Abstract
The protein arginine methyltransferase (PRMT) family of enzymes comprises nine family members in mammals. They catalyze arginine methylation, either monomethylation or symmetric/asymmetric dimethylation of histone and non-histone proteins. PRMT methylation of its substrate proteins modulates cellular processes such as signal transduction, transcription, and mRNA splicing. Recent studies have linked overexpression of PRMT5, a member of PRMT superfamily, to oncogenesis, making it a potential target for cancer therapy. In this study, we developed a highly sensitive (Z’ score = 0.7) robotic high throughput screen (HTS) platform to discover small molecule inhibitors of PRMT5 by adapting the AlphaLISA™ technology. Using biotinylated histone H4 as a substrate, S-adenosyl-l-methionine as a methyl donor, PRMT5 symmetrically dimethylated H4 at arginine (R) 3. Highly specific Acceptor beads for symmetrically dimethylated H4R3 and streptavidin-coated Donor beads bound the substrate, emitting signal that is proportional to the methyltransferase activity. Using this powerful approach, we identified specific PRMT5 inhibitors 1608K04 and P1618J22, and further validated their efficacy and specificity for inhibiting PRMT5. Importantly, these two compounds exhibited much more potent efficacy than the commercial PRMT5 inhibitor EPZ015666 in both pancreatic and colorectal cancer cells. Overall, our work highlights a novel, powerful, and sensitive approach to identify specific PRMT5 inhibitors. The general principle of this HTS screening method can be applied not only to PRMT5 and the PRMT superfamily, but may also be extended to other epigenetic targets. This approach allows us to identify compounds that inhibit the activity of their respective targets, and screening hits like 1608K04 and P1618J22 may serve as basis for novel drug development to treat cancer and/or other diseases.
Keywords: AlphaLISA, HTS, PRMT
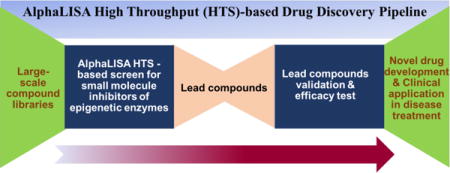
Graphical abstract
Proposed workflow of drug development pipeline with adapted AlphaLISA HTS-based screen to discover inhibitors of epigenetic targets like PRMT5 with possible significance in disease treatment.
1. Introduction
Protein arginine methyltransferases (PRMTs) catalyze the transfer of methyl groups from S-adenosylmethionine (SAM or AdoMet) to guanidino nitrogen of arginine on its substrate protein, resulting in methylated arginine. This posttranslational modification occurs on both histones and non-histone proteins, thereby affecting cellular function and development. Mammalian PRMTs are divided into three groups based on their symmetric or asymmetric arginine methylation pattern: type I PRMTs catalyze the formation of x-NG-monomethylarginine (MMA) and asymmetric x-NG, NG-dimethylarginine (aDMA); type II PRMTs catalyze the formation of MMA and symmetric x-NG, N0G-dimethylarginine (sDMA), and type III PRMTs bring about only monomethylation. Type I PRMTs include PRMT1, 2, 3, 4, 6, and 8, while PRMT5 and 9 are Type II PRMTs. PRMT7 is classified as a type III PRMT.1, 2
Methylation of proteins by PRMTs is involved in a variety of cellular functions related to signal transduction, transcription, DNA repair, and mRNA splicing. Interestingly, recent studies have linked overexpression of these enzymes to a wide variety of cancers.3 Both PRMT1 and PRMT2 have been implicated in epithelial-mesenchymal transition (EMT) in breast cancer cells.4,5 PRMT1 and 6 dysregulations were shown to promote growth of bladder and lung cancer cells.6 Our lab was the first to demonstrate that PRMT5 activates nuclear factor kappa B (NF-κB) by symmetrically dimethylating the p65 subunit of NF-κB at R30.7 Furthermore, PRMT5 is upregulated in several types of cancers including breast, cervix, liver, colon, lung, ovary, bladder, and particularly in pancreatic cancers.7 - 9 Thus, the PRMT family has been widely implicated in cancer development. Since PRMT5 has generated a lot of interest as a potential oncogene in a wide variety of cancers, in this study, we decided to use PRMT5 as the representative PRMT family member as the target for assay development to screen for small molecule inhibitors.
High throughput screening (HTS) techniques can be used to effectively screen for small molecule inhibitors against specific targets. The inhibitors identified using HTS not only serve as invaluable tools to study important targets in cancer such as PRMT5, but also provide prospective candidates for therapeutic development. An ideal HTS screen is reproducible, sensitive, scalable, cost-efficient, and most importantly, effective. In this study, we have adapted the AlphaLISA™ technique, a bead-based Amplified Luminescent Proximity Homogeneous Assay-Linked ImmunoSorbent Assay that satisfies the criteria for an effective HTS approach. Compared to conventional methods like time-resolved fluorescence energy transfer (TR-FRET) assays and enzyme-based assays, the AlphaLISA technology has been proven to have better sensitivity, less variability, and to possess the ability to be automated for large-scale reactions. Additionally, since it is a non-radioactive, proximity-based assay, the hassles and dangers of radioactivity-based approaches can be avoided. Overall, AlphaLISA provides a highly sensitive and scalable approach to perform assay development for an HTS.
We adapted the AlphaLISA technology to develop an effective assay to screen for small molecule inhibitors of PRMT5 (Fig. 1). Symmetric dimethylation of its substrate by PRMT5 brings the AlphaLISA beads in close proximity and a greatly amplified signal can be produced. Upon excitation with 680nm of light, a photosensitizer component in the “Donor” bead converts ambient oxygen to an excited singlet state. The singlet oxygen molecules react with a thioxene derivative in the “Acceptor” bead, emitting light at 370nm that further activates a fluorophore contained in the Acceptor bead itself. The Acceptor beads have a specific antibody tag that recognizes the symmetrically dimethylated PRMT5 substrate. The fluorophore on the Acceptor bead subsequently emits light at 615nm, which can be quantified using an EnVision® reader. If the specific dimethylation tag is absent, the singlet oxygen molecules produced upon the Donor bead excitation will not be transferred to the Acceptor bead, as proximity of the Acceptor bead will not occur in the absence of this specific interaction. As a result, a very low background signal would be produced. This guarantees the specificity of the signal produced and thereby accurate quantification of the methyltransferase activity.
Fig. 1. Principle of AlphaLISA™ screen for quantifying PRMT5 methylation activity.
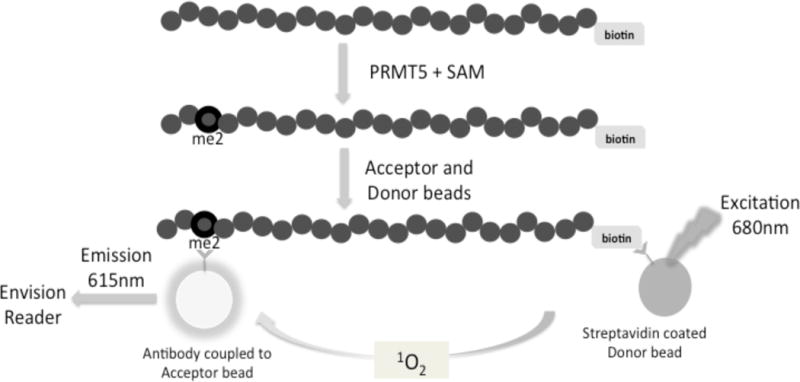
Biotinylated histone H4 is incubated with PRMT5 and a methyl donor, S-adenosyl-l-methionine (SAM). PRMT5 symmetrically dimethylates the third arginine (R3) on Biotin-H4 to form dimethylated Biotin-H4R3 (Biotin-H4R3me2) peptide. This product will further bind to Acceptor beads specific to the methylation site (Biotin-H4R3me2). Streptavidin-coated Donor beads will bind to the biotinylated substrate. Interaction between the Acceptor and Donor beads emits a chemiluminescent signal, which will be detected using an EnVision® reader. The methylation activity of PRMT5 is proportional to the intensity of the signal.
In this paper, we describe the AlphaLISA assay development to the HTS stage with a robust signal to background (S/B) ratio, adapting a PRMT5-specific protocol, such that the efficiency of this approach in an HTS setting is optimized. As a proof-of-principle, we highlight two hits (P1608K04 and P1618J22) from our screening that showed significant inhibition of PRMT5 methyltransferase activity. These hits were identified by screening small molecule compound libraries that contained natural as well as synthetic compounds, and compounds with known pharmacological activity. We confirmed their PRMT5-specific activity using a concentration-dependent AlphaLISA assay and tested the in vitro efficacy by checking their effect on viability of a panel of PRMT5-overexpressing pancreatic ductal adenocarcinoma (PDAC) and colorectal cancer (CRC) cell lines. Further evidence for specificity of these inhibitors was demonstrated by the reduction in methylation of the p65 subunit of NF-κB, subsequent decrease of NF-κB activity, and the reduced expression of NF-κB target genes. Importantly, comparing with previously published data from our lab8 with regards to a commercially available PRMT5 inhibitor EPZ015666,10 we observed much lower IC50s of P1608K04 and P1618J22 than that of EPZ015666 in both PDAC and CRC cells, confirming both the great advantage of the AlphaLISA HTS technique that we adapted and the considerable promise that P1608K04 and P1618J22 hold for further drug development.
