Abstract
Chronic viral infections and cancer often lead to the emergence of dysfunctional or “exhausted” CD8+ T cells, and the restoration of their functions is currently the focus of therapeutic interventions. Here, we detail recent advances in the annotation of the gene modules and the epigenetic landscape associated with T cell dysfunction. Together with analysis of single-cell transcriptomes, these findings have enabled a deeper and more precise understanding of the transcriptional mechanisms that induce and maintain the dysfunctional state and highlight the heterogeneity of CD8+ T cell phenotypes present in chronically inflamed tissue. We discuss the relevance of these findings for understanding the transcriptional and spatial regulation of dysfunctional T cells and for the design of therapeutics.
T cell dysfunction
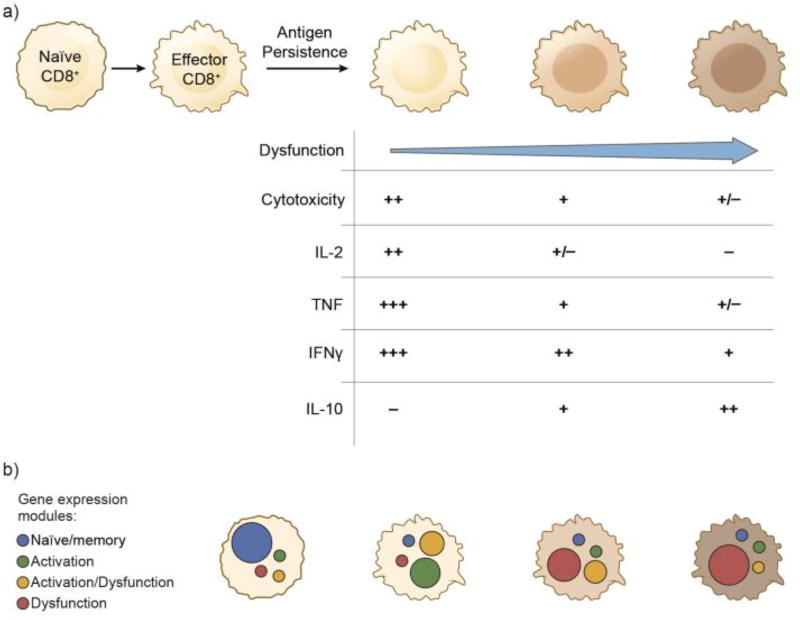
In chronic disease settings, such as in chronic viral infections and cancer, effector CD8+ T cells progressively acquire an “exhausted” or dysfunctional T cell state characterized by variable deficits in their effector functions, including cytotoxicity and the production of pro-inflammatory cytokines (IL-2, TNF-α, IFN-γ) [1] (Figure 1a). Importantly, dysfunctional CD8+ T cells are not completely inert as it has been shown that they can still exert some level of viral control [1]. Moreover, as effector CD8+ T cells lose their ability to produce pro-inflammatory cytokines, they can acquire the ability to produce IL-10, a potent immune suppressive cytokine, indicating a potential role for dysfunctional CD8+ T cells in maintaining immune suppression at sites of chronic inflammation. Thus, dysfunctional CD8+ T cells comprise both an obstacle and a liability for successful anti-viral and anti-tumor immunity. Consequently, achieving a deeper understanding of the mechanisms, both CD8+ T cell extrinsic and intrinsic, which promote the dysfunctional T cell state has the potential to inform the development of therapeutic interventions for both chronic viral infections and cancer.
Figure 1. Spectrum of CD8+ T cell phenotypes and expression of gene modules in chronic disease.
A). In chronic disease settings, antigen persistence and environmental factors drive the variable loss of effector functions, including cytotoxicity and pro-inflammatory cytokine (IL-2, TNF-α, IFN-γ) production, in CD8+ T cells resulting in a spectrum of CD8+ T cell phenotypes. As effector T cells become dysfunctional or “exhausted”, they can also acquire the ability to produce the immunosuppressive cytokine IL-10, suggesting a possible role for dysfunctional CD8+ T cells in maintaining local immune suppression. B) Representation of the expression of the naïve/memory, activation, activation/dysfunction, and dysfunction gene modules in individual CD8+ T cells. The extent to which individual gene modules are expressed in CD8+ T cells will determine their functional phenotype.
T cell dysfunction was first recognized in the setting of chronic lymphocytic choriomeningitis virus (LCMV) infection in mice [2, 3]. Studies in this model showed that the repeated triggering of effector CD8+ T cells as a result of antigen persistence was a key factor leading to the progressive loss of effector functions in CD8+ T cells. Later, it was recognized that other CD8+ T cell extrinsic signals also played important roles in promoting T cell dysfunction. The loss of CD4+ T cell help, particularly in the form of IL-21 [4–6] and signaling downstream of the immunosuppressive cytokines, IL-10 and TGF-β, were shown to promote T cell dysfunction [7, 8]. More recently, IL-6 signaling, alone or in combination with TGF-β, was shown to induce the transcription factor Maf, a potential driver of CD8+ T cell dysfunction in tumor [9]; however the role of IL-6 in driving dysfunctional phenotype in vivo was not addressed. That Maf is driven by Stat3 signaling [10] further raises the possibility that other cytokines that activate Stat3 may also play a role in regulating T cell dysfunction.
Recent advances have provided increased resolution of the cell intrinsic programs associated with CD8+ T cell dysfunction. The ability to measure mRNA expression in individual T cells has revealed not only how these distinct transcriptional programs are expressed at the single-cell level but also the heterogeneity present in CD8+ T cells at sites of chronic inflammation. Although dysfunction has also been described in CD4+ T cells in chronic viral infection [11–20] (see Text Box 1), in this review we will discuss the recent advances in elucidating the gene modules (see glossary) and epigenetic landscape associated with CD8+ T cell dysfunction and discuss a broad framework for understanding the spectrum of CD8+ T cell phenotypes present in chronically inflamed tissue.
