Abstract
Muscular dystrophy is defined as the progressive wasting of skeletal muscles that is caused by inherited or spontaneous genetic mutations. Next-generation sequencing (NGS) has greatly improved the accuracy and speed of diagnosis for different types of muscular dystrophy. Advancements in depth of coverage, convenience, and overall reduced cost, have led to the identification of genetic modifiers that are responsible for phenotypic variability in affected patients. These genetic modifiers have been postulated to explain key differences in disease phenotypes including age of loss of ambulation, steroid-responsiveness, and the presence or absence of cardiac defects in patients with the same form of muscular dystrophy. Here we review and highlight recent findings on genetic modifiers of Duchenne and Facioscapulohumeral muscular dystrophies based on animal and clinical studies. These genetic modifiers hold great promise to be developed into novel therapeutic targets for the treatment of muscular dystrophies.
Keywords: muscular dystrophy, genetic modifier, variant, muscle disease, DMD
Introduction
Duchenne muscular dystrophy (DMD) is the most common form of muscular dystrophy worldwide with estimates of incidence ranging from 1:3500 to more recent estimates of 1:5000 live male births(1, 2). The genetic cause of DMD was identified in 1986 as pathogenic loss-of-function mutations in the dystrophin (DMD) gene that lead to insufficient, and sometimes undetectable, levels of a functional dystrophin protein(3). Conversely, in Becker muscular dystrophy (BMD) a partially-functional, truncated dystrophin protein is produced, typically resulting in a milder clinical pathology(3–6). DMD patients have severe, progressive skeletal muscle wasting and cardiac defects. Many DMD patients lose ambulation by their first decade of life(7). An interesting yet understudied aspect of DMD pathology is that approximately 20–25% of DMD boys develop significant cognitive issues that fall into the Autism spectrum disorder (ASD) scale(7, 8). One explanation for this variation in DMD boys is the alteration in expression levels of neuronal dystrophin protein isoforms, although it remains unclear as to why some DMD boys develop cognitive impairment and others have normal intelligence(9–13). As biochemical and molecular techniques for muscular dystrophy diagnostics improved, additional Dystrophin-interacting proteins were identified(14–16). These proteins are part of a Dystrophin-associated protein complex (DAPC) that bridges the actin cytoskeleton with the extracellular matrix (ECM)(17). With the advancement in genomic sequencing and coverage of the human genome, improved gene panels shifted muscular dystrophy diagnostic methods from predominantly pathology/histology-based to Sanger sequencing and later whole exome/genome sequencing (WES/WGS)(18–24).
The sequencing of increasingly larger numbers of exomes and genomes from healthy and diseased individuals permits large-scale genetic analyses of modifiers of diseases. The Exome Aggregation Consortium (ExAC) and 100,000 genomes project (UK) have banked large datasets of publically available genomic information(25, 26). Genomic analyses of children with Mendelian diseases have revealed that genetic modifiers of diseases have an incomplete penetrance of disease symptoms and pathogenicity(27). Indeed, many predicted loss-of-function (LoF) pathogenic mutations have been identified in healthy children and adults (“human knockouts”), suggesting an incomplete disease penetrance due to protective genetic modifiers(28, 29). As genomic sequencing has become more commonplace, the ability to analyze large amounts of data from different populations of healthy and diseased patients has become easier. Large scale databases and registries have been established to better share genomic and medical data among researchers studying muscle diseases(30, 31). These studies have yielded copious amounts of data on the wide spectrum of phenotypic variation among patients with muscular dystrophies. In conjunction with novel animal model screening platforms, these studies have identified several genetic modifiers of different forms of muscular dystrophy. Whole genomic sequencing among inbred strains of mice, such as the MRL super-healing strain which blocks dystrophic muscle pathology when mated to dystrophin-deficient mdx mice, have yielded valuable genetic modifiers of disease pathology(32). This review will focus on the identification and functional consequences of genetic modifiers in the two most prevalent forms of muscular dystrophy in humans: DMD and Facioscapulohumeral muscular dystrophy (FSHD). We also highlight current advancements in the clinic for these diseases and how genetic modifiers identified from both patient studies and animal models have led to novel therapeutic targets that may affect disease outcomes. These genetic modifiers hold the potential for ameliorating muscular dystrophies, and offer hope for patients with these debilitating diseases.
Genetic modifiers of Duchenne muscular dystrophy
A large number of the Duchenne muscular dystrophy genetic modifiers have been identified in dystrophin-deficient animal models and more recently in genomic analyses of DMD patient cohorts (Table 1). Since the discovery of the first mdx mouse arising from a spontaneous mutation in a mouse colony from the United Kingdom, many additional dystrophin-deficient vertebrate animal models have been generated and phenotypically evaluated(33–36). Mdx mice display progressive skeletal muscle weakness but do not share the same reduced lifespan and early death as observed in DMD patients. This is thought to be due to increased expression of the dystrophin protein analog utrophin, which is expressed during embryonic myogenesis but is silenced during adult myogenesis(37, 38) (Figure 1). An alternative explanation for this phenomenon may be the increased presence in mdx muscle of revertant (dystrophin-positive) myofibers, resulting from undefined RNA-splicing or naturally occurring exon-skipping mechanisms(39–41). Exogenous overexpression of utrophin or compounds that can activate utrophin expression in skeletal muscle remains a promising therapeutic strategy for DMD(42). Transgenic mice that overexpress utrophin on a dystrophin-deficient background have been shown to rescue both the muscle pathology and motor deficits, thereby preventing muscular dystrophy disease progression(43). Drug compounds that induce utrophin transcriptional activation in dystrophin-deficient skeletal muscle (SMT C1100; Ezutromid) are in current clinical trials in DMD boys(44). Ezutromid induces transcription of utrophin mRNA in adult muscle where it is transcriptionally silenced, and thereby results in expression of utrophin protein to compensate for the lack of functional dystrophin protein(45).
Table 1.
Genes with Polymorphic Significance in the Clinical Manifestation of Muscular Dystrophies
| Gene Name | Abbreviation | Normal Function | Disease Modification | Modification-Associated Phenotype |
|---|---|---|---|---|
| Genes with Identified Mutation or Significant Variant Contributing to Pathological Variation | ||||
| α-Actinin-3 | ACTN3 | Skeletal muscle-specific actin binding protein | Homozygosity for SNP R577X in DMD patients | Reduced susceptibility to stretch-induced damage, enhanced fatigue recovery, and slowed dystrophic progression |
| Annexin A6 | ANXA6 | Calcium-dependent membrane and phospholipid binding protein | Exon 11 SNP rs26961431 creates a truncated Annexin A6 protein in mice | ANXA6 truncation results in increased muscle wasting and degeneration in Sgcg mutant mice |
| CD40 Antigen | CD40 | Transmembrane receptor of the tumor necrosis factor receptor family; involved in T helper cell polarization | Minor allele rs1883832 in the 5′ UTR of CD40 associated with diminished CD40 expression | Differential factor for age of loss of ambulation in DMD patients |
| Growth Hormone/Growth Hormone Receptor | GH/GHR | Regulates normal growth and IGF1 signaling in muscle | Growth hormone-deficiency comorbid with DMD in selected case reports | Case reports of DMD patients with dwarfism/short stature having delayed loss of ambulation |
| Latent TGFβ Binding Protein 4 | LTBP4 | Regulator of TGFβ and TGFβ signaling | 1. IAAM haplotype in DMD patients | 1. Reduced inflammation and myofiber breakdown, increased time to loss of ambulation |
| 2. Recessive T allele at rs10880 and IAAM haplotype in DMD patients | 2. Protective against dilated cardiomyopathy (DCM) | |||
| Structural maintenance of chromosomes flexible hinge domain containing 1 | SMCHD1 | Chromatin modification; mediates CpG methylation and chromatin relaxation | In FSHD, allelic variants in SMCHD1 result in hypomethylation and inadvertent expression of DUX4; subsequent diagnosis of FSHD2, genotypically distinct from FSHD1. | FSHD type 2 allows for permissive expression of DUX4 |
| Genes with Protein Expression Differences Contributing to Pathological Variation | ||||
| α-7 Integrin | ITGA7 | Mediates cell-matrix interactions; receptor for basement membrane protein laminin-1 | Increased expression in mdx mice via transgenic cassette or SU9516 compound | Reduced membrane injury, increased muscle fiber size, decreased levels of fibrosis, extended lifespan |
| Biglycan | BGN | Small leucine-rich repeat proteoglycan; ubiquitous structural ECM protein | Increased expression of Biglycan corresponds with stabilization of Utrophin | Decreased diseased muscle pathology |
| Galectin-1 | LGALS1 | Laminin-binding protein; mediates cell-matrix interactions | Increased expression in GRMD dogs | Enhanced muscle repair, reduced immune response and apoptosis |
| Jagged1 | JAG1 | Ligand of the Notch receptor | Increased expression in mdx mice | Improved dystrophic histology and physiology |
| Myostatin | GDF8 | TGF-β superfamily member | Decreased expression in animals and humans | Doubling of muscle mass via myofiber hypertrophy |
| Osteopontin | SPP1 | Diverse matricellular protein | 1. Decreased expression in mice and humans | 1. Decreased immune signaling and fibrosis, increased muscle regeneration |
| 2. Dominant G allele at SPP1 rs28357094 | 2. Protective against dilated cardiomyopathy (DCM) | |||
| Sarcospan | SSPN | A member of the DAPC, which provides a structural anchor of the myocyte cytoskeleton to the ECM | Transgenic overexpression in mdx mice improves muscle function and force. | Increased myofiber membrane stability and reduces muscle tearing |
| Utrophin | UTRN | Autosomal homolog of DMD gene; maintains sarcolemmal integrity during embryonic myogenesis | Overexpression of Utrophin in mice or with Ezutromid compound | Increased lifespan, decreased diseased muscle pathology |
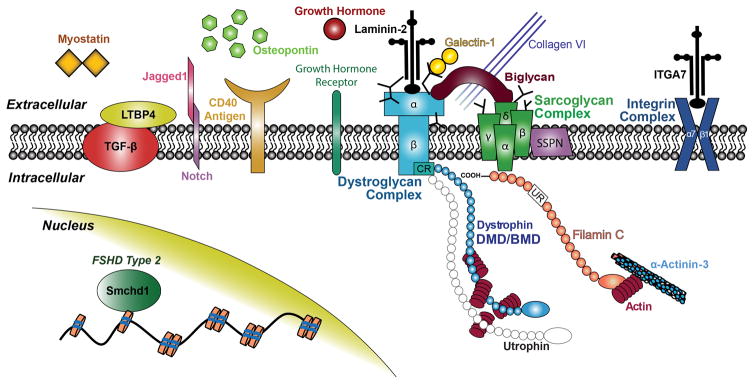
Figure 1. Schematic of significant genetic modifiers of Duchenne and Facioscapulohumeral muscular dystrophies and their sub-cellular localization in skeletal muscle.
