Abstract
Glioma is the most common malignant primary brain tumor which arises from the central nervous system. Our studies reported that an anti-apoptotic factor, activating transcription factor 5 (ATF5), is highly expressed in malignant glioma specimens and cell lines. Downregulation by dominant-negetive ATF5 could repress glioma cell proliferation and accelerate apoptosis. Here, we further investigate the upstream factor which regulates ATF5 expression. Bioinformatic analysis showed that ATF5 was a potential target of miR-141-3p. Luciferase reporter assay verified that miR-141-3p specifically targeted the ATF5 3′-UTR in glioma cells. Functional studied suggested that miR-141-3p overexpression inhibited proliferation and promoted apoptosis of glioma cells (U87MG and U251). Xenograft experiments proved the inhibition of miR-141-3p on glioma growth in vivo. Moreover, exogenous ATF5 without 3′-UTR restored the cell proliferation inhibition triggered by miR-141-3p. Taken together, we put forward that miR-141-3p is a new upstream target towards ATF5. It can serve as a crucial tumor suppressor in regulating the ATF5-regulated growth of malignant glioma.
Keywords: Glioma, miR-141-3p, Activating transcription factor 5, Cell proliferation, Apoptosis
1. Introduction
Glioma is the most common malignant primary brain tumor which arises from the central nervous system [1]. The tumor is highly aggressive with a mean survival time of nine to twelve months from diagnosis to death. According to malignant gradesI-IV distinguished by the World Health Organization (WHO), gliomas were classified into low-grade glioma (LGG) and high-grade glioma consisting of anaplastic glioma and glioblastoma (GBM) [2]. In spite of advances in cancer therapy, treatments towards gliomas remain challenging. Nowadays, increasing knowledge of glioma biology is starting to impact gene therapies towards targeted sites, which has a very exciting prospect [3]. ATF5 is a member of the activating transcription factor (ATF)/cAMP response-element binding protein (CREB) family, which contains a great many of basic leucine zipper proteins [4]. Recently, ATF5 has been emerged as an important regulator of numerous cellular functions, including cell proliferation, survival, and stress response [5–7]. Our previous works proved that ATF5, as an anti-apoptotic factor, was highly expressed in malignant glioma and could activate diverse survival pathways [8–10]. In the latest paper, we also found that ATF5 could serve as an adaptor protein in promoting survival of glioma cells under viruses infection. Although the role of ATF5 as a survival-related protein has been extensively reported, upstream factors which regulate ATF5 expression are only partially understood at present [5].
MicroRNAs (miRNAs) are a kind of endogenous, non-coding, small (19–25 nucleotides) RNAs, which bind to the 3′-UTR of target gene mRNAs to negatively regulate gene expression on a post-transcriptional level [11,12]. Although small, characteristic of regulating downstream oncogenes or tumor suppressors is embodied in cellular processes like development, proliferation and apoptosis [13,14]. Therefore, miRNAs could serve as therapeutic targets and diagnostic tools [15]. Early studies have linked miRNAs to gliomas. Several researchers have found that miRNAs, such as miR-141 [16], miR-152 [17], miR-137 [18], may be involved in the pathogenesis of glioma [19,20]. On the basis of the current understanding on the roles of tumor suppressors in glioma, these miRNAs are attractive for serving as nucleic-acid drugs. Nevertheless, an obstacle in their further clinical development is the potential non-specific toxic side effect of miRNA-based therapeutics.
The overall aim of this article is to explore the upstream molecular base for the regulation of ATF5 in glioma. In the present study, we verified the hypothesis that ATF5 might be a target of miR-141-3p. We provide evidence that miR-141-3p directly binds to ATF5 at its 3′-UTR and dramatically inhibits cell proliferation, promotes apoptosis by the suppression of ATF5. Our data support the conclusion that miR-141-3p can serve as a crucial tumor suppressor modulating ATF5 in malignant glioma.
2. Materials and methods
2.1. Tissues and cell lines
All 36 tissue samples were obtained from the Affiliated Hospital of Qingdao University (Qingdao, Shandong, China) from September 2015 to December 2016. According to the WHO Classification of Nervous System Tumors, the pathological diagnosis and grading for each glioma were assessed by neuropathologists. Hereinto, 33 glioma tissues consist of five cases with low-grade glioma, 11 cases with anaplastic glioma and 17 cases with glioblastoma. None of the patients have accepted radiation or chemotherapy before surgical therapy. In addition, 3 tissue specimens obtained from the normal cortex of patients without glioma were used as the negative control. Our research is received ethical approval from the Institutional Review Board of the Affiliated Hospital of Qingdao University. Human glioblastoma cell lines U87MG and U251 were purchased from Shanghai Cell Bank of the Chinese Academy of Sciences (Shanghai, China). U87MG cells were grown in Minimum Essential Medium (Hyclone, Logan, UT, USA) with 10% fetal bovine serum (FBS; Gibco, Rockville, MD, USA). U251 cells were maintained in Dulbecco’s High Glucose Modified Eagle Medium (Hyclone) with 10% FBS. Human astrocyte cells were separated from fresh fetal tissue of voluntary abortion by mifepristone and cultured in DMEM/F12 1:1 media (Hyclone) with 20% FBS. All cells were adherent cultured at 37 °C with 5% CO2.
