Abstract
Autosomal recessive non-syndromic hearing loss (ARNSHL) is defined as a genetically heterogeneous disorder. The aim of the present study was to screen for pathogenic variants in an Iranian pedigree with ARNSHL. Next-generation targeted sequencing of 127 deafness genes in the proband detected two novel variants, a homozygous missense variant in PTPRQ (c.2599 T>C, p.Ser867Pro and a heterozygous missense variant in MYO1A (c.2804 T>C, p.Ile935Thr), both of which were absent in unaffected sibs and two hundred unaffected controls. Our results suggest that the homozygous PTPRQ variant maybe the pathogenic variant for ARNSHL due to the recessive nature of the disorder. Nevertheless, the heterozygous MYO1A may also be involved in this disorder due to the multigenic pattern of ARNSHL. Our data extend the mutation spectrum of PTPRQ and MYO1A, and have important implications for genetic counseling in unaffected sibs of this family. In addition, PTPRQ and MYO1A pathogenic variants have not to date been reported in the Iranian population.
Keywords: Hearing Loss, MYO1A, Novel Variant, PTPRQ
Introduction
Hearing impairment is one of the most common sensorineural disorders in humans, affecting approximately one in 500-1,000 newborns. Hereditary hearing impairments are mainly transmitted in an autosomal dominant or recessive fashion (1) with mitochondrial (2) or X-linked (3) inheritance reported in frequently. Considering the isolated forms, about 80% of hereditary deafness cases manifest as autosomal recessive non-syndromic hearing loss (ARNSHL) (4). To date, more than 100 genes have been implicated in ARNSHL (5).
Molecular diagnosis plays a key role in clinical management, prognosis evaluation and prenatal diagnosis (PND) for ARNSHL families (6). However, the genetic heterogeneity of hearing impairment had undermined genetic diagnosis in most cases until recently. With the advent of next-generation sequencing (NGS) technology, heterogeneous disorders are now open to routine genetic testing and comprehensive genetic analysis. Targeted NGS of the identified deafness genes (a “gene panel” that generally covers the exons and flanking intronic sequences) can provide a basis for a broad first-step study of pathogenic variants in ARNSHL (7). We thus aimed to screen the deafness gene panel in a proband with ARNSHL and of Iranian origin. Herein, we report two novel missense pathogenic variants in PTPRQ and MYO1A, both of which may explain the ARNSHL phenotype in the proband.
Case report
The proband is a 23-year-old Iranian male with a clinical diagnosis of hearing impairment (Fig .1A). No exact complications have been reported during his perinatal period. However, at age of 21 months, his mother suspected hearing loss because of his poor response to sound. He was born from a consanguineous marriage (first cousin unaffected parents). There was no family history of inherited diseases such as ARNSHL or congenital malformations in his pedigree. Two hundred unrelated subjects of Iranian origin with normal hearing were screened for the pathogenic variants as controls. Written informed consent was obtained from all participants according to the guidelines of the Ethics Committee of the Ministry of Health and Medical Education of Iran.
