Abstract
The long noncoding RNA MSNP1AS (moesin pseudogene 1, antisense) is a functional element that was previously associated with autism spectrum disorder (ASD) with genome-wide significance. Expression of MSNP1AS was increased 12-fold in the cerebral cortex of individuals with ASD and 22-fold in individuals with a genome-wide significantly associated ASD genetic marker on chromosome 5p14.1. Overexpression of MSNP1AS in human neuronal cells caused decreased expression of moesin protein, which is involved in neuronal process stability. In this study, we hypothesize that MSNP1AS knockdown impacts global transcriptome levels. We transfected the human neural progenitor cell line SK-N-SH with constructs that caused a 50% suppression of MSNP1AS expression. After 24 h, cells were harvested for total RNA isolation. Strand-specific RNA sequencing analysis indicated altered expression of 1,352 genes, including altered expression of 318 genes following correction for multiple comparisons. Expression of the OAS2 gene was increased >150-fold, a result that was validated by quantitative PCR. Gene ontology analysis of the 318 genes with altered expression following correction for multiple comparisons indicated that upregulated genes were significantly enriched for genes involved in immune response, and down-regulated genes were significantly enriched for genes involved in chromatin remodeling. These data indicate multiple transcriptional and translational functions of MSNP1AS that impact ASD-relevant biological processes. Chromatin remodeling and immune response are biological processes implicated by genes with rare mutations associated with ASD. Our data suggest that the functional elements implicated by association of common genetic variants impact the same biological processes, suggesting a possible shared common molecular pathway of ASD.
Keywords: RNA sequencing, Autism spectrum disorders, Noncoding RNA, Long noncoding RNA, Small interfering RNA, Small noncoding antisense RNA, Antisense therapy
Introduction
Autism spectrum disorder (ASD) is a neurodevelopmental condition characterized by deficits in social communication and behavioral flexibility [1, 2]. Recent advances in genomics have begun to identify the genetic variants that contribute to ASD risk [3–5]. The important steps are to translate these genetic findings to a biological understanding of ASD pathogenesis and to the development of biologically based treatments [6]. Recently, we identified a long noncoding RNA (lncRNA) that contributes to ASD risk [7]. A genome-wide association study (GWAS) identified a single peak on chromosome 5p14.1 for ASD, indicating a significant association (p = 10−10) for rs4307059 [3]. The same rs4307059 allele was also associated with social communication phenotypes in a general population sample [8]. Directly under the chromosome 5p14.1 ASD GWAS peak, we identified an lncRNA that was almost 4 kb [7]. This transcript is 94% identical and antisense to the X chromosome transcript, MSN, which encodes a neuronal architecture protein and is on the opposite (antisense) strand of moesin pseudogene 1 (MSNP1) [7]. Therefore, we named the newly discovered gene MSNP1AS (moesin pseudogene 1, antisense) [7]. Expression of MSNP1AS was detected in 5 brain regions (frontal cerebral cortex, temporal cerebral cortex, occipital cerebral cortex, cerebellum and spinal cord) of the adult and in the fetal frontal cerebral cortex [7]. In peripheral tissues, MSNP1AS was expressed at high levels in the adult peripheral blood and fetal heart [7]. These data indicate that MSNP1AS is expressed at high levels in both the nervous and circulatory systems. Here we use publicly available databases of gene expression to provide additional details of MSNP1AS expression. MSNP1AS expression is increased 12.7 times in the postmortem temporal cerebral cortex of individuals with ASD compared to controls and increased 22 times in individuals with the ASD-associated rs4307059 allele [7]. Therefore, the highly significant GWAS findings led to the identification of a functional lncRNA that contributes to ASD risk [7].
Jiang et al. [9] have recently reported rescue of cellular phenotypes of Down syndrome by transgenic expression of another lncRNA, XIST (X inactive specific transcript). When XIST, which normally targets the “extra” X chromosome of females for inactivation, was expressed on chromosome 21 in cells derived from Down syndrome patients, the extra copy of chromosome 21 was inactivated [9]. These studies provide a clear path toward translation of manipulating lncRNA expression to rescue disorder phenotypes. Because MSNP1AS is overexpressed in the postmortem cerebral cortex of individuals with ASD [7], we used transcriptional gene silencing (TGS) in human neuronal progenitor cells to examine the potential for rescue. By targeting the MSNP1AS gene promoter, we were able to reduce the expression of the MSNP1AS transcript to half of its normal expression and mimic a potential therapeutic for ASD and social communication deficits. RNA sequencing was used as an unbiased approach to identify genes with altered expression following knockdown of MSNP1AS.
Materials and Methods
Cell Culture
SK-N-SH neuroblastoma neural progenitor cells were demonstrated previously to express levels of MSNP1AS, MSN, and moesin protein consistent with those of developing human brain [7]. The human neural progenitor cell line SK-N-SH (American Type Culture Collection; Manassas, VA, USA) was cultured in minimum essential medium supplemented with 10% heat-inactivated fetal bovine serum, 1% penicillin/streptomycin, nonessential amino acids, and 1.5 g/L sodium bicarbonate in a 75-cm2 flask at 37 ° C and 5% CO2. When cells were 75% confluent, the human neuronal cells were subcultured to a density of 1 × 106 cells/well in a 6-well plate for harvests 24 h after transfection.