Dystrophin and the dystrophin-associated protein complex (DAPC) have an important functional role in the transmission of intercellular force to the extracellular matrix (ECM). Of note, Smchd1 is a chromatin-modifier protein that is believed to allow a permissive state for transcriptional activation of the pathogenic DUX4 transcription factor in FSHD Type 2.
Recent studies in mdx mice have also implicated additional genetic modifiers of dystrophin-deficiency that are found in inbred mouse strains that may also explain the genetic variation in both DMD mice and humans(46–48). In humans, the presence of the R577X (rs1815739) null polymorphism in the α-ACTININ-3 (ACTN3) gene is associated with better overall muscle endurance(49). Male and female athletic sprinters show a higher percentage of the 577R allele over non-sprinters and the general population as a whole(50). In the context of dystrophin-deficiency, the loss of Actn3 in mice blocks muscle wasting and degeneration in mdx mice(51). A Golden Retriever muscular dystrophy (GRMD) dog was identified from a litter of inbred dogs and found to contain a splice acceptor variant resulting in the retention of intron 6 of the canine dystrophin gene(52, 53). Interestingly, in a Brazilian colony of GRMD dogs, an “escaper” dog named Ringo was shown to have no detectable levels of dystrophin protein, but a milder clinical phenotype, normal reproductive capabilities, and a normal lifespan(54, 55). Whole genome sequencing and RNA transcriptome profiling of the muscles from the Golden Retriever muscular dystrophy canine model and the escaper GRMD dog that had a milder phenotype revealed that increased levels of Jagged-1 (Jag1) improved dystrophic symptoms in both zebrafish and dog DMD models(56). With improved genomic editing using clustered regularly interspaced short palindromic repeats (CRISPR) technologies, newer DMD animal models may yield additional genetic modifiers that may lessen or worsen disease symptoms. These dystrophin-deficient models may be beneficial in identifying naturally occurring strain variants that yield additional protective or harmful genetic modifiers of dystrophin deficiency.
Several genetic modifiers of muscular dystrophies have been identified from mouse crosses and are currently being explored for therapeutic targets. Expression levels of two important genetic modifiers (α-7 integrin and laminin-α2) were shown to be elevated in mdx mice and the GRMD dog models of DMD treated with prednisone(57). Loss of α-7 integrin (Itga7) in mice on the mdx background worsens muscle weakness and increases levels of fibrosis(58, 59). Conversely, transgenic overexpression of α-7 integrin reduces muscle weakness and muscle force deficits in both laminin-α2 and mdx/utrn double mutant mice(60, 61). A small molecule SU9516 has been shown to rescue muscular dystrophy phenotypes in mdx mice due to its ability to transcriptionally activate the α-7 integrin promoter(62). Other genetic modifiers such as biglycan (BGN), sarcospan (SSPN), and galectin-1 (Lgals1) overexpression have been demonstrated to act as a muscle membrane “glue” to increase muscle myofiber membrane stability and block muscle tearing in mdx mice(63–65) (Table 1). Biglycan functions to protect against muscle force loss by acting as a protein “anchor” to stabilize the muscle myofibers and their link to the extracellular matrix (ECM) via an interaction with the dystrophin-glycoprotein complex (DGC)(66, 67). (Figure 1). In DMD, Biglycan expression levels are slightly elevated as it is capable of interacting and sequestering TGF-β in the ECM(68, 69). Given the strong role of TGF-β for driving inflammation in DMD, TGF-β antagonists have been effective in blocking some of the dystrophic muscle symptoms associated with DMD(70, 71). Direct intravenously-administered AAV-mediated overexpression of exogenous human biglycan (BGN) was recently shown to ameliorate muscle grip strength deficits and improve overall histology in dystrophin-deficient mice(72). Similar to biglycan, the protein sarcospan (SSPN) also acts as a membrane “glue” to anchor the muscle membrane via an interaction with sarcoglycans, further stabilizing the dystrophin-glycoprotein complex(73). Transgenic overexpression of sarcospan in mdx mice blocks muscle pathology via increasing the levels of utrophin-glycoprotein complex and activates AKT signaling as a compensatory mechanism for the lack of Dystrophin expression(74, 75). Follow-up transgenic mouse studies demonstrated that high levels of sarcospan were sufficient to rescue cardiac and pulmonary defects in mdx mice(76, 77). Together these studies strongly support the notion that overexpression (either naturally-occurring genetic variants or artificially engineered) of selected membrane-associated proteins may be beneficial in blocking or ameliorating dystrophic pathology. As with all of these therapeutic compounds resulting from genetic modification or exogenous delivery, it is possible that human DMD patients may harbor protective or pathogenic variants in these genetic factors that may predict disease progression and outcomes. More whole genome studies of both healthy individuals and the DMD population are needed to test this hypothesis.
Genetic modifiers of DMD identified from study of affected patient populations
Case reports of DMD patients have revealed genetic modifiers that might explain differences in clinical severity. In one case report, a DMD patient presented a milder dystrophic clinical pathology, delayed loss of ambulation, and overall short stature due to a growth hormone (GH)-deficiency(78, 79). A double-blinded controlled study of monozygotic twin DMD boys in which one was administered the growth hormone inhibitor manzindol versus a placebo revealed that the DMD twin receiving growth hormone inhibitor had greater mobility and reduced symptoms compared to his DMD twin on placebo(80). However, a larger double-blinded studies revealed that manzindol-treated DMD boys showed no significant benefit over placebo-treated individuals(81). DMD boys with growth hormone deficiency given growth hormone showed no acceleration of dystrophic disease progression in muscle or cardiopulmonary outputs(82, 83). It has been postulated based on natural history longitudinal studies in DMD boys and dystrophin-deficient animal models that short stature in DMD boys is beneficial for delaying the loss of ambulation(84). Mouse models of growth hormone-deficiency such as the Ames/Dwarf and Growth hormone receptor (GHR) mutant mice have revealed that short stature/growth retardation slows aging via alteration of metabolic responses to insulin/IGF1 signaling(85, 86). It is likely that the regulation of growth hormone may modulate DMD symptoms and influence phenotypic outcomes. The naturally occurring genetic variants resulting in short-stature/dwarfism in DMD patients have been reported to result in overall milder dystrophic pathology and better outcomes(87).
