Abstract
Lentiviral vectors pseudotyped with the baculovirus envelope protein GP64 transduce primary cultures of human airway epithelia (HAE) at their apical surface. Our goal in this study was to harness a directed evolution approach to develop a novel envelope glycoprotein with increased transduction properties for HAE. Using error-prone PCR, a library of GP64 mutants was generated and used to prepare a diverse pool of lentiviral virions pseudotyped with GP64 variants. The library was serially passaged on HAE and three GP64 mutations were recovered. Single-, double- and the triple-combination mutant envelope glycoproteins were compared with wild-type GP64 for their ability to transduce HAE. Our results suggest that lentiviral vectors pseudotyped with evolved GP64 transduced HAE with greater efficiency than wild-type GP64. This effect was not observed in primary cultures of porcine airway epithelial cells, suggesting that the directed evolution protocol was species specific. In summary, our studies indicate that serial passage of a GP64 mutant library yielded specific variants with improved HAE cell tropism, yielding tools with the potential to improve the success of gene therapy for airway diseases.
Introduction
Results from human clinical trials using lentiviral-based vectors are encouraging, and the feasibility of gene therapy for monogenetic diseases is now established.1 Recent promising examples using lentivirus include Wiskott–Aldrich Syndrome,2,3 metachromatic leukodystrophy,4,5 acute lymphoid leukemia6 and multiple primary immunodeficiencies.7 Despite these successes, gene transfer to somatic cells of the airways presents unique challenges. Novel lentiviral vector designs that enhance transduction of airway epithelial cells would improve the chances of a successful gene therapy clinical trial for airway diseases.
We developed gene transfer vectors derived from a nonprimate lentivirus, feline immunodeficiency virus (FIV).8,9 In addition, we screened a wide variety of envelope glycoproteins to determine those with optimal tropism for the airways.10–12 We discovered that the envelope glycoprotein GP64 from baculovirus Autographa californica multinucleocapsid nucleopolyhedrosis virus conferred apical entry into well-differentiated air–liquid interface primary cultures of human airway epithelial cells (HAE). In addition, we observed abundant expression in mouse, ferret and porcine airways in vivo.10,13–16
Neither lentivirus nor baculovirus naturally evolved to enter its host through the airways and, indeed, the natural hosts of A. californica multinucleocapsid nucleopolyhedrosis virus are insects. Similarly, viruses did not evolve to deliver therapeutic genes. In truth, all viral vectors have room for improvement, including their transduction efficiency, cellular tropism, species tropism and vector titer/stability. Directed evolution is an ‘algorithm’ that can be applied to natural viruses or viral proteins to fine-tune their functions for specific cell or tissue applications.
Multiple directed evolution approaches to improve viral vectors have been reported,17,18 and each begins with the generation of large genetic variant libraries followed by selection for enhanced fitness. Our group previously used directed evolution strategies to address challenges with adeno-associated virus and lentivirus vectors.19–24 For example, we evolved AAV libraries with random point mutations, random chimeras of parent AAV serotypes and random peptide library inserted into a defined location of the viral attachment protein.25–28 In addition, we used peptide insertion and error-prone PCR libraries of vesicular stomatitis virus-G.24 We successfully engineered enhanced variants of this envelope protein for lentiviral vector pseudotyping, and the resulting variants exhibited improved resistance to serum resistance.29 This established proof of concept for engineering envelope variants for targeted delivery to specific cells.
In the present study, we implement an analogous approach to modify the baculoviral envelope protein GP64 with a goal of improving the efficiency of lentiviral vector transduction of the airway epithelium. Greater GP64 transduction efficiencies can likely be achieved because the natural evolution of wild-type baculovirus occurs in insect cells rather than mammalian cells. Specifically, we used directed evolution involving a library of GP64 envelope mutants and repeated virus transduction of HAE. The resulting evolved GP64 glycoproteins exhibited improved transduction properties for HAE as compared with wild-type GP64 pseudotyped lentiviral vector.
Results and Discussion
Typically, FIV-based vectors are produced using a standard three-plasmid transfection method.9 For the directed evolution experiments, a two-plasmid transfection was substituted because the transgene plasmid also supplies the GP64 envelope glycoprotein expression cassette (shown schematically in Figure 1). Error-prone PCR (Figure 1, red arrows) was used to generate a library of ∼ 108 GP64 mutants as previously described.22,23 The mutagenesis was focused on amino acids located in the putative GP64 receptor-binding domain that encompasses residues 21 through 294, in which residues 140 to 161 are believed to comprise the core receptor-binding pocket.30 The resulting plasmid pool was then packaged to prepare a viral vector library in which each virion was composed of a GP64 variant presented from the viral envelope and a RNA genome encoding that GP64 variant. We hypothesized that serial passage of a library of GP64 mutants on human airway cells would enable screening and discovery of new GP64 proteins with enhanced cellular transduction properties.