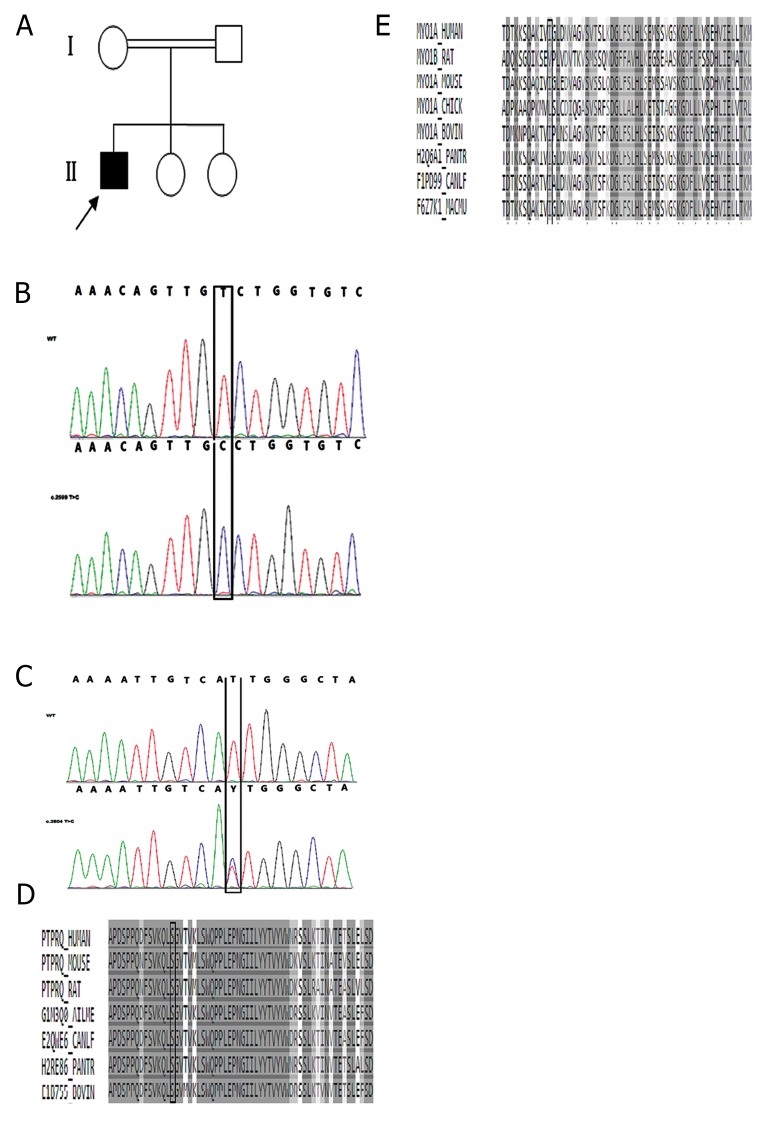
Fig.1.
Genetic analysis of the ARNSHL proband. A. Pedigree of family B with ARNSHL, the proband is denoted in black. Partial sequences of B. PTPRQ, C. MYO1A in the proband showing that homozygous mutation (c.2599T>C) in PTPRQ and the heterozygous mutation (c.2804 T>C) in MYO1A, both co-segregating with the phenotype. Mutated nucleotides are marked with vertical lines (black). Protein alignment shows conservation of residue D. 867 in PTPRQ, and E.935 in MYO1A across seven and eight species respectively. These two novel mutations occur at evolutionarily conserved amino acid positions marked with vertical lines (black).
Blood samples were collected from the proband and his parents. Genomic DNA was extracted from blood samples of all participants using the standard salting out method (8). Targeted NGS was carried out by using a custom designed NimbleGen chip capturing 127 hearing impairment genes including but not limited to PTPRQ, GJB6, MYO1A, MYO7A, SLC26A4, and MTRNR1 (BGI-Clinical Laboratories, China). The genomic region containing the variant were amplified (primer sequences are available upon request) in 25 µL volumes and 35 cycles: 95°C for one minute, 65°C for 40 seconds and 72°C for one minute and then the polymerase chain reaction (PCR) product was sequenced with direct sanger sequencing carried out with automated DNA sequencer (ABI3130, Applied Biosystems, USA) (validation with a second independent sample of DNA) to confirm presence of potential pathogenic variants in the proband and his parents for segregation analysis.
The frequency of the detected variants was checked in the 1000 genomes database (http://WWW.1000genomes. org/.). Next, in silico functional prediction of the missense variants were performed with bioinformatics tools including Sorting Intolerant from Tolerant (SIFT) (9), Polymorphism Phenotyping V2 (PolyPhen2) (10) and Mutation Taster (11).
All genomic data analysis including read alignment, variant calling and novel mutation identification was undertaken by BGI that detected two novel variants in PTPRQ and MYO1A co-segregating in the family. The variant in exon 17 of PTPRQ (c.2599T>C) results in a serine to proline substitution at codon 867 (Ser867Pro) (Fig .1B). The second variant was found in exon 26 of MYO1A (c.2804 T>C) (Fig .1C), leading to an isoleucine to threonine substitution (Ile935Thr). Both missense variants were predicted to be pathogenicity the three prediction tools (Table 1). Reported mutations in PTPRQ and MYO1A are summarized in Table 2, and 3 respectively. Interestingly, no pathogenic variants were identified in the other 125 genes in the proband. The two detected variants were confirmed by sanger sequencing. Both missense variants alter highly evolutionary conserved amino acids (Fig .1D, E). To confirm pathogenicity, presence of the two variants was checked in unaffected individuals in the pedigree. The unaffected parents and one of his sisters (II-3) were heterozygous for the PTPRQ variant while the MYO1A variant was only identified in the mother in a heterozygous state. Both variants were not detected in the other sister (II-2) and the 200 healthy controls of Iranian origin. Figure 2 shows the locations of these variants.