Due to the robust influence of inflammatory and fibrotic signaling in dystrophic muscle pathology, finding ways to alter these signaling pathways remains a promising strategy as more genetic modifiers have been identified. Recent whole exome analyses of the DMD population collected at various clinic sites have revealed additional genetic modifiers of DMD with ties to the TGFβ signaling pathway. Osteopontin is a secreted extracellular matrix protein thought to be essential for normal osteoclast formation and bone mineralization(88). Osteopontin protein was shown to be strongly increased in expression levels in the DMD patient muscle biopsies and the serum of mdx mice(89). The OSTEOPONTIN (SPP1) G allele polymorphism rs28357094 has been shown to be a predictive indicator of loss of ambulation and degree of muscle weakness in DMD patients(90, 91). Indeed, Osteopontin null mice show altered immune signaling and reduced muscle fibrosis in the mdx mice via reducing M1 and M2a macrophage populations to the more pro-regenerative M2c macrophage population subset(92). The Latent TGFβ Binding Protein 4 (LTBP4) gene was initially identified from a screen of gamma-sarcoglycan (Sgcg) null mice for cytoprotective single nucleotide polymorphisms (SNPs) between two interbred mouse strains(93). Studies in DMD patients revealed that four LTBP4 SNPs (V194I, T787A, T820A, and T1140M; VTTT/IAAM haplotype) were predictive of the age at onset of loss of ambulation and of dilated cardiomyopathy (DCM) onset(94, 95) (Table 1). Follow-up studies in mice and cell culture demonstrated that the IAAM residues bound more latent TGFβ compared to the LTBP4 VTTT protein(96). Thus, direct inhibition of LTBP4 via neutralizing antibodies or other RNA/protein antagonistic strategies may lead to a novel therapeutic means of blocking inflammation and myofiber breakdown in dystrophin-deficient skeletal muscles. Whole exome analyses of a large DMD cohort demonstrated that a SNP at rs1883832 of the CD40 gene is predictive of the age at loss of ambulation and further implicated important roles in DMD for both the NFκB and TGFβ signaling pathways(97) (Table 1). CD40 is a cell surface receptor that is predominantly expressed on mature B cells as part of the Tumor Necrosis Factor (TNF) family of proteins(98). CD40 is required for immunoglobulin class switching and CD40 human polymorphisms have also been linked to increased susceptibility to immunological diseases(99, 100). The immune system has long been shown to have an important role in normal muscle growth and regeneration, as well as the progression of muscular dystrophies(101, 102). The CD40 rs1883832 SNP is likely to have functional immunological consequences for the progression of DMD symptoms as the dystrophic muscle begins to deteriorate with the advancing of the disease.
Genetic modifiers of Facioscapulohumeral muscular dystrophy
Facioscapulohumeral muscular dystrophy affects 1:8000 individuals worldwide and is caused by a contraction of D4Z4 expansion repeats in the DUX4 pseudogene resulting in the production of the myotoxic DUX4 protein(103, 104). DUX4 transcriptional activity is thought to activate pro-apoptotic signaling pathways, immune signaling regulators, and retrotransposons resulting in FSHD disease pathology(105). One of the more perplexing aspects of FSHD is the presence of the contracted D4Z4 repeat in asymptomatic familial carriers whom show no disease pathology(106, 107). It has been postulated that single nucleotide polymorphisms (SNPs) affecting DUX4 expression may allow for the permissive state of DUX4 transcriptional activation(108). Interestingly, mutations in the chromatin-modifying gene structural maintenance of chromosomes flexible hinge domain containing 1 (SMCHD1) result in the relaxing of chromatin thereby permitting the expression of the DUX4 protein and affected patients are categorized as FSHD type 2(109) (Figure 1). Additional FSHD epigenetic regulators including a long non-coding RNA (lncRNA) known as DBE-T, have been shown to regulate the expression of DUX4 in skeletal muscles via modulation of transcriptional regulatory complexes(110). The most interesting aspect of FSHD genetics is that many individuals in FSHD families have been identified as having the FSHD permissive allele, but do not have the phenotypic muscle weakness and thus cannot be diagnosed with FSHD(111). As more and more FSHD families and patients have their exomes and genomes fully-sequenced, additional genetic modifiers that may be consequential to disease progression and outcome will likely be identified.
Advances in DMD/FSHD therapeutic strategies and the potential use of corrective genome editing technologies
The identification of protective variants in human patients offers novel therapeutic entry points for the treatment of muscular dystrophies and muscle diseases in general. Improved adeno-associated viral (AAV) vectors for gene therapy and other biologics have been designed and delivered to patients to overexpress key cytoprotective muscle factors(112). As next-generation sequencing (NGS) has become cheaper and more accurate, larger-scale genomic analyses of healthy and diseased populations are now routinely being conducted. These studies are important to demonstrate the presence of genetic variations in specific regions of the human genome, and the function of transcriptional regulatory regions. The ENCODE (Encyclopedia of DNA Elements) project lists additional layers of epigenetic (non-DNA-dependent) regulation of gene expression of many common disease-causing genes(113). Non-coding RNAs (ncRNAs), RNA-splicing factors, and DNA methylation/acetylation factors among other epigenetic elements were shown to be important post-transcriptional regulators of human muscle diseases(114). The cost of whole genome sequencing has dropped significantly, thus resulting in an increased consideration towards newborn screening of infants for diagnostic and epidemiological purposes(115, 116).
CRISPR genomic editing has emerged as a potential method for correcting small DNA mutations via targeting the specific mutation and replacing it with the corrected sequence(117). Several recent studies have shown that AAV-mediated CRISPR genomic editing can correct the dystrophin exon 23 mutation in the mdx mouse(118–121). Another recent study demonstrated up to 70% restoration of dystrophin protein expression in the myogenic area of AAV-CRISPR mediated correction of the mdx4cv (exon 53 mutation) mouse(122). While these therapies may hold promise for direct correction of small DMD point mutations or deletions, the majority of DMD patients have large, multi-exon deletions for which CRISPR-mediated genomic corrective editing is not currently feasible(30). Nevertheless, it may be possible to alter the genomic sequences of patients with muscular dystrophies and insert or remove the functionality of a therapeutic genetic modifier via CRISPR technology.
Recently, the exon-skipping compound eteplirsen (previously referred to as AVI-4658, Sarepta Therapeutics) was conditionally approved by the US Food and Drug Administration (FDA) for the treatment of DMD patients with genetically amenable dystrophin mutations(123). Eteplirsen functions via bypassing the dystrophin mutation (skipping dystrophin exon 51) resulting in the production of a chimeric, partially-functional dystrophin protein that produces a Becker-like phenotype (124). In a clinical trial of DMD boys amenable to skipping dystrophin exon 51, it was shown that DMD boys treated with eteplirsen retained ambulation longer than natural history controls with the same mutations(125). Gene therapies that overexpress a truncated form of dystrophin (micro-dystrophin) have shown efficacy in DMD animal models and are in current clinical trials for DMD(126–128). The naturally-produced myokine myostatin (also called GDF8) has been shown to be a potent negative regulator of muscle mass in mammals(129). Subsequently, naturally occurring loss-of-function genetic mutations in the myostatin gene of Belgian Blue cattle was demonstrated to be the direct cause of the doubling of their muscle mass via muscle hypertrophy(130). Later, human case studies showed that myostatin genetic variants were responsible for the large, hypertrophic muscles in a young German boy(131). Genetic loss-of-function mutations of myostatin have been shown to induce muscle hypertrophy, and protect against muscle force deficits in mdx mice(132). Pharmacological blockade of myostatin or the myostatin receptor Activin IIB (ACVRIIB) similarly demonstrated a physiological benefit in protecting against muscle force loss in dystrophic mice(133, 134). A recent clinical trial involving a soluble form of the human activin receptor type IIB (ACE-031; produced by Acceleron Pharma) given to ambulatory DMD boys showed some benefits over placebo; although the trial was halted due to unforeseen side effects(135). A similar myostatin/TGF-β pathway inhibitory compound (ACE-081) is currently recruiting for a Phase 2 trial for FSHD patients to alleviate symptoms of muscle weakness (Clinical Trials Identifier: NCT02927080). While these compounds may ameliorate muscle weakness and other dystrophic symptoms, they do not correct the underlying cause of the muscle disease. Many additional strategies towards the treatment of DMD and FSHD are currently under development or in clinical trials and many of these therapeutic targets were originally identified from genomic modifiers of muscular dystrophies(136). It is likely that no single treatment for DMD or FSHD will fully cure either disease, but combination treatments targeting multiple factors including those identified as genetic modifiers may improve muscle symptoms and extend the lifespans of affected individuals.
As genomic sequencing data from muscular dystrophies becomes more commonplace, the opportunity and ability to identify protective and pathogenic genetic modifiers of muscular dystrophies will increase dramatically. These novel genetic modifiers may hold biological clues as to why some individuals do not display muscular dystrophy symptoms despite having pathogenic variants. The exciting prospect of exploring these genetic modifiers as therapeutic agents for drug development may lead to novel treatments for treating these debilitating diseases.
