Abstract
Background
Hepatocellular carcinoma (HCC) has the greatest increase in mortality among all solids tumors in the United States related to low rates of early tumor detection. Development of non-invasive biomarkers for early detection of HCC may reduce HCC-related mortality.
Methods
We have developed an algorithm that combines routinely observed clinical values into a single equation that in a study of > 3,000 patients from 5 independent sites, improved detection of HCC as compared to the currently used biomarker, alpha-feto-protein (AFP), by 4–20%. However, this algorithm had limited benefit in those with AFP <20 ng/mL. To that end, we have developed a secondary algorithm that incorporates a marker, fucosylated kininogen, to improve the detection of HCC, especially in those with AFP <20 ng/mL and early stage disease.
Results
The ability to detect early stage AFP negative (AFP< 20 ng/mL) HCC increased from 0% (AFP alone) to 89% (for the new algorithm). Glycan analysis revealed that kininogen has several glycan modifications that have been associated with HCC, but often not with specific proteins, including increased levels of core and outer-arm fucosylation and increased branching.
Conclusions
An algorithm combining fucosylated kininogen, alpha fetoprotein, and clinical characteristics is highly accurate for early HCC detection.
Impact
Our biomarker algorithm could significantly improve early HCC detection and curative treatment eligibility in patients with cirrhosis.
Introduction
Hepatocellular carcinoma (HCC) was once thought to be an uncommon cancer in the United States, but incidence rates have increased dramatically due to increasing number of advanced hepatitis C virus (HCV) and non-alcoholic fatty liver disease (NAFLD) cases. Indeed, in the most recent Annual Report to the Nation on the Status of Cancer, HCC had the second fastest rise in incidence and the greatest increase in mortality among both men and women(1). HCC incidence is predicted to continue rising in the United States and exceed 50,000 cases per year by 2025(2).
Current guidelines by the American Association for the Study of Liver Disease (AASLD) suggest ultrasound for the routine surveillance of those at risk for HCC without the addition of any biomarker(3–5). In practice, many clinicians utilize routine MRI and CT scans for cancer detection, given the low sensitivity of ultrasound alone for early tumor detection(6–8). However, CT scan and MRI have never been studied as surveillance tests for HCC, and modeling studies have shown that these modalities are not cost-effective(9). AFP, while no longer recommended for surveillance in the AASLD guidelines (10, 11), is still widely used in clinical practice despite questionable value as an individual marker(12, 13).
We previously developed an AFP-based algorithm, the Doylestown algorithm (DA), that used standard lab values (age, gender, alanine aminotransferase (ALT), and alkaline phosphatase (ALK))) to improve the sensitivity and specificity of AFP for identifying early stage HCC(14). In a recently published study among 5 independent patient cohorts of > 3000 patients, the DA increased AUROC of AFP by 8% among all patients and by 31% among those with AFP between 10–100 ng/mL. However, the performance of this algorithm in those with AFP <20 ng/mL was poor and thus, we have attempted to determine if we could build on this algorithm to further improve the performance. The logic here is that the Doylestown algorithm would act as a base to add in a limited number of new biomarkers to further increase sensitivity and specificity over both AFP and the base algorithm. As we have previously performed extensive biomarker discovery and identified a number of potential HCC biomarker candidates(15–23), we tested a number of these as potential combinatorial partners with the DA. To that end, we have discovered that the addition of lectin reactive low molecular weight (LMW) kininogen to the algorithm increases performance and allows for a high degree of sensitivity and specificity in the early detection of HCC, especially in those with AFP <20ng/mL. Glycan analysis of kininogen derived from the serum of healthy subjects revealed that this molecule is normally fucosylated but has limited reactivity to the fucose binding Aleuria aurantia lectin (AAL). Advanced glycoproteomic analysis of the site specific glycosylation of kininogen revealed that the glycans on LMW kininogen become further fucosylated in patients with HCC and this is drives the increased reactivity with the core fucose binding lectins and biomarker performance.
Materials and Methods
Identification of Novel Biomarkers for Addition to Doylestown Algorithm
Our base model is an AFP-adjusted algorithm, the DA, as described in a prior study (14). We evaluated the incremental benefit of adding 3 novel HCC biomarkers to the Doylestown algorithm: fucosylated alpha-1-anti-trypsin (A1AT), fucosylated Kininogen or GP73. Fucosylated proteins were detected using a lectin-fluorescently linked immune-absorbent assay (FLISA) method that involves the antibody capture of specific proteins, i.e., kininogen (Abcam, Cambridge, MA, Cat# ab79650) or A1AT (Sigma-Aldrich, St. Louis, MO, Cat #: A0409) followed by detection of specific N-linked glycan with a sugar binding protein called a lectin, in this case the AAL (Vector Laboratories, Burlingame, CA). Details regarding this assay can be found in our previous publications(23, 24). GP73 was measured by western blot as done previously(25).