Text box 1. CD4+ T cell dysfunction.
CD4+ T cells have been reported to show a dysfunctional phenotype during chronic viral and parasitic infections in mice and humans [11–16]. Similar to CD8+ T cells, dysfunctional CD4+ T cells show a loss of the ability to produce cytokines such as IL-2 and TNFα [13] and acquire the expression of co-inhibitory molecules [12, 14]. Prdm1 (transcription factor also known as BLIMP-1) was reported to drive CD4+ T cell dysfunction [12] and several other transcription factors such as Eomes and Helios were shown to be upregulated in dysfunctional CD4+ T cells relative to naïve and memory CD4+ T cells [14]. Overall, several transcription factors associated with CD4+ T cell dysfunction are also implicated in CD8+ T cell dysfunction, suggesting the evolution of shared regulatory pathways to restrict T cell responses. Interestingly, viral persistence during chronic LCMV infection has been reported to redirect CD4+ T cell differentiation towards T follicular helper cells [17]. It was proposed that this allows CD4+ T cells to maintain functionality and host immunity. This observation is in line with the recently described CXCR5+ CD8+ T cells that are present in chronic LCMV infection and also exhibit a T follicular helper-like cell signature [18–20]. Despite the similarities, it has been reported that dysfunctional CD4+ T cells can have distinct features as compared to dysfunctional CD8+ T cells and other CD4+ T cell subsets [14]. This will likely be disease and context dependent. Given the heterogeneity of CD4+ T cells, deep analysis at the single-cell level will greatly enhance our understanding of CD4+ T cell dysfunction.
Molecular signatures of T cell dysfunction
Several gene signatures (see glossary) based on analyses of populations of dysfunctional CD8+ T cells from cancer and chronic viral infections have been published [9, 21–25]. These signatures have shown that there is great similarity between virus- and cancer-associated CD8+ T cell dysfunction and have greatly advanced our understanding of factors that contribute to dysfunctional phenotype. Comparisons of the gene expression profiles of functional versus dysfunctional virus-specific CD8+ T cells taken from mice during acute or chronic infection with LCMV, respectively, led to the discovery of a role for the co-inhibitory receptor PD-1 in promoting dysfunctional phenotype [26], albeit PD-1 is dispensable for the development of T cell dysfunction [27]. Now it is appreciated that high expression of a multitude of co-inhibitory receptors including PD-1, Tim-3, CTLA-4, Lag-3, TIGIT, BTLA, 2B4, and CD160 are associated with CD8+ T cell dysfunction and that the degree of co-expression of these receptors directly correlates with the severity of dysfunctional phenotype (reviewed in [1]). Interference with many of these receptors has been shown to improve CD8+ T cell function as well as viral or tumor growth control in experimental models. Indeed, therapies that block CTLA-4, PD-1, or its ligand PD-L1 have now been harnessed clinically to treat patients with multiple different cancers. Despite their association with dysfunctional CD8+ T cell phenotype, it is important to note that co-inhibitory receptors are upregulated on naïve T cells as they undergo activation and effector differentiation. Indeed, the PD-1+ T cells found in the circulation of healthy humans comprise effector-memory T cells [28]. Moreover, we have observed uncoupling of co-inhibitory receptor expression from dysfunctional phenotype in CD8+ tumor-infiltrating lymphocytes (TILs) [24]. Thus, it is not possible to discern the presence of a dysfunctional versus activated T cell state based on the expression of co-inhibitory receptors alone.
Gene modules for T cell dysfunction and activation
While the dysfunction signatures discussed above have been useful in identifying factors that contribute to T cell dysfunction, it was noted early on that these signatures highly overlap with signatures of healthy T cell activation (reviewed in [29]). For example, co-inhibitory receptors (e.g. PD-1, Tim-3, CTLA4) and effector molecules (e.g. GZMC, IFNγ) present in the dysfunction signatures have been shown to be associated with healthy T cell activation. This overlap between the dysfunction and activation signatures complicates the identification of molecular mechanisms that drive or maintain dysfunction as opposed to programs that drive T cell activation and effector differentiation and ultimately give rise to T cell dysfunction.
Recently, we identified dysregulation of intracellular zinc as having a major role in driving CD8+ T cell dysfunction [24]. We found that metallothioneins (MT1 and MT2), proteins that function as intracellular zinc chaperones and intracellular zinc availability are increased in dysfunctional CD8+ TILs [24]. Indeed, loss of MT1 and MT2 expression resulted in normalization of intracellular zinc, improved CD8+ TIL functionality, and tumor growth control in a mouse model of melanoma. Unsupervised analysis of the RNA profiles of TILs from wildtype and MT1/2 deficient mice, in which CD8+ TILs do not exhibit a dysfunctional phenotype, allowed us to obtain a separate ranking of genes by their association with activated and dysfunctional T cell phenotypes. Using these rankings, we defined four separate modules, consisting of four ranked lists of genes associated with (1) dysfunction (but not activation), (2) activation (but not dysfunction), (3) activation and dysfunction and (4) neither (a “naïve/memory-like” module) [24] (Figure 1b). Our rankings aligned well with expected enrichments from the MSigDB database [24], and were also found to correlate well with gene signatures extracted from a single-cell transcriptomic study of melanoma in humans [30].