Figure 1.
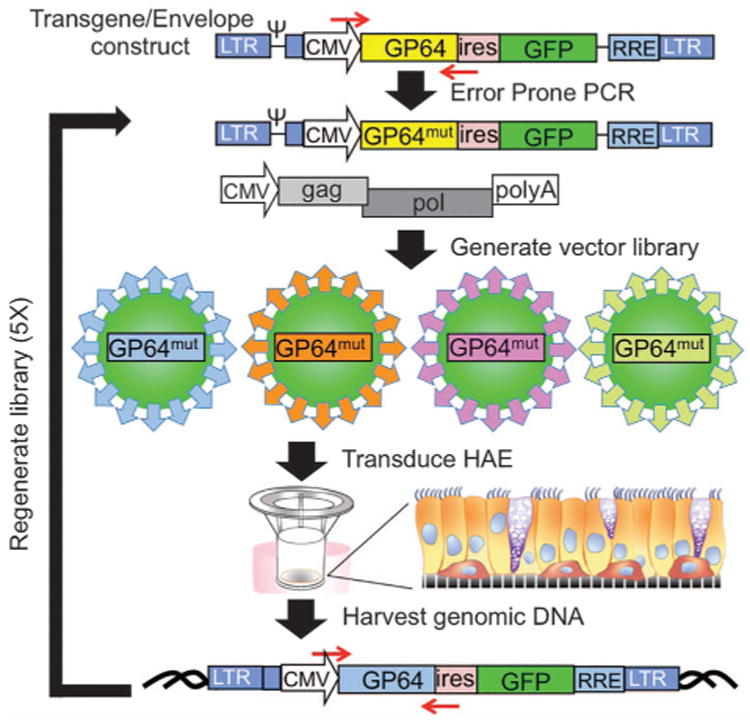
Schematic representation of deriving mutant GP64 libraries. FIV lentiviral vector with a GP64 transgene was subjected to error-prone PCR to generate a mutant GP64 library. Vector libraries were generated by two-plasmid transfection: lentiviral backbone with mutant GP64 cassette and gag/pol construct. Resultant viral particles were applied apically to HAE overnight, and genomic DNA was harvested 4 days post transduction. GP64 sequences were recovered by PCR amplification and cloning into a TA vector. This process was repeated four additional times.
As shown (Figure 1), the library was used to transduce the apical surface of well-differentiated air–liquid interface cultures of HAE. After 4 days, genomic DNA was harvested. GP64 was amplified by PCR and used to generate a new plasmid library. The plasmid library was again packaged to generate a FIV library by two-plasmid transfection. This process was repeated for a total of five passages through HAE. After each passage, clones from the libraries were sequenced to assess convergence of the variant pool. Convergence was not observed until the fifth passage, at which time three single-point mutations or combinations of those mutations were identified (Table 1). G165D was the most commonly recovered mutation. Vector titers among the mutants were slightly reduced from wild-type GP64 (5.6× 108 transducing units (TU) ml−1) and ranged from 0.9 ×108 to 4.1×108 TU ml−1.
Table 1. Summary of GP64 mutations.
| Appearance frequency | Titer (TU ml−1) | |
|---|---|---|
| Wild type | 5.6 ×108 | |
| Single | ||
| E45K | 2 | 1.2 × 108 |
| G165D | 14 | 1.1×108 |
| T259A | 1 | 1.8 × 108 |
| Double | ||
| E45K/G165D | 2 | 1.9 × 108 |
| G165D/T259A | 1 | 0.9 × 108 |
| E45K/T259A | 0 | 1.1 ×108 |
| Triple | ||
| E45K/G165D/T259A | 0 | 4.1 ×108 |
| RR-RR | 1.1 ×108 | |
Three single mutations and two double mutations in GP64 were discovered after library generation. Appearance frequency indicates the number of sequences in which the mutations were observed. Titers (transducing units (TU) ml−1) were an average of three independent viral preparations. All envelope expression plasmids were driven by the CMV promoter.
Site-directed mutagenesis was used to introduce each of the three single mutations (E45K, G165D and T259A) into GP64 expression plasmids that were used to generate FIV-based vectors expressing Gaussia luciferase using a standard three-plasmid production protocol.9,31 The resulting vectors were applied to the apical surface of HAE, and Gaussia luciferase expression was quantified in apical washings 4 days later. Unexpectedly, none of the three single GP64 mutants resulted in increased reporter gene production over wild-type GP64 vector (Figure 2a). In fact, E45K and G165D resulted in significantly reduced transduction.