Based on the DA, we added novel HCC biomarkers, fucosylated A1AT, GP73, or fucosylated LMW-Kininogen into the DA. New models were built from the different combination, and their fitness checked, (deviance, AIC, generalized-R2 (26), Hosmer-Lemeshow GOF test and concordance statistic-c (27)). First order interactions of novel biomarkers with other predictors in the DA were also checked (a p-value less than 5% was considered significant). Testing of model assumptions was applied, included co-linearity among predictors, the linearity of logit, partial residuals plot, etc. as described elsewhere(28).
Comparison of Model Performance
The predictive performances of each model was evaluated by leave one out cross validation (LOOCV), 3-fold cross validation, stratified 3 fold cross validation and bootstrap methods(29). In the case of LOOCV a non-stratified resampling method was adapted (30) to determine statistical difference between models in LOOCV. Statistical difference between models was tested with Wilcoxon sign-rank test or paired t-test based on data distribution (29). In case of the bootstrap cross validation, the Wilcoxon sign rank test or paired t-test was applied to inference statistical difference of prediction between models.
The output equation for the DA is found in another publication (14) and the output equation for the kininogen based DA plus equation is as follows:
In all cases, a P output value of 0.5 was used as a fixed cutoff and patients were classified as being HCC positive when p>=0.5, otherwise they were classified as cirrhotic (p<0.5). The basic methods that were used to develop the different algorithms are described in more detail in our recent publication(14).
Patients
Two sets of serum samples were used. The experiments performed on the two sample sets are shown in Supplementary Figure S1. The first set of samples was from the University of Michigan and this sample set was used for biomarker analysis. Serum samples were obtained via a study protocol approved by the University of Michigan’s Institutional Review Board. In all cases written informed consent was obtained from each subject. Demographic and clinical information was obtained, and a blood sample was collected from each subject. The diagnosis of HCC was made by histopathology, including all T1 lesions, and if histopathology was not available by two imaging modalities (ultrasound [US], magnetic resonance imaging [MRI], or computed tomography [CT]) showing a vascular enhancing mass > 2 cm) (5). Diagnosis of cirrhosis was based on liver histology or clinical, laboratory and imaging evidence of hepatic decompensation or portal hypertension (15). Each of the patients with cirrhosis had a normal US and if serum AFP was elevated a MRI of the liver within 3 months before to enrollment and another one 6 months after enrollment that showed no liver mass. The cirrhotic controls have been followed for a median of 12 months (range 7–18 months) after enrollment, and no one has developed HCC. Tumor staging was determined using the United Network of Organ Sharing-modified TNM staging system for HCC (16). Early HCC was defined as T1 (single lesion < 2 cm in diameter) and T2 (single lesion between 2 and 5 cm in diameter; or < 3 lesions each < 3 cm in diameter) lesions, which met criteria for liver transplantation in the United States. A 20-ml blood sample was drawn from each subject, spun, aliquoted, and serum stored at –80°C until testing. Blood samples were drawn prior to initiation of HCC treatment. AFP was tested using commercially available immunoassays utilizing enhanced chemiluminescence at the University of Michigan Hospital Clinical Diagnostic Laboratory. The upper limit of normal was 8 ng/ml.
The second set of samples from the University of California San Diego (UCSD) was used for kininogen glycomic analysis. These samples were collected under a study protocol approved by the UCSD Institutional Review Board and written informed consent was obtained from each subject. The detailed information regarding patients from the UCSD is found in Supplementary Table S1.
Glycan analysis
Glycan analysis was performed on N-linked glycans enzymatically released from kininogen by treatment with PNGase F (PNGase F PRIME™, Bulldog Bio, Portsmouth, NH), labeled with a fluorescent dye, 2-aminobenzamide (2AB), and resolved via a ultra-high performance liquid chromatography (UPLC) system with a normal phase column complemented with a Waters fluorescence detector and quantified using the Millennium Chromatography Manager software (Waters Corporation, Milford, MA) as described previously (31, 32). Glycans are resolved into discreet chromatographic peaks as has been done previously(33–35) and assigned preliminary glycan identifications based upon retention times that are converted to glucose units (GU) using a glucose homopolymer standard curve. Glycan structures are assigned via treatment with exoglycosidases and alteration in predicted GU values (18, 20, 32, 36–44).
LC-MS/MS analysis of fucosylated site specific N-glycoforms of kininogen
Affinity depletion of abundant serum proteins for the analysis of site specific glycoforms of kininogen was carried out on HP 1290 HPLC system (Agilent, Santa Clara, CA) and Multiple Affinity Removal Column Human 14 (MARS 14, Agilent) according to manufacturer’s protocol. Briefly, human plasma (20 μl) was mixed with buffer A (85 μl) and injected on the column. Affinity buffers A and B were used as mobile phases, 100% A at a flow rate 0.25 ml/min at 0–9 min, 100% B at a flow rate 1 ml/min at 9–15 min, and 100% A at a flow rate 1 ml/min at 15–20 min. The peak of the retained fraction was automatically collected. Amicon microspin column (MilliporeSigma, Darmstadt, Germany) with 3,000 Da cutoff and 0.5 ml volume was used to desalt and concentrate the flow-through fraction (cca 70 μg of protein) and to exchange Agilent buffer A to 50mM ammonium bicarbonate. RapiGest SF anionic surfactant (Waters, Milford, MA) was added to a final concentration of 0.05% and incubated with the proteins at 37°C for 1 hour. Aliquots (20 μg of total proteins) of the samples were reduced with 5 mM DTT and alkylated with 15 mM IAA. Residual IAA was quenched with 5 mM DTT, and the samples were digested overnight using Trypsin Gold (Promega, V5280) in a ratio 1/20 to the total protein.