The shared activation/dysfunction module
The activation/dysfunction module, defined to be genes that scored highly for both activation and dysfunction, was strongly associated with a previously annotated signature of dysfunctional virus-specific CD8+ T cells in chronic LCMV infection (Doering et al., 2012). Included in the activation/dysfunction module were most co-inhibitory receptors (PD-1, CTLA-4, Tim-3, Lag3, etc.), emphasizing the association of these receptors with states of T cell activation and dysfunction and underscoring the need for more precise markers to distinguish between activated and dysfunctional CD8+ T cell states. Interestingly, we found that co-stimulatory receptors (4-1BB, OX-40, GITR) that are members of the TNF superfamily were also included in this gene module, indicating a role for co-stimulatory receptors belonging to the TNF superfamily in counter-balancing the action of co-inhibitory receptors [24].
The dysfunction module
The dysfunction module was defined to be the set of genes that scored high for dysfunction but low for activation, thus specifying a transcriptional program associated with dysfunction that is isolated from CD8+ T cell activation. The dysfunction module significantly overlapped with genes up-regulated in regulatory Ly49+CD8+ cells [31], suggesting possible regulatory features for cells with high expression of this gene module. Within this module, we found that Gata3 was the highest-ranking transcription factor of those consistent across our analyzed datasets. Indeed, transfer of tumor-specific CRISPR-Cas9 deleted Gata3 CD8+ T cells resulted in reduced tumor growth and higher cytokine expression as compared to control CD8+ T cells, validating Gata3 as a driver of CD8+ T cell dysfunction [24]. We further found Helios (Ikzf2) to rank highly among transcription factors in this module. Indeed, Helios is highly expressed in regulatory Ly49+CD8+ T cells and has been shown to be required for the stable inhibitory capabilities of this regulatory population [31]. Interestingly, Helios has been highlighted in association with CD8+ T cell dysfunction in chronic LCMV infection [22] and is up-regulated by NFAT, which has also been implicated in CD8+ T cell dysfunction [25].
That both Gata3 and Helios are zinc finger transcription factors and are high-ranking transcription factors in our dysfunction module is in line with our demonstration that increased zinc availability correlates with the development of dysfunctional phenotype in CD8+ T cells [24]. Indeed, gene signatures of dysfunctional CD8+ T cells from chronic LCMV infection have also shown enrichment for C2H2-like zinc finger transcription factors [22], which includes Helios. Zinc is an essential metal that is required for the structure and function of over 1,000 zinc-finger containing proteins including not only transcription factors but also serine-threonine kinases, ubiquitin ligases, and matrix metallopeptidases. Thus, one can envision how alterations in zinc availability can have a profound effect on T cell behavior.
The activation module
The activation module was defined to be the set of genes that scored high for activation, but low for dysfunction, thus specifying a transcriptional program associated with CD8+ T cell activation that is isolated from dysfunction. Indeed, highly significant enrichment was observed for this gene module with signatures of naïve CD8+ T cells activated in vitro and weaker but significant enrichments for signatures of CD8+ T cells activated in vivo [24]. Included in this module are genes involved in metabolism (ACAT2), double strand break repair (RAD50), and mitochondrial ribosome assembly (MTG1).
Heterogeneity of CD8+ T cell states within tumor
The discovery of refined gene modules for T cell dysfunction, activation/dysfunction, and activation raises the question of how these modules are expressed in individual CD8+ T cells and how this relates to their functional phenotype (Figure 1b). For example, do the dysfunctional CD8+ T cells previously defined based on surface expression of co-inhibitory receptors uniformly express the activation/dysfunction and dysfunction modules or are there subpopulations that express these modules to different extents? If so, what would be the functional phenotype of cells that predominantly express the dysfunction module or the activation/dysfunction module? The ability to profile the transcriptome at single-cell resolution allows for investigation of how these gene modules are either mutually or exclusively expressed in CD8+ T cells.
Clustering of single-cells by their mRNA transcriptional profiles enables the identification of distinct groups of cells that share similar transcriptional states, forming a basis for exploring the functional heterogeneity within the system [32]. Clustering of high-dimensional data (e.g. single-cell mRNA measurements) can be done with a variety of methods (e.g. hierarchical clustering). Graph-based clustering approaches, such as K-nearest-neighbor (KNN) graph construction followed by community grouping with the infomap algorithm are widely used with single-cell RNA-seq data and have enabled successful identification of distinct cellular states [33]. Using this approach, we found seven separate transcriptionally-defined populations of CD8+ TILs present in B16F10 melanoma tumors [24], implying complexity of CD8+ T cell states in tumor tissue. Intriguingly, among these populations, we found that expression of the activation/dysfunction module correlates strongly with expression of the activation module, and that while some populations showed diverse expression of numerous modules, others predominantly expressed either the activation and activation/dysfunction or the dysfunction modules. Functional annotation of these novel populations, along with their driving regulators, will greatly advance our understanding of T cell heterogeneity and function in tumor tissue. Thus, we have discovered a molecular framework that will enable study of the regulation of distinct CD8+ T cell populations in tumor tissue. Understanding the heterogeneity of CD8+ TILs at such resolution and how the spectrum of CD8+ TILs shifts in response to different perturbations will be highly beneficial for both the evaluation and design of immunotherapies that target the CD8+ T cell response.