Table 1.
Results of in silico prediction tools for functional effect of the novel missense mutations
| Gene/Variant | SIFT score | PolyPhen score | Mutation taster |
|---|---|---|---|
| ENST00000614701, S867P | 0.787 (possibly damaging) | disease causing | |
| ENST00000300119, I935T | 0.908 (possibly damaging) | disease causing | |
Table 2.
Reported mutations in PTPRQ
| Origin | Pathogenic variant | Protein effect | Domain | Exon | Type of mutation | Inheritance pattern | Zygosity |
|---|---|---|---|---|---|---|---|
| Palestinian | c.1285C>T | p.Gln429Stop | EC | 9 | Nonsense | AR | Homozygous |
| Dutch | c.1491T>A | p.Tyr497Stop | EC | 10 | Nonsense | AR | Homozygous |
| Moroccan | c.1369A>G | p.Ala457Gly | EC | 10 | Missense | AR | Homozygous |
| Chinese | c.3125A>G | p.Asp1042Gly | EC | 20 | Missense | AR | Homozygous |
| Chinese | c.5981A>G | p.Glu1994Gly | EC | 37 | Missense | AR | Homozygous |
| Japanese | c.166C>G | p.Pro56Ala | EC | 2 | Missense | AR | Compoundheterozygous |
| Japanese | c.1261C>T | p.Arg421Stop | EC | 9 | Nonsense | AR | Homozygous |
| Japanese | c.4046T>C | p.Met1349Thr | EC | 25 | Missense | AR | Compoundheterozygous |
| Japanese | c.6453+3delA | - | CP | 41 | Splice site | AR | Compoundheterozygous |
| Iranian | c.2599T>C | Ser867Pro | EC | 17 | Missense | AR | Homozygous |
CP; Cytoplasmic domain, EC; Extracellular domain, and AR; Autosomal recessive.
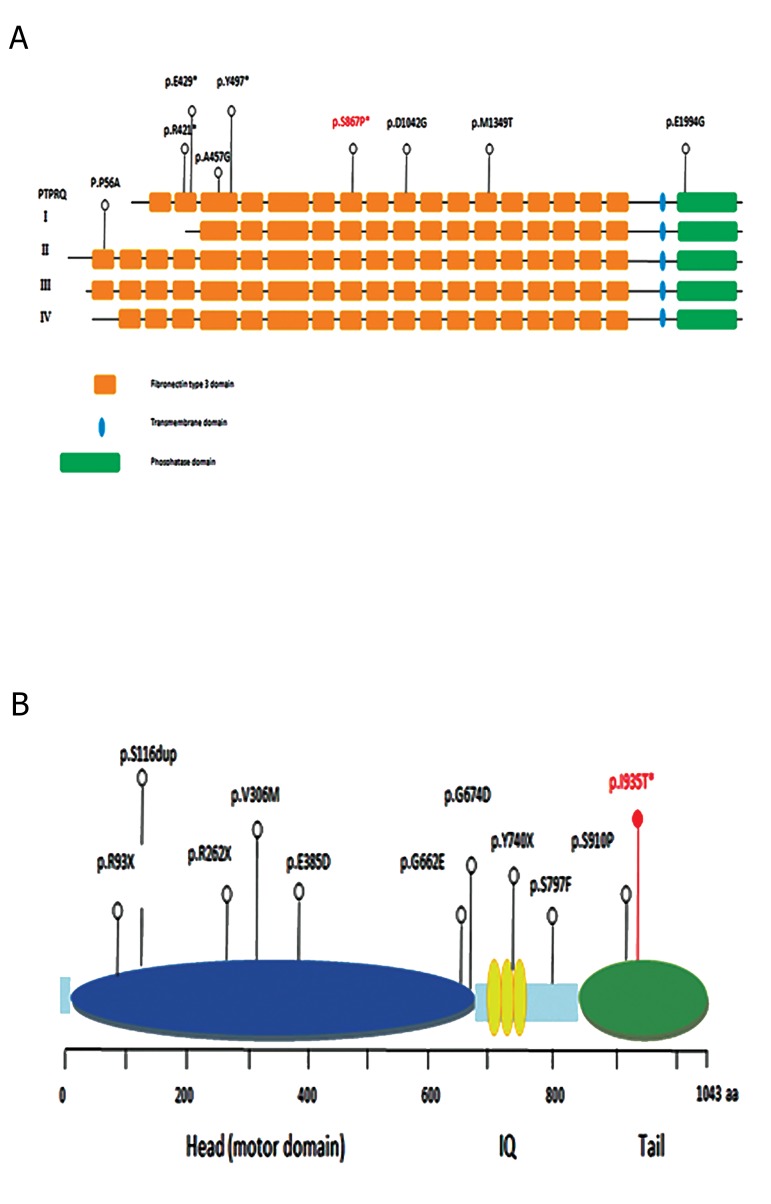
Fig.2.