Acknowledgments
The authors wish to thanks Drs. Louis Kunkel, Janelle Spinazzola, Emanuela Gussoni, and Glenn Rowe for critical discussions of this manuscript. The authors wish to thank Paige Souder for assistance with figure preparation. The authors also wish to thank Anete Rozkalne for assistance with formatting and editing. The authors wish to apologies to those researchers whose works were omitted due to space constraints. R.M.H. is a member of the RoadMap Scholars Program at UAB (NIH Grant Number: 5R25NS089463) and is also supported by a T32 training fellowship sponsored by the University of Alabama at Birmingham Center for Exercise Medicine. Research reported in this publication was supported by the Eunice Kennedy Shriver National Institute of Child Health and Human Development of the National Institutes of Health under Award Number 2T32HD071866-06. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. M.S.A. is supported by a grant from the Kaul Pediatrics Research Institute (Award Number: 000514637).
List of Abbreviations
- AAV
Adeno-associated viral vector
- ASD
Autism spectrum disorder
- BMD
Becker muscular dystrophy
- CNV
Copy number variant
- CRISPR
Clustered regularly interspaced short palindromic repeats
- DAPC
Dystrophin-associated protein complex
- DCM
Dilated cardiomyopathy
- DGC
Dystrophin-glycoprotein complex
- DM
Myotonic dystrophy
- DMD
Duchenne muscular dystrophy
- DUX4
Double homeobox 4
- ECM
Extracellular matrix
- ENCODE
Encyclopedia of DNA Elements
- ExAC
Exome Aggregation Consortium
- FSHD
Facioscapulohumeral muscular dystrophy
- GH/GHR
Growth hormone/Growth hormone receptor
- GRMD
Golden Retriever muscular dystrophy
- JAG1
Jagged-1
- lncRNA
Long non-coding RNA
- LoF
Loss-of-function
- LTBP4
Latent TGFβ Binding Protein 4
- mdx
X chromosome-linked muscular dystrophy
- ncRNA
Non-coding RNAs
- NGS
Next-generation sequencing
- SMCHD1
Structural maintenance of chromosomes flexible hinge domain containing 1
- SNP
Single nucleotide polymorphism
- SPP1
Osteopontin
- WES
Whole exome sequencing
- WGS
Whole genome sequencing
Footnotes
Statement of Financial Disclosures and Conflicts of Interest
The authors (R.M.H. and M.S.A.) declare no financial holdings or conflicts of interest for all work presented in this manuscript.
Ethical Publication Statement
We confirm that we have read the Journal’s position on issues involved in ethical publication and affirm that this report is consistent with those guidelines.
Author Contributions
R.M.H. and M.S.A. together wrote, edited, and approved the final version of this manuscript.
References Cited
- 1.Emery AEH. Population frequencies of inherited neuromuscular diseases--A world survey. Neuromuscular Disorders. 1991;1(1):19–29. doi: 10.1016/0960-8966(91)90039-u. [DOI] [PubMed] [Google Scholar]
- 2.Mendell JR, Shilling C, Leslie ND, Flanigan KM, al-Dahhak R, Gastier-Foster J, et al. Evidence-based path to newborn screening for duchenne muscular dystrophy. Annals of Neurology. 2012;71(3):304–13. doi: 10.1002/ana.23528. [DOI] [PubMed] [Google Scholar]
- 3.Monaco AP, Neve RL, Colletti-Feener C, Bertelson CJ, Kurnit DM, Kunkel LM. Isolation of candidate cDNAs for portions of the Duchenne muscular dystrophy gene. Nature. 1986;323(6089):646–50. doi: 10.1038/323646a0. [DOI] [PubMed] [Google Scholar]
- 4.Koenig M, Hoffman EP, Bertelson CJ, Monaco AP, Feener C, Kunkel LM. Complete cloning of the duchenne muscular dystrophy (DMD) cDNA and preliminary genomic organization of the DMD gene in normal and affected individuals. Cell. 1987;50(3):509–17. doi: 10.1016/0092-8674(87)90504-6. [DOI] [PubMed] [Google Scholar]
- 5.Hoffman EP, Brown RH, Kunkel LM. Dystrophin: the protein product of the Duchenne muscular dystrophy locus. Cell. 1987;51:919–28. doi: 10.1016/0092-8674(87)90579-4. [DOI] [PubMed] [Google Scholar]
- 6.Hoffman EP, Fischbeck KH, Brown RH, Johnson M, Medori R, Loire JD, et al. Characterization of Dystrophin in Muscle-Biopsy Specimens from Patients with Duchenne’s or Becker’s Muscular Dystrophy. New England Journal of Medicine. 1988;318(21):1363–8. doi: 10.1056/NEJM198805263182104. [DOI] [PubMed] [Google Scholar]
- 7.Connolly AM, Florence JM, Cradock MM, Malkus EC, Schierbecker JR, Siener CA, et al. Motor and cognitive assessment of infants and young boys with Duchenne Muscular Dystrophy: results from the Muscular Dystrophy Association DMD Clinical Research Network. Neuromuscular Disorders. 2013;23(7):529–39. doi: 10.1016/j.nmd.2013.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Anderson JL, Head SI, Rae C, Morley JW. Brain function in Duchenne muscular dystrophy. Brain. 2002;125(1):4–13. doi: 10.1093/brain/awf012. [DOI] [PubMed] [Google Scholar]
- 9.Bardoni A, Felisari G, Sironi M, Comi G, Lai M, Robotti M, et al. Loss of Dp140 regulatory sequences is associated with cognitive impairment in dystrophinopathies. Neuromuscular Disorders. 2000;10(3):194–9. doi: 10.1016/s0960-8966(99)00108-x. [DOI] [PubMed] [Google Scholar]
- 10.Chamova T, Guergueltcheva V, Raycheva M, Todorov T, Genova J, Bichev S, et al. Association Between Loss of Dp140 and Cognitive Impairment in Duchenne and Becker Dystrophies. Balkan Journal of Medical Genetics: BJMG. 2013;16(1):21–30. doi: 10.2478/bjmg-2013-0014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pane M, Lombardo ME, Alfieri P, D’Amico A, Bianco F, Vasco G, et al. Attention Deficit Hyperactivity Disorder and Cognitive Function in Duchenne Muscular Dystrophy: Phenotype-Genotype Correlation. The Journal of Pediatrics. 2012;161(4):705–9. e1. doi: 10.1016/j.jpeds.2012.03.020. [DOI] [PubMed] [Google Scholar]
- 12.Taylor PJ, Betts GA, Maroulis S, Gilissen C, Pedersen RL, Mowat DR, et al. Dystrophin Gene Mutation Location and the Risk of Cognitive Impairment in Duchenne Muscular Dystrophy. PLoS ONE. 2010;5(1):e8803. doi: 10.1371/journal.pone.0008803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wingeier K, Giger E, Strozzi S, Kreis R, Joncourt F, Conrad B, et al. Neuropsychological impairments and the impact of dystrophin mutations on general cognitive functioning of patients with Duchenne muscular dystrophy. Journal of Clinical Neuroscience. 2011;18(1):90–5. doi: 10.1016/j.jocn.2010.07.118. [DOI] [PubMed] [Google Scholar]
- 14.Campbell KP, Kahl SD. Association of dystrophin and an integral membrane glycoprotein. Nature. 1989;338(6212):259–62. doi: 10.1038/338259a0. [DOI] [PubMed] [Google Scholar]
- 15.Ervasti JM, Kahl SD, Campbell KP. Purification of dystrophin from skeletal muscle. Journal of Biological Chemistry. 1991;266(14):9161–5. [PubMed] [Google Scholar]
- 16.Ervasti J, Campbell K. A role for the dystrophin-glycoprotein complex as a transmembrane linker between laminin and actin. The Journal of Cell Biology. 1993;122(4):809–23. doi: 10.1083/jcb.122.4.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guiraud S, Aartsma-Rus A, Vieira NM, Davies KE, Ommen G-JBv, Kunkel LM. The Pathogenesis and Therapy of Muscular Dystrophies. Annual Review of Genomics and Human Genetics. 2015;16(1):281–308. doi: 10.1146/annurev-genom-090314-025003. [DOI] [PubMed] [Google Scholar]
- 18.Ghaoui R, Cooper ST, Lek M, et al. Use of whole-exome sequencing for diagnosis of limb-girdle muscular dystrophy: Outcomes and lessons learned. JAMA Neurology. 2015;72(12):1424–32. doi: 10.1001/jamaneurol.2015.2274. [DOI] [PubMed] [Google Scholar]
- 19.Aartsma-Rus A, Ginjaar IB, Bushby K. The importance of genetic diagnosis for Duchenne muscular dystrophy. Journal of Medical Genetics. 2016;53(3):145–51. doi: 10.1136/jmedgenet-2015-103387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nallamilli BRR, Ankala A, Hegde M. Current Protocols in Human Genetics. John Wiley & Sons, Inc; 2001. Molecular Diagnosis of Duchenne Muscular Dystrophy. [DOI] [PubMed] [Google Scholar]