Tryptic digests (20 μg) were de-sialylated with neuraminidase (New England BioLabs, Ipswich, MA) at 37°C using pressurized accelerated digestion (barocycler) technology (Pressure Biosciences, South Easton, MA). Completion of the de-sialylation reaction was confirmed by LC-MS/MS analysis. De-sialylated glycopeptides were evaporated in a vacuum concentrator (Labconco, Kansas City, MO) and dissolved in mobile phase A (2% acetonitrile (ACN), 0.1% formic acid (FA)) for LC-MS/MS measurements. Neuraminidase treated tryptic digests of the MARS 14 depleted samples were separated using a 90 min gradient elution. A 5 min trapping step using 2% ACN, 0.1% formic acid at 15 μl/min was followed by chromatographic separation at 0.4 μl/min as follows: starting conditions 1% ACN, 0.1% FA; 0–1 min 1–5% ACN, 1–60 min, 5–40% ACN, 0.1% FA; 60–65 min, 40–98% ACN, 0.1% FA; 65–70 min 98% ACN, 0.1% FA followed by equilibration to starting conditions for additional Samples were measured under optimized DIA conditions using rolling collision energy and a 15 Da SWATH window step. Glycopeptide specific Y-ion isotope cluster with isolation of a window of 1.2 Da, extracted from the SWATH MS/MS with a 15 Da step window, was used for analysis of the glycopeptide intensities. Areas of the fucosylated glycopeptides were normalized to the areas of corresponding non-fucosylated analytes for the analysis of the degree (%) of fucosylation.
Results
Identification of Biomarkers for Algorithm
The previously reported DA had limited benefit in those with AFP < 20 ng/mL(14), highlighting the need for new biomarkers to help identify those with AFP negative disease. To that end, we initiated a program to identify novel biomarkers required to improve the detection of HCC, especially in those with AFP <20 ng/mL (See Supplementary Table S2 and Supplementary Figure S2 and S3). In our testing, the DA was considered the base marker and additional previously identified biomarkers were incorporated to improve performance. The markers tested included fucosylated A1AT, fucosylated kininogen and GP73(18, 20, 45, 46). Analysis was performed in 115 HCC and 93 cirrhotic cases (See Table 1). While GP73 was measured directly by immunoblotting, fucosylated A1AT and fucosylated kininogen were measured by a lectin-FLISA based method that measures the lectin reactivity of captured protein with a specific fucose binding lectin (AAL). GP73, fucosylated A1AT and fucosylated kininogen data were available in 93 of the HCC samples and 73 of the cirrhotic samples and model development used only in the sub-set of patients where all biomarker and clinical variables were available (distribution of these markers is found in Supplementary Figure S2). In our testing, the addition of fucosylated A1AT had limited benefit in this model, while the incorporation of GP73 and fucosylated kininogen resulted in the greatest increase in AUC, as compared to AFP alone (or each marker alone, Supplementary Tables S2–S3). However, the algorithm using GP73 plus fucosylated kininogen was not statistically different than an algorithm that contained only fucosylated kininogen (see Supplementary Table S4). Indeed, the best performance with a single biomarker was obtained with a fucosylated kininogen based algorithm. This algorithm is referred to as the DA Plus and the predictive performance of this model was evaluated using leave-one-out cross validation (LOOCV) and three-fold cross validation (3CV) and bootstrap cross validation (Supplementary Tables S3–S4).
Table 1.
Sample population characteristics
| Diagnostic state | HCC2 | Cirrhosis3 | p-value |
|---|---|---|---|
| Male:Female(%)4 | 73%:27% | 51%:49% | 0.0033 |
| Etiology of liver disease (HBV:HCV:Other) 5 | 6:61:34 | 10:48:42 | 0.1991 |
| % Early HCC | 60% | – | |
| AGE (mean ±sd)6 | 58.4 ± 13.0 | 50.5 ± 8.6 | <0.0001 |
| ALT (mean ±sd)7 | 74.7 ± 47.1 | 65.7 ± 44.0 | 0.1051 |
| AST (mean ±sd)8 | 105.4 ± 80.3 | 121.3 ± 85.9 | 0.2401 |
| ALK (mean ±sd)9 | 239.2 ± 200.9 | 94.2 ± 45.4 | <0.0001 |
| Bilirubin (mean ±sd)10 | 1.43 ± 1.05 | 1.16 ± 0.76 | 0.2208 |
| AFP (mean ±sd)11 | 9657.6 ±3975.3 | 11.8 ± 34.6 | <0.0001 |
| log (AFP) (mean ±sd)11 | 2.061 ±1.3257 | 0.714 ± 0.4271 | <0.0001 |
| Sample size | 115 | 93 | N/A |
Samples obtained from the clinical collaborators from the University of Michigan.