Design of Antisense RNA to the MSNP1AS Proximal Gene Promoters
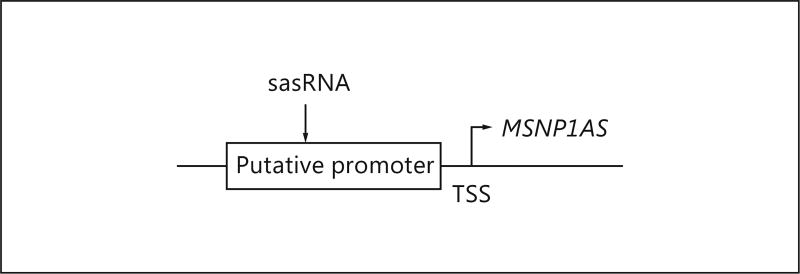
Small noncoding antisense RNAs (sasRNA) to silence gene transcription were designed with proprietary software [10–13]. The program was used to identify the best sasRNA to silence the lncRNA MSNP1AS gene transcript (Fig. 1). We cloned sasRNAs directed to the putative gene promoter into the pCDNA3-U6M2 expression vector with the following target sequence for MSNP1AS knockdown: AATTCTAGAAATGTGCCAACA (Fig. 1).
Fig. 1.
sasRNAs were designed to silence transcription by targeting the MSNP1AS gene promoter.
Transfection of sasRNAs
Targets for TGS of MSNP1AS were defined in human neuronal cell lines. Eight sasRNAs were designed directed to MSNP1AS TGS targets. The TGS targets were screened for effectiveness in U87 or Lan 6 cells. The TGS sasRNAs were screened for efficacy, and 3 were chosen for use in SK-N-SH cells. Each transfection set was considered an independent experiment. The 3 sasRNAs were tested at 5 different vector concentrations (online suppl. Table 1; see www.karger.com/doi/10.1159/000453258 for all online suppl. material). Cell harvest occurred 24 h after transfection. Based on optimization experiments, we used 1 µg of sasRNA/million cells to achieve ~50% suppression of MSNP1AS in further studies.
Cells were transfected using Amaxa Nucleofector (Lonza; Walkersville, MD, USA) technology and subcultured into 6-well plates. One milliliter of fresh prewarmed medium was added to each well, and the cells were centrifuged twice at 130 g for 10 min for the SK-N-SH cells with PBS washes of 7 and 4 mL, respectively. The cell pellet was resuspended in Nucleofector solution containing supplement using 1 × 106 cells/well. One hundred microliters of cell/nucleofector solution were added to a new cuvette and placed in the Amaxa Nucleofector using the T-16 program. Five hundred microliters of fresh prewarmed medium were added to the cuvette, mixed, and transferred to the well with a disposable pipet. Two milliliters of culture medium were added to the transfected cells in the 6-well plate. The cells were incubated at 37 °C in 5% CO2 until harvest. Each experiment was repeated 4 times.
Neural Progenitor Cell Harvest
After 24 h, the culture medium was removed and discarded. The cells were washed with 1 mL of PBS wash, and the PBS wash was aspirated. One milliliter of trypsin/EDTA solution was added to each well, and the cells were incubated for 4 min. Five milliliters of cell type-specific medium were added, and the cells were triturated and transferred to 15-mL conical tubes. SK-N-SH cells were centrifuged at 130 g for 10 min. The supernatant was aspirated and 5 mL of PBS added. The cell pellet was triturated and centrifuged at 130 g for 10 min. The PBS wash was aspirated, and 1 mL of fresh PBS was added. Half of the solution was transferred to each of two 1.5-mL Eppendorf tubes. The tubes were centrifuged at 4 °C at 130 g for 10 min. The supernatant was decanted, and the cell pellets were frozen at −80 °C.
RNA Purification
The Qiagen RNEasy kit was used to isolate total RNA using vacuum technology according to the manufacturer’s protocol (Qiagen; Valencia, CA, USA). The RNA was eluted with 35 µL of RNAse-free water and quantified using the NanoDrop ND-1000 spectrophotometer (v3.1.2; Thermo Fisher Scientific, Waltham, MA, USA). The RNA was stored at −80 °C.
Quantitative RT-PCR to Confirm MSNP1AS Knockdown
To confirm knockdown of MSNP1AS, cDNA was synthesized using the SuperScript III First-Strand Synthesis System for qRT-PCR protocol (Invitrogen/Thermo Fisher Scientific, Waltham, MA, USA). Five hundred nanograms of RNA were used to make one and a half reaction volumes (35 µL) of cDNA. The cDNA was stored at −20 °C. The qRT-PCR protocol described in Kerin et al. [7] was used to validate knockdown of MSNP1AS.
Construction of Strand-Specific, Ribosomal RNA-Depleted RNA Sequencing Libraries
Directional RNA sequencing libraries were prepared for Illumina HiSeq 2000 sequencing using the Stranded Total TruSeq RNA Sample Preparation kit with Ribo-Zero Gold (Illumina) using the manufacturer’s protocol using the Hamilton Starlet Liquid Handling robot. One nanogram of RNA was used for each sample. Ribo-Zero was used to deplete cytoplasmic and mitochondrial ribosomal RNA from total RNA. The depleted RNA was fragmented and primed with random hexamers to synthesize first-strand cDNA using Superscript II (Life Technologies). Next, the second strand was synthesized, incorporating dUTP in place of dTTP. A single “A” base was added to the 3 ′-ends of the fragments, and the indexed adaptors were ligated to the ends of the double-stranded cDNA to prepare them for hybridization onto the flow cell. PCR was used to selectively enrich the fragments with ligated adaptors and to amplify the amount of DNA in the library. The libraries were produced in a 96-well format and quality controlled using the Agilent Technologies 2200 TapeStation Instrument. Libraries were pooled (4 samples/lane) and sequenced on Illumina HiSeq 2000 to a targeted depth generating an average of 20 million paired-end 50-cycle reads for each sample (online suppl. Table 2).