2.2. Plasmids and siRNA
MiRNA control (miR-control), miR-141-3p mimic and its corresponding miR-141-3p inhibitor (anti-miR-141-3p) were purchased from Ribobio (Guangzhou, China). Plasmid of ATF5 without 3′-UTR (pEF1α-ATF5) was recombined by Applied Biological Materials (Nanjing, China). Cell transfection was performed with Lipofectamine 2000 Reagent (Invitrogen, Carlsbad, CA, USA) according to manufacturer’s protocol. After incubating for 48 or 72 h, cells were harvested then experiments were performed. Loss of ATF5 expression achieved using small interfering RNA (siRNA) of ATF5. To establish U87MG cell lines with a stable knock-down of ATF5, a GV113 control plasmid and a GV113 plasmid containing ATF5 lentivirus short hairpin RNAs, LV-ATF5-RNAi (8842-1; GCGAGATCCAGTACGTCAA) (Shanghai GeneChem Co. Ltd. Shanghai, China), targeting the ATF5 coding sequence were separately transduced into U87MG.
2.3. RNA extraction and reverse transcription PCR
Total RNA was extracted with TRIzol Reagent (Invitrogen). Reverse transcription of total RNA was performed using Prime-Script RT reagent Kit (TaKaRa, Dalian, China). Quantitative real time PCR to measure ATF5 expression was carried out with SYBR® Green PCR Kit (Qiagen, Germany). Micro-RNA was isolated by miRcute miRNA isolation kit (Tiangen, Beijing, China). Reverse transcription of miRNA was performed using miRcute miRNA First-Strand cDNA Synthesis (Tiangen). Quantitative real time PCR to measure miRNA expression was carried out with miRcute miRNA qRT-PCR Detection Kit (Tiangen). Primers used were shown as following: ATF5 forward 5′-AAGTCGGCGGCTCTGAGGTA-3′ and reverse 5′-GGACTCTGCCCGTTCCTTCA-3′, human β-actin forward 5′-CCCAGCACAATGAAGATCAA-3′ and reverse 5′-GATCCACACGGAGTACTTG-3′, GAPDH forward 5′-GAAGGTGAAGGTCGGAGTC-3′ and reverse 5′-GAAGATGGTGATGGGATTTC-3′. In addition, primers of miR-141-3p and U6 were purchased from Tiangen. The mRNA expression of ATF5 was normalized with β-actin and GAPDH, miR-141 expression was normalized with U6. qRT-PCR was performed on iQ5 Real-Time PCR Detection system (Bio-Rad, USA), the results were analyzed by the 2−ΔΔCt method.
2.4. CCK8 assay
Cell proliferation was detected by Cell Counting Kit (7 sea biotech, Shanghai, China). Cells were grown in 96-well plate with 1 × 104 per well and incubated in 37 °C with 5% CO2 until cell confluent rate reached 70%. After transfected with plasmid for 48 h, cells were still incubated for 24, 48, 72 and 96 h 10 μL CCK8 solution was seed into each well. The absorbance at 450 nm was measured with SUNRISE Microplate Reader (Tecan, Switzerland).
2.5. Flow cytometry
After 48-hr transfection, the cells were harvested, centrifuged, and washed with phosphate-buffered saline for three times. 100 μL 5%MEM was added to the resuspended cells in a 1.5 mL centrifuge tube. Every tube was incubated at room temperature for 20 min without light with 100 μL of Guava Nexin Reagant (Millipore Guava technologies, Billerica, MA, USA). Samples were analyzed by a Guava Easy Cyte Mini Flow cytometry instrument (Millipore Guava), and each sample was repeated 3 times.
2.6. Luciferase reporter assay
The primers used to amplify 3′-UTR of ATF5 mRNA from cDNA were 5′-GAGTGGCGACAGGATAGAGC-3′ (forward) and 5′-CCCCTCTCCAACACCTTCAC-3′ (reverse). Mutation in the miR-141 binding sites of ATF5 was performed using QuickChange Site-Directed Mutagenesis kit (Stratagene, La Jolla, CA, USA). The PCR products of wild-type 3′-UTR and mutant 3′-UTR were cloned into pMIR-REPORT vector (Ambion, Austin, TX, USA) within Spe I and Hind III restriction sites. Then cells were transfected with miR-141-3p mimics and pMIR-3′-UTRWt or pMIR-3′-UTRMut using Lipofectamine 2000. The relative luciferase activity was detected by Luciferase Assay Kit (Promega, Fitchburg, WI, USA). Firefly activity was normalized to Renilla activity to control the transfection efficiency. Renilla luciferase activity acts as a normalized control.
2.7. Western blot
Total proteins were extracted using RIPA (Solarbio), which contains 10 μL PMSF (Solarbio) per milliliter at 4 °C for 20 min. Lysates were collected by centrifugation at 13000g for 15 min. The proteins were separated on 12% SDS-PAGE gel and transferred onto polyvinylidene fluoride membranes. The antibodies used were rabbit anti-ATF5 (Abcam, UK), rabbit anti-β-actin (Bioss, Beijing, China) and hosrseradish peroxidase-conjugated goat anti-rabbit IgG (Abgent, USA). The membranes were developed using ECL regents (Thermo Fisher, USA) and visualized using the BIO-PRINT ST4 gel imaging system (Vilber Lourmat, Marne-la-Vallée, France). The bands of gray intensity were analyzed by Image J software.