Given the similarity between T cell dysfunction in the tumor and viral settings, it is of interest to determine whether our gene modules could exist in CD8+ T cells during chronic viral infections. Indeed, different populations of dysfunctional CD8+ T cells have been previously described in chronic LCMV infection. Progenitor and terminal subsets of dysfunctional T cells have been described, marked by the expression of differential levels of PD-1 and distinct transcription factors [34]. Moreover, a novel CD8+ T cell population was identified recently in chronic LCMV and HIV infection [18–20, 35]. This population expresses Cxcr5, Tcf7, Bcl6, has high proliferative capacity, and gives rise to effector cells in response to PD1 blockade [20, 35]. A recent study leveraged single-cell CD8 T cell transcriptomic data along a time course of acute viral infection (LCMV Armstrong) to discover two distinct transcriptional populations early in the time course [36]. Of the populations defined, one transcriptionally resembled terminal effectors and the other a memory-like state. The authors then combine their findings with epigenetic data to show that the early transcriptional programs may predict the cells’ differentiation trajectories. Expanding such single-cell analyses to the chronic viral settings as well as to additional tumor models, and evaluating how the activation, activation/dysfunction, and dysfunction gene modules are expressed in these cells and whether the different CD8+ T cell populations present in chronic viral infection mirror populations present in cancer, will greatly advance our understanding of the dysfunctional phenotype (e.g. by defining precise gene signatures), its heterogeneity across tissues, and its underlying regulatory circuitry.
Regulation of CD8+ T cell dysfunction
Transcription factors
In addition to Gata3 [24], several other transcription factors have been shown to play a role in promoting CD8+ T cell dysfunction. The leucine zipper-containing transcription factor Maf was reported to be up-regulated in dysfunctional CD8+ TILs in murine melanoma tumors and in TILs from melanoma patients when compared to naïve and effector CD8+ T cells [9]. Loss of Maf resulted in better tumor control while overexpression resulted in dampened TIL accumulation and anti-tumor activity. Martinez et al. showed that nuclear factor of activated T cells (NFAT), which is mostly known for its role in T cell activation, can promote CD8+ T cell dysfunction in both tumor and viral infection models when it is not bound to activator protein (AP)-1 transcription factors [25]. Interestingly, Batf, a transcription factor in the AP-1 family, was shown to drive T cell dysfunction during HIV infection in a pathway downstream of PD-1 [37]. In line with the connection of zinc regulation to T cell dysfunction, high expression of the zincfinger transcription factor Prdm1 (Blimp-1) has been shown to drive dysfunctional phenotype in chronic LCMV infection [38].
Some studies have started to delineate how the balance of transcription factor activity impacts on CD8+ T cell function. Eomesedermin (Eomes) was predicted in a computational analysis to be a key transcription factor involved in CD8+ T cell dysfunction during chronic LCMV infection (Doering et al., 2012). In both murine chronic LCMV infection and human HIV infection, Eomes and T-bet (Tbx21) expression were found to be anti-correlated, with Eomes expression marking a terminal subset of dysfunctional CD8+ T cells with reduced proliferative capacity and T-bet expression marking a progenitor subset with high proliferative capacity [34, 39]. Interestingly, IRF4 has been shown to regulate the ratio of T-bet to Eomes in chronic LCMV [40]. Indeed, IRF4 is required for sustaining anti-viral CD8+ T cell responses in chronic LCMV [41]. These observations provide a glimpse of the complex regulatory network that determines CD8+ T cell phenotype. It is important to note that expression of a single transcription factor is not sufficient to mark the dysfunctional state as transcription factors are often recruited to play context-dependent roles. For example, T-bet and Eomes have different roles at different stages of CD8+ T cell differentiation (reviewed in [42]).
In line with the balance of Eomes and T-bet being an important determinant of dysfunctional phenotype, Eomes is associated with the dysfunction module that we have defined, while T-bet is associated with the activation module. Additionally, both NFATc1 and Prdm1 are associated with the activation/dysfunction module. Thus, it is possible that other transcription factors in this module (e.g. Bhlhe40 and Hif1a, Table S4 in [24]) will have functional roles in determining dysfunctional phenotype.
The identification of transcription factors that drive dysfunctional phenotype and how they are expressed in the settings of chronic viral infection and cancer is essential to understanding how cells enter and maintain the dysfunctional state. Additionally, constructing the regulatory networks in which these transcription factors take part will be instrumental for elucidating nodes for intervention. To achieve this, data regarding the gene targets of relevant transcription factors in the setting of T cell dysfunction must be collected and combinatorial knockout experiments of key regulators will be helpful. Lastly, deeper characterization of the mechanisms by which identified transcription factors are regulated will inform as to cues that push cells to adopt dysfunctional phenotype.
Epigenetics
Beyond transcriptional programs per se, several studies have highlighted the importance of understanding the epigenetic changes associated with CD8+ T cell dysfunction [43–45]. These studies showed that the epigenetic landscape of CD8+ T cells significantly varies between the dysfunctional, memory, and effector CD8+ T cell states [44, 45]. Sen et al showed that differential regulatory regions in CD8+ T cells between acute and chronic LCMV infection were found mostly away from transcription start sites (TSS) and gene promoters, and that the dysfunction-specific epigenetic profiles observed in experimental mouse models are conserved in CD8+ T cells from human HIV infection. Importantly, Pauken et al showed using ATAC-seq in the LCMV viral model that open chromatin regions in dysfunctional CD8+ T cells are not significantly altered following PD-1 blockade, indicating that dysfunctional cells are “locked in” to a state that is not reversed upon co-inhibitory receptor blockade, thereby limiting its efficacy. Thus, characterizing the heterogeneity across subpopulations of dysfunctional CD8+ T cells with respect to their epigenetic states and their plasticity in both the chronic viral infection and tumor settings will be crucial for improving therapeutic targeting and diagnosis.
