArT
diversity arrays technology
- AIP
AhMITE1 insertion polymorphism
- PTEMD
polymorphic TEs and their movement detection
- QTL
quantitative trait loci
- CTAB
cetyl trimethyl ammonium bromide
- PCR
polymerase chain reaction
- PIC
polymorphic information content
- RIL
recombinant inbred line
Footnotes
Electronic supplementary material
The online version of this article (10.1186/s13104-017-3121-8) contains supplementary material, which is available to authorized users.
Contributor Information
M. Gayathri, Email: gaya1293@gmail.com
Kenta Shirasawa, Email: shirasaw@kazusa.or.jp.
R. K. Varshney, Email: R.K.Varshney@cgiar.org
M. K. Pandey, Email: M.Pandey@cgiar.org
R. S. Bhat, Email: bhatrs@uasd.in
References
- 1.Kochert G, Halward T, Branch WD, Simpson CE. RFLP variability in peanut (Arachis hypogaea L.) cultivars and wild species. Theor Appl Genet. 1991;81(5):565–570. doi: 10.1007/BF00226719. [DOI] [PubMed] [Google Scholar]
- 2.Halward TM, Stalker TH, Larue EA, Kochert G. Use of single-primer DNA amplifications in genetic studies of peanut (Arachis hypogaea L.) Plant Mol Biol. 1992;18(2):315–325. doi: 10.1007/BF00034958. [DOI] [PubMed] [Google Scholar]
- 3.Paik-Ro OG, Smith RL, Knauft DA. Restriction fragment length polymorphism evaluation of six peanut species within the Arachis section. Theor Appl Genet. 1992;84(1):201–208. doi: 10.1007/BF00224001. [DOI] [PubMed] [Google Scholar]
- 4.Khera P, Upadhyaya HD, Pandey MK, Roorkiwal M, Sriswathi M, Janila P, et al. Single nucleotide polymorphism-based genetic diversity in the reference set of peanut (spp.) by developing and applying cost-effective kompetitive allele specific polymerase chain reaction genotyping assays. Plant Genome. 2013 [Google Scholar]
- 5.Casa AM, Brouwer C, Nagel A, Wang L, Zhang Q, Kresovich S, et al. The MITE family Heartbreaker (Hbr): molecular markers in maize. Proc Natl Acad Sci. 2000;97(18):10083–10089. doi: 10.1073/pnas.97.18.10083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Queen R, Gribbon B, James C, Jack P, Flavell A. Retrotransposon-based molecular markers for linkage and genetic diversity analysis in wheat. Mol Genet Genomics. 2004;271(1):91–97. doi: 10.1007/s00438-003-0960-x. [DOI] [PubMed] [Google Scholar]
- 7.Hui W, ShuangLing L, Yan R, MengQi X, YanMao S, Mei Y, et al. Genetic relationship of peanut (Arachis hypogaea L.) varieties and advanced generation lines evaluated by AhMITE transposable markers. J Agric Biotechnol. 2013;21(10):1176–1184. [Google Scholar]
- 8.Huo H, Conner J, Ozias-Akins P. Genetic mapping of the apospory-specific genomic region in Pennisetum squamulatum using retrotransposon-based molecular markers. Theor Appl Genet. 2009;119(2):199. doi: 10.1007/s00122-009-1029-y. [DOI] [PubMed] [Google Scholar]
- 9.Bhat RS, Patil VU, Chandrashekar TM, Sujay V, Gowda MVC, Kuruvinashetti MS. Recovering flanking sequence tags of miniature inverted-repeat transposable element by thermal asymmetric interlaced-PCR in peanut. Curr Sci. 2008;95(4):452–453. [Google Scholar]
- 10.Gowda MVC, Bhat RS, Motagi BN, Sujay V. Varshakumari, Bhat S. Association of high-frequency origin of late leaf spot resistant mutants with AhMITE1 transposition in peanut. Plant Breed. 2010;129(5):567–569. [Google Scholar]
- 11.Gowda MVC, Bhat RS, Sujay V, Kusuma P, Varshakumari Bhat S, et al. Characterization of AhMITE1 transposition and its association with the mutational and evolutionary origin of botanical types in peanut (Arachis spp.) Plant Syst Evol. 2011;291(3–4):153–158. [Google Scholar]