Figure 2.
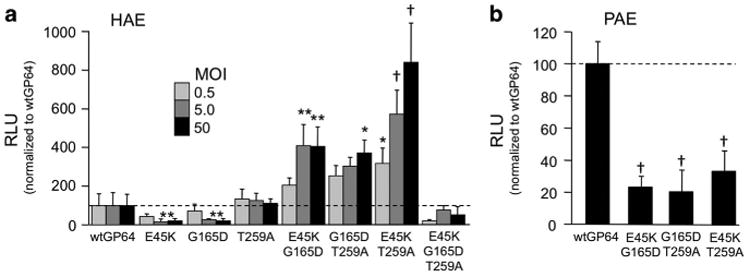
Double mutant GP64 vectors exhibit dose-dependent entry. wtGP64-FIV-gaussia luciferase (gLuc), single mutant GP64-FIV-gLuc and all mutant combinations were individually applied to the apical side of HAE overnight at a multiplicity of infection (MOI) of 0.5, 5 and 50 (n =9). Gaussia luciferase was measured 4 days post transduction as relative light units (RLUs). Expression for each MOI was normalized to the respective wild-type GP64 control (a). wtGP64 and GP64 double mutants were applied to the apical side of pig airway epithelia (PAE) overnight at an MOI of 50 (n = 9). At 4 days post transduction, Gaussia luciferase expression was measured as RLU and normalized to wtGP64 (b). *P<0.05, **P<0.01, †P<0.001.
Using the same delivery protocol, we next tested combinatorial mutations. The double mutants E45K/G165D, G165D/T259A and E45K/T259A all resulted in transduction efficiencies that outperformed wild-type GP64 (Figure 2a). The highest Gaussia luciferase expression in HAE was observed with FIV pseudotyped with the E45K/T259A mutant GP64 that reached levels ∼8 times that of wild-type GP64 at a multiplicity of infection of 50. However, the triple mutant E45K/G165K/T259A resulted in reduced trans-gene expression. Interestingly, when these mutant envelopes were applied to well-differentiated primary cultures of porcine airway epithelia, reduced reporter gene expression was observed as compared with wild-type GP64 (Figure 2b). Similarly, we compared transduction efficiency in the nasal epithelia of mice and observed reduced reporter gene expression as compared with wild-type GP64 (Supplementary Figure 1). These results suggest that directed evolution by serial passage through human cells yielded enrichment of species-specific mutations.
Recently, O'Flynn et al.32 reported strategies to modify a basic loop contained within GP64 residues 268–281 to increase entry efficiency into different cell types. They hypothesized that increasing the number of charged residues around the acidic tripeptide VEH in the center of the basic loop would enhance fusion activity, and insertion of four arginines (271_272insRR, 274_275insRR) was shown to enhance transduction of three different mammalian cell lines.32 We used this mutant GP64 (referred to as RR-RR) to pseudotype FIV-based vector, and its titer was similar to wild-type GP64 and the evolved GP64 mutants (Table 1). FIV pseudotyped with wild-type GP64 or the RR-RR mutant GP64, both encoding Gaussia luciferase, was applied to the apical surface of HAE. We observed an approximately two-fold increase in reporter gene expression using the RR-RR vector; however, the trend did not reach statistical significance (Figure 3a). This level of enhancement is consistent with the previously reported results in other cell types.32 Interestingly, in porcine airway epithelia no increase in reporter gene expression, as compared with wild-type GP64, was observed (Figure 3b). A future direction will be to compare the RR-RR mutations in combination with the evolution-derived mutations.
Figure 3.
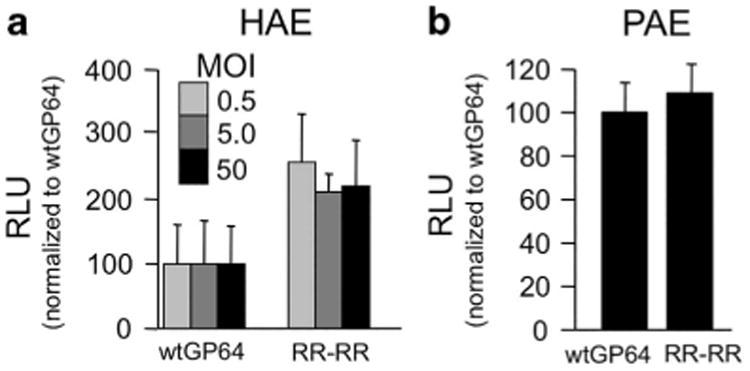
RR-RR GP64 mutant enhances entry into HAE. wtGP64-FIV-gaussia luciferase (gLuc) and RR-RR GP64-FIV-gLuc were applied to the apical surface of polarized HAE at multiplicity of infection (MOI) of 0.5, 5 and 50 (n = 9) (a) and pig airway epithelia at an MOI of 50 (n =9) (b) overnight. Apical washes were collected 4 days post transduction, and relative light unit (RLU) intensity of Gaussia luciferase was measured and normalized to wtGP64.