Overall, this study illustrates a systematic approach to design, optimize and execute a HTS approach for targets of interest such as PRMT5 using AlphaLISA. This study is significant as it describes a highly effective (Z’ factor=0.7) HTS system to screen for PRMT5 inhibitors with a robotic system. Importantly, it can serve as a template for other studies involving small molecule inhibitors for other epigenetic enzymes. Guided protocol development based on this study will allow researchers to follow similar considerations and develop HTS screening protocols to achieve their scientific goals. Notably, we identified two novel PRMT5 small molecule inhibitors that are more potent than the commercial inhibitor EPZ015666 in PDAC and CRC cells. In the broader sense, compounds identified in such HTS studies are critical tools to study the underlying mechanism of the targets of interest in disease models, thus, may serve as basis for novel drug development in the future.
2. Materials and methods
2.1 Reagents and peptides
The methyl group donor SAM was purchased from New England Biolabs (Ipswich, MA). Unmethylated biotinylated histone H4 peptide substrate at arginine (R) 3 (unmeH4R3) was obtained from AnaSpec (Fremont, CA). The 23-amino acid sequence of H4R3 peptide was as follows: SGRGKGGKGLGKGGAKRHRKVLRGG-K(biotin)-NH2, with the third arginine site available for dimethylation as per the assay protocol. For screening, dimethyl sulfoxide (DMSO) stock of library compounds comprising of approximately 10,000 pure natural products, semi-synthetic natural products and reported bioactives were purchased from Analyticon Discovery (Rockville, MD), MilliporeSigma (St. Louis, MO) and Microsource Discovery Systems Inc (Gaylordsville, CT). The compound libraries were stored at −80°C. Anti-methyl-H4R3 AlphaLISA beads, Streptavidin-tagged Donor beads, 1× Epigenetics buffer, TopSeal™-A film, OptiPlate™-384 white opaque plates, and EnVision® Multilabel Reader were obtained from PerkinElmer (Waltham, MA).
2.2 Cell lines
PDAC cell lines (PANC1, MiaPaCa2 and AsPC1) were kindly provided by Dr. Murray Korc (Indiana University School of Medicine). CRC cell lines (HT29, HCT116, and DLD1) were purchased from American Type Culture Collection (ATCC). PANC1 and MiaPaCa2 cell lines were grown in HyClone™ Dulbecco’s Modified Eagle Medium (DMEM) (GE Healthcare, Logan, UT), with addition of 1% of penicillin/streptomycin and 5% fetal bovine serum (FBS). CRC cells were maintained in HyClone™ Roswell Park Memorial Institute Medium (RPMI 1640) (GE Healthcare, Logan, UT), with 1% penicillin/streptomycin and 5% FBS. 293 cells overexpressing Flag-PRMT5 were generated as described previously7 and cultured in HyClone™ DMEM with 1% penicillin/streptomycin and 5% FBS. The cells were grown at 37°C and 5% CO2 conditions. Authentication of cell lines was carried out using 9Marker STR Profile.
2.3 Enzyme purification
Flag-tagged PRMT5 enzyme was purified from 293 cells overexpressing Flag-PRMT5 by co-immunoprecipitating with Flag beads from MilliporeSigma (St. Louis, MO). 293-Flag-PRMT5 cells were cultured in 15cm plates, ~90% confluence per plate. After washing the cells with 1× phosphate buffered saline (PBS), the cells were spun down at 1,200rpm for 10 min at 4°C. The pellets were lysed by adding lysis buffer (50mM Tris-HCl, pH7.4, 150mM NaCl, 1mM EDTA, 100nM Phenylmethanesulfonyl fluoride, 0.5M Na3NO4, 1% Triton X-100 and protease inhibitor tablets) and vortexing 4-5 times over a period of 20 min. The lysed cells were spun at 3,400rpm for 30 min, and the supernatant was transferred to the prewashed Flag beads suspended in 1× cold PBS. The lysed cells/beads mixture was rotated at 4°C. After overnight incubation, the beads were spun down and washed 4 times using 1× wash buffer (50mM Tris-HCl, pH 7.4, 150mM NaCl, 1mM EDTA, 0.5% Triton X-100). Post-washing, the beads were mixed with Flag peptide and rotated for 1 h at 4°C, then spun down. Supernatant (the PRMT5 enzyme product) was added with storage buffer (40mM Tris, 110mM NaCl, 2.2mM KCl, 3mM Dithiothreitol (DTT), 20% glycerol), and used immediately for the AlphaLISA assay or aliquoted at −80°C until further use.
2.4 AlphaLISA-based H4R3me2 detection assay
The AlphaLISA reaction was run in 384-well white opaque plates with a total reaction volume of 30μl. All the parameters of the assay were determined using standardized conditions by testing 10-100nM of unmeH4R3 and of 0.2-4000μM SAM. In the pilot experiments as well as the Z’ test, the following protocol was used: the biotinylated unmeH4R3 substrate stock as well as the SAM stock was diluted using milliQ water with final concentrations of 30nM and 100nM respectively in the reaction well. The PRMT5 enzyme prep was diluted at 1:10 ratio in Assay buffer (30mM Tris, pH 8.0, 1mM DTT, 0.01% Bovine serum albumin, 0.01% Tween-20) before being used. 10μl of unmeH4R3:SAM mixture and 10μl of diluted enzyme stock were mixed in each well and incubated at R.T. for 1h. Following this, the Acceptor bead stock (with an antibody tag specific for H4R3me2) was diluted 1:50 using 1× Epigenetics buffer to a final concentration of 20μg/ml. 5μl of the diluted bead stock was dispensed in each of the reaction wells. After incubation with the Acceptor beads for 1h at room temperature (R.T.), the streptavidin-coated Donor bead stock (specific for the biotinylated substrate) was diluted at 1:50 using 1× Epigenetics buffer to a final concentration of 20μg/ml, and 5μl was dispensed in each of the reaction wells (Table 1). Following incubation with Donor beads for 30 min in the dark, the plates were immediately read using the “Alphascreen” filter in the EnVision® Reader. To assess background of the reaction in the negative control wells, all the conditions were same except 10μl of 1× assay buffer was added instead of enzyme stock.
Table 1.
Protocol for High Throughput Screening for PRMT5 AlphaLISA Assay
| Step | Parameter | Value | Description |
|---|---|---|---|
| 1 | Dispense substrate & cofactor | 10μl | 2× mix of SAM/peptide |
| 2 | Dispense compounds | 250nL | 80× compound (1.25% (v/v) DMSO) |
| 3 | Dispense enzyme | 10μl | 2× PRMT5 enzyme |
| 4 | Incubation time | 1h | RT, sealed |
| 5 | AlphaLISA Acceptor beads | 5μl | 1:42 dilution of Acceptor beads mix |
| 6 | Incubation time | 1h | RT, sealed |
| 7 | AlphaLISA Donor beads | 5μl | 1:42 dilution of Donor beads mix |
| 8 | Incubation time | 0.5h | RT, sealed |
| 9 | Assay readout | 615nm | EnVision plate reader; Alphascreen mode |
Step Notes
1. Final concentrations are 30nM Biotin-unmeH4R3, 100μM SAM prepared in MilliQ water.
2. Final compound concentration: 12.5μM; columns 21–24 do not have compound and have only DMSO/water with a final DMSO concentration at 1.25%.
3. Columns 23 and 24 only has no enzyme, prepared in enzyme reaction buffer.
4. Plates sealed with adhesive film and covered with aluminum foil.
5. Final concentration is 20μg/ml for Acceptor beads mix, prepared in the dark using 1× Epigenetics buffer.
6. Plates sealed with adhesive film, covered with aluminum foil and kept in dark.
7. Final concentration is 20μg/ml for Donor beads mix, prepared in the dark using 1× Epigenetics buffer.
8. Plates sealed with adhesive film, covered with aluminum foil and kept in dark.
9. Alphascreen (570nm) emission filter
PRMT5, protein arginine methyltransferase 5; H4R3, arginine 3 of histone H4; SAM, S-adenosyl-l-methionine
2.5 High-throughput screening
For the HTS, we used the optimized protocol as described in Table 1. Briefly, 10μL of the substrate 2× mix (60nM unmethylated peptide, 200μM SAM in MilliQ water) was dispensed using Multidrop into each well of the assay plates. The final reaction concentration was 30nM unmeH4R3 peptide and 100μM SAM. Next, 250nL of 1mM DMSO stock of the library compounds was transferred to columns 1-20, with a final compound concentration of 12.5μM and final DMSO of 1.25% in each well. 5% DMSO/water (1% DMSO final) was added to column 21-24 to keep the DMSO concentration consistent with the wells in which the compounds were added. Next, 10μL enzyme (ten-fold dilution of the enzyme prep in assay buffer) was dispensed to columns 1-22 using MultiFlo in each plate. At the same time, 10μL of the 1× assay buffer was added to columns 23-24 in each plate. Thus, columns 21-22 serve as positive “maximum signal” controls (no compound, with PRMT5 enzyme added) and columns 23-24 act as “background signal” controls (no compound, no enzyme added). Columns 1-20 in each plate had the different compounds added, with 1 replicate for each compound in the library. The plates were incubated at R.T. for 1h. 5μL anti-H4R3me2 Acceptor beads (1:50 dilution of the 5mg/mL stock in 1× Epigenetics Buffer) were then dispensed using MultiFlo into each well of the assay plates. The plates were incubated at R.T. for 1 h. 5μL Donor beads (1:50 dilution of the 5mg/mL stock in 1× Epigenetics Buffer) were dispensed using MultiFlo into each well of the assay plates. The plates were incubated at R.T. for 30 min followed by Alpha signal reading on EnVision® reader.