- 21.Bennett R, den Dunnen J, O’Brien K, Darras B, Kunkel L. Detection of mutations in the dystrophin gene via automated DHPLC screening and direct sequencing. BMC Genetics. 2001;2(1):17. doi: 10.1186/1471-2156-2-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schmutz J, Wheeler J, Grimwood J, Dickson M, Yang J, Caoile C, et al. Quality assessment of the human genome sequence. Nature. 2004;429(6990):365–8. doi: 10.1038/nature02390. [DOI] [PubMed] [Google Scholar]
- 23.O’Grady GL, Lek M, Lamande SR, Waddell L, Oates EC, Punetha J, et al. Diagnosis and etiology of congenital muscular dystrophy: We are halfway there. Annals of Neurology. 2016;80(1):101–11. doi: 10.1002/ana.24687. [DOI] [PubMed] [Google Scholar]
- 24.Reddy HM, Cho K-A, Lek M, Estrella E, Valkanas E, Jones MD, et al. The sensitivity of exome sequencing in identifying pathogenic mutations for LGMD in the United States. J Hum Genet. 2017;62(2):243–52. doi: 10.1038/jhg.2016.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536(7616):285–91. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peplow M. The 100 000 Genomes Project. BMJ. 2016:353. doi: 10.1136/bmj.i1757. [DOI] [PubMed] [Google Scholar]
- 27.Chen R, Shi L, Hakenberg J, Naughton B, Sklar P, Zhang J, et al. Analysis of 589,306 genomes identifies individuals resilient to severe Mendelian childhood diseases. Nat Biotec. 2016 doi: 10.1038/nbt.3514. advance online publication. [DOI] [PubMed] [Google Scholar]
- 28.Narasimhan VM, Hunt KA, Mason D, Baker CL, Karczewski KJ, Barnes MR, et al. Health and population effects of rare gene knockouts in adult humans with related parents. Science. 2016;352(6284):474–7. doi: 10.1126/science.aac8624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Saleheen D, Natarajan P, Armean IM, Zhao W, Rasheed A, Khetarpal SA, et al. Human knockouts and phenotypic analysis in a cohort with a high rate of consanguinity. Nature. 2017;544(7649):235–9. doi: 10.1038/nature22034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bladen CL, Salgado D, Monges S, Foncuberta ME, Kekou K, Kosma K, et al. The TREAT-NMD DMD Global Database: Analysis of More than 7,000 Duchenne Muscular Dystrophy Mutations. Human Mutation. 2015;36(4):395–402. doi: 10.1002/humu.22758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang RT, Silverstein Fadlon CA, Ulm JW, Jankovic I, Eskin A, Lu A, et al. Online Self-Report Data for Duchenne Muscular Dystrophy Confirms Natural History and Can Be Used to Assess for Therapeutic Benefits. PLoS Currents. 2014;6 doi: 10.1371/currents.md.e1e8f2be7c949f9ffe81ec6fca1cce6a. ecurrents.md.e1e8f2be7c949f9ffe81ec6fca1cce6a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Demonbreun AR, Allen MV, Warner JL, Barefield DY, Krishnan S, Swanson KE, et al. Enhanced Muscular Dystrophy from Loss of Dysferlin Is Accompanied by Impaired Annexin A6 Translocation after Sarcolemmal Disruption. The American Journal of Pathology. 2016;186(6):1610–22. doi: 10.1016/j.ajpath.2016.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McGreevy JW, Hakim CH, McIntosh MA, Duan D. Animal models of Duchenne muscular dystrophy: from basic mechanisms to gene therapy. Disease Models & Mechanisms. 2015;8(3):195–213. doi: 10.1242/dmm.018424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Whitmore C, Morgan J. What do mouse models of muscular dystrophy tell us about the DAPC and its components? International Journal of Experimental Pathology. 2014;95(6):365–77. doi: 10.1111/iep.12095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bulfield G, Siller WG, Wight PA, Moore KJ. X chromosome-linked muscular dystrophy (mdx) in the mouse. Proceedings of the National Academy of Sciences. 1984;81(4):1189–92. doi: 10.1073/pnas.81.4.1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chapman VM, Miller DR, Armstrong D, Caskey CT. Recovery of induced mutations for X chromosome-linked muscular dystrophy in mice. Proceedings of the National Academy of Sciences of the United States of America. 1989;86(4):1292–6. doi: 10.1073/pnas.86.4.1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Matsumura K, Ervasti JM, Ohlendieck K, Kahl SD, Campbell KP. Association of dystrophin-related protein with dystrophin-associated proteins in mdx mouse muscle. Nature. 1992;360(6404):588–91. doi: 10.1038/360588a0. [DOI] [PubMed] [Google Scholar]
- 38.Mizuno Y, Yoshida M, Nonaka I, Hirai S, Ozawa E. Expression of utrophin (dystrophin-related protein) and dystrophin-associated glycoproteins in muscles from patients with Duchenne muscular dystrophy. Muscle & Nerve. 1994;17(2):206–16. doi: 10.1002/mus.880170212. [DOI] [PubMed] [Google Scholar]
- 39.Thanh LT, Nguyen TM, Helliwell TR, Morris GE. Characterization of revertant muscle fibers in Duchenne muscular dystrophy, using exon-specific monoclonal antibodies against dystrophin. American Journal of Human Genetics. 1995;56(3):725–31. [PMC free article] [PubMed] [Google Scholar]
- 40.Sherratt TG, Vulliamy T, Dubowitz V, Sewry CA, Strong PN. Exon skipping and translation in patients with frameshift deletions in the dystrophin gene. American Journal of Human Genetics. 1993;53(5):1007–15. [PMC free article] [PubMed] [Google Scholar]
- 41.Dickson G, Hill V, Graham IR. Screening for antisense modulation of dystrophin pre-mRNA splicing. Neuromuscular Disorders. 2002;12:S67–S70. doi: 10.1016/s0960-8966(02)00085-8. [DOI] [PubMed] [Google Scholar]
- 42.Fairclough RJ, Wood MJ, Davies KE. Therapy for Duchenne muscular dystrophy: renewed optimism from genetic approaches. Nat Rev Genet. 2013;14(6):373–8. doi: 10.1038/nrg3460. [DOI] [PubMed] [Google Scholar]
- 43.Tinsley J, Deconinck N, Fisher R, Kahn D, Phelps S, Gillis J-M, et al. Expression of full-length utrophin prevents muscular dystrophy in mdx mice. Nat Med. 1998;4(12):1441–4. doi: 10.1038/4033. [DOI] [PubMed] [Google Scholar]
- 44.Ricotti V, Spinty S, Roper H, Hughes I, Tejura B, Robinson N, et al. Safety, Tolerability, and Pharmacokinetics of SMT C1100, a 2-Arylbenzoxazole Utrophin Modulator, following Single- and Multiple-Dose Administration to Pediatric Patients with Duchenne Muscular Dystrophy. PLoS ONE. 2016;11(4):e0152840. doi: 10.1371/journal.pone.0152840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guiraud S, Squire SE, Edwards B, Chen H, Burns DT, Shah N, et al. Second generation compound for the modulation of utrophin in the therapy of DMD. Human Molecular Genetics. 2015 doi: 10.1093/hmg/ddv154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Calyjur PC, Almeida CdF, Ayub-Guerrieri D, Ribeiro AFJ, Fernandes SdA, Ishiba R, et al. The mdx Mutation in the 129/Sv Background Results in a Milder Phenotype: Transcriptome Comparative Analysis Searching for the Protective Factors. PLoS ONE. 2016;11(3):e0150748. doi: 10.1371/journal.pone.0150748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Coley WD, Bogdanik L, Vila MC, Yu Q, Van Der Meulen JH, Rayavarapu S, et al. Effect of genetic background on the dystrophic phenotype in mdx mice. Human Molecular Genetics. 2016;25(1):130–45. doi: 10.1093/hmg/ddv460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Heydemann A, Swaggart KA, Kim GH, Holley-Cuthrell J, Hadhazy M, McNally EM. The superhealing MRL background improves muscular dystrophy. Skeletal Muscle. 2012;2(1):26. doi: 10.1186/2044-5040-2-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.North KN, Yang N, Wattanasirichaigoon D, Mills M, Easteal S, Beggs AH. A common nonsense mutation results in [alpha]-actinin-3 deficiency in the general population. Nat Genet. 1999;21(4):353–4. doi: 10.1038/7675. [DOI] [PubMed] [Google Scholar]
- 50.Yang N, MacArthur DG, Gulbin JP, Hahn AG, Beggs AH, Easteal S, et al. ACTN3 Genotype Is Associated with Human Elite Athletic Performance. The American Journal of Human Genetics. 2003;73(3):627–31. doi: 10.1086/377590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hogarth MW, Houweling PJ, Thomas KC, Gordish-Dressman H, Bello L, Pegoraro E, et al. Evidence for ACTN3 as a genetic modifier of Duchenne muscular dystrophy. Nature Communications. 2017;8:14143. doi: 10.1038/ncomms14143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kornegay JN, Tuler SM, Miller DM, Levesque DC. Muscular dystrophy in a litter of golden retriever dogs. Muscle & Nerve. 1988;11(10):1056–64. doi: 10.1002/mus.880111008. [DOI] [PubMed] [Google Scholar]