Using the published crystal structure of post-fusion GP64,33 we mapped the location of the evolved mutations (Figure 4), and each is surface exposed. E45K and T259A reside in close proximity in three dimensions. Mutation of the negatively charged Glu-45 (top inset; blue sticks) to positively charged Lys (magenta sticks) (E45K) likely disrupts the hydrogen bond interaction network that involves Lys-317 and Lys-44 (Figure 4, top inset), thereby potentially facilitating interactions between these basic residues and the negatively charged cell surface. Thr-259 lies in the vicinity of the disordered loop (residues 272–287) within the crystal structure of the closed form of GP64, but the effect of the Thr → Ala mutation (T259A) on the structure, stability or loop dynamics is not discernable from this single static model (Figure 4, middle inset). Finally, Gly-165 lies in the middle of a β-sheet above a disulfide bond, and its mutation to the much larger Asp likely clashes with the neighboring Tyr-68 (G165D). This would be expected to significantly affect the structure and stability of this region (Figure 4, bottom inset). The G165D mutation appeared 14 times in the sequences acquired from the GP64 library generation, more than E45K or T259A (Table 1). Based on this frequency, we anticipated that this mutation would enhance cell entry better than the others. In particular, residues 21–159 comprise a putative receptor-binding domain,34 and thus the proximity of residue 165 may be important for cell entry. However, the addition of a bulky aspartic acid at 165 seems to disrupt the stability of GP64, potentially preventing critical residues from interacting with the cell membrane and thus not increasing efficiency. Yet based on functional experimentation, we show that when G165D is combined with either E45K or T259A, transduction of HAE was substantially increased.
Figure 4.
Location of the mutations affecting transduction in the baculovirus fusion protein GP64. Cartoon representation of the post-fusion trimeric GP64 protein based off a crystal structure (PDB ID 3DUZ), with each chain of the trimer colored from blue (N terminus) to red (C terminus). The disordered loop (residues 272–287) missing in the crystal structure was modeled using Yasara. Mutations affecting transduction (Glu-45, Thr-259 and Gly-165) are highlighted in the full-length protein in magenta on the left and in zoomed-in insets on the right. The cartoon coloring scheme for the insets is the same as in the full view on the left. Wild-type residues Glu-45 (blue sticks), Thr-259 (cyan sticks) and Gly-165 (green spheres) were respectively mutated in silico to Lys-45 (magenta sticks), Ala-259 (magenta sticks) and Asp-165 (magenta spheres) to determine local effects of the mutations on the structure. Residues within 5 Å of the mutations are shown as white sticks.
Materials and Methods
Library generation of mutant GP64
Error-prone PCR protocol was used to amplify the GP64 region encoding amino acids 21–294, using primers (5′-AGTGAA CCGTCAGATCCGCTAGCATG-3′ and 5′-GCATCAGGTCGCCTTTGGTG GC-3) that flank that region. Specifically, 25 cycles of Taq polymerase-based amplification was performed in the presence of 5 mM MnCl2, and elevated levels of MgCl2 and unequal quantities of deoxynucleotides produce error-prone products35 with the goal of introducing 1–3 amino acid changes per product. Four independent amplifications were performed and pooled to provide greater heterogeneity.36 The C-terminal region (295–512) of GP64 was amplified using primers (5′-GCCACCAAA GGCGACCTGATGC-3′ and 5′-AGGGAGAGGGGCGGATCCTAAGC-3′) through normal PCR protocol by Taq polymerase. The complete GP64 cDNA was amplified using primers (5′-AGTGAAC CGTCAGATCCGCTAGCATG-3′ and 5′-AGGGAGAGGGGCGGATCC TAAGC-3′) and the PCR fragments by overlap PCR. The resulting library of GP64 PCR fragments with errors concentrated in the receptor binding domain were digested with the unique restriction enzymes (NheI and HindIII) and ligated into the FIV vector plasmid.9
Vector production
The FIV vector system utilized in this study37,38 expressed Gaussia luciferase in the FIV3.3RSV backbone.8 GP64-pseudotyped FIV vector particles were generated by transient transfection, concentrated 250-fold by centrifugation and titered using real-time PCR as previously described.8 The library of plasmids containing GP64 was used to package FIV vector in the University of Iowa Viral Vector Core. The envelope plasmid was diluted during packaging such that each packaging cell received on average only one envelope plasmid.20,22–24 In this case, a 15 cm plate of 293T cells was transfected with 25 μg of FIV packaging plasmid and 7 ng of the pooled transgene/envelope plasmid library. The supernatant containing the vector particle library was collected 24–48 h later after transfection.