2.8. Xenograft experiments
All animal experiments were conducted in accordance with the China animal welfare law and approved by local authorities. The BALB/c mice (4 weeks old) were obtained from the Institute of Laboratory Animal Science, Chinese Academy of Medical Sciences and kept under specific pathogen-free conditions. U87MG cells were overexpressed miR-control or miR-141-3p for 48 h before inoculating and maintained in serum free MEM medium. U87MG cells (3 × 106) in 200 μL of PBS were inoculated subcutaneously into the armpits of the mice. Mice were weighed and tumors were measured with external calipers every 4 days and each tumor volume in cubic millimeters was calculated by the following formula: V (mm3) = 0.5 × D × d2 (V, volume; D, longitudinal diameter; d, latitudinal diameter). After 24 days, the mice were euthanized using a subcutaneous injection with sodium pentobarbital (50 mg/kg), and the tumor tissues were striped and weighted.
2.9. Statistics
All data are shown as mean ± standard deviation (SD). Statistical significance was determined using Student’s t-test by SPSS 13.0 and Graphpad Prism 6. P < 0.05 was considered statistically significant.
3. Result
3.1. ATF5 was highly expressed in gliomas and was essential for cell proliferation and apoptosis
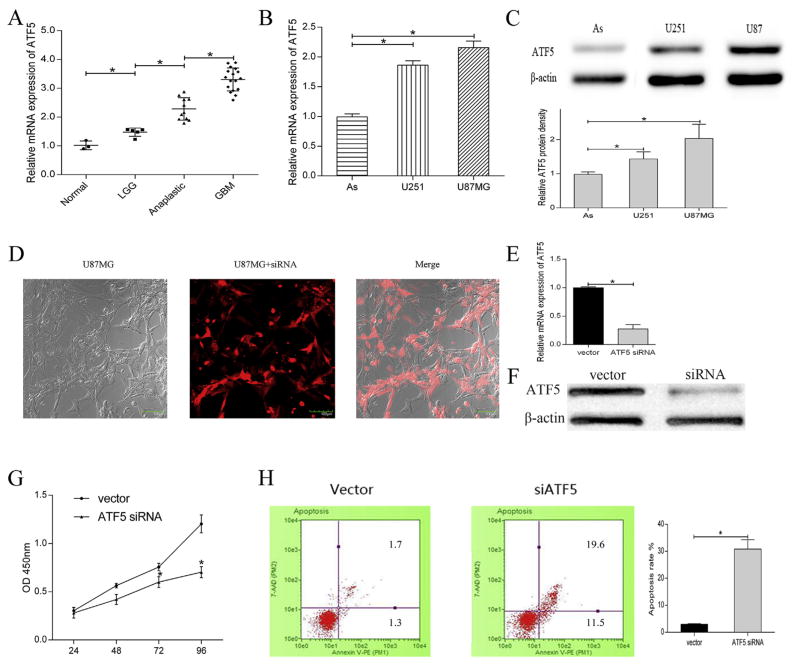
To confirm the upregulation of ATF5 in different grade glioma as suggested in previous study, quantitative RT-PCR was used to detect the mRNA expression level of ATF5 in glioma tissues (Fig. 1A). The results showed that the mRNA expression of ATF5 was gradually increased along with the malignant grade of glioma. Similarly, the high mRNA (Fig. 1B) and protein (Fig. 1C) expression of ATF5 was verified in glioma cell lines. Several studies proved that ATF5 is an anti-apoptosis regulator of tumors and plays an important role in glioma proliferation. In our study, we verify the functions of ATF5 on cell proliferation and apoptosis. The efficiency of short interfering RNA towards ATF5 was shown in Fig. 1D. Parallel results demonstrated the ATF5 siRNA successfully suppress the mRNA and protein expression of ATF5 in U87MG (Fig. 1E and F). In addition, siRNA-mediated ATF5 downregulation inhibited cell proliferation and accelerated apoptosis compared with vector group (Fig. 1G and H).
Fig. 1. ATF5 was highly expressed in gliomas and was essential for cell proliferation and apoptosis.
(A)Relative mRNA expression of ATF5 in different grades of glioma tissues detecting by qRT-PCR. Normal cortex tissues (n = 3) low-grade glioma (LGG, n = 5), anaplastic glioma (n = 11), glioblastoma (GBM, n = 17).
(B,C) Relative mRNA expression (B) and protein expression (C) of ATF5 in glioma cell lines.
(D, E,F) Images of U87MG (D) transfected with ATF5 siRNA. Transfection of ATF5 siRNA reduced the ATF5 mRNA and protein level.
(G,H) The inhibition on cell proliferation (G) and the acceleration on apoptosis (H) by siRNA-ATF5 were shown. All experiments were repeated three times. All data are shown as mean ± SD, *P < 0.05.