HCC as determined by either MRI or CT.
Cirrhosis confirmed by biopsy.
The gender breakdown of patient samples. P value determine by the Chi-squared test.
Etiology of liver disease. HBV, hepatitis B virus; HCV, hepatitis C virus, P value determine by the Chi-squared test.
Mean Age of patients. Students t-test
Alanine transaminase, ALT. P value determined by Wilcoxon rank sum test.
Aspartate transaminase, AST. P value determined by Wilcoxon rank sum test.
Alkaline phosphatase, ALK. P value determine by Wilcoxon rank sum test.
Serum bilirubin levels. P value determined by Wilcoxon rank sum test.
Alpha-feto protein levels in ng/mL. P value determined by Wilcoxon rank sum test.
Performance of Models
In the full cohort of 208 patients (115 HCC cases, 93 non-HCC controls), AFP alone was able to distinguish these groups with an AUC of 0.828 (Figure 1). The DA had a mean AUC of 0.951, which was increased to 0.9777 with the addition of fucosylated kininogen in the DA plus algorithm (Figure 1A). As Table 2A shows, at a fixed false positive rate (FPR) of 5%, the true positive rate (TPR) of AFP alone, the DA and the DA Plus algorithm were 60% (95% CI: 48–71%), 85% (95% CI: 72–93%), and 90% (95% CI: 80–97%), respectively.
Figure 1.
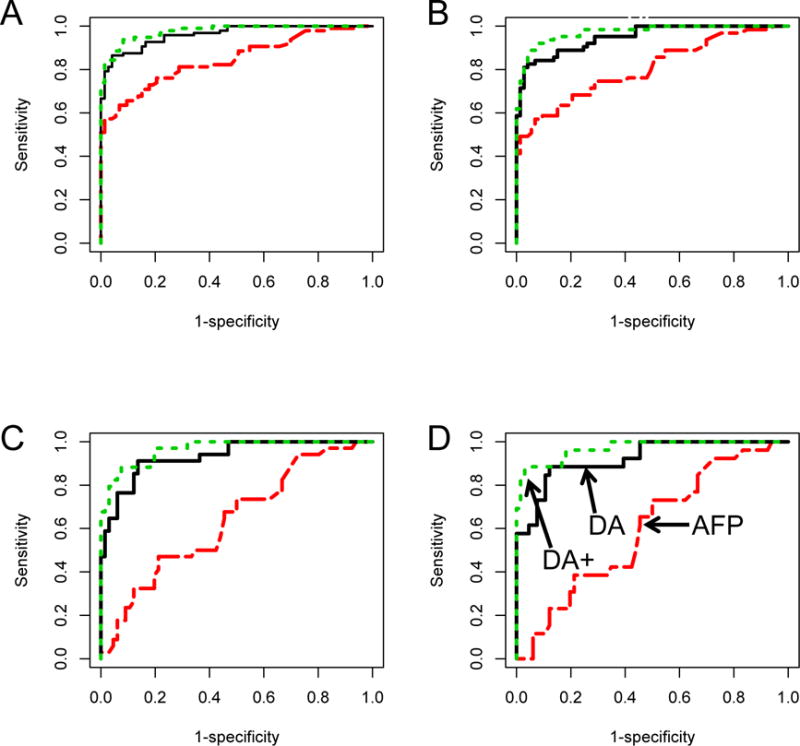
AUROC for AFP, the DA or the DA Plus (DA+) algorithm in A) All patients; B) Patients with early stage HCC; C) patients with AFP<20 ng/mL and D) patients with early stage HCC and AFP<20 ng/mL.
Table 2A.
Sensitivity and Specificity values for the DA plus algorithm, the DA and AFP following cross validation.
| All HCC(HCC=115,Cir=93)1 | Early HCC(T1/2)(HCC=69,Cir=93) 2 | ||||||
|---|---|---|---|---|---|---|---|
| AFP | DA | DA plus | AFP | DA | DA plus | ||
|
AUC 95%CI |
0.844 0.787–0.902 |
0.951 0.922–0.980 |
0.977 0.960–0.994 |
0.803 0.723–0.878 |
0.934 0.893–0.976 |
0.971 0.947–0.994 |
|
| TPR at fixed FPR |
Fixed FPR | TPR | TPR | TPR | TPR | TPR | TPR |
|
5% 95% CI |
60% 48–71 |
85% 72–93 |
90% 80–97 |
53% 38–67 |
83% 67–92 |
86% 68–97 |
|
|
10% 95% CI |
66% 53–77 |
88% 80–94 |
94% 86–99 |
59% 44–73 |
84% 75–94 |
92% 81–98 |
|
|
15% 95% CI |
69% 57–80 |
89% 81–95 |
96% 90–99 |
62% 48–76 |
84% 75–94 |
95% 86–100 |
|
Either all patients in Table 1 or only those with early HCC
were analyzed. For table, AUC is area under the curve, AFP is alpha-fetal protein, DA is Doylestown algorithm, DA plus is the Doylestown plus algorithm. FPR is false positive rate, TPR is true positive rate. The 95% confidence interval is given for each TPR at each fixed FPR.