Temporal and spatial variables
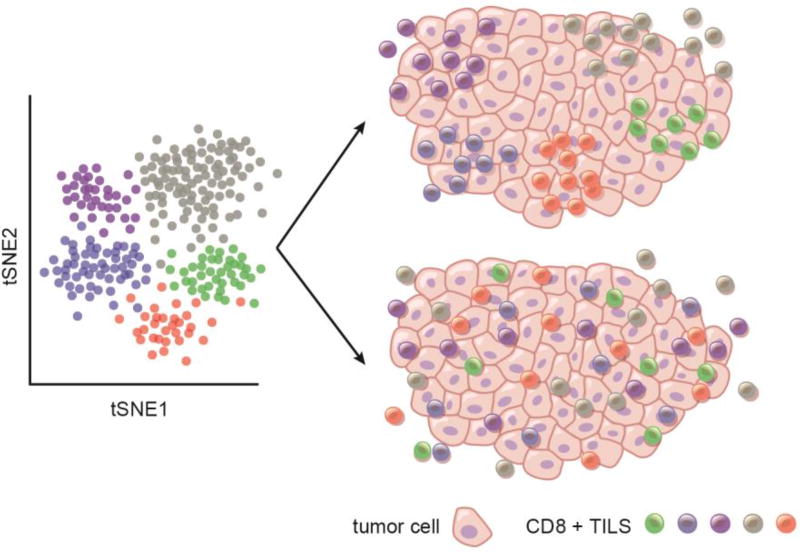
The existence of multiple CD8+ T cell populations in tumor tissue that express distinct combinations of our refined gene modules raises important questions with regard to the possible temporal and spatial regulation of CD8+ T cell functional states in tumor tissue. Are distinct CD8+ TILs populations localized to discrete tissue niches that direct their transcriptional and functional phenotype? Or, are CD8+ TILs randomly scattered in tumor tissue, indicating that anatomic localization has no bearing on their transcriptional/functional phenotype (Figure 2)? Do the more functional CD8+ T cells represent those newly recruited to the tumor and/or are they clustered and localized in a more favorable environment for T cell activation, potentially tertiary lymphoid structures (TLS)?
Figure 2. Spatial organization of CD8+ T cells.
Representation of CD8+ TILs populations as defined by clustering analysis (tSNE plot). Two models of spatial organization of CD8+ T cells are being proposed: CD8+ TILs expressing similar transcriptomes either occupy distinct tissue niches that direct their transcriptional and functional phenotype or are driven by cell-intrinsic programs and are randomly located in tumor tissue.
The localization of lymphocytes in tumor tissue is likely regulated through a chemokine network. It is possible that CD8+ T cells that express a distinct chemokine receptor profile will traffic to and reside in specific niches, which in turn will direct their acquisition of a specific functional state. For example, TLS have been identified in a number of tumors and their presence is linked to better patient prognosis [46, 47]. Follicular helper T cells are a major constituent of TLS and were identified as a major cellular source of CXCL13 [48]. Interestingly, CXCR5, the receptor for CXCL13, was found recently to mark a subpopulation of CD8+ T cells that bears a follicular helper T cell-like signature during chronic LCMV infection [18, 20]. A similar population of CXCR5+ CD8+ T cells that inversely correlates with viral load was also identified in HIV patients [18]. While it is unclear whether such a population of CD8+ T cells exists in tumor tissue, the presence of CXCR5 mRNA in tumor tissue has been detected and associated with better breast cancer patient survival [49]. Thus, should a similar population of CD8+ T cells exist in tumor tissue, one might speculate that CXCR5+ CD8 T cells would be recruited to TLS where they would receive a better T cell activation signal and thus better sustain functionality.
Implications for therapy
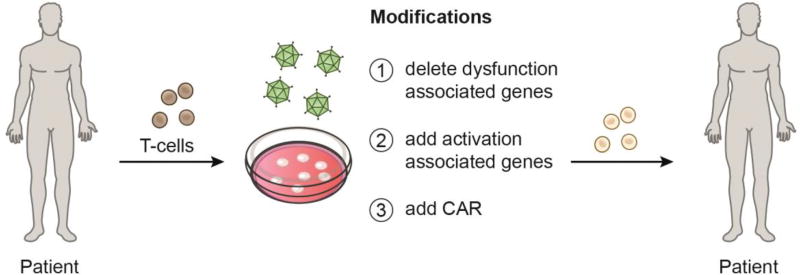
Immunotherapies, such as co-inhibitory receptor blockade and adoptive T cell transfer (ACT), are transforming the treatment for advanced cancer [50]. Understanding how these therapies work and finding ways to improve them is an active area of research [51–55]. Technological advances such as in-depth genomic analyses will be instrumental in this regard. Indeed, such analyses have shown that CTLA-4 and PD-1 blockade therapies achieve efficacy through distinct mechanisms [56]. Moreover, transcriptome analyses of responders and non-responders to PD-1 blockade in cancer patients have begun to elucidate distinct pathways that may be targeted to improve this therapy [57–59]. Furthermore, the identification of distinct gene modules associated with different functions in CD8+ TIL has laid the groundwork for discovery of novel targets that have important implications for current therapeutic approaches [24]. Despite the promising results of PD-1 and CTLA-4 blockade, many patients don’t respond or fail to develop durable responses. Examination of surface receptors in the activation/dysfunction module could lead to the discovery of novel co-inhibitory receptors or co-stimulatory receptors that can complement these therapies. The identification of distinct genes that drive CD8+ T cell dysfunction and activation can enable engineering of T cells that are resistant to dysfunction for use in ACT (Figure 3). Such approaches can be combined with expression of a chimeric antigen receptor (CAR) to engineer powerful anti-tumor T cells.
Figure 3. Engineering potent anti-tumor T cells.
The identification of genes specifically associated with activation versus dysfunction enables more precise engineering of T cells for use in adoptive T cell therapies. Deletion of dysfunction-associated genes and/or addition of activation-associated genes can further be combined with addition of a chimeric antigen receptor (CAR) to engineer T cells with desired functions against tumors.