3.2. MiR-141-3p specifically targeted the ATF5 3′-UTR in glioma cells
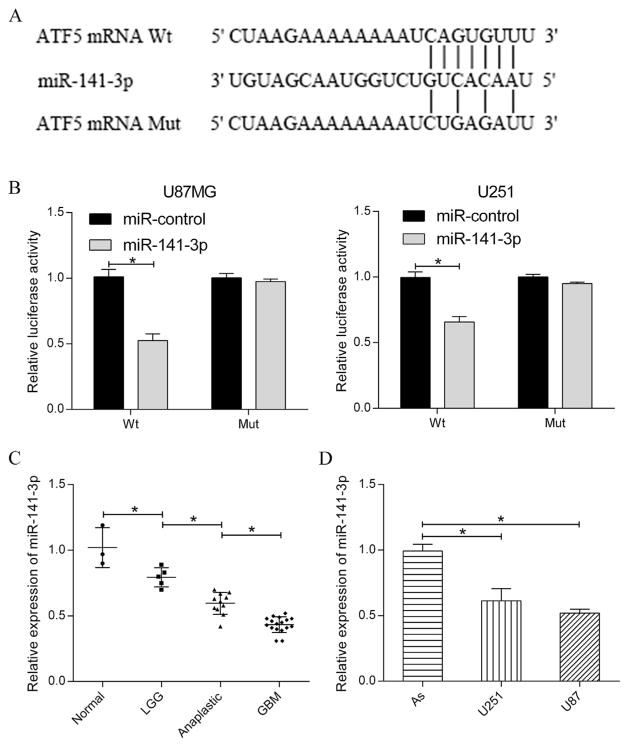
To explore the upstream factor which would modulate ATF5 expression, TargetScan (http://www.targetscan.org) and PicTar (http://www.pictar.org/) were employed to predict target of miR-141-3p. The results showed that miR-141-3p can directly bind to the 3′-UTR of ATF5 mRNA (Fig. 2A). In order to confirm this prediction, 3′-UTR wild type (Wt) and 3′-UTR mutation type (Mut) of ATF5 were cloned into the pMIR-REPORT vector. As expected, dual luciferase assay proved that the relative luciferase activity was significantly lower (~50%) in the group which co-transfected with pMIR-3′-UTRWt and miR-141-3p than the group co-transfected with pMIR-3′-UTRWt and miR-cotrol. In contrast, the effect of miR-141-3p on luciferase activity observed in pMIR-3′-UTRWt was absent in pMIR-3′-UTRMut (Fig. 2B). These results indicated that ATF5 mRNA 3′-UTR is a specific functional target of miR-141-3p in glioma cells.
Fig. 2. MiR-141-3p was significantly downregulated in glioma and specifically targeted ATF5 3′-UTR.
(A) Predicted binding sites of miR-141-3p in ATF5 mRNA 3′-UTR and the mutant type was shown in the schematic diagram.
(B) Dual luciferase assay was performed in U87MG and U251 at 48 h after transfection with pMIR-3′-UTR or pMIR-3′-UTRMut together with miR-control or miR-141-3p.
(C) Relative expression of miR-141-3p in different grades of glioma tissues.
(D) Relative expression of miR-141-3p in glioma cell lines (U87MG and U251) and human astrocyte cell line (As). U6 was used as internal control. All experiments were repeated three times. All data are shown as mean ± SD, *P < 0.05.
3.3. MiR-141-3p was significantly downregulated in glioma tissues and cell lines
The expression of miR-141-3p in 33 glioma patient tissues and two glioma cell lines was detected by quantitative RT-PCR. Analysis of the relative expression (Fig. 2C) revealed that miR-141-3p expression in GBM tissues was evidently decreased compared with the expression in anaplastic glioma tissues, miR-141-3p expression in anaplastic glioma was significantly decreased compared with the level in low-grade glioma and the expression of miR-141-3p was apparently reduced in low-grade glioma compared with normal brain tissues. In a word, it indicated that the expression of miR-141-3p is related to the different malignant grade of glioma. Similarly, compared with human astrocyte cell, miR-141-3p expression was significantly decreased in both U251 and U87MG cell lines (Fig. 2D).