Data Analysis
Data analysis was performed using TopHat (version 2.0.10) [14] to align the Illumina short reads against the reference human genome ENSEMBL GrCH38 version 81. Sequence alignments were generated as BAM files [15], and then Cuffdiff (version 2.2.1) [16] was used to summarize the gene expression values as FPKM measures. The gene expression of samples with MSNP1AS knocked down was compared to the gene expression of the negative control experiment samples to find other differentially expressed genes. Cuffdiff was also used to calculate the expression n-fold change, p values and false discovery rate (FDR) values. Genes with FDR q < 0.05 were used as the input for DAVID (version 6.7) functional annotation [17, 18].
Quantitative PCR to Confirm Altered Gene Expression Observed in RNA Sequencing
Ten RNAs with q < 0.05 in the RNA sequencing results were selected to validate by quantitative PCR (qPCR) using Taqman gene expression assays from Life Technologies: ABCA1 (assay ID Hs01059118_m1), ANGPTL4 (assay ID Hs01101127_m1), ATF3 (assay ID Hs00231069_m1), CXCL8 (assay ID H s00174103_m1), GADD45B (assay ID Hs04188837_g1), ID1 (assay ID Hs03676575_s1), INSM2 (assay ID Hs00261625_s1), MIR320A (assay ID Hs04233529_s1), OAS2 (assay ID Hs00942643_m1), and TMEM215 (assay ID Hs01942498_s1).
Analysis of BrainSpan Data
Normalized expression data representing reads per kilobase per million mapped reads (RPKM) were downloaded from the BrainSpan Atlas of the Developing Human Brain database. Details on the tissues, materials, and sequencing protocols are available on the BrainSpan website (www.brainspan.org). Data used in this analysis were restricted to neocortical brain tissues given the importance of these regions in ASD pathology as well as the altered expression of MSNP1AS in these regions of the brain (primary auditory cortex, dorsolateral prefrontal cortex, inferior parietal cortex, inferolateral temporal cortex, primary motor cortex, medial prefrontal cortex, orbital frontal cortex, primary somatosensory cortex, superior temporal cortex, primary visual cortex, ventrolateral prefrontal cortex). The data were also partitioned into 10 age groups to identify the expression pattern of MSNP1AS across age. Only genes showing a RPKM >1 in at least 1 sample were used in this analysis to reduce the amount of noise generated from genes showing extremely low expression values in all samples. All RPKM values were then transformed to the log2 + 1 as suggested when constructing a weighted gene coexpression network.
Results
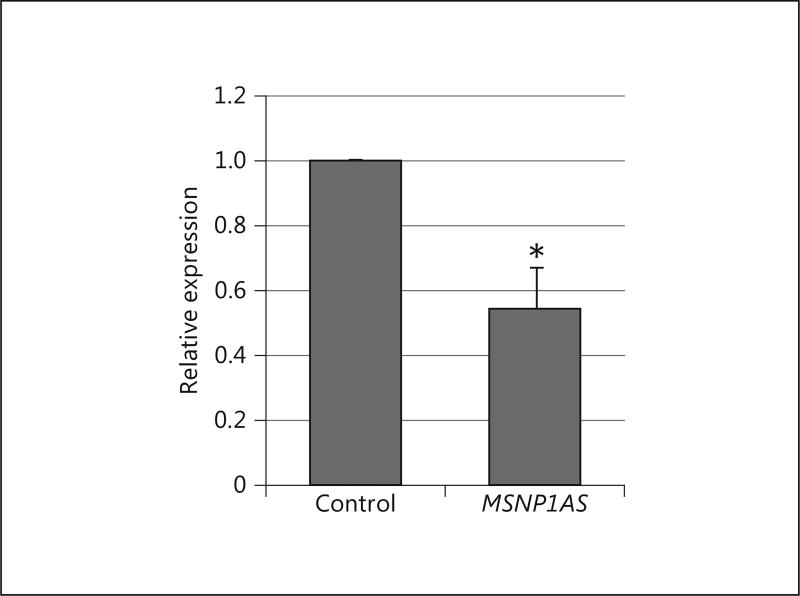
Expression of the ASD-associated lncRNA MSNP1AS is increased 12 times in the cerebral cortex of individuals with ASD compared to controls [7]. We used TGS to knock down MSNP1AS and thus mimic a potential therapeutic for ASD. To quantitatively model the consequences of MSNP1AS knockdown, sasRNA-mediated knockdown of MSNP1AS was performed in the human neural progenitor cell line SK-N-SH. The knockdown efficiency of MSNP1AS transcript levels was confirmed via qPCR (Fig. 2).
Fig. 2.
Quantitation of M SNP1AS knockdown. Quantitative PCR showed that a 1-µg transfection of MSNP1AS sasRNA resulted in a ~45% reduction of MSNP1AS transcript (* p < 0.05 by the Student t test).