Understanding how the state of CD8+ T cells that express the gene modules associated with different functions shift over time and in response to immunotherapy could have prognostic value. In this regard it is important to note that not only does it matter what transcriptional and functional phenotype a T cell may express but where it is located. Thus, the use of integrative analysis that combines platforms that assess spatial information such as the immunoscore [60, 61] together with novel computational approaches (eg. CIBERSORT [62]) that assess cellular composition in tissue will provide even more accurate assessment of immune correlation with clinical outcome.
Concluding Remarks and Future Perspectives
Single-cell analysis has the potential to identify new and more refined gene modules in CD8+ T cells. It will be important to discern transient from more stable states, potentially with epigenetic analysis coupled with perturbations (see Outstanding Questions). Testing whether the different transcriptional populations identified by single-cell transcriptomics are distinct in cytotoxicity, cytokine secretion, proliferative capacity, and T cell receptor expression will resolve their functional relevance. Connecting the transcriptional state of a T cell to its functional role holds great promise to advance our understanding of CD8+ T cell function and regulation within different systems. Various technologies (e.g. drop-seq, 10×, in-drop, and plate-based technology) currently enable the fast and cost-effective generation of single-cell transcriptomic data at high capture rates. By leveraging these technologies with appropriate computational frameworks and biological validation, it is now possible to transcriptionally characterize in a per-system and per-condition manner the T cell transcriptional landscape across a multitude of environments (e.g. cancer, chronic virus infection) and conditions (e.g. immunotherapy). Thus, connecting the transcriptional heterogeneity observed in T cells with functional phenotype will enable significant advances in our understanding of CD8+ T cell dysfunction and functional potential.
Outstanding Questions Box.
-
-
Is the spectrum of CD8+ T cell phenotypes (transcriptional and functional) in chronic viral infection similar to that in cancer?
-
-
How do spatial versus temporal variables impact on transcriptional programs and functional CD8+ T cell states in tissue?
-
-
What is the transcription factor network that gives rise to dysfunctional CD8+ T cells?
-
-
What cell surface markers can reliably distinguish different CD8+ T cell states?
-
-
How does immunotherapy change the spectrum of CD8+ T cell states observed in chronic viral infection and cancer?
Trends Box.
-
-
Recent advances have identified novel CD8+ T cell functional states in chronic inflammatory conditions, associated with distinct transcriptional programs.
-
-
Single cell analysis has revealed extensive transcriptional heterogeneity in the CD8+ T cell response in cancer
-
-
CRISPR/Cas9 genome-editing in mature CD8+ T cells has enabled testing of candidate regulators in vivo
-
-
Analysis of the chromatin landscape in CD8+ T cells has revealed distinct epigenetic changes associated with distinct functional states
Glossary
- Gene Signature
a set of genes that defines a specific cell state. Typically, gene signatures include tens to hundreds of genes. Example: an activated CD8+ T cell signature would consist of a set of genes upregulated when CD8+ T cells are activated, and downregulated otherwise. Importantly, the identification of optimal and precise signatures for various cell states is the subject of ongoing research.
- Gene Module
a set of genes that have similar expression profiles across conditions tested. Gene modules may include hundreds or even thousands of genes. Unlike a gene signature, a module does not necessarily define a specific cell state. Example: A dysfunction/activation module for CD8+ T cells consists of genes that are upregulated in both the dysfunction and activation states.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Wherry EJ, Kurachi M. Molecular and cellular insights into T cell exhaustion. Nat Rev Immunol. 2015;15(8):486–99. doi: 10.1038/nri3862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zajac AJ, et al. Viral immune evasion due to persistence of activated T cells without effector function. J Exp Med. 1998;188(12):2205–13. doi: 10.1084/jem.188.12.2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moskophidis D, et al. Virus persistence in acutely infected immunocompetent mice by exhaustion of antiviral cytotoxic effector T cells. Nature. 1993;362(6422):758–61. doi: 10.1038/362758a0. [DOI] [PubMed] [Google Scholar]
- 4.Elsaesser H, Sauer K, Brooks DG. IL-21 is required to control chronic viral infection. Science. 2009;324(5934):1569–72. doi: 10.1126/science.1174182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Frohlich A, et al. IL-21R on T cells is critical for sustained functionality and control of chronic viral infection. Science. 2009;324(5934):1576–80. doi: 10.1126/science.1172815. [DOI] [PubMed] [Google Scholar]
- 6.Yi JS, Du M, Zajac AJ. A vital role for interleukin-21 in the control of a chronic viral infection. Science. 2009;324(5934):1572–6. doi: 10.1126/science.1175194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tinoco R, et al. Cell-intrinsic transforming growth factor-beta signaling mediates virus-specific CD8+ T cell deletion and viral persistence in vivo. Immunity. 2009;31(1):145–57. doi: 10.1016/j.immuni.2009.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wilson EB, Brooks DG. The role of IL-10 in regulating immunity to persistent viral infections. Curr Top Microbiol Immunol. 2011;350:39–65. doi: 10.1007/82_2010_96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Giordano M, et al. Molecular profiling of CD8 T cells in autochthonous melanoma identifies Maf as driver of exhaustion. EMBO J. 2015;34(15):2042–58. doi: 10.15252/embj.201490786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang Y, et al. IL-6 plays a unique role in initiating c-Maf expression during early stage of CD4 T cell activation. J Immunol. 2005;174(5):2720–9. doi: 10.4049/jimmunol.174.5.2720. [DOI] [PubMed] [Google Scholar]