3.4. MiR-141-3p inhibited growth through repressing ATF5 expression in vitro and in vivo
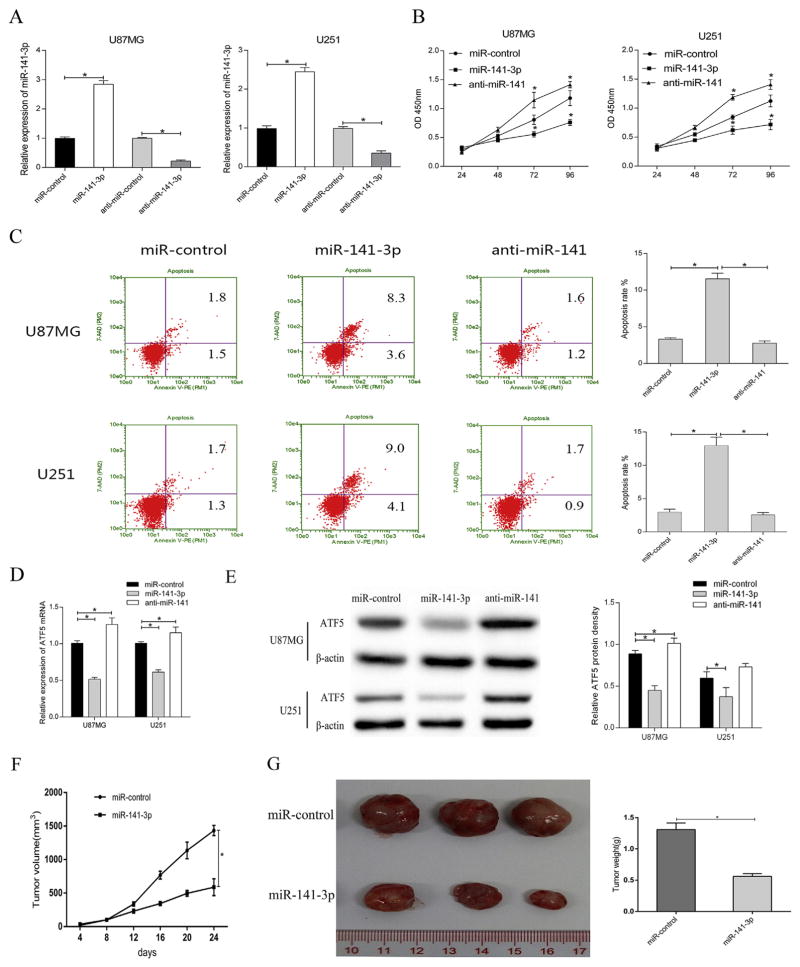
To evaluate the impact of miR-141-3p on glioma cell proliferation, U87MG and U251 were transfected with miR-control, miR-141-3p mimics, anti-miR-control and anti-miR-141-3p separately. At 48 h after transfection of miR-141-3p, miR-141-3p expression was increased compared with miR-control group (Fig. 3A). Reversely, anti-miR-141-3p, which specifically binds to mature miR-141-3p, weakened endogenous miR-141-3p expression and then impacted on target gene expression. CCK8 assay was carried out to analyze cell proliferation at 24, 48, 72 and 96 h following transfection (Fig. 3B). Overexpression of miR-141-3p observably inhibited cell proliferation in both U87MG and U251 compared with control. Reversely, down regulation by anti-miR-141-3p promoted the cell proliferation. In addition, Flow Cytometry using Annexin V/7-AAD staining (Fig. 3C) showed that apoptotic ratio was significantly higher in miR-141-3p-transfected cells compared with negative control. Nevertheless, as endogenous miR-141-3p was inhibited, the apoptosis rate was obviously decreased. In addition, we analyzed the effect of miR-141-3p on ATF5 expression using qRT-PCR and western blot. The result showed that ATF5 mRNA and protein expression were inhibited by over-expressed miR-141-3p whereas anti-miR-141-3p obviously up-regulate ATF5. It indicated that miR-141-3p may affect glioma cell proliferation and apoptosis through ATF5 (Fig. 3D and E).
Fig. 3. MiR-141-3p inhibited glioma growth in vitro and in vivo.
(A)qRT-PCR was used to detect the expression of miR-141-3pin U87MG and U251 after transfection with miR-control or miR-141-3p or anti-miR-141-3p.
(B) Effect of miR-141-3p on cell proliferation of glioma cell lines was examined by CCK8 assay.
(C) Effect of miR-141-3p on cell apoptosis of glioma cell lines was detected by Flow Cytometry.
(D) The mRNA expression level of ATF5 was inhibited by miR-141-3p in U87MG and U251 cells.
(E) ATF5 protein levels were evaluated by western blot assay after transient transfection in U87MG and U251 cells.
(F) Growth curve of U87MG xenograft tumors were measured volume every four days.
(G) Tumor tissues image and weight at the end of experiments. All experiments were repeated three times. All data are shown as mean ± SD, *P < 0.05.
To further confirm the correlation of miR-141 expression with the growth of glioma in vivo, we examined tumor growth in nude mice. U87MG cells expressed miR-control or miR-141-3p were subcutaneously inoculated into the armpits of the mice. During the 24 days, tumors of miR-141-3p overexpression group grew much smaller and slower than control transfected group (Fig. 3F). At the endpoint of the experiments, the tumor volume and weight of miR-141-3p group was significantly lower than that of control group (Fig. 3G).