MSNP1AS Knockdown Results in Genome-Wide Transcriptional Changes
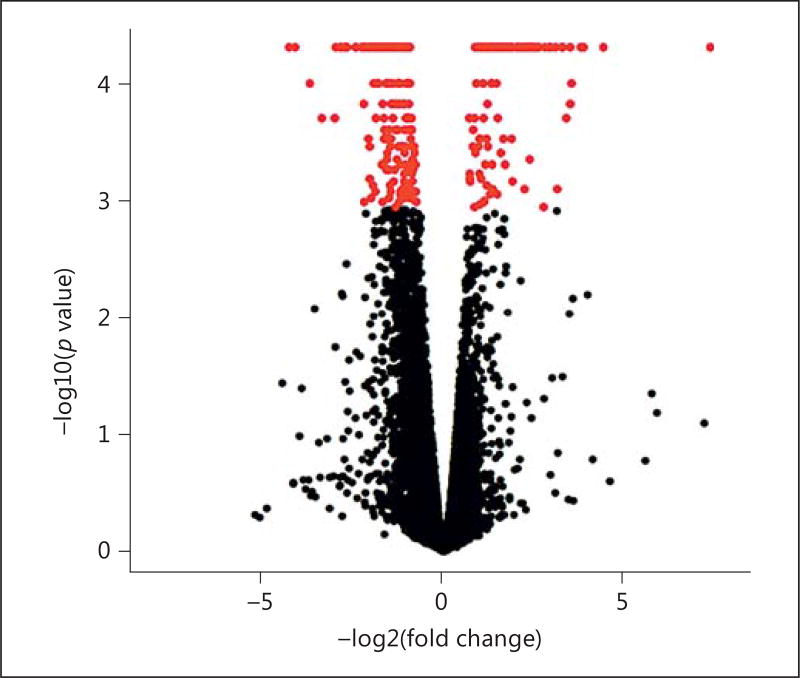
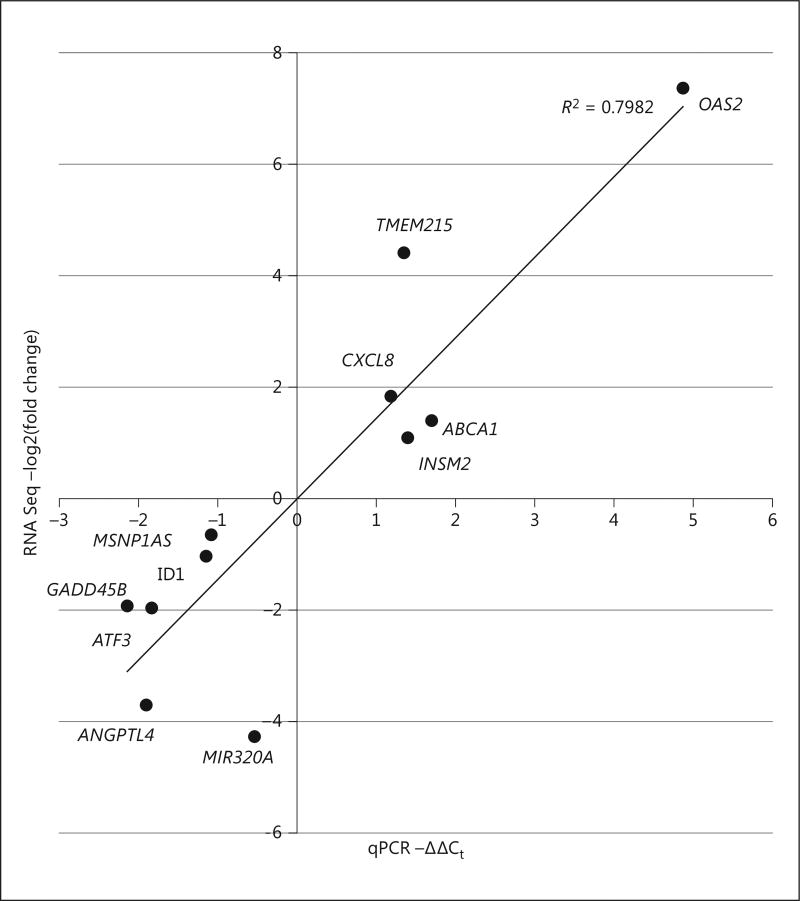
Changes in the transcriptome caused by MSNP1AS knockdown may be able to reveal the impact of biologically based therapeutic approaches on human neural progenitor cell differentiation. The transcriptional changes resulting from the knockdown were evaluated using genome-wide transcriptome profiling of human neuronal progenitor cells SK-N-SH, via RNA sequencing. Differential gene expression analysis using the Ensembl GrCH38 genome annotation revealed 1,352 differentially expressed genes (p < 0.05) (online suppl. Table 3). When a more stringent test for multiple comparisons was applied, a significance level of q < 0.05 showed 318 differentially expressed genes (Table 1, Fig. 3), with 65% of the genes downregulated. Subsequent qPCR validation was performed for genes with large changes in expression following MSNP1AS knockdown. Comparison of gene expression levels measured by RNA sequencing and qPCR indicated a highly significant Pearson correlation coefficient (Fig. 4), validating the magnitude of gene expression changes identified by RNA sequencing.
Table 1.
Top ten differentially expressed genes following knockdown of MSNP1AS
| Gene | Locus | n-fold change | Protein or noncoding | p value | q value |
|---|---|---|---|---|---|
| OAS2 | 12:112906776 – 113017751 | 164.90 | protein coding | 5.00E–05 | 4.12E–03 |
| TMEM215 | 9:32783498 – 32787399 | 21.24 | protein coding | 5.00E–05 | 4.12E–03 |
| IFI44L | 1:78619921 – 78646145 | 14.59 | protein coding | 5.00E–05 | 4.12E–03 |
| LRRC55 | 11:57181746 – 57191717 | 13.67 | protein coding | 5.00E–05 | 4.12E–03 |
| LAMP3 | 3:183122212 – 183163839 | 11.53 | protein coding | 1.00E–04 | 7.33E–03 |
| ITIH4 | 3:52812974 – 52897596 | −8.11 | protein coding | 2.00E–04 | 1.28E–02 |
| CDC42EP1 | 22:37560446 – 37569405 | −10.38 | protein coding | 2.00E–04 | 1.28E–02 |
| ANGPTL4 | 19:7958578 – 8489114 | −13.10 | protein coding | 1.00E–04 | 7.33E–03 |
| HIST1H4J | 6:27824107 – 27824480 | −17.32 | protein coding | 5.00E–05 | 4.12E–03 |
| MIR320A | 8:22244961 – 22245043 | −19.42 | noncoding | 5.00E–05 | 4.12E–03 |
Gene and locus refer to ENSEMBL human genome GrCH38 build 81.