2.6 Quenching experiment
In this assay, the incubation and assay conditions were relatively similar to the HTS assay described above. The major differences were: (1) histone H4 pre-methylated at arginine 3 (Biotin-H4R3me2) (AnaSpec, Fremont, CA) was used as a substrate at a final concentration of 15nM; (2) No PRMT5 enzyme or unmethylated substrate was included in the experiment, and (3) The plate design differed as follows: the first two columns did not have any compound (positive control). Columns 1-22 had pre-methylated Biotin-H4R3me2 substrate. The last two columns did not have this pre-methylated substrate (negative control; background). Diluted AlphaLISA beads (Acceptor and Donor) were added to each well.
2.7 MTT [(3-(4, 5-dimethylthiazolyl-2)-2, 5-diphenyltetrazolium bromide)] assay
Cells were seeded at 60% confluence in Corning® Costar® clear flat-bottom 96-well plates and increasing concentrations of P1608K04 and P1618J22 were added to the wells respectively. After incubation for 4 days, 10μl of MTT dye (Sigma-Aldrich, St. Louis, MO) was added directly to each well. The dye was incubated with the cells for 2 h at 37°C. The media was then aspirated and 10μl of DMSO was then added to each well. The colorimetric dye was quantified using Synergy H1 Multi-Mode Reader from BioTek Instruments Inc. (Winooski, VT).
2.8 Western blotting
10% polyacrylamide gel was loaded with protein samples and transferred overnight at 4°C to a polyvinylidene fluoride (PVDF) membrane (Thermo Fisher Scientific, Waltham, MA). Symmetric demethylation at R30 subunit of p65 was detected by generating a customized polyclonal primary antibody (in collaboration with GenScript Inc. Piscataway, NJ) used at 1:750 dilution in 5% bovine serum albumin. β-actin was detected using its corresponding antibody with 1:5000 dilution in 5% milk (MilliporeSigma, St. Louis, MO). Corresponding secondary antibodies were used and the protein signal was detected using enhanced chemiluminescent (ECL) detection method (PerkinElmer, Waltham, MA).
2.9 NF-κB Luciferase assay
NF-κB luciferase construct p5XIP10 containing five tandem copies of NF-κB binding site from one of its target genes, IP10 was used.7, 8 The p5XIP10 construct was transfected transiently in cells using Lipofectamine™ LTX Reagent with PLUS™ Reagent (Thermo Fisher Scientific, Waltham, MA). Cells were lysed 48 h post transfection with or without PRMT5 small molecule inhibitors treatment. Luciferase activity was measured using the Luciferase Assay System with Reporter Lysis Buffer kit (Promega, Madison, WI) by using a manufacturer provided protocol and the Synergy H1 Multi-Mode Reader.
2.10 Quantitative PCR
Total RNA was isolated from ~90% confluent plate of cells using Trizol as per previously described protocol.7 cDNA was then synthesized using the SuperScript III First-Strand Synthesis System (Invitrogen, Carlsbad, CA) by reverse-transcriptase (RT) PCR. Primers for the final step were designed using Primer Express 3.0 software and quantitative PCR (qPCR) was carried out using the FastStart Universal SYBR Green Master ROX kit (Roche, Indianapolis, IN).
2.11 Structural analysis and docking
For docking, chain A of PRMT5 protein in 4×61.pdb from the Protein Data Bank was used. All docking experiments were performed by Glide program (version 7.5) of the Schrodinger suite in standard precision mode.11–14 Control docking experiment was performed by deleting the ligand EPZ015666 from the crystal structure while keeping the ligand SAM and could reproduce the binding pose of EPZ015666 seen in the crystal structure. Compounds P1608K04 and P1618J22 were docked into the PRMT5 active site under two conditions. In one experiment, the compound was docked into the PRMT5 active site in which SAM was kept. In the other experiment, compounds were docked into the completely empty active site in which SAM was deleted. Figures were prepared with Maestro version 11.2.14
2.12 Statistical analysis
The Z’ factor was calculated using the formula described in Zhang et al.: [Z’ = 1− {3 × SD positive control + 3 × SD negative control} / (mean positive control – mean negative control}]. Percent inhibition for the compound library was calculated as follows: [({Avg. maximum reading – compound reading}/ Avg. maximum reading) × 100].15 Statistical software GraphPad Prism (San Diego, CA) was used. The p-values <0.05 were considered statistically significant.
3. Results
3.1 Development and optimization of AlphaLISA protocol
As shown in Fig. 1, AlphaLISA protocol requires several critical components, including substrate H4R3, the methyl donor SAM, PRMT5 enzyme as well as Acceptor and Donor beads. We purified PRMT5 enzyme using co-immunoprecipitation experiments with Flag beads from 293-PRMT5-Flag cells. The manufacturer-recommended dilutions for the Acceptor as well as the Donor beads helped us to obtain the desired signal. The major optimization was necessary for the unmethylated H4R3 substrate (unmeH4R3) and SAM concentrations to be used in the experiment. First, we optimized the substrate concentration by running the AlphaLISA assay with 10-100nM final concentration of the unmeH4R3 substrate in the reaction well. We observed that with increasing concentrations of substrate there was a predictable increase in the AlphaLISA signal (Fig. 2A). Since using the lower concentrations provided quite a robust signal in the pilot experiment, we decided to use 30nM of unmeH4R3 to avoid unnecessary use of substrate.
Fig. 2. Optimization of the AlphaLISA protocol.
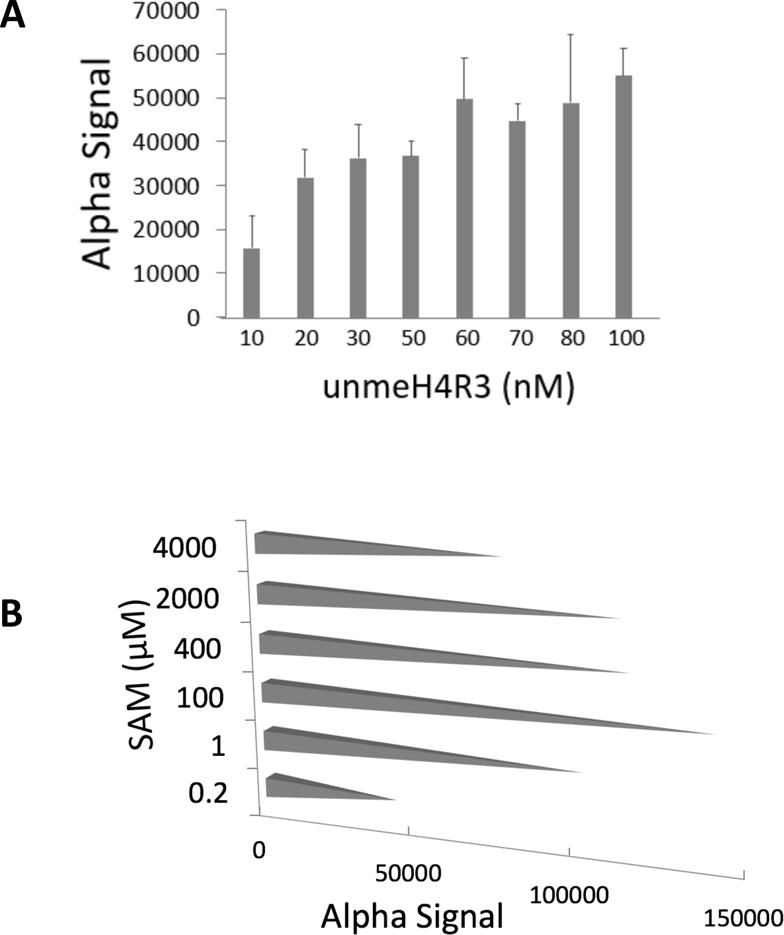
(A) Alpha signal graph, showing that the Alpha signal increased with increased substrate (Biotin-unmeH4R3) concentrations (10–100nM). (B) Alpha signal graph, showing that increased concentrations of the methyl donor SAM (0.2–4000μM) led to bell-shaped distribution of Alpha signal strength. We observed an increase in the Alpha signal, with a peak at 100μM of SAM, followed by a subsequent decrease with further increased SAM concentrations.
Next, with the aim to maximize the AlphaLISA signal, we determined if a specific concentration for methyl donor SAM worked best in this assay. We tested a wide range of SAM concentrations (0.2μM to 2000μM) while keeping all the other components same, including the substrate concentration at 30nM as described above. We observed a steady increase in AlphaLISA signal, with a peak at 100μM SAM, and then decreasing with the higher concentrations (Fig. 2B). Thus, we decided to choose a final concentration of 100μM SAM in our HTS assay. These assays also allowed us to adjust and test volumes and concentrations that were permissible for the robotic approach, thus helping us to automate the addition of these reagents in the actual HTS.
3.2 Determination of Z’ factor
After successfully determining the important parameters for assay development, we executed the Z’ test to check the robustness of the assay. Calculation of the Z’ factor allows for evaluation and validation of HTS assays .15 We used a 384-well white opaque plate to run the Z’ test. Briefly, half of the plate was used as “maximum signal” wells (with the PRMT5 enzyme added) and the other half was used as “background wells” (with no PRMT5 enzyme, only assay buffer added). We used a 30μl reaction volume as described in the Materials and Methods section. The graph for Z’ factor determination is illustrated in Fig. 3. The Z’ factor was calculated to be 0.7 and a signal/background ratio of ~8.3, which is indicative of the robustness and reproducibility of the assay. Once the assay was proven HTS-compatible, the next step was to scale it up to run a HTS screen using large-scale small compound libraries.