Diagram structure of PTPRQ and Myosin-IA proteins. Schematic of A. PTPRQ and B. Myosin-IA proteins show the locations of the pathogenic variants in humans. The two novel mutations reported in this study are shown in red font (p.Ser867Pro and p.Ile935Thr).
Table 3.
Reported mutations in MYO1A
| Origin | Pathogenic variant | Protein effect | Exon | Domain | Type of mutation | Inheritance pattern | Zygosity |
|---|---|---|---|---|---|---|---|
| Italian | 277C/T | R93X | 3 | Myosin motor | Nonsence | AD | Heterozygous |
| Italian | 349-350A | 349-350insCTT | 4 | Myosin motor | Insertion | AD | Heterozygous |
| Italian | 916G/A | V306M | 10 | Myosin motor | Missence | AD | Heterozygous |
| Italian | 1155G/T | E385D | 12 | Myosin motor | Missence | AD | Heterozygous |
| Italian | 1985G/A | G662E | 18 | Myosin motor | Missence | AD | Heterozygous |
| Italian | 2021G/A | G674D | 18 | Myosin motor | Missence | AD | Heterozygous |
| Italian | 2390C/T | S797F | 22 | - | Missence | AD | Heterozygous |
| Italian | 2728T/C | S910P | 25 | TH1 | Missence | AD | Heterozygous |
| Pakistani | c.784C>T | p.Arg262∗ | 10 | Myosin motor | nonsense | AD | Heterozygous |
| German | c.2220T>G | p.Tyr740∗ | 21 | IQ 2 | nonsense | AD | Heterozygous |
| Iranian | c.2804T>C | I935T | 26 | TH1 | Missence | AR/ compound heterozygous | Heterozygous |
TH1; Class I myosin tail homology, AD; Autosomal dominant, and AR; Autosomal recessive.
Conclusion
Here we report two novel missense variants in PTPRQ and MYO1A in an Iranian family displaying hearing loss. Protein Tyrosine Phosphatase, Receptor Type Q (PTPRQ) is a stereociliar membrane protein, composed of three domains which include an extracellular domain (containing 18 fibronectin III repeats), a membrane spanning domain (trans membrane domain) and a cytoplasmic domain (phosphatase domain) (12-14). It plays key roles in cell shape changes, regulation of actin filament organization and formation of stereocilia in hair cells of the inner ear (15) with its loss or malfunction resulting in shaft connector malformation of hair cell stereocilia (16).
The novel homozygous PTPRQ variant detected in the proband is located in the fibronectin type III-9 domain (extracellular domain). This extracellular domain is able to bind ligands including extracellular proteins, collagen and heparin as well as ligands on the cell (17-19). The wild-type residue is polar while the mutant residue is non-polar, thus likely to affect PTPRQ interactions with ligands.
Additionally, this is the first PTPRQ variant found in an Iranian population. To date, 9 variants in PTPRQ have been reported. All PTPRQ variants previously reported were detected in prelinguistic or congenital hearing loss patients (20). The proband in this study had congenital hearing loss, consistent with previous reports. Of the 9 reported PTPRQ variants, five were missense variants in the extracellular (EC) domain of which three were found in a homozygous state [p.A457G in Morocco (12), and p.D1042G and p.E1994G in China (21)] and two in a heterozygous state [p.P56A and p.M1349T in Japan (20)]. Three were also nonsense variants in the EC domain that were found in homozygous [p.Q429X in Palestine (13) and p.Y497X in Holland (112)] or heterozygous [p.R421X in Japan (20)] state. The ninth variant was a heterozygous splice site variant (c.6453+3delA) detected in a Japanese family (20).
We also identified a novel heterozygous variant in MYO1A as a potentially causative variant of congenital ARNSHL in the proband. MYO1A encodes Myosin- IA, a protein with 1043 amino acids, belonging to the myosin super family (22, 23). MYO1A contains three core domains, an N-terminal motor domain, a central neck region made up of IQ motifs and a tail region. MYO1A functions as an actin-based molecular motor and is implicated in directing the movement of organelles along the actin filaments (24).