Cell culture
Primary cultures of HAE were prepared from trachea and bronchi by enzymatic dispersion using established methods.39 Briefly, epithelial cells were dissociated and seeded onto collagen-coated, semipermeable membranes with a 0.4 μm (no. 3470, Corning, Lowell, MA, USA) or 3 μm (no. 3472, Corning) pore size (surface area 0.33 cm2). HAE cultures were maintained in Ultroser G media at 37 °C, 5% CO2. Polyester transwell inserts were placed into 24-well plastic cell culture plates (Costar, Cambridge, MA, USA). At 24 h after seeding, the mucosal medium was removed and the cells were allowed to grow at the air-liquid interface as reported previously.39 Only well-differentiated cultures (>3 weeks old) were used in these studies. We note that choice of insert was important because substantially reduced MeV infection was observed through alternative membrane materials (no. 3413, Corning). The presence of tight junctions was confirmed by measuring the transepithelial resistance using a volt-ohm meter (World Precision Instruments, Sarasota, FL, USA; resistance > 500 Ω•cm2).
Serial passaging vector in vitro
Virions were transduced into HAE cell cultures at low multiplicity of infection (∼0.005). A low multiplicity of infection helps to ensure that transduced epithelial cells contain a single virion. After 4 days, the cell cultures were lysed, genomic DNA was purified using QIAamp genomic DNA kit from Qiagen (Hilden, Germany) and the purified genomic DNA was digested by XmaI. PCR using primers (5′-AGA TCCGCTAGCATGGTAAGCGCTATTGTTTTATATGTGCTTTTGG-3′ and 5′-GATCCTAAGCTTTTAATATTGTCTATTACGGTTTCTAATCATACAGTAC-3′) and the digested genomic DNA as a template was used to recover the mutant GP64 clones. These PCR fragments were recloned and repackaged as described above, and the resulting vector was again used to infect epithelial cell cultures. We repeated the transduction process for four additional selection steps. After the five selection steps, the resulting PCR fragments were cloned into a TA vector, and ∼100 clones were sequenced. Evolved GP64 sequences were determined, and the consensus sequences were cloned using site-directed mutagenesis. Evolved clones were compared with wild-type GP64 for its ability to transduce primary cultures of HAE cells.
Protein modeling
YASARA Structure (version 16.7.22; www.yasara.org) was used to build and optimize the missing loop (residues 272–287) in the post-fusion monomer form of the GP64 crystal structure (Protein Data Bank (PDB) ID 3DUZ). PyMOL (Version 1.8; Schrödinger, LLC, New York, NY, USA) was used to generate the GP64 trimer, generate in silico mutations based on the most prevalent rotamer in the PDB and for preparing all the structural figures.
Delivery to mouse nasal epithelia and bioluminescence imaging This study was approved by the institutional animal care and use committee at the University of Iowa. Female 6–8-week-old Balb/c mice (National Cancer Institute (NCI) mouse repository) were used in this study. No randomization was used. The exact sample size (n) for each experimental group is indicated in Supplementary Figure 1. Sample sizes were chosen based on prior experience with similar studies. Mice were first anesthetized by intraperitoneal injection of ketamine/xylazine (87.5+2.5 mg kg−1). Approximately 1×107 TU of FIV vector formulated with 1% methylcellulose (1:1) in a total volume of 50 μl was delivered to the nasal epithelia via direct instillation. At 1 week following vector delivery, mice were injected intraperitoneally with 200 μl of D-luciferin (15 mg ml−1 in 1× phosphate-buffered saline; Xenogen, Alameda, CA, USA) using a 25-gauge needle. After 5 min, mice were imaged using the Xenogen IVIS charge-coupled device camera while under 1–3% isoflurane anesthetization. Imaging data were analyzed unblinded and signal intensity was quantified using Xenogen Living Image software. The negative controls in all in vivo bioluminescent assays were naive strain-matched mice. We included negative controls in every assay and report only background subtracted levels of experimental groups.
Statistics
All numerical data are presented as the mean ± s.e. Statistical analyses were performed using GraphPad Prism software (Graph-Pad Software, Inc., La Jolla, CA, USA) and two-tailed Student's t-test with unequal variance. F-tests were performed in parallel to determine that variance was similar between groups. For nonparametric data, Mann–Whitney U-test was used.