Fig. 3. Determination of Z’ factor for PRMT5-specific AlphaLISA assay.
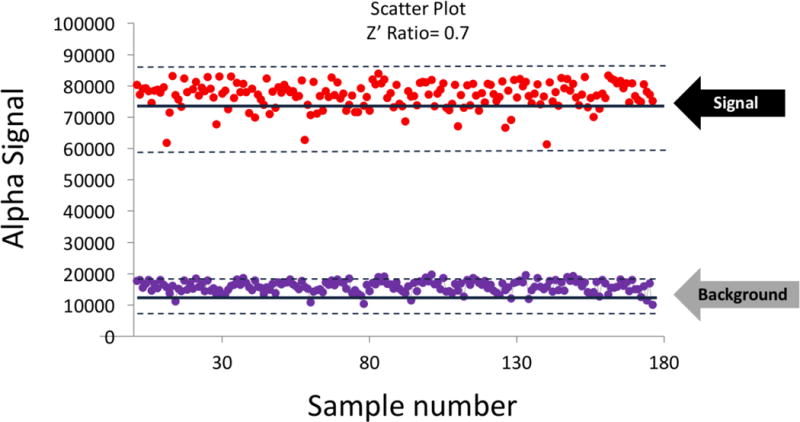
Scatter plot, representing the data points for the 384-well plate, with the red points indicating the “maximum signal” wells and the purple points representing the “background” wells. The solid lines at the center represent the average values and the dotted lines at the top and bottom represent the +/− 3 × SD (standard deviation) values for the respective groups. The S/B (signal vs. background) ratio was calculated to be ~8.3 and the Z’ factor was determined to be 0.7.
3.3 Execution of the HTS screening to identify specific inhibitors of PRMT5 methyltransferase activity
A major advantage of AlphaLISA technology is the ability to be scaled to a HTS setting upon validation. After the Z’ test confirmed the robustness of the assay, we used this assay to screen small-molecule compound libraries (as described in the Materials and methods sections 2.1 and 2.5) to identify hits with PRMT5 methyltransferase inhibition activity. 384-well white opaque plates were used like our pilot experiments for assay development. By adapting our pilot AlphaLISA assay to the MultiFlo robotic system to dispense reagents, we automated the lengthy process of testing each of the 10,000 compounds and reduced the volume of reagents required, as the system could dispense low microliter volumes, thus making the overall process time-efficient and cost-effective. Unmethylated histone H4 substrate:methyl donor SAM were added to all wells of the plate. Each of these wells was also added with individual compounds in columns 1-20. An equivalent amount of DMSO:water was pipetted into columns 21-24 to equalize the final DMSO concentration across all wells in the plate. Enzyme was dispensed to columns 1-22 and 1× assay buffer was added to columns 23-24 in each plate. Thus, columns 21-22 served as positive “maximum signal” controls (no compound, with PRMT5 enzyme added) and columns 23-24 acted as “background signal” controls (no compound, no enzyme added). Acceptor beads, specific for the dimethylated H4R3 and Donor beads were dispensed in all the wells.
Several compounds showed the capacity to inhibit PRMT5 methyltransferase activity. These hits were ranked based on their percent inhibition values. Next, in order to obtain an even more accurate list of positive hits, it was imperative to rule out the false positive hits. To achieve this goal, we conducted the Alpha signal quenching experiment. False hits can result from interference with the AlphaLISA assay itself rather than inhibiting the enzymatic reaction, such as compounds that absorb light at 620 nm or the compounds that absorb singlet O2, resulting in quenching of the Alpha signal. One way to eliminate these “Alpha signal quenchers” was to screen those compounds in a control assay, in which only histone H4 pre-methylated at arginine 3 (Biotin-H4R3me2) was used without adding the PRMT5 enzyme and running the enzymatic reaction. In this way, the Biotin-H4R3me2 peptide will bring the Donor beads and Acceptor beads in close proximity to produce the Alpha signal. In this setting, if any compound reduced the Alpha signal, it would be a quencher instead of a real inhibitor. This is because this reduction in signal is unrelated to PRMT5 activity which itself is absent from this reaction. Fig. 4 illustrates the experimental design and plate layout for this filtering experiment among 320 top hits from the HTS screening to eliminate any Alpha signal quenchers. The first and last two columns did not contain compounds, but the first two columns had the pre-methylated peptide substrate which resulted in a high signal; the last two columns did not have me-H4R3 substrate and provided the readings for the background signal. Every well of the plate had both the Acceptor and Donor beads. We identified 64 potential hits that were not false positives and repeated the AlphaLISA to reconfirm their inhibition activity. Several of the top hits identified after this rigorous screening process have been illustrated in Fig. 5A, with a cut-off value of over 60% inhibition of PRMT5 methyltransferase activity. We conducted preliminary testing using concentration-dependent AlphaLISA assay for all these compounds to check the corresponding effect on PRMT5 methyltransferase activity. P1608K04 and P1618J22 (Fig. 5B) showed the most promise. In a confirmation experiment, we observed that increased concentrations of both P1608K04 and P1618J22 led to a concurrent decrease in AlphaLISA signal with low IC50 values of ~1.5μM and 16.5μM, respectively (Fig. 6A and B). Thus, we employed a stepwise filtering protocol for screening ~10,000 compounds using an AlphaLISA-based approach, and identified two promising hits that are specific for reducing the PRMT5 methyltransferase activity (Fig. 5B).
Fig. 4. Plate design for the quenching experiment to identify false hits from the AlphaLISA screen top hits.
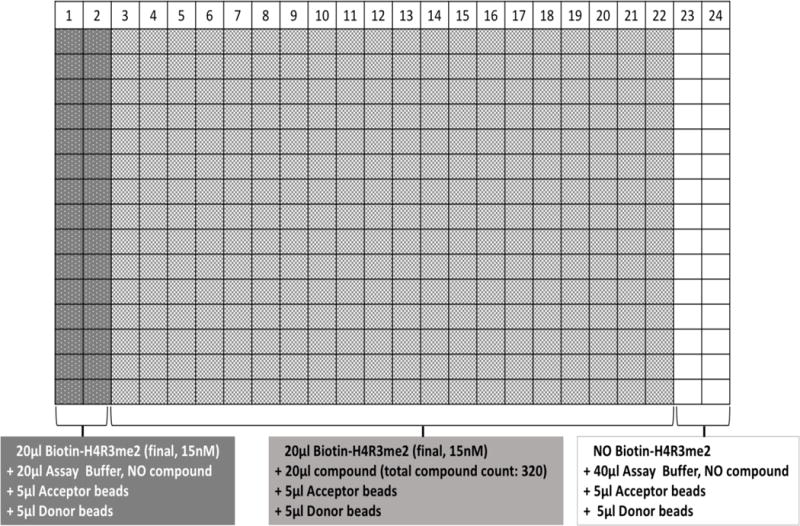
The first two columns (white dots; grey background) do not contain any compound and act as positive control, signifying “maximum signal”. Columns 1–22 had Biotin-H4R3me2 substrate (dimethylated Biotin-H4R3) added at a final concentration of 15nM in each well. The last two columns do not have the Biotin-H4R3me2, and act as the “negative control”. Acceptor and Donor beads were added to each well. No PRMT5 enzyme was included in this experiment. Compounds that exhibited quenching of the Alpha signal were “false hits” and not true PRMT5 inhibitors, thus were removed from further testing.
Fig. 5. Identification of top hits P1608K04 and P1618J22.
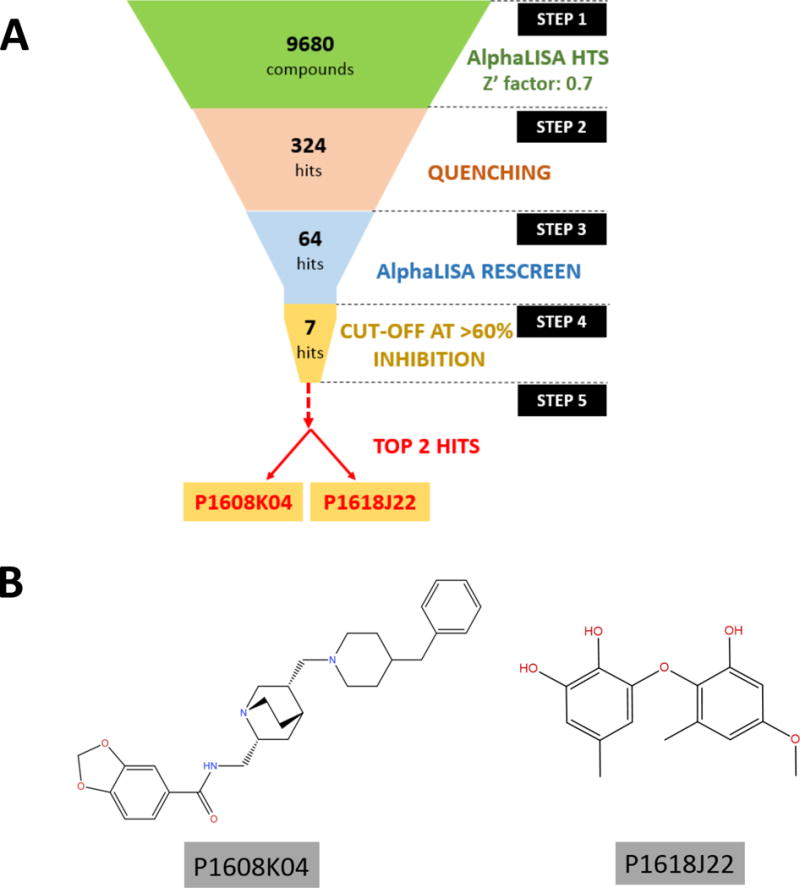
(A) Workflow for the stepwise procedure that eventually led to the shortlisting of two top PRMT5 inhibitors, P1608K04 and P1618J22, among ~10,000 compounds from small compound libraries. Each step represents the experimental approach and the number of compounds to start with. (B) Structures of the 2 top hits: P1608K04 (Left panel) and P1618J22 (Right panel).