Fig. 3.
Volcano plot generated to show the differentially expressed genes resulting from the MSNP1AS knockdown. Each dot represents a change in gene expression, with genes above the FDR-corrected significance threshold shown in red (color in the online version only).
Fig. 4.
Confirmation of altered expression of genes identified by RNA sequencing (RNA Seq). Ten genes with large changes in gene expression following MSNP1AS knockdown were assayed via qPCR to establish the validity of the RNA sequencing differential expression analysis. Knockdown of MSNP1AS was also validated. qPCR and RNA sequencing values showed a statistically significant correlation with both Pearson (p = 0.0002) and Spearman (p = 0.009) methods.
Transcriptional Consequences of MSNP1AS Knockdown
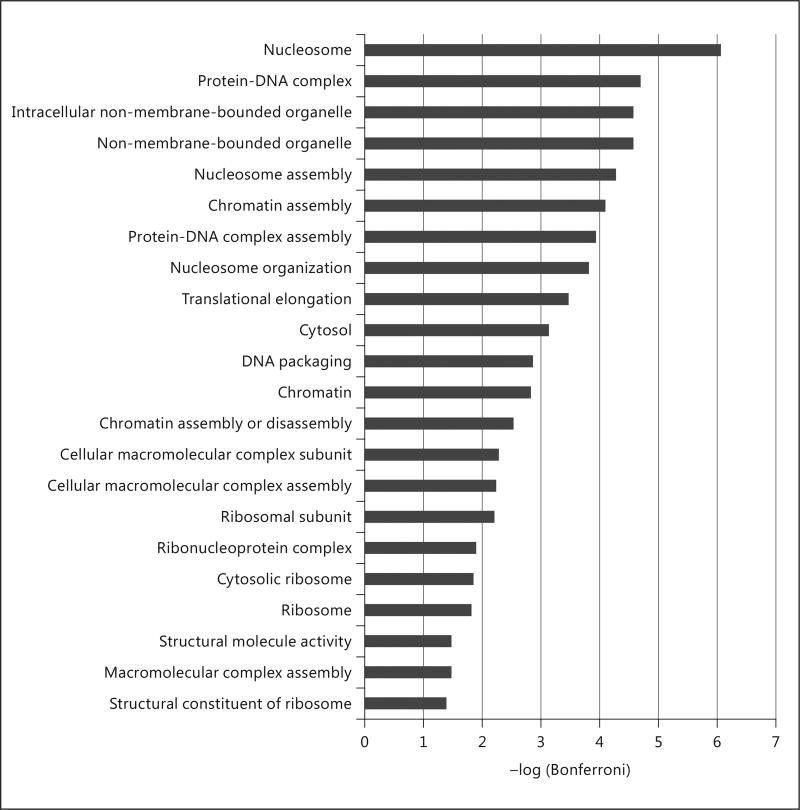
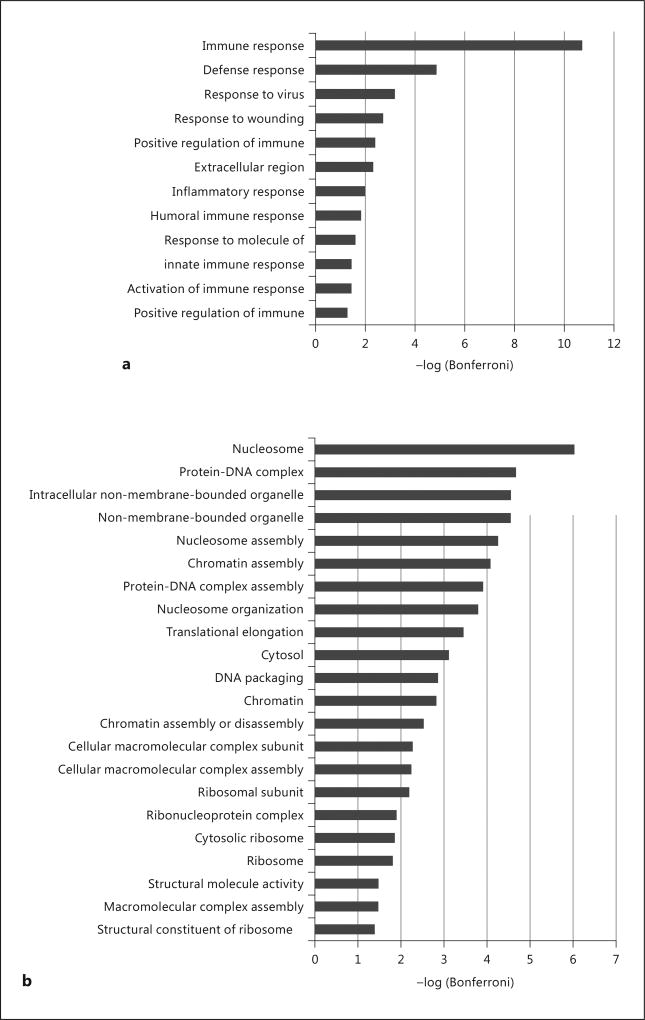
Gene ontology analysis using the DAVID web server of the MSNP1AS knockdown revealed functions important in chromatin assembly and nucleosome organizaton (Fig. 5). We also performed gene ontology analysis grouping the differentially expressed genes (q < 0.05) by their direction of differential expression. Genes that are upregulated by MSNP1AS knockdown were enriched for immune response (Fig. 6a). Genes that are downregulated by MSNP1AS knockdown were enriched for nucleosome assembly and chromatin assembly (Fig. 6b). DAVID analysis indicates that genes encoding proteins involved in protein synthesis and chromatin regulation were altered in expression following MSNP1AS transcriptional gene silencing. There is no evidence of enrichment of genes in the moesin pathway, suggesting that inhibition of moesin protein translation is independent of transcriptional regulation. Comparison of genes with altered expression following MSNP1AS TGS did not reveal enrichment of genes previously implicated in ASD [19].