Variants within this gene have been reported to cause ARNSHL (25). To date, 10 recessive variants in MYO1A have been shown to be associated with ARNSHL in patients of Italian, German and Pakistani descent. However, variants in MYO1A have not to date been reported in the Iranian population.
The c.2804 T>C variant located in the C-terminal tail homology-1 (TH1) domain, which is responsible for membrane binding (26). Therefore, missense variants that alter a nonpolar aliphatic amino acid to polar amino acids with a hydroxyl group may modify the interaction of the tail domain with membranous compartments and alter its movement. Therefore this novel variant is likely to negatively affect the function of the TH1 domain. ARNSHL has an autosomal recessive inheritance pattern and since neither parents nor the proband are homozygous, it is unlikely to be causal in this case. However, this variant might cause pathogenicity in case another variant is acquired in future generations and result in compound heterozygosity.
Our findings confirm that two novel variants in PTPRQ and MYO1A may be causative of ARNSHL in a consanguineous Iranian family. In conclusion, by using NGS in this study, we show that this method can be useful for detecting rare causative genetic variants in ARNSHL patients, such as those detected in MYO1A and PTPRQ.
Acknowledgments
The authors thank the Milad Genetic Counseling Center and all of the individuals for their participation in this study. There is no financially support and conflict of interest in this study.
Author’s Contributions
F.T.; Study conception and design. F.Gh.M.; Acquisition of data, analysis and interpretation of data, drafting of manuscript, critical revision. S.T., M.N.Z., J.M.A.; Analysis and interpretation of data. All authors read and approved the final manuscript.
References
- 1.Duman D, Tekin M. Autosomal recessive nonsyndromic deafness genes: a review. Front Biosci (Landmark Ed) 2012;17:2213–2236. doi: 10.2741/4046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cengiz FB, Duman D, Sırmacı A, Tokgöz-Yilmaz S, Erbek S, Öztürkmen-Akay H, et al. Recurrent and private MYO15A mutations are associated with deafness in the Turkish population. Genet Test Mol Biomarkers. 2010;14(4):543–550. doi: 10.1089/gtmb.2010.0039. [DOI] [PubMed] [Google Scholar]
- 3.Wang Q, Li R, Zhao H, Peters JL, Liu Q, Yang L, et al. Clinical and molecular characterization of a Chinese patient with auditory neuropathy associated with mitochondrial 12S rRNA T1095C mutation. Am J Med Genet A. 2005;133(1):27–30. doi: 10.1002/ajmg.a.30424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Babanejad M, Fattahi Z, Bazazzadegan N, Nishimura C, Meyer N, Nikzat N, et al. A comprehensive study to determine heterogeneity of autosomal recessive nonsyndromic hearing loss in Iran. Am J Med Genet A. 2012;158A(10):2485–2492. doi: 10.1002/ajmg.a.35572. [DOI] [PubMed] [Google Scholar]
- 5.Taghizadeh SH, Kazeminezhad SR, Sefidgar SA, Yazdanpanahi N, Tabatabaeifar MA, Yousefi A, et al. Investigation of LRTOMT gene (locus DFNB63) mutations in Iranian patients with autosomal recessive non-syndromic hearing loss. Int J Mol Cell Med. 2013;2(1):41–45. [PMC free article] [PubMed] [Google Scholar]
- 6.Chang MY, Choi BY. Strategy for the customized mass screening of genetic sensorineural hearing loss in koreans. Korean J Audiol. 2014;18(2):45–49. doi: 10.7874/kja.2014.18.2.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rehm HL. Disease-targeted sequencing: a cornerstone in the clinic. Nat Rev Genet. 2013;14(4):295–300. doi: 10.1038/nrg3463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988;16(3):1215–1215. doi: 10.1093/nar/16.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kumar P, Henikoff S, Ng PC. Predicting the effects of coding nonsynonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009;4(7):1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- 10.Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7(4):248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schwarz JM, Cooper DN, Schuelke M, Seelow D. MutationTaster2: mutation prediction for the deep-sequencing age. Nat Methods. 2014;11(4):361–362. doi: 10.1038/nmeth.2890. [DOI] [PubMed] [Google Scholar]