Fig. 6. Determination of IC50 for using AlphaLISA assay.
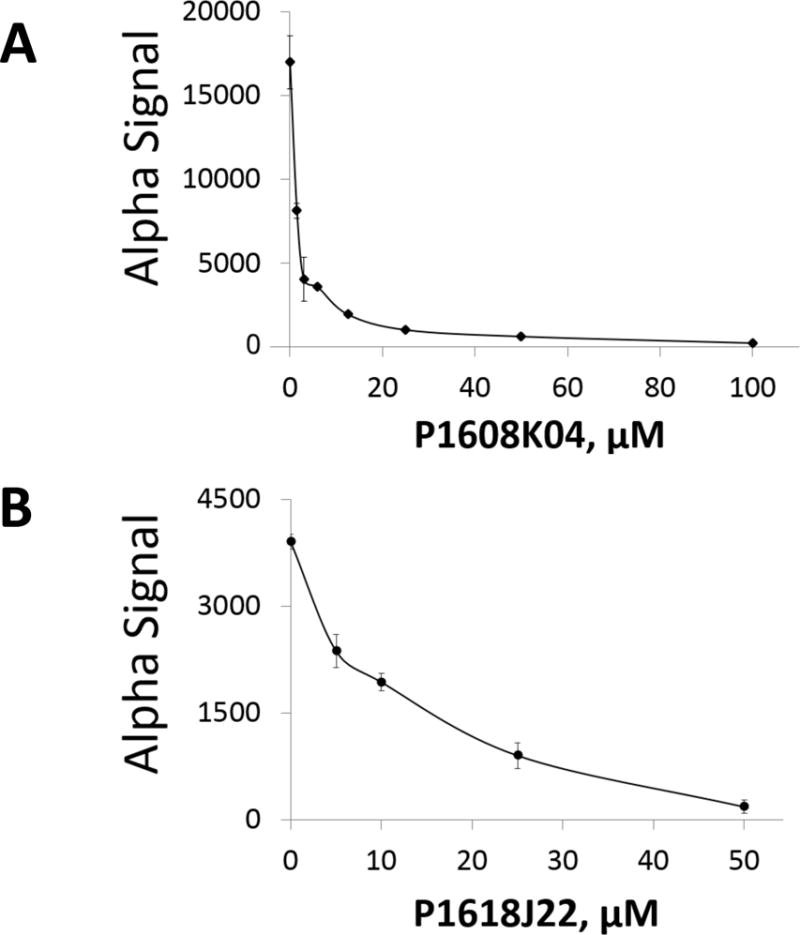
(A) Calculation of IC50 of P1608K04 using AlphaLISA, showing concentration-dependent decrease of Alpha signal. The IC50was~1.5μM. (B) The IC50of P1618J22 was calculated to be 16.5μM, using the same protocol as (A). The data represent the means ± SD for three independent experiments.
3.4 P1608K04 and P1618J22 are potent inhibitors of PRMT5 activity in vitro
In order to test the effectiveness of these promising PRMT5 inhibitors in the context of PDAC and CRC, we used the MTT assay with increasing concentrations of P1608K04 and P1618J22 in PDAC and CRC cell lines. Both P1608K04 (Fig. 7A and C) and P1618J22 (Fig. 7B and C) showed great efficacy in decreasing the cell viability in PDAC (PANC1, MiaPaCa2, and AsPC1) and CRC cells (HT29, HCT116, and DLD1). Importantly, both compounds were much more potent (have lower IC50) than the commercial PRMT5 inhibitor EPZ015666 in PDAC and CRC cells (Fig. 7C and Supplementary Table 1). Therefore, our data suggested that P1608K04 and P1618J22 are powerful PRMT5 inhibitors in PDAC and CRC cells. Furthermore, the inhibitors identified using our PRMT5-specific AlphaLISA assay could be used as basis for novel drug development and tools to further study PRMT5-driven mechanisms in these cancers.
Fig. 7. P1608K04 and P1618J22 are potent inhibitors for viability in PDAC and CRC cells.
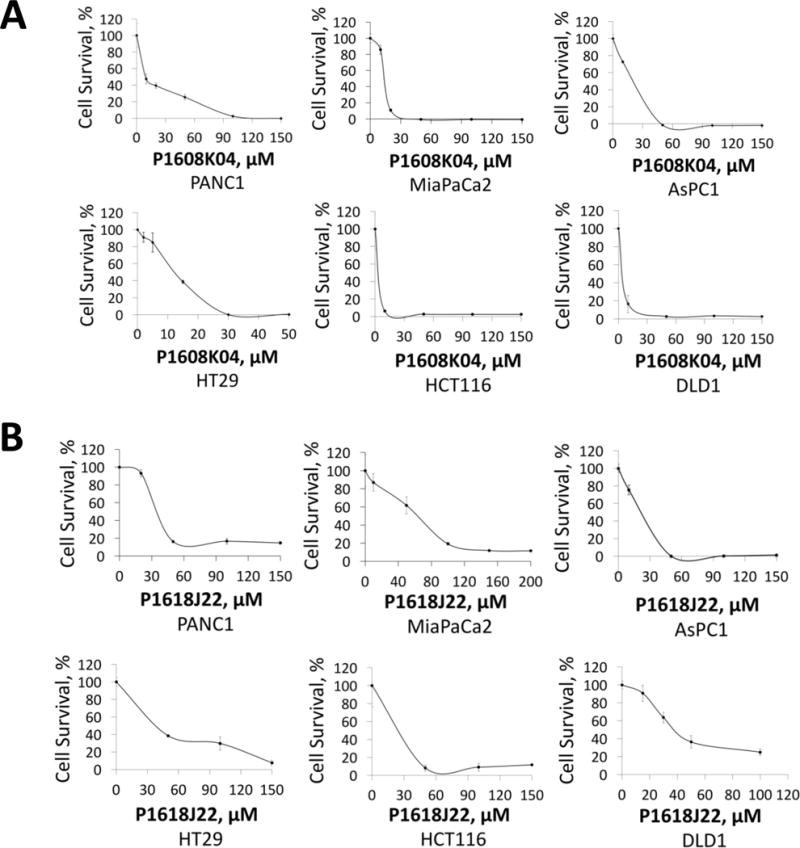
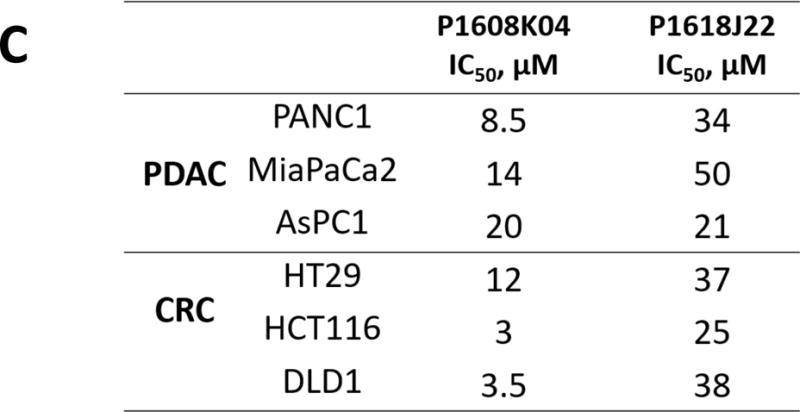
(A) MTT assay with the treatment of P1608K04 in both PDAC (PANC1, MiaPaCa2, and AsPC1) and CRC cells (HT29, HCT116, and DLD1), showing that cell viability dramatically decreased in the presence of increasing concentrations of P1608K04. (B) MTT assay with the treatment of P1618J22 in both PDAC and CRC. The data represent the means ± SD for three independent experiments. (C) Table, summarizing the IC50 values for P1608K04 and P1618J22 in PDAC and CRC cells, respectively.
3.5 P1608K04 and P1618J22 inhibited PRMT5-mediated NF-κB methylation, activation and target gene expression
A previous study in our lab identified p65 subunit of NF-κB as a novel substrate of PRMT5 via methylation of its R30 residue.7 To test the specificity of PRMT5 inhibitors, we examined their effect on p65 methylation in both PANC1 and HT29 cells. Western analysis with a site-specific antibody against dimethylated R30 residue of p65 (Materials and method section 2.8) showed decreased R30 methylation on p65 after the treatment of P1608K04 or P1618J22 (Fig. 8A and B), confirming that these inhibitors specifically attenuated PRMT5-mediated NF-κB methylation in these cells.
Figure 8. P1608K04 and P1618J22 decreased methylation at R30 on the p65 subunit of NF-κB.