Fig. 5.
Gene ontology enrichment analysis of differentially expressed genes (q < 0.05) in SK-N-SH cells after 24 h of MSNP1AS knockdown revealed changes in chromatin assembly.
Fig. 6.
Gene ontology enrichment analysis of differentially expressed genes (q < 0.05) in SK-N-SH cells after 24 h of MSNP1AS knockdown. a Analysis of upregulated genes revealed enrichment of genes involved in immune response following knockdown of MSNP1AS. b Analysis of downregulated genes revealed changes in chromatin and nucleosome assembly following knockdown of MSNP1AS.
Expression of MSNP1AS in Developing Human Brain
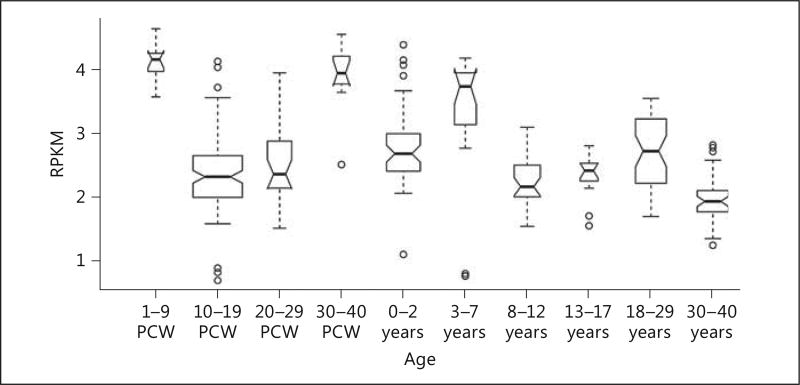
Analysis of BrainSpan data indicate high levels of MSNP1AS in the cerebral cortex during early embryogenesis and again immediately after birth (Fig. 7). GTex indicates levels of expression of MSNP1AS at low to undetectable levels in 12 regions of the adult brain. However, GTex indicates high levels of MSNP1AS expression in peripheral blood, consistent with our previous report [7].
Fig. 7.
Boxplot representing the expression of MSNP1AS through human cerebral cortex development. MSNP1AS exhibits a dynamic expression pattern across development with the highest expression being evident during early and late prenatal development. PCW, postconception weeks.
Discussion
Our findings indicate that MSNP1AS knockdown disrupted the expression of 318 genes (q < 0.05), many of which are involved in chromatin organization and immune response. MSNP1AS is the functional element revealed by genome-wide significant association with ASD [3, 7] and is increased 12.7-fold in the postmortem cerebral cortex of individuals with ASD [7]. We test the effect of a rescue of this phenotype by TGS of MSNP1AS in human neurons. After knocking down the lncRNA in SK-N-SH cells, we evaluated changes in the transcriptome using RNA sequencing to elucidate the consequences of suppressing MSNP1AS expression. These experiments seek to define the role of MSNP1AS in the pathways that lead to ASD pathogenesis and suggest that MSNP1AS acts at the transcriptional level to affect immune response and chromatin organization.
The biological function of the MSNP1AS lncRNA revealed by these data, chromatin organization, is similar to that of genes with rare de novo mutations associated with ASD [20, 21]. The etiology of ASD seems to be affected by both rare and common variants in similar ways. lncRNAs are regulated through spliceosome machinery and post-translational modifications, much like protein-coding transcripts [22–25]. However, lncRNAs are more loosely regulated, are produced in relatively lower abundance, and have fewer exons than protein-coding genes. In spite of this, lncRNA expression is extremely specific to cell type and tissues [22, 25, 26]. lncRNAs can regulate proteins in a variety of ways, including as a framework or scaffold to facilitate interaction with other proteins or DNA, as enhancers or silencers to regulate transcription, and as decoys to bind proteins or other RNA [24, 27–35]. In addition, lncRNAs are being evaluated for epigenetic modifications and chromatin accessibility to reveal their contribution to psychiatric disease etiology [36].