3.5. Exogenous expression of ATF5 rescued cells from proliferation inhibition triggered by miR-141-3p
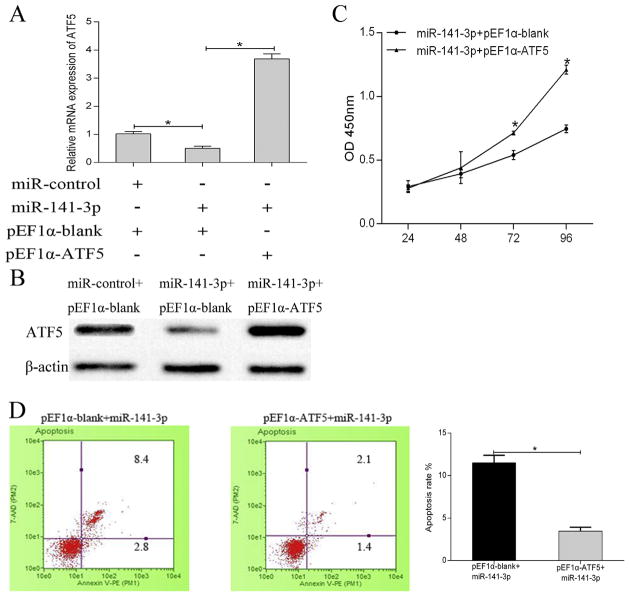
To determine the mechanism underlying the inhibitory effects of miR-141-3p on glioma, we constructed pEF1α-ATF5 which embedded the segment of ATF5 without specific miR-141-3p binding sequences in the 3′-UTR to avoid the miRNA interference. Then qRT-PCR and western blot were performed after cell transfection in U87MG. The mRNA expression of ATF5 was decreased in cells transfected with miR-141-3p, while the ATF5 expression was obviously restored in cells co-transfected with miR-141-3p and pEF1α-ATF5 (Fig. 4A). Western blot has shown the ATF5 protein level (Fig. 4B). Moreover, exogenous ATF5 reverses the inhibitory effect on cell proliferation (Fig. 4C) and the stimulative effect on apoptosis (Fig. 4D). Similar results were also verified in U251 cell line. These data illustrated that miR-141-3p suppressed glioma cell proliferation through binding to ATF5 3′-UTR.
Fig. 4.
Exogenous expression of ATF5 rescued cells from proliferation inhibition triggered by miR-141-3p.
(A,B) ATF5 mRNA (A) and protein (B) expression in U87MG transfected with pEF1α-ATF5 or pEF1α-blank.
(C) Overexpressed ATF5 reversed the cell proliferation suppression caused by miR-141-3p in CCK8 assay.
(D) Exogenous ATF5 expression reversed the stimulative effect caused by miR-141-3p on apoptosis. All experiments were repeated three times. All data are shown as mean ± SD, *P < 0.05.
4. Discussion
Gliomas are the most common clinical primary brain tumors. ATF5, an anti-apoptotic protein, is highly expressed in malignant gliomas. Angelastro et al. [21] reported ATF5 mRNA and protein are down-regulated in PC12 and neuroprogenitor cells by nerve growth factor. Persengiev et al. [22] confirmed withdrawal of interleukins result in an obvious loss of ATF5 transcripts in lymphocytic and bone marrow cells. Although many results including our previous findings have showed that ATF5 is a mark of gliomas, especially in glioblastoma, there is still no clear mechanism of how ATF5 be regulated. To explore the upstream factor regulated ATF5 expression, TargetScan, and PicTar were employed to identify ATF5 as a putative target of miR-141-3p. This hypothesis was further confirmed by luciferase assay analysis.
Aberrant miRNAs expression, regulating transcription of targeted oncogenes or tumor suppressor genes, is closely related to various types of tumors [23]. Nowadays, quite a few studies have reported miR-141, a member of miR-200 family, is down-regulated and has an effect on various tumors. For example, miR-141, acting as an oncogene, accelerates non-small cell lung cancer cell proliferation in vitro and tumor growth [24]. However, it functions as a tumor suppressor in gastric cancer [25], hepatocellular carcinoma [26] and breast cancer [27]. In addition, Guo et al. [28] previously demonstrated miR-141, decreasing in glioma tissues and cell lines, inhibited glioma cell growth and migration by targeting ZEB1. Similarly, Peng et al. [16] reported miR-141, which is also down-regulated in glioma tissues and cell lines, inhibited glioma cells growth and metastasis by targeting TGF-β2. In this study, we have detected the low expression of miR-141-3p in 33 different malignant glioma tissues (low-grade glioma, anaplastic glioma and glioblastoma) for the first time. Results indicate that miR-141-3p expression is gradually decreased with the increased malignancy of glioma.
MiRNA inhibitor is a length of antisense oligonucleotides which is considered as the best and possibly the only practical approach for specific pharmacological inhibition of miRNAs function [29]. It complementarily forms a high affinity duplex with seed region of miRNA within the RNA induced silencing complex and functionally blocks miRNA activity leading to de-repression of miRNA target mRNAs [30]. In 2005, it has reported that locked nucleic acid modified oligonucleotides could be used to inhibit miRNA function in cells [31]. Recent study proved that inhibition of miR-9 by a systemically delivered antimiR-9 in a xenograft model of Hodgkin lymphoma results in decreased tumour outgrowth, confirming that miR-9 actively participates in Hodgkin lymphoma pathogenesis [32]. Therefore, miRNA inhibition gives insight into a more physiological response pattern of miRNA regulation and biological targets. In our study, we employed miR-141-3p inhibitor to validate the function of miR-141-3p on cell proliferation and apoptosis. We put forward that overexpression of miR-141-3p was related to the inhibition of cell proliferation as well as the promotion of apoptosis in U87MG and U251. Xenograft experiments indicated miR-141-3p significantly inhibited glioma growth in vivo.