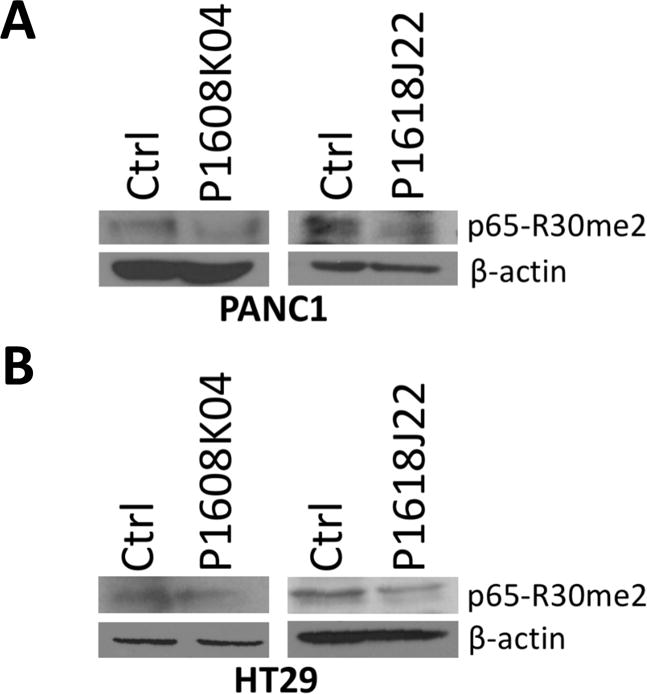
Western blot analysis, (A) PANC1 and (B) HT29 cells were treated with P1608K04 (Left panels) or P1618J22 (Right panels) at their IC50values, respectively. Samples were probed with specific anti-p65-R30me2 antibody. Decreased p65-R30me2 levels were observed in the compounds treated groups as compared with the control (ctrl) groups that are without compounds treatment. β-actin was used as a loading control.
Since NF-κB R30 methylation leads to its activation,7 we wondered whether P1608K04 and P1618J22 may inhibit NF-κB activation and its target gene expression. NF-κB luciferase assay was conducted to determine the effect of compound treatment on PRMT5-mediated NF-κB activation in PDAC and CRC cells. We confirmed that treatment of P1608K04 and P1618J22 for 48 h at the indicated concentrations significantly decreased NF-κB activity in both PANC1 (Fig. 9A) and HT29 (Fig. 9B), respectively. It is important to note that it requires a much higher concentration of the commercial PRMT5 inhibitor EPZ015666 (~85-200μM in PANC1, and 200-300μM in HT29 cells)8 to observe a similar effect per our previously published data,8 demonstrating that both P1608K04 and P1618J22 are more effective than EPZ015666 in decreasing PRMT5-mediated NF-κB activation in PDAC and CRC cells.8
Fig. 9. P1608K04 and P1618J22 inhibited NF-κB activation and its target gene expression in PDAC and CRC cells.
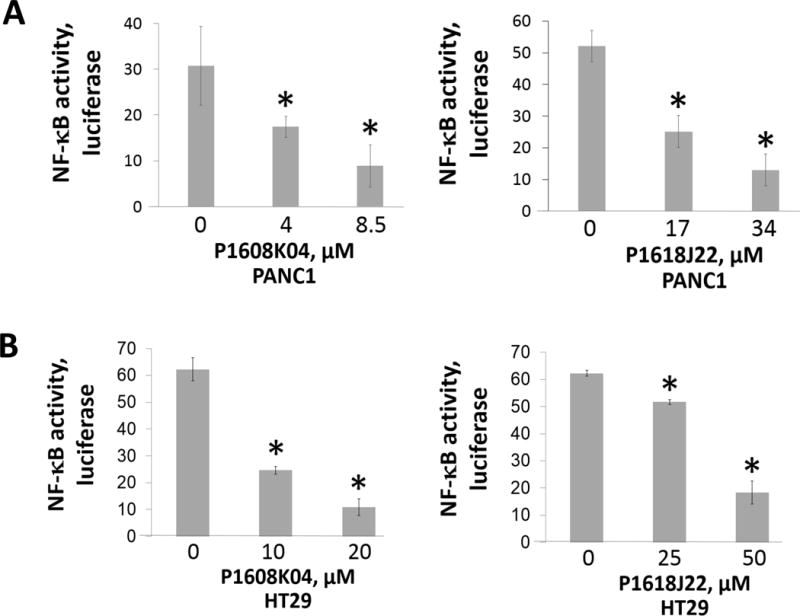
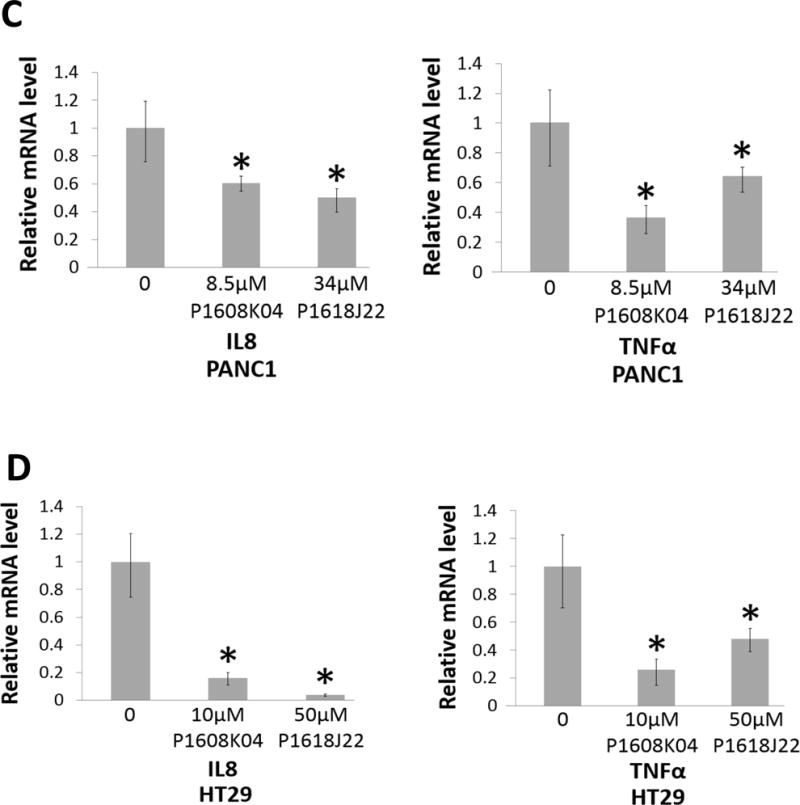
NF-κB luciferase assay in (A) PANC1 and (B) HT29 cells, indicating that treatment with increasing concentrations of P1608K04 or P1618J22 respectively for 24h led to a corresponding decrease in NF-κB activation. The data represent the means ± SD for three independent experiments. *P < 0.05 vs. 0μM. (C) and (D) Quantitative PCR (qPCR) analysis, showing the expression of the typical NF-κB target genes, IL8 and TNFα, was decreased after the treatment of P1608K04 or P1618J22 respectively in PANC1 (C) and HT29 cells (D). The data represent the means ± SD for three independent experiments. *P < 0.05 vs. 0μM.
In addition to the above evidence, since PRMT5-mediated NF-κB activation leads to the induction of well-known cancer-promoting NF-κB target genes, such as interleukin8 (IL8) and tumor necrosis α (TNFα) in PDAC and CRC cells,8 we conducted quantitative PCR to determine the effect of P1608K04 and P1618J22 on the expression of these two genes. As shown in Fig. 9, treatment of PANC1 (Fig. 9C) or CRC (Fig. 9D) cells with these two compounds led to significantly decreased expression of both IL8 and TNFα.
Collectively, the above evidence affirmed that P1608K04 and P1618J22 are PRMT5 specific inhibitors that function through inhibiting PRMT5-mediated NF-κB methylation, activation, and its downstream target gene expression.
3.6 Predicted structural binding of P1608K04 and P1618J22 to PRMT5
To better understand the potential binding mechanism of P1608K04 and P1618J22 to PRMT5, we used a directed computer-based docking approach of these inhibitors to the PRMT5 crystal structure. Comparing the crystal structure of commercial PRMT5 inhibitor EPZ015666 with the predicted binding poses of the two inhibitors we identified, we explored how our inhibitors bind in conjunction with EPZ015666. As illustrated in Fig. 10A and B, the best-scoring binding pose for P1608K04 has been depicted in the presence (SAM-bound PRMT5) and absence (Apo-PRMT5) of the methyl donor SAM, respectively. As shown in Fig. 10A, P1608K04 binds to a similar location to that of EPZ015666 in the presence of SAM. Interestingly, in the absence of SAM (Fig. 10B), P1608K04 partially occupies the SAM binding site, thereby potentially blocks the SAM-PRMT5 interaction. Blocking SAM binding could be a potential avenue through which PRMT5 methyltransferase activity is affected. Based on the docking scores listed in Fig. 10E, P1608K04 is more likely to bind in the absence of SAM.
Fig. 10. In silico prediction of P1608K04 and P1618J22 binding to PRMT5. (A)–(D).