Importantly, if only the patients with early tumors were examined, the mean AUC of the DA Plus algorithm was 0.971 (0.95–0.99) with an 86% TPR at a 5% FPR. In contrast, AFP was reduced to an AUC of 0.803 with a TPR of 53% at a 5% FPR. Significantly, if only those patients with AFP <20 ng/mL and early stage HCC were examined, the DA Plus algorithm maintained an AUC of 0.970 (95% CI: 0.936–1.0) with a TPR of 89% (95% CI: 70–100%). In contrast, the AUC of AFP was reduced to 0.597 (95% CI: 0.47 to 0.72) with a TPR of 0% (95% CI: 0–24%) at a 5% FPR. In these patients the DA was limited to 65% TPR (95% CI: 42–81%) at a 5% FPR. In all cases, the increased performance of the DA Plus over both AFP and the DA was statistically significant (Supplementary Table S4).
Structural Analysis of Kininogen
The lectin used in patient analysis binds to fucose bound to the parent glycan in α-1,2, α-1,3, α-1,4 outer arm linkages or an α 1,6 core linkage. The specific types of fucose molecules found on kininogen are not known, although kininogen was previously implicated as a candidate biomarker of HCC through a glycoproteomic method that involved extraction of proteins with a fucose binding lectin (Lens culinaris agglutinin), with greater affinity for α 1,6 linked fucosylated glycan (19, 20, 46). However, while this method suggested that the protein contains glycan with fucose moieties, it does not provide any true structural glycan information. To address this, we have performed glycan analysis of human LMW-kininogen purified from normal human serum, from patients with liver cirrhosis, and from patients with HCC in the background of liver cirrhosis. Analysis of normal human kininogen was first analyzed by chromatographic N-linked glycan analysis. These results are shown in Figure 2. Using sequential exoglycosidase digestion of 2AB labeled glycan and resolution by hydrophilic interaction chromatography (HILAC) we have identified the major structures to contain, a bi-antennary glycan, a tri-antennary glycan and a tri-antennary glycan with a single outer-arm linked α-1,3 fucose residue. Minor N-linked glycan included those with core α-1,6 fucose and tetra- antennary glycan.
Figure 2. Characterization of low molecular weight kininogen glycosylation.
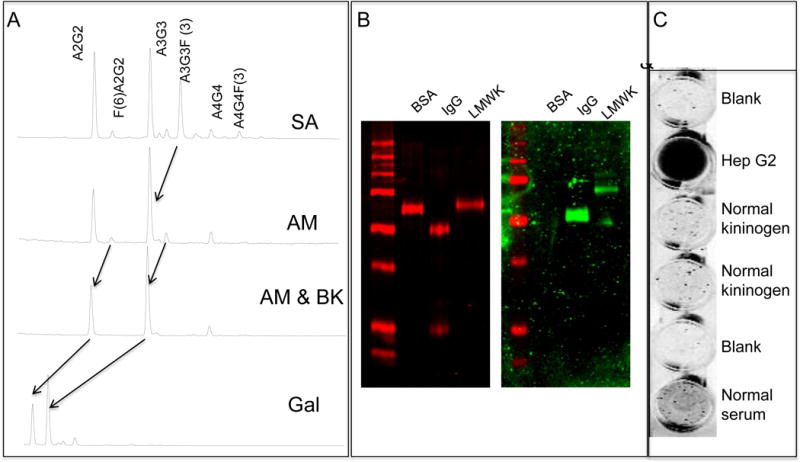
A) Normal phase UPLC analysis of low molecular weight kininogen following sequential exoglycosidase digestion. B) Lectin blotting of low molecular weight kininogen. The left panel is stained with colloidal Coomassie brilliant blue (red channel) and the right panel (green channel) is a lectin blot with the AAL lectin. Proteins are Bovine serum albumin (BSA, as negative control, human IgG as a positive control) and low molecular weight kininogen (LMWK). C) Lectin-FLISA of captured kininogen. Supernatant from Hep G2 cells served as the positive control.