In summary, miR-141-3p, which targets ATF5 mRNA 3′-UTR, was down-regulated ATF5 mRNA and protein expression leading to cell proliferation inhibition and cell apoptosis promotion in glioma. It interprets the molecular mechanism of miR-141-3p-induced cell behaviors. Moreover, we tentatively put forward that a new factor influenced transcription of ATF5 has been found. MiR-141-3p provides a novel insight into the upstream mechanism of regulating ATF5 function.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (NO.81601754 and NO.81471958); the Qingdao Postdoctoral Science Foundation (NO.2015160); and the Scientific Technology Plan Projects of Shandong, China (NO.J12LL11).
Abbreviations
- ATF5
activating transcription factor 5
- miRNAs
microRNAs
- WHO
the World Health Organization
- LGG
low-grade glioma
- GBM
glioblastoma
- 3′-UTR
3′-Untranslated Regions
- qRT-PCR
quantitative Reverse Transcription PCR
Footnotes
Conflict of interest
The authors declare that they have no competing interest.
Transparency document
Transparency document related to this article can be found online at http://dx.doi.org/10.1016/j.bbrc.2017.05.174.
References
- 1.Ilkanizadeh S, Lau J, Huang M, Foster DJ, Wong R, Frantz A, Wang S, Weiss WA, Persson AI. Glial progenitors as targets for transformation in glioma. Adv Cancer Res. 2014;121:1–65. doi: 10.1016/B978-0-12-800249-0.00001-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Castro MG, Candolfi M, Kroeger K, King GD, Curtin JF, Yagiz K, Mineharu Y, Assi H, Wibowo M, Ghulam MA, Foulad D, Puntel M, Lowenstein PR. Gene therapy and targeted toxins for glioma. Curr Gene Ther. 2011;11:155–180. doi: 10.2174/156652311795684722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rizzo D, Ruggiero A, Martini M, Rizzo V, Maurizi P, Riccardi R. Molecular biology in pediatric high-grade glioma: impact on prognosis and treatment. Biomed Res Int. 2015;2015:215135. doi: 10.1155/2015/215135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sheng Z, Li L, Zhu LJ, Smith TW, Demers A, Ross AH, Moser RP, Green MR. A genome-wide RNA interference screen reveals an essential CREB3L2-ATF5-MCL1 survival pathway in malignant glioma with therapeutic implications. Nat Med. 2010;16:671–677. doi: 10.1038/nm.2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Greene LA, Lee HY, Angelastro JM. The transcription factor ATF5: role in neurodevelopment and neural tumors. J Neurochem. 2009;108:11–22. doi: 10.1111/j.1471-4159.2008.05749.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li G, Xu Y, Guan D, Liu Z, Liu DX. HSP70 protein promotes survival of C6 and U87 glioma cells by inhibition of ATF5 degradation. J Biol Chem. 2011;286:20251–20259. doi: 10.1074/jbc.M110.211771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu DX, Qian D, Wang B, Yang JM, Lu Z. P300-Dependent ATF5 acetylation is essential for Egr-1 gene activation and cell proliferation and survival. Mol Cell Biol. 2011;31:3906–3916. doi: 10.1128/MCB.05887-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Angelastro J, Canoll P, Kuo J, Weicker M, Costa A, Bruce J, Greene LA. Selective destruction of glioblastoma cells by interference with the activity or expression of ATF5. Oncogene. 2006;25:907–916. doi: 10.1038/sj.onc.1209116. [DOI] [PubMed] [Google Scholar]
- 9.Huang R, Qian D, Hu M, Zhang X, Song J, Li L, Chen H, Wang B. Association between human cytomegalovirus infection and histone acetylation level in various histological types of glioma. Oncol Lett. 2015;10:2812–2820. doi: 10.3892/ol.2015.3638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang T, Qian D, Hu M, Li L, Zhang L, Chen H, Yang R, Wang B. Human cytomegalovirus inhibits apoptosis by regulating the activating transcription factor 5 signaling pathway in human malignant glioma cells. Oncol Lett. 2014;8:1051–1057. doi: 10.3892/ol.2014.2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.He L, He X, Lim LP, de Stanchina E, Xuan Z, Liang Y, Xue W, Zender L, Magnus J, Ridzon D, Jackson AL, Linsley PS, Chen C, Lowe SW, Cleary MA, Hannon GJ. A microRNA component of the P53 tumour suppressor network. Nature. 2007;447:1130–1134. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Slezak-Prochazka I, Durmus S, Kroesen BJ, van den Berg A. MicroRNAs, macrocontrol: regulation of miRNA processing. Rna. 2010;16:1087–1095. doi: 10.1261/rna.1804410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Novakova J, Slaby O, Vyzula R, Michalek J. MicroRNA involvement in glioblastoma pathogenesis. Biochem Biophys Res Commun. 2009;386:1–5. doi: 10.1016/j.bbrc.2009.06.034. [DOI] [PubMed] [Google Scholar]