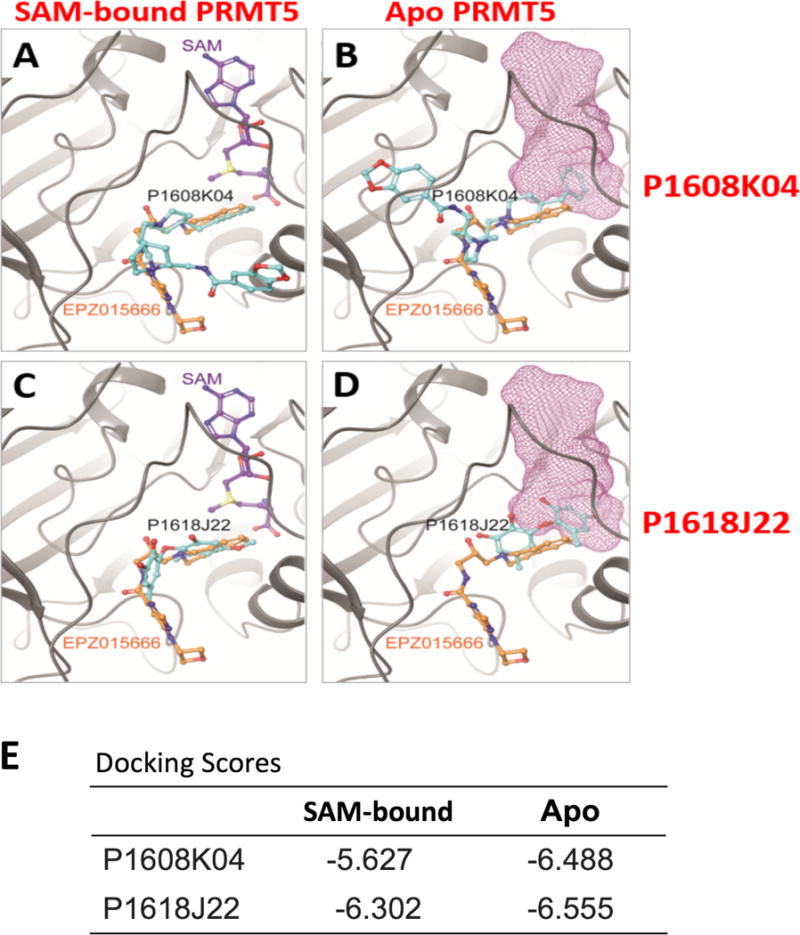
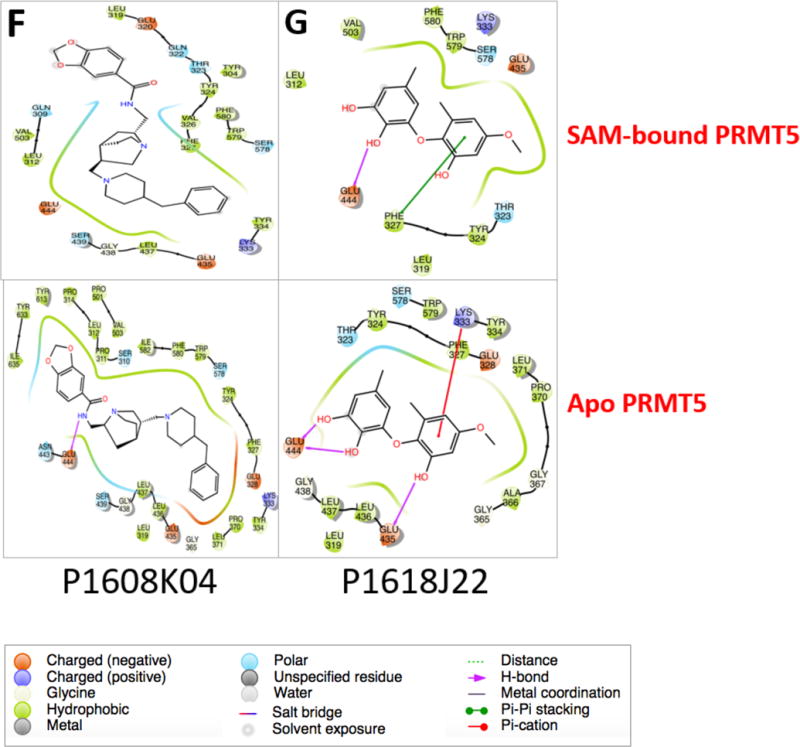
Binding poses predicted for P1608K04 and P1618J22. (A) and (C), in the presence or (B) and (D) in the absence of SAM, respectively. PRMT5 is shown in gray ribbons, while all ligands are shown in balls and sticks and colored with respect to atom type (0 atoms in red, N atoms in blue, S atoms in yellow. C atoms are colored in purple, orange, and cyan in SAM, EPZ015666, and the P1608O04 or P1618J22, respectively.) In panels (B) and (D), the position of SAM is depicted with a purple wireframe surface to indicate if the docked ligand overlaps. (E) Docking scores for the docking poses depicted in (A)-(D). (F) and (G) Ligand affinity maps, depicting the residues of PRMT5 that interact with P1608K04 (F) or P1618J22 (G) in the docking poses in (A)–(D).
In the case of P1618J22, we also observed a similar binding pose to that of EPZ015666 in the presence of SAM, but a preferential binding to the SAM binding site in the absence of SAM (Fig. 10C and D). However, for P1618J22, binding affinities are very close in the presence and absence of SAM, thus it is impossible to predict which pose the ligand would prefer solely based on structural docking studies.
The ligand affinity maps in Fig. 10F and G illustrate the PRMT5 residues that can potentially interact with P1608K04 (Left panels) or P1618J22 (Right panels), in the presence (upper panels) or absence (lower panels) of SAM, respectively. Key residues unique to the PRMT5 protein, such as Glutamine (E) 444 residue that is part of the catalytic cleft of PRMT5 and Phenylalanine (F) 327 critical for PRMT5 product specificity8 are involved in the interactions with these inhibitors as well. The interactions also differ from EPZ015666,8 possibly leading to the different efficacy between them. In the future, it will be interesting to explore with the site mutagenesis approach in determining how important these residues and others (Fig. 10F and G) are for the binding mechanism of these inhibitors to PRMT5.
Collectively, our data support the hypothetical model (Fig. 11) that inhibitors of PRMT5, such as P1608K04 and P1618J22, bind to PRMT5, inhibit PRMT5-mediated NF-κB activation and its downstream target gene expression, therefore, resulting in the alleviation of cancer phenotype in PDAC and CRC.
Fig. 11. Hypothetical model.
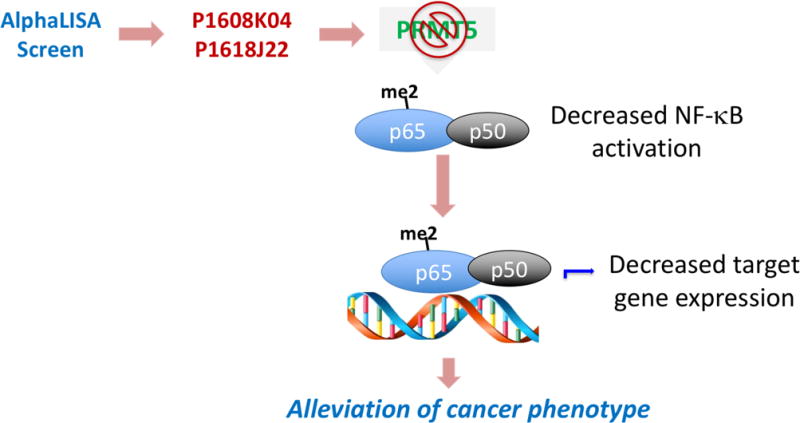
Optimized AlphaLISA screen for PRMT5 inhibitors identified and confirmed P1608K04 and P1618J22 as two top hits. This inhibitory effect could partly be via inhibiting PRMT5-mediated NF-κB methylation and subsequent activation, resulting in decreased cancer associated target gene expression. This process would ultimately lead to the alleviation of cancer phenotype.
4. Discussion
Epigenetic modifications play an important role in normal cellular function and development in nearly every aspect of biology, making it one of the most important fields in scientific research. Dysregulation of epigenetic modifications leads to serious disruptions of regular functioning in humans and is the underlying cause of promoting a wide range of disorders including cancers. A plethora of studies have linked overexpression of epigenetic enzymes with promotion and metastasis of varied cancers. Thus, a great deal of interest has been generated in the field to exploit these epigenetic enzymes as potential therapeutic targets. To date, successful attempts at developing inhibitors for epigenetic targets have been made, with the histone deacetylases (HDAC) family being the most prominent example. Vorinostat and romidepsin are HDAC inhibitors that are already FDA approved, and many others are currently under clinical trials.16 It is imperative to also explore other critical epigenetic enzyme families like PRMTs by developing HTS screens and identifying potential inhibitors of clinical significance.
Various assays for HTS that have been used in recent times were considered to screen for PRMT family member inhibitors. Among the current available technologies, AlphaLISA is proved to be an excellent choice for this purpose for various reasons. For instance, in the case of radiometric assays, the cost and danger associated with its usage and the generation of radioactive waste are a huge deterrent. Enzyme-based assays like ELISA have been quite popular for screening in the past. However, with these assays, the scaling-up approach can prove to be quite expensive and cumbersome. As the HTS field grows, there is a dire need to develop economical and scalable approaches to screen for active compounds against potential targets. AlphaLISA is a viable option because it provides a straightforward protocol for assay development, and its robustness, sensitivity, cost-effectiveness, and ease of use. We conducted extensive troubleshooting and adapted all the loading volumes and concentrations to be compatible with a robotic system. A big limitation of the non-robotic approach is the cumbersome and expensive nature of the study, with addition of 10,000 compounds and reagents involved as part of the screening protocol. The ease of automation provided by our technique will help to drive high scale screening studies. Additionally, the robotic system allows for higher accuracy with its state-of-the-art pipetting system as well as cost efficiency as it enables the use of very low volumes of reagents. If using the appropriate biotinylated substrate, methyl donor, epigenetic-tag specific Acceptor beads, and Streptavidin-coated Donor beads, this assay can be customized for other epigenetic targets as well (Fig. 1).