Accumulating data support the role of the immune system in the pathogenesis of ASD [37, 38]. Our previously published data showed that MSNP1AS binds moesin protein, which is involved in neuronal process stability and immune response [7]. Our data show that after MSNP1AS has been knocked down, MSN transcript levels remain unchanged (online suppl. Table 4). This lack of expression change suggests that, while MSNP1AS regulates moesin protein, suppression of MSNP1AS does not alter MSN transcription. The lack of MSN suppression, combined with the decrease in moesin protein expression shown [7], suggests that MSNP1AS binds to the MSN transcript to prevent translation. Further experiments will be necessary to determine the mechanisms of moesin protein regulation. The biological function revealed by genes upregulated after MSNP1AS knockdown points to the immune response as well. Since MSN transcript levels are not perturbed by MSNP1AS knockdown, these immune response genes may be acting independently of MSN. Our results indicate a 150-fold increase in OAS2 expression in SK-N-SH human neural progenitor cells following MSNP1AS transcriptional gene silencing. OAS2 is 1 of 3 genes that make up the OAS gene cluster. These genes regulate viral infection resistance and are involved in cell growth, differentiation and death. The role of the OAS gene cluster as an ASD susceptibility locus is currently under investigation. Genetic variants in OAS2 are associated with another neurodevelopmental disorder, attention deficit hyperactivity disorder [39].
Because the ASD phenotype is highly heterogeneous, a network of many genes is likely contributing to abnormal regulation. A variety of genes may contribute to several different ASD phenotypes; however, strong evidence suggests that many of the pathways involved may overlap to form an interconnected network. The results presented here give some insight into the interconnected network and possible molecular pathology of ASD. Translation of these findings to patients will be a challenge due to technical delivery concerns. However, these first steps are important to identify potential biomarkers and treatment targets.
Supplementary Material
References
- 1.Levitt P, Campbell DB. The genetic and neurobiologic compass points toward common signaling dysfunctions in autism spectrum disorders. J Clin Invest. 2009;119:747–754. doi: 10.1172/JCI37934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wilkinson B, Campbell DB. Contribution of long noncoding RNAs to autism spectrum disorder risk. Int Rev Neurobiol. 2013;113:35–59. doi: 10.1016/B978-0-12-418700-9.00002-2. [DOI] [PubMed] [Google Scholar]
- 3.Wang K, Zhang H, Ma D, Bucan M, Glessner JT, Abrahams BS, Salyakina D, Imielinski M, Bradfield JP, Sleiman PM, Kim CE, Hou C, Frackelton E, Chiavacci R, Takahashi N, Sakurai T, Rappaport E, Lajonchere CM, Munson J, Estes A, Korvatska O, Piven J, Sonnenblick LI, Alvarez Retuerto AI, Herman EI, et al. Common genetic variants on 5p14.1 associate with autism spectrum disorders. Nature. 2009;459:528–533. doi: 10.1038/nature07999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barnard RA, Pomaville MB, O’Roak BJ. Mutations and modeling of the chromatin remodeler CHD8 define an emerging autism etiology. Front Neurosci. 2015;9:477. doi: 10.3389/fnins.2015.00477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wilkinson B, Grepo N, Thompson BL, Kim J, Wang K, Evgrafov OV, Lu W, Knowles JA, Campbell DB. The autism-associated gene chromodomain helicase DNA-binding protein 8 (CHD8) regulates noncoding RNAs and autism-related genes. Transl Psychiatry. 2015;5:e568. doi: 10.1038/tp.2015.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.DeWitt J, Campbell DB. Targeting noncoding RNA for treatment of autism spectrum disorders. In: Hu V, editor. Frontiers in Autism Research, Diagnosis, and Treatment. Singapore: World Scientific Publishing; 2014. pp. 203–228. [Google Scholar]
- 7.Kerin T, Ramanathan A, Rivas K, Grepo N, Coetzee GA, Campbell DB. A noncoding RNA antisense to moesin at 5p14.1 in autism. Sci Transl Med. 2012;4:128ra140. doi: 10.1126/scitranslmed.3003479. [DOI] [PubMed] [Google Scholar]
- 8.St Pourcain B, Wang K, Glessner JT, Golding J, Steer C, Ring SM, Skuse DH, Grant SF, Hakonarson H, Davey Smith G. Association between a high-risk autism locus on 5p14 and social communication spectrum phenotypes in the general population. Am J Psychiatry. 2010;167:1364–1372. doi: 10.1176/appi.ajp.2010.09121789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jiang J, Jing Y, Cost GJ, Chiang JC, Kolpa HJ, Cotton AM, Carone DM, Carone BR, Shivak DA, Guschin DY, Pearl JR, Rebar EJ, Byron M, Gregory PD, Brown CJ, Urnov FD, Hall LL, Lawrence JB. Translating dosage compensation to trisomy 21. Nature. 2013;500:296–300. doi: 10.1038/nature12394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Turner AM, Ackley AM, Matrone MA, Morris KV. Characterization of an HIV-targeted transcriptional gene-silencing RNA in primary cells. Hum Gene Ther. 2012;23:473–483. doi: 10.1089/hum.2011.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hawkins PG, Morris KV. Transcriptional regulation of OCT4 by a long non-coding RNA antisense to OCT4-pseudogene 5. Transcription. 2010;1:165–175. doi: 10.4161/trns.1.3.13332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hawkins PG, Santoso S, Adams C, Anest V, Morris KV. Promoter targeted small RNAs induce long-term transcriptional gene silencing in human cells. Nucleic Acids Res. 2009;37:2984–2995. doi: 10.1093/nar/gkp127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Knowling S, Morris KV. Non-coding RNA and antisense RNA. Nature’s trash or treasure? Biochimie. 