- 12.Schraders M, Oostrik J, Huygen PL, Strom TM, van Wijk E, Kunst HP, et al. Mutations in PTPRQ are a cause of autosomal-recessive nonsyndromic hearing impairment DFNB84 and associated with vestibular dysfunction. Am J Hum Genet. 2010;86(4):604–610. doi: 10.1016/j.ajhg.2010.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shahin H, Rahil M, Abu Rayan A, Avraham KB, King MC, Kanaan M, et al. Nonsense mutation of the stereociliar membrane protein gene PTPRQ in human hearing loss DFNB84. J Med Genet. 2010;47(9):643–645. doi: 10.1136/jmg.2009.075697. [DOI] [PubMed] [Google Scholar]
- 14.Oganesian A, Poot M, Daum G, Coats SA, Wright MB, Seifert RA, et al. Protein tyrosine phosphatase RQ is a phosphatidylinositol phosphatase that can regulate cell survival and proliferation. Proc Natl Acad Sci USA. 2003;100(13):7563–7568. doi: 10.1073/pnas.1336511100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Takenawa T, Itoh T. Phosphoinositides, key molecules for regulation of actin cytoskeletal organization and membrane traffic from the plasma membrane. Biochim Biophys Acta. 2001;1533(3):190–206. doi: 10.1016/s1388-1981(01)00165-2. [DOI] [PubMed] [Google Scholar]
- 16.Goodyear RJ, Legan PK, Wright MB, Marcotti W, Oganesian A, Coats SA, et al. A receptor-like inositol lipid phosphatase is required for the maturation of developing cochlear hair bundles. J Neurosci. 2003;23(27):9208–9219. doi: 10.1523/JNEUROSCI.23-27-09208.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chi-Rosso G, Gotwals PJ, Yang J, Ling L, Jiang K, Chao B, et al. Fibronectin type III repeats mediate RGD-independent adhesion and signaling through activated beta1 integrins. J Biol Chem. 1997;272(50):31447–31452. doi: 10.1074/jbc.272.50.31447. [DOI] [PubMed] [Google Scholar]
- 18.Napper CE, Drickamer K, Taylor ME. Collagen binding by the mannose receptor mediated through the fibronectin type II domain. Biochem J. 2006;395(3):579–586. doi: 10.1042/BJ20052027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weber P, Zimmermann DR, Winterhalter KH, Vaughan L. TenascinC binds heparin by its fibronectin type III domain five. J Biol Chem. 1995;270(9):4619–4623. doi: 10.1074/jbc.270.9.4619. [DOI] [PubMed] [Google Scholar]
- 20.Sakuma N, Moteki H, Azaiez H, Booth KT, Takahashi M, Arai Y, et al. Novel PTPRQ mutations identified in three congenital hearing loss patients with various types of hearing loss. Ann Otol Rhinol Laryngol. 2015;124(1):184S–192S. doi: 10.1177/0003489415575041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gao X, Su Y, Chen YL, Han MY, Yuan YY, Xu JC, et al. Identification of two novel compound heterozygous PTPRQ mutations associated with autosomal recessive hearing loss in a chinese family. PLoS One. 2015;10(4):e0124757–e0124757. doi: 10.1371/journal.pone.0124757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hasson T, Skowron JF, Gilbert DJ, Avraham KB, Perry WL, Bement WM, et al. Mapping of unconventional myosins in mouse and human. Genomics. 1996;36(3):431–439. doi: 10.1006/geno.1996.0488. [DOI] [PubMed] [Google Scholar]
- 23.Tyska MJ, Mooseker MS. MYO1A (brush border myosin I) dynamics in the brush border of LLC-PK1-CL4 cells. Biophys J. 2002;82(4):1869–1883. doi: 10.1016/S0006-3495(02)75537-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eisenberger T, Di Donato N, Baig SM, Neuhaus C, Beyer A, Decker E, et al. Targeted and genomewide NGS data disqualify mutations in MYO1A, the “DFNA48 gene”, as a cause of deafness. Hum Mutat. 2014;35(5):565–570. doi: 10.1002/humu.22532. [DOI] [PubMed] [Google Scholar]
- 25.Donaudy F, Ferrara A, Esposito L, Hertzano R, Ben-David O, Bell RE, et al. Multiple mutations of MYO1A, a cochlear-expressed gene, in sensorineural hearing loss. Am J Hum Genet. 2003;72(6):1571–1577. doi: 10.1086/375654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nambiar R, McConnell RE, Tyska MJ. Myosin motor function: the ins and outs of actin-based membrane protrusions. Cell Mol Life Sci. 2010;67(8):1239–1254. doi: 10.1007/s00018-009-0254-5. [DOI] [PMC free article] [PubMed] [Google Scholar]