Based on the UPLC based glycan sequencing, approximately 30% of the N-linked glycan on kininogen contained fucose residues (~5% core fucose; 25% outer-arm fucose). Orthogonal conformation of the fucosylation was performed by lectin blotting using the fucose specific AAL, by commercial glycan analysis via MALDI-TOF in combination with exoglycosidase digestion of core fucosylated glycan (Supplementary Figure S4) and by the lectin-FLISA based method as used in Figure 1 for patient analysis. As Figure 2B shows, kininogen, when denatured and resolved via SDS-PAGE, is highly lectin reactive, consistent with the UPLC based glycan analysis. Similarly, MALDI-TOF glycan analysis in combination with exoglycosidase digestion of core fucosylated glycan also confirmed that normal kininogen was 30% fucosylated (Supplementary Figure S4). Surprisingly, this material is not lectin reactive when analyzed via the lectin-FLISA method used for patient analysis. That is, as Figure 2C shows, when kininogen is left in a native state following capture with an anti-kininogen antibody and fucose detected with AAL (Figure 2C), there is no lectin reactivity. This is in contrast to the results observed when kininogen is analyzed via lectin blotting. This result suggests that the conformation of native kininogen inhibits lectin binding and that either a change in glycosylation or potentially a change in protein structure could be responsible for the increased lectin reactivity observed in HCC patients.
Glycoproteomic Analysis of Patient Derived Kininogen
To determine if the glycans on kininogen change with the development of HCC, we performed glycoproteomics on kininogen at the site-specific level. Kininogen contains four potential N-linked glycosylation sites, and we performed glycoproteomics to identify the site specific glycoforms on the kininogen molecule (47). Table 3 presents the detected glycoforms on the four N-linked glycosylation sites and shows that complex N-linked glycan structures were detectable at all the sites of N-linked glycan attachment. We were able to detect bi- and tri-antennary glycoforms with at most one fucose on asparagine (N) 57. In contrast, the glycoforms at sites N169, N205 and N294 contained bi- to tetra- antennary glycans with varying degree of fucosylation. Site N294 was the only site where we detected up to four fucoses associated with the tetra-antennary glycoform. Figure 3 shows the extracted ion chromatograms of select glycoforms at the N294 glycan site; peak areas of the glycoforms were used to quantify the site specific glycoforms of kininogen in patients with liver cirrhosis (10) to patients with liver cirrhosis and HCC (20). Clinical information regarding these patients is found in Supplementary Table S1 and chromatograms for glycopeptides containing sites N57, N205 and N294 are shown in Supplementary Figures S5–S7. Even though the structures detected at each site differ, fucosylation of the glycans at all the four peptides increased in HCC compared to cirrhosis. This is documented in Figure 4 on the N294 glycosylation site that shows, as compared to the cirrhotic control, a greater level of fucosylation for all glycoforms from bi- to tetra- antennary. Thus, this data suggests that the increased reactivity with fucose binding lectin observed with the development of HCC is the result of increased levels of fucosylation, both core and outer-arm.
Table 3.
All detected N-glycoforms of kininogen in samples of healthy, cirrhotic, and HCC participants.
| A2G2 | A2G2F1 | A3G3 | A3G3F1 | A3G3F2 | A3G3F3 | A4G4 | A4G4F1 | A4G4F2 | A4G4F3 | A4G4F4 | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| YNSQNQSNNQFVLYR1 | Q | Q | Q | Q | ND | ND | ND | ND | ND | ND | ND |
| HGIQYFNNN TQHSSLFMLNEVK2 | Q | D | Q | Q | Q | D | Q | Q | Q | D | ND |
| ITYSIVQTNCSK3 | Q | Q | Q | Q | Q | ND | Q | D | D | ND | ND |
| LNAENNATFYFK4 | Q | Q | Q | Q | Q | D | Q | Q | Q | D | D |
The quantifiable glycoforms (Q) are detected in all patient samples with signal to noise greater than 10; the detectable glycoforms (D) were measurable in a subset of participants only; some glycoform were not detectable (ND) on all peptides.
The site of glycosylation is at asparagine (N) 57.
The site of glycosylation is at N169.
The site of glycosylation is at N205.
The site of glycosylation is at N294.
Figure 3. Glycopeptide analysis of tryptic glycopeptide 290–300 highlighting the presence of branched and fucosylated glycan.
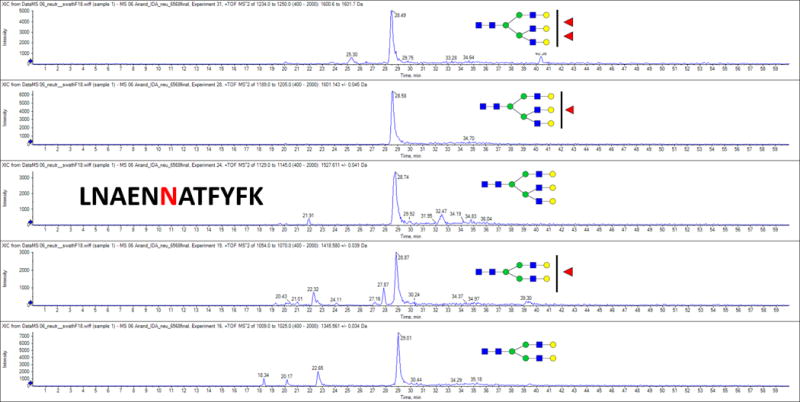
For glycan structures, blue squares represent N-acetylglucosamine monosaccharides (GlcNAc); green circles represent mannose residues; yellow circles represent galactose residues and red triangles represent fucose residues.