- 53.Sharp NJH, Kornegay JN, Van Camp SD, Herbstreith MH, Secore SL, Kettle S, et al. An error in dystrophin mRNA processing in golden retriever muscular dystrophy, an animal homologue of Duchenne muscular dystrophy. Genomics. 1992;13(1):115–21. doi: 10.1016/0888-7543(92)90210-j. [DOI] [PubMed] [Google Scholar]
- 54.Ambrósio CE, Valadares MC, Zucconi E, Cabral R, Pearson PL, Gaiad TP, et al. Ringo, a Golden Retriever Muscular Dystrophy (GRMD) dog with absent dystrophin but normal strength. Neuromuscular Disorders. 2008;18(11):892–3. doi: 10.1016/j.nmd.2008.06.385. [DOI] [PubMed] [Google Scholar]
- 55.Zatz M, Vieira NM, Zucconi E, Pelatti M, Gomes J, Vainzof M, et al. A normal life without muscle dystrophin. Neuromuscular Disorders. 2015;25(5):371–4. doi: 10.1016/j.nmd.2015.02.007. [DOI] [PubMed] [Google Scholar]
- 56.Vieira Natassia M, Elvers I, Alexander Matthew S, Moreira Yuri B, Eran A, Gomes Juliana P, et al. Jagged 1 Rescues the Duchenne Muscular Dystrophy Phenotype. Cell. 2015;163(5):1204–13. doi: 10.1016/j.cell.2015.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wuebbles RD, Sarathy A, Kornegay JN, Burkin DJ. Levels of α7 integrin and laminin-α2 are increased following prednisone treatment in the mdx mouse and GRMD dog models of Duchenne muscular dystrophy. Disease Models & Mechanisms. 2013;6(5):1175–84. doi: 10.1242/dmm.012211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Guo C, Willem M, Werner A, Raivich G, Emerson M, Neyses L, et al. Absence of α7 integrin in dystrophin-deficient mice causes a myopathy similar to Duchenne muscular dystrophy. Human Molecular Genetics. 2006;15(6):989–98. doi: 10.1093/hmg/ddl018. [DOI] [PubMed] [Google Scholar]
- 59.Rooney JE, Welser JV, Dechert MA, Flintoff-Dye NL, Kaufman SJ, Burkin DJ. Severe muscular dystrophy in mice that lack dystrophin and α7 integrin. Journal of Cell Science. 2006;119(11):2185–95. doi: 10.1242/jcs.02952. [DOI] [PubMed] [Google Scholar]
- 60.Doe JA, Wuebbles RD, Allred ET, Rooney JE, Elorza M, Burkin DJ. Transgenic overexpression of the α7 integrin reduces muscle pathology and improves viability in the dy(W) mouse model of merosin-deficient congenital muscular dystrophy type 1A. Journal of Cell Science. 2011;124(13):2287–97. doi: 10.1242/jcs.083311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Burkin DJ, Wallace GQ, Milner DJ, Chaney EJ, Mulligan JA, Kaufman SJ. Transgenic Expression of α7β1 Integrin Maintains Muscle Integrity, Increases Regenerative Capacity, Promotes Hypertrophy, and Reduces Cardiomyopathy in Dystrophic Mice. The American Journal of Pathology. 2005;166(1):253–63. doi: 10.1016/s0002-9440(10)62249-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sarathy A, Wuebbles RD, Fontelonga TM, Tarchione AR, Mathews Griner LA, Heredia DJ, et al. SU9516 increases alpha7beta1 integrin and ameliorates disease progression in the mdx mouse model of Duchenne Muscular Dystrophy. Molecular Therapy. 2017 doi: 10.1016/j.ymthe.2017.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Amenta AR, Yilmaz A, Bogdanovich S, McKechnie BA, Abedi M, Khurana TS, et al. Biglycan recruits utrophin to the sarcolemma and counters dystrophic pathology in mdx mice. Proceedings of the National Academy of Sciences. 2011;108(2):762–7. doi: 10.1073/pnas.1013067108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Marshall JL, Oh J, Chou E, Lee JA, Holmberg J, Burkin DJ, et al. Sarcospan integration into laminin-binding adhesion complexes that ameliorate muscular dystrophy requires utrophin and α7 integrin. Human Molecular Genetics. 2015;24(7):2011–22. doi: 10.1093/hmg/ddu615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Van RyPM, Wuebbles RD, Key M, Burkin DJ. Galectin-1 Protein Therapy Prevents Pathology and Improves Muscle Function in the mdx Mouse Model of Duchenne Muscular Dystrophy. Molecular Therapy. 2015;23(8):1285–97. doi: 10.1038/mt.2015.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Casar JC, McKechnie BA, Fallon JR, Young MF, Brandan E. Transient up-regulation of biglycan during skeletal muscle regeneration: delayed fiber growth along with decorin increase in biglycan-deficient mice. Developmental Biology. 2004;268(2):358–71. doi: 10.1016/j.ydbio.2003.12.025. [DOI] [PubMed] [Google Scholar]
- 67.Bowe MA, Mendis DB, Fallon JR. The Small Leucine-Rich Repeat Proteoglycan Biglycan Binds to α-Dystroglycan and Is Upregulated in Dystrophic Muscle. The Journal of Cell Biology. 2000;148(4):801–10. doi: 10.1083/jcb.148.4.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zanotti S, Negri T, Cappelletti C, Bernasconi P, Canioni E, Di Blasi C, et al. Decorin and biglycan expression is differentially altered in several muscular dystrophies. Brain. 2005;128(11):2546–55. doi: 10.1093/brain/awh635. [DOI] [PubMed] [Google Scholar]
- 69.Hildebrand A, Romarís M, Rasmussen LM, Heinegård D, Twardzik DR, Border WA, et al. Interaction of the small interstitial proteoglycans biglycan, decorin and fibromodulin with transforming growth factor beta. Biochemical Journal. 1994;302(Pt 2):527–34. doi: 10.1042/bj3020527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Acuña MJ, Pessina P, Olguin H, Cabrera D, Vio CP, Bader M, et al. Restoration of muscle strength in dystrophic muscle by angiotensin-1-7 through inhibition of TGF-β signalling. Human Molecular Genetics. 2014;23(5):1237–49. doi: 10.1093/hmg/ddt514. [DOI] [PubMed] [Google Scholar]
- 71.Kemaladewi DU, Pasteuning S, van der Meulen JW, van Heiningen SH, van Ommen G-J, ten Dijke P, et al. Targeting TGF-β Signaling by Antisense Oligonucleotide-mediated Knockdown of TGF-β Type I Receptor. Molecular Therapy Nucleic Acids. 2014;3(4):e156. doi: 10.1038/mtna.2014.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ito MEY, Li J, Inada K, Ohno K. Protein-Anchoring Therapy of Biglycan for Mdx Mouse Model of Duchenne Muscular Dystrophy. Hum Gene Ther. 2017;28(5):428–36. doi: 10.1089/hum.2015.088. [DOI] [PubMed] [Google Scholar]
- 73.Lebakken CS, Venzke DP, Hrstka RF, Consolino CM, Faulkner JA, Williamson RA, et al. Sarcospan-Deficient Mice Maintain Normal Muscle Function. Molecular and Cellular Biology. 2000;20(5):1669–77. doi: 10.1128/mcb.20.5.1669-1677.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Peter AK, Marshall JL, Crosbie RH. Sarcospan reduces dystrophic pathology: stabilization of the utrophin–glycoprotein complex. The Journal of Cell Biology. 2008;183(3):419–27. doi: 10.1083/jcb.200808027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Marshall JL, Holmberg J, Chou E, Ocampo AC, Oh J, Lee J, et al. Sarcospan-dependent Akt activation is required for utrophin expression and muscle regeneration. The Journal of Cell Biology. 2012;197(7):1009–27. doi: 10.1083/jcb.201110032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Parvatiyar MS, Marshall JL, Nguyen RT, Jordan MC, Richardson VA, Roos KP, et al. Sarcospan Regulates Cardiac Isoproterenol Response and Prevents Duchenne Muscular Dystrophy–Associated Cardiomyopathy. Journal of the American Heart Association. 2015;4(12) doi: 10.1161/JAHA.115.002481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gibbs EM, Marshall JL, Ma E, Nguyen TM, Hong G, Lam JS, et al. High levels of sarcospan are well tolerated and act as a sarcolemmal stabilizer to address skeletal muscle and pulmonary dysfunction in DMD. Human Molecular Genetics. 2016;25(24):5395–406. doi: 10.1093/hmg/ddw356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zatz M, Betti RTB, Levy JA. Begnign duchenne muscular dystrophy in a patient with growth hormone deficiency. American Journal of Medical Genetics. 1981;10(3):301–4. doi: 10.1002/ajmg.1320100313. [DOI] [PubMed] [Google Scholar]