Conclusions
For cystic fibrosis gene therapy, our goal is to develop an efficient airway delivery vector for cystic fibrosis transmembrane conductance regulator (CFTR). Such a gene replacement approach would be efficacious regardless of underlying CFTR mutation, and if a population of progenitor cells is targeted, it offers the possibility of a single-dose, life-long curative therapeutic strategy for a devastating disease. Current lentiviral vector technology has made considerable progress toward this goal, but improvements are needed. Future studies will determine whether CFTR delivered by FIV pseudotyped with the E45K/T259A GP64 double mutant confers improved anion transport properties in HAE derived from cystic fibrosis patients.
In this study, we improved the gene transfer efficiency of the GP64 envelope glycoprotein through directed evolution. We performed serial passage on cultured primary HAE to evolve an airway-adapted viral vector, and sequencing of the evolved GP64 coding region revealed three-point mutations. Individually the mutations were detrimental, but combinatorial mutations showed enhanced transduction of airway epithelia. The dose-dependent effect translated to a two–eight-fold increase in transduction efficiency, significantly greater than wild-type GP64 in HAE. The E45K/T259A double mutant outperformed the single mutants, E45K/G165D, G165D/T259A, and the triple mutant. E45K and T259A are located in close proximity, and are both surface exposed. Inexplicably, the E45K and T259A single mutants were recovered in our library sequencing but the double mutant was not; thus, inclusion of this comparison was a serendipitous choice. The enhanced entry of the E45K/T259A GP64 mutant could be due to enhanced accessibility to interact laterally with known host cell factors, such as phospholipids, before fusion with the cell membrane.
Interestingly, multiple combinatorial GP64 mutants improved transduction in human cultures but not in pig airway epithelia. This observation supports our central hypothesis that directed evolution is a strategy to improve gene transfer efficiency in a species or cell type for which the virus did not naturally evolve, and different mutations would likely arise if the libraries were serially passaged through pig primary epithelia. Determining the mechanism of improved gene transfer, such as binding assays and integration analyses, will require further study. Ultimately, our goal is to incorporate these vector design modifications into a clinical therapeutic for cystic fibrosis gene therapy.
Supplementary Material
Acknowledgments
We thank Kimberly Bekas and Mayumi Oakland for their technical assistance. Lokesh Gakhar of the Protein Crystallography Core Facility assisted with the protein modeling. The In Vitro Models and Cell Culture Core provided the primary cell cultures. The Viral Vector Core produced and titered the viral vector. This work was supported by the National Institutes of Health: P01 HL-51670, P01 HL-091842 and R01 HL-105821, and the Center for Gene Therapy of Cystic Fibrosis: P30 DK-54759.
Footnotes
Conflict of Interest: PBM is a founder of and holds equity in Talee Bio. DVS is a co-inventor and has a patent related to directed evolution of lentiviral vectors. The other authors declare no conflict of interest.
References
- 1.McGarrity GJ, Hoyah G, Winemiller A, Andre K, Stein D, Blick G, et al. Patient monitoring and follow-up in lentiviral clinical trials. J Gene Med. 2013;15:78–82. doi: 10.1002/jgm.2691. [DOI] [PubMed] [Google Scholar]
- 2.Aiuti A, Bacchetta R, Seger R, Villa A, Cavazzana-Calvo M. Gene therapy for primary immunodeficiencies: Part 2. Curr Opin Immunol. 2012;24:585–591. doi: 10.1016/j.coi.2012.07.012. [DOI] [PubMed] [Google Scholar]
- 3.Aiuti A, Biasco L, Scaramuzza S, Ferrua F, Cicalese MP, Baricordi C, et al. Lentiviral hematopoietic stem cell gene therapy in patients with Wiskott-Aldrich syndrome. Science. 2013;341:1233151. doi: 10.1126/science.1233151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cartier N, Hacein-Bey-Abina S, Bartholomae CC, Veres G, Schmidt M, Kutschera I, et al. Hematopoietic stem cell gene therapy with a lentiviral vector in X-linked adrenoleukodystrophy. Science. 2009;326:818–823. doi: 10.1126/science.1171242. [DOI] [PubMed] [Google Scholar]
- 5.Biffi A, Montini E, Lorioli L, Cesani M, Fumagalli F, Plati T, et al. Lentiviral hema-topoietic stem cell gene therapy benefits metachromatic leukodystrophy. Science. 2013;341:1233158. doi: 10.1126/science.1233158. [DOI] [PubMed] [Google Scholar]