Figure 4. Glycopeptide analysis of kininogen tryptic glycopeptide 290–300 containing site N294.
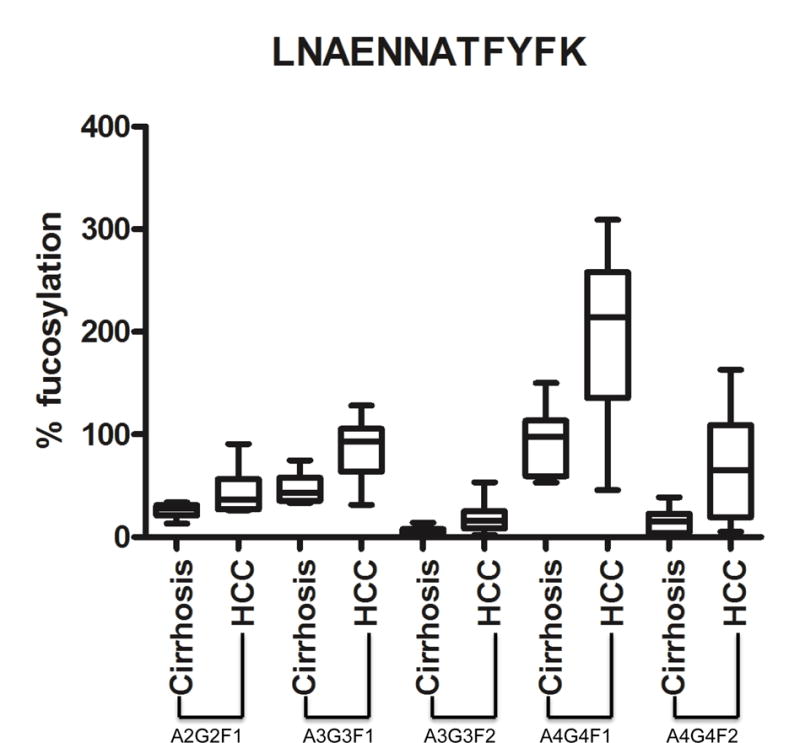
The median, 25%, 75% and standard deviation are shown for each fucosylated glycan observed. Results are from an analysis of 10 individual cirrhotic and 20 individual HCC patients. For structures, A2G2F1, biantennary N-glycan glycan with a single fucose residue; A3G3F1, tri-antennary N-glycan glycan with a single fucose residue; A3G3F2, tri-antennary N-glycan glycan with two fucose residues; A4G4F1, tetra-antennary N-glycan glycan with a single fucose residue; A4G4F2, tetra-antennary N-glycan glycan with two fucose residues.
Discussion
We have recently reported the usefulness of incorporating biomarkers and relevant clinical variables into a statistical model for predicting the incidence of HCC. In that prior work, we investigated the predictive performance of AFP alone or after the inclusion of clinical factors such as age, gender, and serum ALK and ALT levels. However, the major issue with AFP and the DA is the poor discriminatory ability when AFP is <20 ng/mL. This is a major issue as 50% of all cases of HCC can fall into this category. In this situation, new biomarkers, which can identify early cancer in patients with low AFP, are of great value. Thus, we wanted to determine if any of our previously identified markers could increase the AUC of the DA and specifically in those with early HCC. Surprisingly, fucosylated kininogen, which, by itself only has limited discriminatory ability (AUC of 0.829), was the marker to best complement AFP and the clinical factors in this cohort. GP73, in contrast, had better overall individual performance (AUC of 0.870) but offered no better complement to the DA then fucosylated kininogen.
N-linked glycan analysis of kininogen revealed a high level of fucosylation, with roughly 25% of the glycan containing outer-arm fucosylation with a small degree of core fucosylation (5%). Surprisingly, when analyzed by the plate based lectin FLISA, this molecule has very limited lectin reactivity with the fucose specific AAL (figure 2C). Analysis of kininogen from patients with HCC revealed an increased amount of both core and outer-arm fucosylation as compared to the cirrhotic controls. Interestingly, the alteration in glycosylation appeared to be site specific with one particular site of N-linked glycosylation modified by fucosylation. The potential influence of the different sites in the lectin FLISA and in the detection of HCC will have to be determined.
It is noted that alterations in N-linked glycosylation have been associated with the development of cancer. In our recent study, we showed that increased core fucosylation was directly associated with de-differentiation of hepatocytes and the transition of cells from an epithelial to a mesenchymal state (EMT)(48). However, this modification was not present in the all patients and may be directly related to the underlying genetic abnormalities associated with the cancer. This is currently under investigation.