- 79.Zatz M, Betti RTB, Opitz JM, Reynolds JF. Benign Duchenne muscular dystrophy in a patient with growth hormone deficiency: A five years follow-up. American Journal of Medical Genetics. 1986;24(3):567–72. doi: 10.1002/ajmg.1320240323. [DOI] [PubMed] [Google Scholar]
- 80.Zatz M, Betti RTB, Frota-Pessoa O, Opitz JM, Reynolds JF. Treatment of duchenne muscular dystrophy with growth hormone inhibitors. American Journal of Medical Genetics. 1986;24(3):549–66. doi: 10.1002/ajmg.1320240322. [DOI] [PubMed] [Google Scholar]
- 81.Griggs RC, Moxley RT, Mendell JR, Fenichel GM, Brooke MH, Miller PJ, et al. Randomized, double-blind trial of mazindol in Duchenne dystrophy. Muscle & Nerve. 1990;13(12):1169–73. doi: 10.1002/mus.880131212. [DOI] [PubMed] [Google Scholar]
- 82.Rutter MM, Collins J, Rose SR, Woo JG, Sucharew H, Sawnani H, et al. Growth hormone treatment in boys with Duchenne muscular dystrophy and glucocorticoid-induced growth failure. Neuromuscular Disorders. 2012;22(12):1046–56. doi: 10.1016/j.nmd.2012.07.009. [DOI] [PubMed] [Google Scholar]
- 83.Frank GR, Smith RE. Effective Growth Hormone Therapy in a Growth Hormone Deficient Patient with Duchenne Muscular Dystropy Without Evidence of Acceleration of the Dystrophic Process. Journal of Pediatric Endocrinology and Metabolism. 2001:211. doi: 10.1515/jpem.2001.14.2.211. [DOI] [PubMed] [Google Scholar]
- 84.Bodor M, McDonald CM. Why short stature is beneficial in duchenne muscular dystrophy. Muscle & Nerve. 2013;48(3):336–42. doi: 10.1002/mus.23793. [DOI] [PubMed] [Google Scholar]
- 85.Robertson K, Kopchick JJ, Liu J-L. Growth hormone receptor gene deficiency causes delayed insulin responsiveness in skeletal muscles without affecting compensatory islet cell overgrowth in obese mice. American Journal of Physiology - Endocrinology And Metabolism. 2006;291(3):E491–E8. doi: 10.1152/ajpendo.00378.2005. [DOI] [PubMed] [Google Scholar]
- 86.Miquet JG, Muñoz MC, Giani JF, González L, Dominici FP, Bartke A, et al. Ames dwarf (Prop1df/Prop1df) mice display increased sensitivity of the major GH-signaling pathways in liver and skeletal muscle. Growth Hormone & IGF Research. 2010;20(2):118–26. doi: 10.1016/j.ghir.2009.11.003. [DOI] [PubMed] [Google Scholar]
- 87.Zatz M, Pavanello RCM, Lazar M, Yamamoto GL, Lourenço NCV, Cerqueira A, et al. Milder course in Duchenne patients with nonsense mutations and no muscle dystrophin. Neuromuscular Disorders. 2014;24(11):986–9. doi: 10.1016/j.nmd.2014.06.003. [DOI] [PubMed] [Google Scholar]
- 88.Rittling SR, Matsumoto HN, McKee MD, Nanci A, An X-R, Novick KE, et al. Mice Lacking Osteopontin Show Normal Development and Bone Structure but Display Altered Osteoclast Formation In Vitro. Journal of Bone and Mineral Research. 1998;13(7):1101–11. doi: 10.1359/jbmr.1998.13.7.1101. [DOI] [PubMed] [Google Scholar]
- 89.Vetrone SA, Montecino-Rodriguez E, Kudryashova E, Kramerova I, Hoffman EP, Liu SD, et al. Osteopontin promotes fibrosis in dystrophic mouse muscle by modulating immune cell subsets and intramuscular TGF-β. The Journal of Clinical Investigation. 2009;119(6):1583–94. doi: 10.1172/JCI37662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bello L, Kesari A, Gordish-Dressman H, Cnaan A, Morgenroth LP, Punetha J, et al. Genetic modifiers of ambulation in the cooperative international Neuromuscular research group Duchenne natural history study. Annals of Neurology. 2015;77(4):684–96. doi: 10.1002/ana.24370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Pegoraro E, Hoffman EP, Piva L, Gavassini BF, Cagnin S, Ermani M, et al. SPP1 genotype is a determinant of disease severity in Duchenne muscular dystrophy. Neurology. 2011;76(3):219–26. doi: 10.1212/WNL.0b013e318207afeb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Capote J, Kramerova I, Martinez L, Vetrone S, Barton ER, Sweeney HL, et al. Osteopontin ablation ameliorates muscular dystrophy by shifting macrophages to a pro-regenerative phenotype. The Journal of Cell Biology. 2016 doi: 10.1083/jcb.201510086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Heydemann A, Ceco E, Lim JE, Hadhazy M, Ryder P, Moran JL, et al. Latent TGF-β–binding protein 4 modifies muscular dystrophy in mice. The Journal of Clinical Investigation. 2009;119(12):3703–12. doi: 10.1172/JCI39845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Flanigan KM, Ceco E, Lamar K-M, Kaminoh Y, Dunn DM, Mendell JR, et al. LTBP4 genotype predicts age of ambulatory loss in duchenne muscular dystrophy. Annals of Neurology. 2013;73(4):481–8. doi: 10.1002/ana.23819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Barp A, Bello L, Politano L, Melacini P, Calore C, Polo A, et al. Genetic Modifiers of Duchenne Muscular Dystrophy and Dilated Cardiomyopathy. PLoS ONE. 2015;10(10):e0141240. doi: 10.1371/journal.pone.0141240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lamar K-M, Miller T, Dellefave-Castillo L, McNally EM. Genotype-Specific Interaction of Latent TGFβ Binding Protein 4 with TGFβ. PLOS ONE. 2016;11(2):e0150358. doi: 10.1371/journal.pone.0150358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Bello L, Flanigan Kevin M, Weiss Robert B, Dunn Diane M, Swoboda Kathryn J, Gappmaier E, et al. Association Study of Exon Variants in the NFkB and TGFBeta; Pathways Identifies CD40 as a Modifier of Duchenne Muscular Dystrophy. The American Journal of Human Genetics. 2016 doi: 10.1016/j.ajhg.2016.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.van Kooten C, Banchereau J. CD40-CD40 ligand. Journal of Leukocyte Biology. 2000;67(1):2–17. doi: 10.1002/jlb.67.1.2. [DOI] [PubMed] [Google Scholar]
- 99.Kawabe T, Naka T, Yoshida K, Tanaka T, Fujiwara H, Suematsu S, et al. The immune responses in CD40-deficient mice: Impaired immunoglobulin class switching and germinal center formation. Immunity. 1994;1(3):167–78. doi: 10.1016/1074-7613(94)90095-7. [DOI] [PubMed] [Google Scholar]
- 100.Peters AL, Plenge RM, Graham RR, Altshuler DM, Moser KL, Gaffney PM, et al. A novel polymorphism of the human CD40 receptor with enhanced function. Blood. 2008;112(5):1863–71. doi: 10.1182/blood-2008-02-138925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Tidball JG. Regulation of muscle growth and regeneration by the immune system. Nat Rev Immunol. 2017;17(3):165–78. doi: 10.1038/nri.2016.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Rosenberg AS, Puig M, Nagaraju K, Hoffman EP, Villalta SA, Rao VA, et al. Immune-mediated pathology in Duchenne muscular dystrophy. Science Translational Medicine. 2015;7(299):299rv4-rv4. doi: 10.1126/scitranslmed.aaa7322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Deenen JCW, Arnts H, van der Maarel SM, Padberg GW, Verschuuren JJGM, Bakker E, et al. Population-based incidence and prevalence of facioscapulohumeral dystrophy. Neurology. 2014;83(12):1056–9. doi: 10.1212/WNL.0000000000000797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Lemmers RJLF, van der Vliet PJ, Klooster R, Sacconi S, Camaño P, Dauwerse JG, et al. A Unifying Genetic Model for Facioscapulohumeral Muscular Dystrophy. Science. 2010;329(5999):1650–3. doi: 10.1126/science.1189044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Geng Linda N, Yao Z, Snider L, Fong Abraham P, Cech Jennifer N, Young Janet M, et al. DUX4 Activates Germline Genes, Retroelements, and Immune Mediators: Implications for Facioscapulohumeral Dystrophy. Developmental Cell. 2012;22(1):38–51. doi: 10.1016/j.devcel.2011.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Tonini MMO, Pavanello RCM, Gurgel-Giannetti J, Lemmers RJ, van der Maarel SM, Frants RR, et al. Homozygosity for autosomal dominant facioscapulohumeral muscular dystrophy (FSHD) does not result in a more severe phenotype. Journal of Medical Genetics. 2004;41(2):e17-e. doi: 10.1136/jmg.2003.010637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Tonini MMO, Passos-Bueno MR, Cerqueira A, Matioli SR, Pavanello R, Zatz M. Asymptomatic carriers and gender differences in facioscapulohumeral muscular dystrophy (FSHD) Neuromuscular Disorders. 2004;14(1):33–8. doi: 10.1016/j.nmd.2003.07.001. [DOI] [PubMed] [Google Scholar]