- 6.Grupp SA, Kalos M, Barrett D, Aplenc R, Porter DL, Rheingold SR, et al. Chimeric antigen receptor-modified T cells for acute lymphoid leukemia. N Engl J Med. 2013;368:1509–1518. doi: 10.1056/NEJMoa1215134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Farinelli G, Capo V, Scaramuzza S, Aiuti A. Lentiviral vectors for the treatment of primary immunodeficiencies. J Inherit Metab Dis. 2014;37:525–533. doi: 10.1007/s10545-014-9690-y. [DOI] [PubMed] [Google Scholar]
- 8.Sinn PL, Goreham-Voss JD, Arias AC, Hickey MA, Maury W, Chikkanna-Gowda CP, et al. Enhanced gene expression conferred by stepwise modification of a non-primate lentiviral vector. Hum Gen Ther. 2007;18:1244–1252. doi: 10.1089/hum.2006.127. [DOI] [PubMed] [Google Scholar]
- 9.Johnston JC, Gasmi M, Lim LE, Elder JH, Yee JK, Jolly DJ, et al. Minimum requirements for efficient transduction of dividing and nondividing cells by feline immunodeficiency virus vectors. J Virol. 1999;73:4991–5000. doi: 10.1128/jvi.73.6.4991-5000.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sinn PL, Burnight ER, Hickey MA, Blissard GW, McCray PB., Jr Persistent gene expression in mouse nasal epithelia following feline immunodeficiency virus-based vector gene transfer. J Virol. 2005;79:12818–12827. doi: 10.1128/JVI.79.20.12818-12827.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kang Y, Stein CS, Heth JA, Sinn PL, Penisten AK, Staber PD, et al. In vivo gene transfer using a nonprimate lentiviral vector pseudotyped with ross river virus glycoproteins. J Virol. 2002;76:9378–9388. doi: 10.1128/JVI.76.18.9378-9388.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sinn PL, Hickey MA, Staber PD, Dylla DE, Jeffers SA, Davidson BL, et al. Lentivirus pseudotyped with filoviral envelope glycoproteins transduce airway epithelia from the apical surface independent of folate receptor-alpha. J Virol. 2003;77:5902–5910. doi: 10.1128/JVI.77.10.5902-5910.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sinn PL, Cooney AL, Oakland M, Dylla DE, Wallen TJ, Pezzulo AA, et al. Lentiviral vector gene transfer to porcine airways. Mol Ther Nucleic Acids. 2012;1:e56. doi: 10.1038/mtna.2012.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oakland M, Maury W, McCray PB, Jr, Sinn PL. Intrapulmonary versus nasal transduction of murine airways with GP64-pseudotyped viral vectors. Mol Ther Nucleic Acids. 2013;2 doi: 10.1038/mtna.2012.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yan Z, Stewart ZA, Sinn PL, Olsen JC, Hu J, McCray PB, Jr, et al. Ferret and pig models of cystic fibrosis: prospects and promise for gene therapy. Hum Gene Ther Clin Dev. 2015;26:38–49. doi: 10.1089/humc.2014.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cooney AL, Abou Alaiwa MH, Shah VS, Bouzek DC, Stroik MR, Powers LS, et al. Lentiviral-mediated phenotypic correction of cystic fibrosis pigs. JCI Insight. 2016;1:e88730. doi: 10.1172/jci.insight.88730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Buchholz CJ, Friedel T, Buning H. Surface-engineered viral vectors for selective and cell type-specific gene delivery. Trends Biotechnol. 2015;33:777–790. doi: 10.1016/j.tibtech.2015.09.008. [DOI] [PubMed] [Google Scholar]
- 18.Ojala DS, Amara DP, Schaffer DV. Adeno-associated virus vectors and neurological gene therapy. Neuroscientist. 2015;21:84–98. doi: 10.1177/1073858414521870. [DOI] [PubMed] [Google Scholar]
- 19.Koerber JT, Jang JH, Schaffer DV. DNA shuffling of adeno-associated virus yields functionally diverse viral progeny. Mol Ther. 2008;16:1703–1709. doi: 10.1038/mt.2008.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koerber JT, Schaffer DV. Transposon-based mutagenesis generates diverse adeno-associated viral libraries with novel gene delivery properties. Methods Mol Biol. 2008;434:161–170. doi: 10.1007/978-1-60327-248-3_10. [DOI] [PubMed] [Google Scholar]
- 21.Koerber JT, Jang JH, Yu JH, Kane RS, Schaffer DV. Engineering adeno-associated virus for one-step purification via immobilized metal affinity chromatography. Hum Gene Ther. 2007;18:367–378. doi: 10.1089/hum.2006.139. [DOI] [PubMed] [Google Scholar]
- 22.Koerber JT, Maheshri N, Kaspar BK, Schaffer DV. Construction of diverse adeno-associated viral libraries for directed evolution of enhanced gene delivery vehicles. Nat Protoc. 2006;1:701–706. doi: 10.1038/nprot.2006.93. [DOI] [PubMed] [Google Scholar]