- 12.Shirasawa K, Hirakawa H, Tabata S, Hasegawa M, Kiyoshima H, Suzuki S, et al. Characterization of active miniature inverted-repeat transposable elements in the peanut genome. Theor Appl Genet. 2012;124(8):1429–1438. doi: 10.1007/s00122-012-1798-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shirasawa K, Koilkonda P, Aoki K, Hirakawa H, Tabata S, Watanabe M, et al. In silico polymorphism analysis for the development of simple sequence repeat and transposon markers and construction of linkage map in cultivated peanut. BMC Plant Biol. 2012;12(1):80. doi: 10.1186/1471-2229-12-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shirasawa K, Bertioli DJ, Varshney RK, Moretzsohn MC, Leal-Bertioli SC, Thudi M, et al. Integrated consensus map of cultivated peanut and wild relatives reveals structures of the A and B genomes of Arachis and divergence of the legume genomes. DNA Res. 2013;20(2):173–184. doi: 10.1093/dnares/dss042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kolekar RM, Sujay V, Shirasawa K, Sukruth M, Khedikar YP, Gowda MVC, et al. QTL mapping for late leaf spot and rust resistance using an improved genetic map and extensive phenotypic data on a recombinant inbred line population in peanut (Arachis hypogaea L.) Euphytica. 2016;209(1):147–156. [Google Scholar]
- 16.Hake AA, Shirasawa K, Yadawad A, Sukruth M, Patil M, Nayak SN, et al. Mapping of important taxonomic and productivity traits using genic and non-genic transposable element markers in peanut (Arachis hypogaea L.) PLoS One. 2017;12(10):e0186113. doi: 10.1371/journal.pone.0186113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Macko A, Grzebelus D. DcMaster transposon display markers as a tool for diversity evaluation of carrot breeding materials and for hybrid seed purity testing. J Appl Genet. 2008;49(1):33–39. doi: 10.1007/BF03195246. [DOI] [PubMed] [Google Scholar]
- 18.Kwon S-J, Park K-C, Kim J-H, Lee JK, Kim N-S. Rim 2/Hipa CACTA transposon display; a new genetic marker technique in Oryza species. BMC Genet. 2005;6(1):15. doi: 10.1186/1471-2156-6-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bouck A, Peeler R, Arnold ML, Wessler SR. Genetic mapping of species boundaries in Louisiana irises using IRRE retrotransposon display markers. Genetics. 2005;171(3):1289–1303. doi: 10.1534/genetics.105.044552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sato K, Mukainari Y, Naito K, Fukunaga K. Construction of a foxtail millet linkage map and mapping of spikelet-tipped bristles1 (stb1) by using transposon display markers and simple sequence repeat markers with genome sequence information. Mol Breed. 2013;31(3):675–684. [Google Scholar]
- 21.Gao D-Y, Vallejo VA, He B, Gai Y-C, Sun L-H. Detection of DNA changes in somaclonal mutants of rice using SSR markers and transposon display. Plant Cell Tissue Organ Cult. 2009;98(2):187–196. [Google Scholar]
- 22.Yaakov B, Ceylan E, Domb K, Kashkush K. Marker utility of miniature inverted-repeat transposable elements for wheat biodiversity and evolution. Theor Appl Genet. 2012;124(7):1365–1373. doi: 10.1007/s00122-012-1793-y. [DOI] [PubMed] [Google Scholar]
- 23.Kang H, Zhu D, Lin R, Opiyo SO, Jiang N, Shiu S-H, et al. A novel method for identifying polymorphic transposable elements via scanning of high-throughput short reads. DNA Res. 2016;23(3):241–251. doi: 10.1093/dnares/dsw011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou X, Xia Y, Liao J, Liu K, Li Q, Dong Y, et al. Quantitative trait locus analysis of late leaf spot resistance and plant-type-related traits in cultivated peanut (Arachis hypogaea L.) under multi-environments. PLoS ONE. 2016;11(11):e0166873. doi: 10.1371/journal.pone.0166873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhao Y, Zhang C, Chen H, Yuan M, Nipper R, Prakash C, et al. QTL mapping for bacterial wilt resistance in peanut (Arachis hypogaea L.) Mol Breed. 2016;36(2):1–11. doi: 10.1007/s11032-015-0432-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shirasawa K, Kuwata C, Watanabe M, Fukami M, Hirakawa H, Isobe S. Target amplicon sequencing for genotyping genome-wide single nucleotide polymorphisms identified by whole-genome resequencing in peanut. Plant Genome. 2016 doi: 10.3835/plantgenome2016.06.0052. [DOI] [PubMed] [Google Scholar]