- 108.Lemmers RJLF, Wohlgemuth M, van der Gaag KJ, van der Vliet PJ, van Teijlingen CMM, de Knijff P, et al. Specific Sequence Variations within the 4q35 Region Are Associated with Facioscapulohumeral Muscular Dystrophy. The American Journal of Human Genetics. 2007;81(5):884–94. doi: 10.1086/521986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Lemmers RJLF, Tawil R, Petek LM, Balog J, Block GJ, Santen GWE, et al. Digenic inheritance of an SMCHD1 mutation and an FSHD-permissive D4Z4 allele causes facioscapulohumeral muscular dystrophy type 2. Nat Genet. 2012;44(12):1370–4. doi: 10.1038/ng.2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Cabianca Daphne S, Casa V, Bodega B, Xynos A, Ginelli E, Tanaka Y, et al. A Long ncRNA Links Copy Number Variation to a Polycomb/Trithorax Epigenetic Switch in FSHD Muscular Dystrophy. Cell. 2012;149(4):819–31. doi: 10.1016/j.cell.2012.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Jones TI, Chen JCJ, Rahimov F, Homma S, Arashiro P, Beermann ML, et al. Facioscapulohumeral muscular dystrophy family studies of DUX4 expression: evidence for disease modifiers and a quantitative model of pathogenesis. Human Molecular Genetics. 2012;21(20):4419–30. doi: 10.1093/hmg/dds284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Hollinger K, Chamberlain JS. Viral Vector Mediated Gene Therapies. Current opinion in neurology. 2015;28(5):522–7. doi: 10.1097/WCO.0000000000000241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.The EPC. An Integrated Encyclopedia of DNA Elements in the Human Genome. Nature. 2012;489(7414):57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Sincennes M-C, Brun CE, Rudnicki MA. Concise Review: Epigenetic Regulation of Myogenesis in Health and Disease. STEM CELLS Translational Medicine. 2016;5(3):282–90. doi: 10.5966/sctm.2015-0266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Friedman JM, Cornel MC, Goldenberg AJ, Lister KJ, Sénécal K, Vears DF. Genomic newborn screening: public health policy considerations and recommendations. BMC Medical Genomics. 2017;10(1):9. doi: 10.1186/s12920-017-0247-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Berg JS, Agrawal PB, Bailey DB, Beggs AH, Brenner SE, Brower AM, et al. Newborn Sequencing in Genomic Medicine and Public Health. Pediatrics. 2017;139(2) doi: 10.1542/peds.2016-2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Hsu Patrick D, Lander Eric S, Zhang F. Development and Applications of CRISPR-Cas9 for Genome Engineering. Cell. 2014;157(6):1262–78. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Nelson CE, Hakim CH, Ousterout DG, Thakore PI, Moreb EA, Rivera RMC, et al. In vivo genome editing improves muscle function in a mouse model of Duchenne muscular dystrophy. Science. 2016;351(6271):403–7. doi: 10.1126/science.aad5143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Long C, Amoasii L, Mireault AA, McAnally JR, Li H, Sanchez-Ortiz E, et al. Postnatal genome editing partially restores dystrophin expression in a mouse model of muscular dystrophy. Science. 2016;351(6271):400–3. doi: 10.1126/science.aad5725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Tabebordbar M, Zhu K, Cheng JKW, Chew WL, Widrick JJ, Yan WX, et al. In vivo gene editing in dystrophic mouse muscle and muscle stem cells. Science. 2016;351(6271):407–11. doi: 10.1126/science.aad5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Xu L, Park KH, Zhao L, Xu J, El Refaey M, Gao Y, et al. CRISPR-mediated Genome Editing Restores Dystrophin Expression and Function in mdx Mice. Molecular Therapy. 2016;24(3):564–9. doi: 10.1038/mt.2015.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Bengtsson NE, Hall JK, Odom GL, Phelps MP, Andrus CR, Hawkins RD, et al. Muscle-specific CRISPR/Cas9 dystrophin gene editing ameliorates pathophysiology in a mouse model for Duchenne muscular dystrophy. Nature Communications. 2017;8:14454. doi: 10.1038/ncomms14454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Lim KRQ, Maruyama R, Yokota T. Eteplirsen in the treatment of Duchenne muscular dystrophy. Drug Design, Development and Therapy. 2017;11:533–45. doi: 10.2147/DDDT.S97635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Mendell J, Rodino-Klapac LR, Sahenk Z, Roush K, Bird L, Lowes LP, et al. Eteplirsen for the treatment of duchenne muscular dystrophy. Annals of Neurology. 2013 doi: 10.1002/ana.23982. n/a-n/a. [DOI] [PubMed] [Google Scholar]
- 125.Mendell JR, Goemans N, Lowes LP, Alfano LN, Berry K, Shao J, et al. Longitudinal effect of eteplirsen versus historical control on ambulation in Duchenne muscular dystrophy. Annals of Neurology. 2016;79(2):257–71. doi: 10.1002/ana.24555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Zhang Y, Yue Y, Li L, Hakim CH, Zhang K, Thomas GD, et al. Dual AAV therapy ameliorates exercise-induced muscle injury and functional ischemia in murine models of Duchenne muscular dystrophy. Human Molecular Genetics. 2013;22(18):3720–9. doi: 10.1093/hmg/ddt224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Bostick B, Yue Y, Lai Y, Long C, Li D, Duan D. Adeno-Associated Virus Serotype-9 Microdystrophin Gene Therapy Ameliorates Electrocardiographic Abnormalities in mdx Mice. Human Gene Therapy. 2008;19(8):851–6. doi: 10.1089/hum.2008.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Yue Y, Pan X, Hakim CH, Kodippili K, Zhang K, Shin J-H, et al. Safe and bodywide muscle transduction in young adult Duchenne muscular dystrophy dogs with adeno-associated virus. Human Molecular Genetics. 2015;24(20):5880–90. doi: 10.1093/hmg/ddv310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.McPherron AC, Lawler AM, Lee S-J. Regulation of skeletal muscle mass in mice by a new TGF-p superfamily member. Nature. 1997;387(6628):83–90. doi: 10.1038/387083a0. [DOI] [PubMed] [Google Scholar]
- 130.McPherron AC, Lee SJ. Double muscling in cattle due to mutations in the myostatin gene. Proc Natl Acad Sci USA. 1997:94. doi: 10.1073/pnas.94.23.12457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Schuelke M, Wagner KR, Stolz LE, Hübner C, Riebel T, Kömen W, et al. Myostatin Mutation Associated with Gross Muscle Hypertrophy in a Child. New England Journal of Medicine. 2004;350(26):2682–8. doi: 10.1056/NEJMoa040933. [DOI] [PubMed] [Google Scholar]
- 132.Wagner KR, McPherron AC, Winik N, Lee S-J. Loss of myostatin attenuates severity of muscular dystrophy in mdx mice. Annals of Neurology. 2002;52(6):832–6. doi: 10.1002/ana.10385. [DOI] [PubMed] [Google Scholar]
- 133.Bogdanovich S, Krag TOB, Barton ER, Morris LD, Whittemore L-A, Ahima RS, et al. Functional improvement of dystrophic muscle by myostatin blockade. Nature. 2002;420(6914):418–21. doi: 10.1038/nature01154. [DOI] [PubMed] [Google Scholar]
- 134.Morine KJ, Bish LT, Selsby JT, Gazzara JA, Pendrak K, Sleeper MM, et al. ACTIVIN IIB RECEPTOR BLOCKADE ATTENUATES DYSTROPHIC PATHOLOGY IN A MOUSE MODEL OF DUCHENNE MUSCULAR DYSTROPHY. Muscle & nerve. 2010;42(5):722–30. doi: 10.1002/mus.21743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Campbell C, McMillan HJ, Mah JK, Tarnopolsky M, Selby K, McClure T, et al. Myostatin inhibitor ACE-031 treatment of ambulatory boys with Duchenne muscular dystrophy: Results of a randomized, placebo-controlled clinical trial. Muscle & Nerve. 2017;55(4):458–64. doi: 10.1002/mus.25268. [DOI] [PubMed] [Google Scholar]
- 136.Guiraud S, Davies KE. Pharmacological advances for treatment in Duchenne muscular dystrophy. Current Opinion in Pharmacology. 2017;34:36–48. doi: 10.1016/j.coph.2017.04.002. [DOI] [PubMed] [Google Scholar]