- 23.Maheshri N, Koerber JT, Kaspar BK, Schaffer DV. Directed evolution of adeno-associated virus yields enhanced gene delivery vectors. Nat Biotechnol. 2006;24:198–204. doi: 10.1038/nbt1182. [DOI] [PubMed] [Google Scholar]
- 24.Yu JH, Schaffer DV. Selection of novel vesicular stomatitis virus glycoprotein variants from a peptide insertion library for enhanced purification of retroviral and lentiviral vectors. J Virol. 2006;80:3285–3292. doi: 10.1128/JVI.80.7.3285-3292.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kwon I, Schaffer DV. Designer gene delivery vectors: molecular engineering and evolution of adeno-associated viral vectors for enhanced gene transfer. Pharm Res. 2008;25:489–499. doi: 10.1007/s11095-007-9431-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jang JH, Koerber JT, Kim JS, Asuri P, Vazin T, Bartel M, et al. An evolved adeno-associated viral variant enhances gene delivery and gene targeting in neural stem cells. Mol Ther. 2011;19:667–675. doi: 10.1038/mt.2010.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Drapkin PT, O'Riordan CR, Yi SM, Chiorini JA, Cardella J, Zabner J, et al. Targeting the urokinase plasminogen activator receptor enhances gene transfer to human airway epithelia. J Clin Invest. 2000;105:589–596. doi: 10.1172/JCI8858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kotterman MA, Schaffer DV. Engineering adeno-associated viruses for clinical gene therapy. Nat Rev Genet. 2014;15:445–451. doi: 10.1038/nrg3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hwang BY, Schaffer DV. Engineering a serum-resistant and thermostable vesicular stomatitis virus G glycoprotein for pseudotyping retroviral and lentiviral vectors. Gene Therapy. 2013;20:807–815. doi: 10.1038/gt.2013.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dong S, Blissard GW. Functional analysis of the Autographa californica multiple nucleopolyhedrovirus GP64 terminal fusion loops and interactions with membranes. J Virol. 2012;86:9617–9628. doi: 10.1128/JVI.00813-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sinn PL, Goreham-Voss JD, Arias AC, Hickey MA, Maury W, Chikkanna-Gowda CP, et al. Enhanced gene expression conferred by stepwise modification of a non-primate lentiviral vector. Hum Gene Ther. 2007;18:1244–1252. doi: 10.1089/hum.2006.127. [DOI] [PubMed] [Google Scholar]
- 32.O'Flynn NM, Patel A, Kadlec J, Jones IM. Improving promiscuous mammalian cell entry by the baculovirus Autographa californica multiple nuclear polyhedrosis virus. Biosci Rep. 2012;33:23–36. doi: 10.1042/BSR20120093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kadlec J, Loureiro S, Abrescia NG, Stuart DI, Jones IM. The postfusion structure of baculovirus gp64 supports a unified view of viral fusion machines. Nat Struct Mol Biol. 2008;15:1024–1030. doi: 10.1038/nsmb.1484. [DOI] [PubMed] [Google Scholar]
- 34.Zhou J, Blissard GW. Identification of a GP64 subdomain involved in receptor binding by budded virions of the baculovirus Autographica californica multi-capsid nucleopolyhedrovirus. J Virol. 2008;82:4449–4460. doi: 10.1128/JVI.02490-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wilson DS, Keefe AD. Random mutagenesis by PCR. Curr Protoc Mol Biol. 2001;Chapter 8 doi: 10.1002/0471142727.mb0803s51. Unit 8.3. [DOI] [PubMed] [Google Scholar]
- 36.Koerber JT, Klimczak R, Jang JH, Dalkara D, Flannery JG, Schaffer DV. Molecular evolution of adeno-associated virus for enhanced glial gene delivery. Mol Ther. 2009;17:2088–2095. doi: 10.1038/mt.2009.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Johnston J, Power C. Productive infection of human peripheral blood mono-nuclear cells by feline immunodeficiency virus: implications for vector development. J Virol. 1999;73:2491–2498. doi: 10.1128/jvi.73.3.2491-2498.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang G, Slepushkin V, Zabner J, Keshavjee S, Johnston JC, Sauter SL, et al. Feline immunodeficiency virus vectors persistently transduce nondividing airway epithelia and correct the cystic fibrosis defect. J Clin Invest. 1999;104:R55–R62. doi: 10.1172/JCI8390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Karp PH, Moniger TO, Weber SP, Nesselhauf TS, Launspach JL, Zabner J, et al. In: Methods in Molecular Biology. Wise C, editor. Vol. 188. Humana Press Inc.; 2002. pp. 115–137. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