- 14.Zhang Y, Dutta A, Abounader R. The role of microRNAs in glioma initiation and progression. Front Biosci (Landmark Ed) 2012;17:700–712. doi: 10.2741/3952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Luo JW, Wang X, Yang Y, Mao Q. Role of micro-RNA (miRNA) in pathogenesis of glioblastoma. Eur Rev Med Pharm. 2015;19:1630–1639. [PubMed] [Google Scholar]
- 16.Peng T, Zhang S, Li W, Fu S, Luan Y, Zuo L. MicroRNA-141 inhibits glioma cells growth and metastasis by targeting TGF-beta2. Am J Transl Res. 2016;8:3513–3521. [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 17.Ma J, Yao Y, Wang P, Liu Y, Zhao L, Li Z, Li Z, Xue Y. MiR-152 functions as a tumor suppressor in glioblastoma stem cells by targeting kruppel-like factor 4. Cancer Lett. 2014;355:85–95. doi: 10.1016/j.canlet.2014.09.012. [DOI] [PubMed] [Google Scholar]
- 18.Sun G, Cao Y, Shi L, Sun L, Wang Y, Chen C, Wan Z, Fu L, You Y. Overexpressed miRNA-137 inhibits human glioma cells growth by targeting Rac1. Cancer Biother Radiopharm. 2013;28:327–334. doi: 10.1089/cbr.2012.1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Silber J, James CD, Hodgson JG. MicroRNAs in gliomas: small regulators of a big problem. Neuromolecular Med. 2009;11:208–222. doi: 10.1007/s12017-009-8087-9. [DOI] [PubMed] [Google Scholar]
- 20.Wong JW. MicroRNA-induced silencing of glioma progression. J Neurosci. 2010;30:3868–3869. doi: 10.1523/JNEUROSCI.0442-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Angelastro JM, Mason JL, Ignatova TN, Kukekov VG, Stengren GB, Goldman JE, Greene LA. Downregulation of activating transcription factor 5 is required for differentiation of neural progenitor cells into astrocytes. J Neurosci. 2005;25:3889–3899. doi: 10.1523/JNEUROSCI.3447-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Persengiev SP, Devireddy LR, Green MR. Inhibition of apoptosis by ATFx: a novel role for a member of the ATF/CREB family of mammalian bZIP transcription factors. Genes Dev. 2002;16:1806–1814. doi: 10.1101/gad.992202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jansson MD, Lund AH. MicroRNA and cancer. Mol Oncol. 2012;6:590–610. doi: 10.1016/j.molonc.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mei Z, He Y, Feng J, Shi J, Du Y, Qian L, Huang Q, Jie Z. MicroRNA-141 promotes the proliferation of non-small cell lung cancer cells by regulating expression of PHLPP1 and PHLPP2. FEBS Lett. 2014;588:3055–3061. doi: 10.1016/j.febslet.2014.06.020. [DOI] [PubMed] [Google Scholar]
- 25.Chen B, Huang T, Jiang J, Lv L, Li H, Xia S. MiR-141 suppresses proliferation and motility of gastric cancer cells by targeting HDGF. Mol Cell Biochem. 2014;388:211–218. doi: 10.1007/s11010-013-1912-3. [DOI] [PubMed] [Google Scholar]
- 26.Liu Y, Ding Y, Huang J, Wang S, Ni W, Guan J, Li Q, Zhang Y, Ding Y, Chen B, Chen L. MiR-141 suppresses the migration and invasion of HCC cells by targeting Tiam1. PLoS One. 2014;9:e88393. doi: 10.1371/journal.pone.0088393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Abedi N, Mohammadi-Yeganeh S, Koochaki A, Karami F, Paryan M. MiR-141 as potential suppressor of beta-catenin in breast cancer. Tumour Biol. 2015;36:9895–9901. doi: 10.1007/s13277-015-3738-y. [DOI] [PubMed] [Google Scholar]
- 28.Guo E, Wang Z, Wang S. MiR-200c and miR-141 inhibit ZEB1 synergistically and suppress glioma cell growth and migration. Eur Rev Med Pharmacol Sci. 2016;20:3385–3391. [PubMed] [Google Scholar]
- 29.Esau CC. Inhibition of microRNA with antisense oligonucleotides. Methods. 2008;44:55–60. doi: 10.1016/j.ymeth.2007.11.001. [DOI] [PubMed] [Google Scholar]
- 30.Wen J, Leucci E, Vendramin R, Kauppinen S, Lund AH, Krogh A, Parker BJ. Transcriptome dynamics of the microRNA inhibition response. Nucleic Acids Res. 2015;43:6207–6221. doi: 10.1093/nar/gkv603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ørom UA, Kauppinen S, Lund AH. LNA-modified oligonucleotides mediate specific inhibition of microRNA function. Gene. 2006;372:137–141. doi: 10.1016/j.gene.2005.12.031. [DOI] [PubMed] [Google Scholar]
- 32.Leucci E, Zriwil A, Gregersen LH, Jensen KT, Obad S, Bellan C, Leoncini L, Kauppinen S, Lund AH. Inhibition of miR-9 de-represses HuR and DICER1 and impairs Hodgkin lymphoma tumour outgrowth in vivo. Oncogene. 2012;31:5081–5089. doi: 10.1038/onc.2012.15. [DOI] [PubMed] [Google Scholar]