- 27.Gowda MVC, Nadaf HL, Sheshagiri R. The role of mutations in intraspecific differentiation of groundnut (Arachis hypogaea L.) Euphytica. 1996;90(1):105–113. [Google Scholar]
- 28.Clevenger J, Chu Y, Chavarro C, Agarwal G, Bertioli DJ, Leal-Bertioli SC, et al. Genome-wide SNP genotyping resolves signatures of selection and tetrasomic recombination in peanut. Mol Plant. 2017;10(2):309–322. doi: 10.1016/j.molp.2016.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gowda MVC, Motagi BN, Naidu GK, Diddimani SB, Sheshagiri R. GPBD 4: a spanish bunch groundnut genotype resistant to rust and late leaf spot. Int Arachis News Lett. 2002;22:29–32. [Google Scholar]
- 30.Kale D, Badigannavar A, Murty G. Groundnut variety, TAG 24, with potential for wider adaptability. Int Arachis News Lett. 1999;19:12–13. [Google Scholar]
- 31.Varman PV. A foliar disease resistant line developed through interspecific hybridization in groundnut (Arachis hypogaea) Indian J Agril Sci. 1999;69(1):67–68. [Google Scholar]
- 32.You FM, Huo N, Gu YQ, M-c Luo, Ma Y, Hane D, et al. BatchPrimer3: a high throughput web application for PCR and sequencing primer design. BMC Bioinform. 2008;9(1):253. doi: 10.1186/1471-2105-9-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cuc LM, Mace ES, Crouch JH, Quang VD, Long TD, Varshney RK. Isolation and characterization of novel microsatellite markers and their application for diversity assessment in cultivated groundnut (Arachis hypogaea) BMC Plant Biol. 2008;8(1):55. doi: 10.1186/1471-2229-8-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu K, Muse SV. PowerMarker: an integrated analysis environment for genetic marker analysis. Bioinformatics. 2005;21(9):2128–2129. doi: 10.1093/bioinformatics/bti282. [DOI] [PubMed] [Google Scholar]
- 35.Bhat RS, Sujay V, Mukri G, Khedikar YP, Sarvamangala C, Yeri SB et al. Utility of peanut mapping populations developed at UAS Dharwad, India. In: VI international Conference on legume genetics and genomics (ICLGG); 2–7 October 2012; Hyderabad, India; 2012.
- 36.Yeri SB. Validation of late leaf spot and rust resistance-linked markers and transfer of associated QTL to JL 24 in groundnut (Arachis hypogaea L.) Dharwad: University of Agricultural Sciences; 2015. [Google Scholar]
- 37.Kolekar RM. Transfer of resistance to late leaf spot and rust in groundnut through marker assisted backcrossing. Dharwad: University of Agricultural Sciences; 2017. [Google Scholar]
- 38.Shirasawa K, Hirakawa H, Isobe S. Analytical workflow of double-digest restriction site-associated DNA sequencing based on empirical and in silico optimization in tomato. DNA Res. 2016;23:145–153. doi: 10.1093/dnares/dsw004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wishart DS, Stothard P, Van Domselaar GH. PepTool™ and GeneTool™: platform-independent tools for biological sequence analysis. In: Misener S, Krawetz SA, editors. Bioinformatics Methods and Protocols, Methods in Molecular Biology. Totowa, NJ: Humana Press; 1999. p. 93–113. [DOI] [PubMed]
- 40.Bertioli DJ, Cannon SB, Froenicke L, Huang G, Farmer AD, Cannon EK, et al. The genome sequences of Arachis duranensis and Arachis ipaensis, the diploid ancestors of cultivated peanut. Nat Genet. 2016;48(4):438–446. doi: 10.1038/ng.3517. [DOI] [PubMed] [Google Scholar]
- 41.Chen X, Li H, Pandey MK, Yang Q, Wang X, Garg V, et al. Draft genome of the peanut A-genome progenitor (Arachis duranensis) provides insights into geocarpy, oil biosynthesis, and allergens. Proc Natl Acad Sci. 2016;113(24):6785–6790. doi: 10.1073/pnas.1600899113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roy NS, Choi J-Y, Lee S-I, Kim N-S. Marker utility of transposable elements for plant genetics, breeding, and ecology: a review. Genes Genom. 2015;37(2):141–151. [Google Scholar]