- 11.Yi JS, Cox MA, Zajac AJ. T-cell exhaustion: characteristics, causes and conversion. Immunology. 2010;129(4):474–81. doi: 10.1111/j.1365-2567.2010.03255.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hwang S, et al. Blimp-1-mediated CD4 T cell exhaustion causes CD8 T cell dysfunction during chronic toxoplasmosis. J Exp Med. 2016;213(9):1799–818. doi: 10.1084/jem.20151995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brooks DG, et al. Intrinsic functional dysregulation of CD4 T cells occurs rapidly following persistent viral infection. J Virol. 2005;79(16):10514–27. doi: 10.1128/JVI.79.16.10514-10527.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Crawford A, et al. Molecular and transcriptional basis of CD4(+) T cell dysfunction during chronic infection. Immunity. 2014;40(2):289–302. doi: 10.1016/j.immuni.2014.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oxenius A, Zinkernagel RM, Hengartner H. Comparison of activation versus induction of unresponsiveness of virus-specific CD4+ and CD8+ T cells upon acute versus persistent viral infection. Immunity. 1998;9(4):449–57. doi: 10.1016/s1074-7613(00)80628-7. [DOI] [PubMed] [Google Scholar]
- 16.Schulze Zur Wiesch J, et al. Broadly directed virus-specific CD4+ T cell responses are primed during acute hepatitis C infection, but rapidly disappear from human blood with viral persistence. J Exp Med. 2012;209(1):61–75. doi: 10.1084/jem.20100388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fahey LM, et al. Viral persistence redirects CD4 T cell differentiation toward T follicular helper cells. J Exp Med. 2011;208(5):987–99. doi: 10.1084/jem.20101773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.He R, et al. Follicular CXCR5-expressing CD8+ T cells curtail chronic viral infection. Nature. 2016;537(7620):412–428. doi: 10.1038/nature19317. [DOI] [PubMed] [Google Scholar]
- 19.Leong YA, et al. CXCR5(+) follicular cytotoxic T cells control viral infection in B cell follicles. Nat Immunol. 2016;17(10):1187–96. doi: 10.1038/ni.3543. [DOI] [PubMed] [Google Scholar]
- 20.Im SJ, et al. Defining CD8+ T cells that provide the proliferative burst after PD-1 therapy. Nature. 2016;537(7620):417–421. doi: 10.1038/nature19330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wherry EJ, et al. Molecular signature of CD8+ T cell exhaustion during chronic viral infection. Immunity. 2007;27(4):670–84. doi: 10.1016/j.immuni.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 22.Doering TA, et al. Network analysis reveals centrally connected genes and pathways involved in CD8+ T cell exhaustion versus memory. Immunity. 2012;37(6):1130–44. doi: 10.1016/j.immuni.2012.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baitsch L, et al. Exhaustion of tumor-specific CD8(+) T cells in metastases from melanoma patients. J Clin Invest. 2011;121(6):2350–60. doi: 10.1172/JCI46102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Singer M, et al. A Distinct Gene Module for Dysfunction Uncoupled from Activation in Tumor-Infiltrating T Cells. Cell. 2016;166(6):1500–1511 e9. doi: 10.1016/j.cell.2016.08.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martinez GJ, et al. The transcription factor NFAT promotes exhaustion of activated CD8(+) T cells. Immunity. 2015;42(2):265–78. doi: 10.1016/j.immuni.2015.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barber DL, et al. Restoring function in exhausted CD8 T cells during chronic viral infection. Nature. 2006;439(7077):682–7. doi: 10.1038/nature04444. [DOI] [PubMed] [Google Scholar]
- 27.Odorizzi PM, et al. Genetic absence of PD-1 promotes accumulation of terminally differentiated exhausted CD8+ T cells. J Exp Med. 2015;212(7):1125–37. doi: 10.1084/jem.20142237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Duraiswamy J, et al. Phenotype, function, and gene expression profiles of programmed death-1(hi) CD8 T cells in healthy human adults. J Immunol. 2011;186(7):4200–12. doi: 10.4049/jimmunol.1001783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Speiser DE, Ho PC, Verdeil G. Regulatory circuits of T cell function in cancer. Nat Rev Immunol. 2016;16(10):599–611. doi: 10.1038/nri.2016.80. [DOI] [PubMed] [Google Scholar]
- 30.Tirosh I, et al. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science. 2016;352(6282):189–96. doi: 10.1126/science.aad0501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim HJ, et al. Stable inhibitory activity of regulatory T cells requires the transcription factor Helios. Science. 2015;350(6258):334–9. doi: 10.1126/science.aad0616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wagner A, Regev A, Yosef N. Revealing the vectors of cellular identity with single-cell genomics. Nat Biotechnol. 2016;34(11):1145–1160. doi: 10.1038/nbt.3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shekhar K, et al. Comprehensive Classification of Retinal Bipolar Neurons by Single-Cell Transcriptomics. Cell. 2016;166(5):1308–1323 e30. doi: 10.1016/j.cell.2016.07.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Paley MA, et al. Progenitor and terminal subsets of CD8+ T cells cooperate to contain chronic viral infection. Science. 2012;338(6111):1220–5. doi: 10.1126/science.1229620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Utzschneider DT, et al. T Cell Factor 1-Expressing Memory-like CD8(+) T Cells Sustain the Immune Response to Chronic Viral Infections. Immunity. 2016;45(2):415–27. doi: 10.1016/j.immuni.2016.07.021. [DOI] [PubMed] [Google Scholar]