2011;93:1922–1927. doi: 10.1016/j.biochi.2011.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Trapnell C, Pachter L, Salzberg SL. Tophat: discovering splice junctions with RNA-seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. Genome Project Data Processing S: the sequence alignment/map format and samtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. Transcript assembly and quantification by RNA-seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 18.Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Abrahams BS, Arking DE, Campbell DB, Mefford HC, Morrow EM, Weiss LA, Menashe I, Wadkins T, Banerjee-Basu S, Packer A. SFARI gene 2.0: a community-driven knowledgebase for the autism spectrum disorders (ASDs) Mol Autism. 2013;4:36. doi: 10.1186/2040-2392-4-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee H, Lin MC, Kornblum HI, Papazian DM, Nelson SF. Exome sequencing identifies de novo gain of function missense mutation in KCND2 in identical twins with autism and seizures that slows potassium channel inactivation. Hum Mol Genet. 2014;23:3481–3489. doi: 10.1093/hmg/ddu056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.De Rubeis S, He X, Goldberg AP, Poultney CS, Samocha K, Cicek AE, Kou Y, Liu L, Fromer M, Walker S, Singh T, Klei L, Kosmicki J, Shih-Chen F, Aleksic B, Biscaldi M, Bolton PF, Brownfeld JM, Cai J, Campbell NG, Carracedo A, Chahrour MH, Chiocchetti AG, Coon H, Crawford EL, Curran SR, Dawson G, et al. Synaptic, transcriptional and chromatin genes disrupted in autism. Nature. 2014;515:209–215. doi: 10.1038/nature13772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A, Rinn JL. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011;25:1915–1927. doi: 10.1101/gad.17446611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, Guernec G, Martin D, Merkel A, Knowles DG, Lagarde J, Veeravalli L, Ruan X, Ruan Y, Lassmann T, Carninci P, Brown JB, Lipovich L, Gonzalez JM, Thomas M, Davis CA, Shiekhattar R, Gingeras TR, Hubbard TJ, Notredame C, Harrow J, Guigo R. The Gencode v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22:1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guttman M, Rinn JL. Modular regulatory principles of large non-coding RNAs. Nature. 2012;482:339–346. doi: 10.1038/nature10887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ravasi T, Suzuki H, Pang KC, Katayama S, Furuno M, Okunishi R, Fukuda S, Ru K, Frith MC, Gongora MM, Grimmond SM, Hume DA, Hayashizaki Y, Mattick JS. Experimental validation of the regulated expression of large numbers of non-coding RNAs from the mouse genome. Genome Res. 2006;16:11–19. doi: 10.1101/gr.4200206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mercer TR, Dinger ME, Sunkin SM, Mehler MF, Mattick JS. Specific expression of long noncoding RNAs in the mouse brain. Proc Natl Acad Sci USA. 2008;105:716–721. doi: 10.1073/pnas.0706729105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Batista PJ, Chang HY. Long noncoding RNAs: cellular address codes in development and disease. Cell. 2013;152:1298–1307. doi: 10.1016/j.cell.2013.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Karreth FA, Pandolfi PP. ceRNA cross-talk in cancer: when ce-bling rivalries go awry. Cancer Discov. 2013;3:1113–1121. doi: 10.1158/2159-8290.CD-13-0202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee JT. Epigenetic regulation by long noncoding RNAs. Science. 2012;338:1435–1439. doi: 10.1126/science.1231776. [DOI] [PubMed] [Google Scholar]
- 30.Mattick JS, Rinn JL. Discovery and annotation of long noncoding RNAs. Nat Struct Mol Biol. 2015;22:5–7. doi: 10.1038/nsmb.2942. [DOI] [PubMed] [Google Scholar]
- 31.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 32.Morris KV, Mattick JS. The rise of regulatory RNA. Nat Rev Genet. 2014;15:423–437. doi: 10.1038/nrg3722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Orom UA, Shiekhattar R. Long noncoding RNAs usher in a new era in the biology of enhancers. Cell. 2013;154:1190–1193. doi: 10.1016/j.cell.2013.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Prensner JR, Chinnaiyan AM. The emergence of lncRNAs in cancer biology. Cancer Discov. 2011;1:391–407. doi: 10.1158/2159-8290.CD-11-0209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ulitsky I, Bartel DP. lincRNAs: genomics, evolution, and mechanisms. Cell. 2013;154:26–46. doi: 10.1016/j.cell.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Akbarian S, Liu C, Knowles JA, Vaccarino FM, Farnham PJ, Crawford GE, Jaffe AE, Pinto D, Dracheva S, Geschwind DH, Mill J, Nairn AC, Abyzov A, Pochareddy S, Prabhakar S, Weissman S, Sullivan PF, State MW, Weng Z, Peters MA, White KP, Gerstein MB, Amiri A, Armoskus C, Ashley-Koch AE, Bae T, et al. The PsychENCODE project. Nat Neurosci. 2015;18:1707–1712. doi: 10.1038/nn.4156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bourgeron T. From the genetic architecture to synaptic plasticity in autism spectrum disorder. Nat Rev Neurosci. 2015;16:551–563. doi: 10.1038/nrn3992. [DOI] [PubMed] [Google Scholar]
- 38.Ashwood P, Wills S, Van de Water J. The immune response in autism: a new frontier for autism research. J Leukoc Biol. 2006;80:1–15. doi: 10.1189/jlb.1205707. [DOI] [PubMed] [Google Scholar]
- 39.De Jong S, Newhouse SJ, Patel H, Lee S, Dempster D, Curtis C, Paya-Cano J, Murphy D, Wilson CE, Horder J, Mendez MA, Asherson P, Rivera M, Costello H, Maltezos S, Whitwell S, Pitts M, Tye C, Ashwood KL, Bolton P, Curran S, McGuffin P, Dobson R, Breen G. Immune signatures and disorder-specific patterns in a cross-disorder gene expression analysis. Br J Psychiatry. 2016;209:202–208. doi: 10.1192/bjp.bp.115.175471. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.