- 43.Kwon S, Hong S, Son J, Lee J, Cha Y, Eun M, et al. CACTA and MITE transposon distributions on a genetic map of rice using F15 RILs derived from Milyang 23 and Gihobyeo hybrids. Mol Cells. 2006;21(3):360. [PubMed] [Google Scholar]
- 44.Sampath P, Murukarthick J, Izzah NK, Lee J, Choi HI, Shirasawa K, et al. Genome-wide comparative analysis of 20 miniature inverted-repeat transposable element families in Brassica rapa and B oleracea. PLoS ONE. 2014;9(4):e94499. doi: 10.1371/journal.pone.0094499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yadav CB, Bonthala VS, Muthamilarasan M, Pandey G, Khan Y, Prasad M. Genome-wide development of transposable elements-based markers in foxtail millet and construction of an integrated database. DNA Res. 2015;22(1):79–90. doi: 10.1093/dnares/dsu039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lu C, Chen J, Zhang Y, Hu Q, Su W, Kuang H. Miniature inverted–repeat transposable elements (MITEs) have been accumulated through amplification bursts and play important roles in gene expression and species diversity in Oryza sativa. Mol Biol Evol. 2012;29(3):1005–1017. doi: 10.1093/molbev/msr282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Korswagen HC, Durbin RM, Smits MT, Plasterk RH. Transposon Tc1-derived, sequence-tagged sites in Caenorhabditis elegans as markers for gene mapping. Proc Natl Acad Sci. 1996;93(25):14680–14685. doi: 10.1073/pnas.93.25.14680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chu Y, Holbrook CC, Ozias-Akins P. Two alleles of ahFAD2B control the high oleic acid trait in cultivated peanut. Crop Sci. 2009;49(6):2029–2036. [Google Scholar]
- 49.Jung S, Powell G, Moore K, Abbott A. The high oleate trait in the cultivated peanut [Arachis hypogaea L.]. II. Molecular basis and genetics of the trait. Mol Gen Genet. 2000;263(5):806–811. doi: 10.1007/s004380000243. [DOI] [PubMed] [Google Scholar]
- 50.Yeri SB, Bhat RS. Development of late leaf spot and rust resistant backcross lines in JL 24 variety of groundnut (Arachis hypogaea L.) Electron J Plant Breed. 2016;7(1):37–41. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. Details on the genotypes used for marker discovery.
Additional file 2: Table S2a. PCR components for AhMITE1 marker assay. Table S2b. PCR temperature profile used for AhMITE1 markers. Table S2c. PCR components for CAPS marker assay. Table S2d. PCR temperature profile used for CAPS markers. Table S2e. Restriction digestion components for CAPS assay.
Additional file 3: Table S3. Genotype-specific AhMITE1 markers.
Additional file 4. Details on the newly developed AhMITE1 markers.
Additional file 5: Figure S1. Polymorphism survey for AhTE1131 among the parents of RIL and backcross populations of peanut. [M: 100 bp ladder, 1: DER, 2: VL 1, 3: 110, 4: 110(S), 5: TAG 24, 6: GPBD 4, 7: JL 24, 8: TMV 2, 9: ICGV 86699, 10: ICGV 99005, 11: IL 1 and 12: IL 2].
Additional file 6. Details on the potential CAPS sites for 25 restriction enzymes on A03, B01, B02 and B03 chromosomes of peanut.
Additional file 7. Details on the newly developed CAPS markers.
Additional file 8: Figure S2. Validation of selected CAPS markers in peanut. [M: 100 bp ladder, 1: DER, 2: VL 1, 3: 110 and 4: 110(S)].
Data Availability Statement
The sequences have been submitted and the GenBank accession numbers DRA005803 (WGRS of DER and VL 1), DRA005805 [WGRS of 110 and 110(S)], and DRA006239 (WGRS of ICGV 86855 and VG 9514) have been obtained. The newly developed markers have been given under Additional file 1: Table S1, Additional file 2: Table S2, Additional file 3: Table S3, Additional file 4: Table S4, Additional file 6: Table S5, Additional file 7: Table S6. The genotypes used in this study are the released varieties or the breeding lines.