As a prominent member in its superfamily, PRMT5 was chosen as our representative enzyme in this screen system. As described in Figs. 2–4, we employed a multi-step approach to identify top hits from small compound libraries. We then optimized the assay and tested its robustness using the reliable Z’ test, which was determined to be 0.7. Upon completion of the screen of libraries containing natural products and biologically active compounds, we then sorted through the top hits. Our results indicated that P1608K04 and P1618J22 are the most effective PRMT5 inhibitors from our screen. Both compounds demonstrated considerable efficacy in concentration-dependent AlphaLISA assays (Fig. 6), and in MTT assays in PDAC and CRC cells (Fig. 7), respectively.
The identification of novel PRMT5 inhibitors is of great importance. Several years ago, our lab demonstrated that PRMT5 symmetrically dimethylates NF-κB to trigger its activation.7 Since NF-κB is a pivotal culprit in PDAC and CRC,8 inhibition of PRMT5-mediated NF-κB activity can ultimately lead to the impediment of its tumorigenic properties in PDAC and CRC.
In this study, we confirmed the significant inhibitory effect of P1608K04 and P1618J22 on PRMT5-mediated NF-κB methylation, activation, and its downstream target gene expression (Figs. 8 and 9). Overall, our findings support the specific activity of these inhibitors against PRMT5.
To understand how P1608K04 and P1618J22 may bind to PRMT5, we delved deeper into the mechanism of the inhibitor-enzyme interaction using structural docking analyses. As shown in Fig. 10, we identified specific residues in the binding interaction between these two compounds and PRMT5. This analysis suggested that both inhibitors can potentially inhibit PRMT5 through certain similar but also varied mechanisms. For instance, both seem to bind to residues that intersect with the SAM binding site in the Apo states, suggesting that one possible way these inhibitors work is by interfering with SAM binding to its consensus site on PRMT5 structure. Ligand affinity map analysis (Fig. 10F and G) suggests that both P1608K04 and P1618J22 interact with two key residues (E444 and F327) that are unique to the PRMT5 structure,8 also alluding to their specificity. E444 is a key residue in the catalytic cleft of PRMT5, while F327 interacts with the substrate and plays an important role in determining product specificity of PRMT5.17 Both P1608K04 and P168J22 could contribute to PRMT5 inhibition via the interaction with these two key residues. This binding poses of P1608K04 and P168J22 are also different from that of EPZ015666, suggesting that there is a distinction between our compounds and EPZ015666, and could possibly contribute to the different efficacies in inhibiting PRMT5 (Figs. 10A–D). In the future, mutating some of these residues can help us locate critical sites on PRMT5 for the inhibitor-enzyme interactions and elucidate the mechanism of action of P1608K04 and P1618J22 for PRMT5 inhibition. Furthermore, several unique residues to each type of interaction in the ligand affinity maps (Fig. 10F and G) can be further pursued and analyzed in details. Additionally, it would be of great interest to employ structure activity relationship (SAR) analyses to design derivatives of the top hits. This would assist in maximizing the efficacy of these compounds as would also validate anti-tumor efficacy in in vivo mouse cancer xenograft models. From a broader perspective, epigenetic enzymes are proving to be increasingly critical in a wide variety of diseases, and the approach described here can accelerate the development of important tools required for this purpose.
5. Conclusion
In summary, our study allows for rapid optimization of a robot-based automated AlphaLISA protocol that can be modified for other epigenetic targets of interest. Here, we described PRMT5 as a representative target and adapted the AlphaLISA screen to build a HTS screen platform to identify PRMT5 inhibitors. The adaptability and high throughput nature of this approach can be exploited to pursue a screening protocol for any enzymatic target of interest. Importantly, the two compounds that we identified, P1608K04 and P1618J22, are novel PRMT5 inhibitors that exhibited much higher efficacy than the commercial PRMT5 inhibitor EPZ015666 in PDAC and CRC cells. These exciting studies can lead to the development of more effective therapeutic options targeting PRMT5 in PDAC and CRC. In a broader sense, the adapted AlphaLISA HTS approach we reported here may serve as a powerful tool for studying a wide variety of critical targets in cancers and other diseases.
Supplementary Material
Acknowledgments
AlphaLISA technology is a registered trademark of the PerkinElmer Corporation. We would like to thank Ms. Andrea Gunawan from Chemical Genomics Core at Indiana University School of Medicine for her technical assistance with Z’ test and HTS. We would like to thank Ms. Lisa King from the Department of Pharmacology and Toxicology at Indiana University School of Medicine for helping to edit this manuscript. The research is supported by grants from Indiana Center for Technology and Science Innovation (CTSI) and Indiana Drug Discovery Alliance (IDDA) (Grants 2286230 and 2286233 to TL), Indiana University Pancreatic Cancer Signature Center (Grant 4189911 MUR1 to TL), V foundation Kay Yow Cancer Fund (Grant 4486242 to TL), NIH-NIGMS Grant (# 1R01GM120156-01A1 to TL), NIH-NCI Grant (# 1 R03 CA223906-01 to TL), and 100 VOH Grant (# 2987613 to TL). This work was also supported by NIH-NIGMS Grant (#P41-GM103426 and DP2OD007237 to REA), and the NSF Grant (#CHE060073N to REA).
Footnotes
Disclosure statement
The use of P1608K04 and P1618J22 in cancer treatment was filed as US Provisional Patent application 62/534,969 on July 20, 2017.
References
- 1.Bedford MT, Clarke SG. Mol Cell. 2009;33:1–13. doi: 10.1016/j.molcel.2008.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zurita-Lopez CI, Sandberg T, Kelly R, Clarke SG. J Biol Chem. 2012;287:7859–7870. doi: 10.1074/jbc.M111.336271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wei H, Mundade R, Lange KC, Lu T. Cell Cycle. 2014;13:32–41. doi: 10.4161/cc.27353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Oh TG, Bailey P, Dray E, Smith AG, Goode J, Eriksson N, Funder JW, Fuller PJ, Simpson ER, Tilley WD, Leedman PJ, Clarke CL, Grimmond S, Dowhan DH, Muscat GE. Mol Endocrinol. 2014;28:1166–1185. doi: 10.1210/me.2013-1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gao Y, Zhao Y, Zhang J, Lu Y, Liu X, Geng P, Huang B, Zhang Y, Lu J. Sci Rep. 2016;6:19874. doi: 10.1038/srep19874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yoshimatsu M, Toyokawa G, Hayami S, Unoki M, Tsunoda T, Field HI, Kelly JD, Neal DE, Maehara Y, Ponder BA, Nakamura Y, Hamamoto R. Int J Cancer. 2011;128:562–573. doi: 10.1002/ijc.25366. [DOI] [PubMed] [Google Scholar]
- 7.Wei H, Wang B, Miyagi M, She Y, Gopalan B, Huang DB, Ghosh G, Stark GR, Lu T. Proc Natl Acad Sci U S A. 2013;110:13516–13521. doi: 10.1073/pnas.1311784110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Prabhu L, Wei H, Chen L, Demir Ö, Sandusky G, Sun E, Wang J, Mo J, Zeng L, Fishel M, Safa A, Amaro R, Korc M, Zhang ZY, Lu T. Oncotarget. 2017;8:39963–39977. doi: 10.18632/oncotarget.18102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang L, Pal S, Sif S. Mol Cell Biol. 2008;28:6262–6277. doi: 10.1128/MCB.00923-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chan-Penebre E, Kuplast KG, Majer CR, Boriack-Sjodin PA, Wigle TJ, Johnston LD, Rioux N, Munchhof MJ, Jin L, Jacques SL, West KA, Lingaraj T, Stickland K, Ribich SA, Raimondi A, Scott MP, Waters NJ, Pollock RM, Smith JJ, Barbash O, Pappalardi M, Ho TF, Nurse K, Oza KP, Gallagher KT, Kruger R, Moyer MP, Copeland RA, Chesworth R, Duncan KW. Nat Chem Biol. 2015;11:432–437. doi: 10.1038/nchembio.1810. [DOI] [PubMed] [Google Scholar]; Quinn AM, Allali-Hassani A, Vedadi M, Simeonov A. Mol Biosyst. 2010;6:782–788. doi: 10.1039/b921912a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Halgren TA, Murphy RB, Friesner RA, Beard HS, Frye LL, Pollard WT, Banks JL. J Med Chem. 2004;47:1750–1759. doi: 10.1021/jm030644s. [DOI] [PubMed] [Google Scholar]
- 12.Sastry GM, Adzhigirey M, Day T, Annabhimoju R, Sherman W. J Comput Aided Mol Des. 2013;27:221–234. doi: 10.1007/s10822-013-9644-8. [DOI] [PubMed] [Google Scholar]
- 13.Small-Molecule Drug Discovery Suite 2017-2: Glide, version 7.5. Schrödinger, LLC; New York, NY: 2017. [Google Scholar]
- 14.Schrödinger Release 2017-2: Maestro, version 11.2. Schrödinger, LLC; New York, NY: 2017. [Google Scholar]
- 15.Zhang JH, Chung TD, Oldenburg KR. J Biomol Screen. 1999;4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- 16.Yu XR, Tang Y, Wang WJ, Ji S, Ma S, Zhong L, Zhang CH, Yang J, Wu XA, Fu ZY, Li LL, Yang SY. Bioorg Med Chem Lett. 2015;25:5449–5453. doi: 10.1016/j.bmcl.2015.06.095. [DOI] [PubMed] [Google Scholar]
- 17.Schapira M, Ferreira de Freitasa R. Medchemcomm. 2014;5:1779–1788. doi: 10.1039/c4md00269e. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