- 36.Kakaradov B, et al. Early transcriptional and epigenetic regulation of CD8+ T cell differentiation revealed by single-cell RNA sequencing. Nat Immunol. 2017;18(4):422–432. doi: 10.1038/ni.3688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Quigley M, et al. Transcriptional analysis of HIV-specific CD8+ T cells shows that PD-1 inhibits T cell function by upregulating BATF. Nat Med. 2010;16(10):1147–51. doi: 10.1038/nm.2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shin H, et al. A role for the transcriptional repressor Blimp-1 in CD8(+) T cell exhaustion during chronic viral infection. Immunity. 2009;31(2):309–20. doi: 10.1016/j.immuni.2009.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Buggert M, et al. T-bet and Eomes are differentially linked to the exhausted phenotype of CD8+ T cells in HIV infection. PLoS Pathog. 2014;10(7):e1004251. doi: 10.1371/journal.ppat.1004251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nayar R, et al. IRF4 Regulates the Ratio of T-Bet to Eomesodermin in CD8+ T Cells Responding to Persistent LCMV Infection. PLoS One. 2015;10(12):e0144826. doi: 10.1371/journal.pone.0144826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Grusdat M, et al. IRF4 and BATF are critical for CD8(+) T-cell function following infection with LCMV. Cell Death Differ. 2014;21(7):1050–60. doi: 10.1038/cdd.2014.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kaech SM, Cui W. Transcriptional control of effector and memory CD8+ T cell differentiation. Nat Rev Immunol. 2012;12(11):749–61. doi: 10.1038/nri3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Youngblood B, et al. Chronic virus infection enforces demethylation of the locus that encodes PD-1 in antigen-specific CD8(+) T cells. Immunity. 2011;35(3):400–12. doi: 10.1016/j.immuni.2011.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sen DR, et al. The epigenetic landscape of T cell exhaustion. Science. 2016;354(6316):1165–1169. doi: 10.1126/science.aae0491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pauken KE, et al. Epigenetic stability of exhausted T cells limits durability of reinvigoration by PD-1 blockade. Science. 2016;354(6316):1160–1165. doi: 10.1126/science.aaf2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dieu-Nosjean MC, et al. Tertiary lymphoid structures in cancer and beyond. Trends Immunol. 2014;35(11):571–80. doi: 10.1016/j.it.2014.09.006. [DOI] [PubMed] [Google Scholar]
- 47.Hiraoka N, Ino Y, Yamazaki-Itoh R. Tertiary Lymphoid Organs in Cancer Tissues. Front Immunol. 2016;7:244. doi: 10.3389/fimmu.2016.00244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gu-Trantien C, et al. CD4(+) follicular helper T cell infiltration predicts breast cancer survival. J Clin Invest. 2013;123(7):2873–92. doi: 10.1172/JCI67428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Razis E, et al. Improved outcome of high-risk early HER2 positive breast cancer with high CXCL13-CXCR5 messenger RNA expression. Clin Breast Cancer. 2012;12(3):183–93. doi: 10.1016/j.clbc.2012.03.006. [DOI] [PubMed] [Google Scholar]
- 50.Khalil DN, et al. The future of cancer treatment: immunomodulation, CARs and combination immunotherapy. Nat Rev Clin Oncol. 2016;13(5):273–90. doi: 10.1038/nrclinonc.2016.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Klebanoff CA, Rosenberg SA, Restifo NP. Prospects for gene-engineered T cell immunotherapy for solid cancers. Nat Med. 2016;22(1):26–36. doi: 10.1038/nm.4015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Restifo NP, Smyth MJ, Snyder A. Acquired resistance to immunotherapy and future challenges. Nat Rev Cancer. 2016;16(2):121–6. doi: 10.1038/nrc.2016.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lim WA, June CH. The Principles of Engineering Immune Cells to Treat Cancer. Cell. 2017;168(4):724–740. doi: 10.1016/j.cell.2017.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mahoney KM, Rennert PD, Freeman GJ. Combination cancer immunotherapy and new immunomodulatory targets. Nat Rev Drug Discov. 2015;14(8):561–84. doi: 10.1038/nrd4591. [DOI] [PubMed] [Google Scholar]
- 55.Sharma P, Allison JP. Immune checkpoint targeting in cancer therapy: toward combination strategies with curative potential. Cell. 2015;161(2):205–14. doi: 10.1016/j.cell.2015.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gubin MM, et al. Checkpoint blockade cancer immunotherapy targets tumour-specific mutant antigens. Nature. 2014;515(7528):577–81. doi: 10.1038/nature13988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ascierto ML, et al. The Intratumoral Balance between Metabolic and Immunologic Gene Expression Is Associated with Anti-PD-1 Response in Patients with Renal Cell Carcinoma. Cancer Immunol Res. 2016;4(9):726–33. doi: 10.1158/2326-6066.CIR-16-0072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hugo W, et al. Genomic and Transcriptomic Features of Response to Anti-PD-1 Therapy in Metastatic Melanoma. Cell. 2016;165(1):35–44. doi: 10.1016/j.cell.2016.02.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zaretsky JM, et al. Mutations Associated with Acquired Resistance to PD-1 Blockade in Melanoma. N Engl J Med. 2016;375(9):819–29. doi: 10.1056/NEJMoa1604958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mlecnik B, et al. Integrative Analyses of Colorectal Cancer Show Immunoscore Is a Stronger Predictor of Patient Survival Than Microsatellite Instability. Immunity. 2016;44(3):698–711. doi: 10.1016/j.immuni.2016.02.025. [DOI] [PubMed] [Google Scholar]
- 61.Mlecnik B, et al. The tumor microenvironment and Immunoscore are critical determinants of dissemination to distant metastasis. Sci Transl Med. 2016;8(327):327ra26. doi: 10.1126/scitranslmed.aad6352. [DOI] [PubMed] [Google Scholar]
- 62.Newman AM, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12(5):453–7. doi: 10.1038/nmeth.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]