The data presented in this paper also has several limitations that will have to be addressed in future studies. The first is a lack of external validation. One patient cohort was used for the examination of the enhanced DA plus, although the basic components minus the one biomarker have been examined in much larger sets. In addition this study was done with the clinical information collected either at the time of HCC detection or close to it. Thus, a follow-up longitudinal follow-up study will have to be performed to truly determine how this algorithm would be used in the management of patients at risk of developing HCC. Additionally, it is possible that other markers that we have identified, such as fucosylated hemopexin, fucosylated fetuin-A and others, including A1AT shown here, may complement AFP and kininogen in other patient populations. It is noted that the use of patient aspartate aminotransferase (AST) level in place of ALT actually lead to the greatest AUC for the kininogen based algorithm (0.9875), but not when kininogen was excluded. This is currently being investigated as an advanced algorithm. These markers do not appear to impart any negative attributes to the model and one could imagine their inclusion to broaden to potential biomarker coverage.
In conclusion, we present a biomarker algorithm that uses existing clinical information along with a single novel biomarker that dramatically improves the detection of early stage HCC and in the detection of HCC in patients who lack the currently used biomarker, AFP. If this system can be further validated and commercialized, it could potentially be used as a surveillance test to identify those with HCC and to select patients for further imaging and management. Additionally, such a system may be useful in patient monitoring after surgical resection or other treatment modalities.
Supplementary Material
Table 2B.
Sensitivity and Specificity values for the DA plus algorithm, the DA and AFP following cross validation in only those with low AFP
| AFP negative (<20ng/ml)(HCC=39,Cir=84)1 | Early HCC and AFP negative (HCC=29,Cir=84) 2 | ||||||
|---|---|---|---|---|---|---|---|
| AFP | DA | DA plus | AFP | DA | HCC panel | ||
| AUC 95%CI |
0.637 0.523–0.751 |
0.896 0.830–0.962 |
0.965 0.935–0.996 |
0.597 0.473–0.721 |
0.875 0.794–0.956 |
0.970 0.936–1 |
|
| TPR at fixed FPR |
Fixed FPR | TPR | TPR | TPR | TPR | TPR | TPR |
|
5% 95% CI |
9% 0–35 |
71% 50–85 |
82% 62–97 |
0% 0–24 |
65% 42–81 |
89% 70–100 |
|
|
10% 95% CI |
24% 3–44 |
71% 53–88 |
88% 74–97 |
14% 0–35 |
65% 46–85 |
89% 77–100 |
|
|
15% 95% CI |
33% 12–53 |
74% 56–91 |
91% 77–100 |
19% 4–46 |
69% 46–89 |
92% 77–100 |
|
Only those patients with AFP <20ng/mL 1 or only those with early HCC and AFP < HCC
were analyzed. For table, as above.
Acknowledgments
Financial Support: This work was supported by grants R01 CA120206 (A. Mehta), U01 CA168856 (A. Mehta), UO1 CA171146 (R. Goldman and M. Sanda) and RO1 CA135069 (R. Goldman and M. Sanda) from the National Cancer Institute (NCI), the Hepatitis B Foundation, an appropriation from The Commonwealth of Pennsylvania and a sponsored research agreement from Glycotest, INC. to Drexel University (A. Mehta and T.M Block).
Footnotes
Conflict of Interest: M. Wang, MA. Comunale, T.M. Block and A. Mehta have filed a patent regarding the markers in this manuscript (through Drexel University). They have been licensed to Glycotest, Inc. A. Mehta, T.M. Block and C. Swindell have an ownership stake in Glycotest, Inc. T.M Block receives a sponsored research grant from Arbutus bioPharma and is a member of the Advisory board for ContraVir Pharmaceuticals Inc. A. Singal acts as consultant for Instadiagnostics, Bayer, Wako Diagnostics, Eisai Co., Ltd., EMD Serano and Abbott. A. Singal has also received an Honoraria from Bayer.
References
- 1.Ryerson AB, Eheman CR, Altekruse SF, Ward JW, Jemal A, Sherman RL, et al. Annual Report to the Nation on the Status of Cancer, 1975–2012, featuring the increasing incidence of liver cancer. Cancer. 2016;122:1312–37. doi: 10.1002/cncr.29936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mehta A, Herrera H, Block T. Glycosylation and liver cancer. Adv Cancer Res. 2015;126:257–79. doi: 10.1016/bs.acr.2014.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bruix J, Sherman M, American Association for the Study of Liver D Management of hepatocellular carcinoma: an update. Hepatology. 2011;53:1020–2. doi: 10.1002/hep.24199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bruix J, Sherman M, Practice Guidelines Committee AAftSoLD Management of hepatocellular carcinoma. Hepatology. 2005;42:1208–36. doi: 10.1002/hep.20933. [DOI] [PubMed] [Google Scholar]
- 5.Bruix J, Sherman M, Llovet JM, Beaugrand M, Lencioni R, Burroughs AK, et al. Clinical management of hepatocellular carcinoma. Conclusions of the Barcelona-2000 EASL conference. European Association for the Study of the Liver. J Hepatol. 2001;35:421–30. doi: 10.1016/s0168-8278(01)00130-1. [DOI] [PubMed] [Google Scholar]
- 6.Singal AG, Nehra M, Adams-Huet B, Yopp AC, Tiro JA, Marrero JA, et al. Detection of hepatocellular carcinoma at advanced stages among patients in the HALT-C trial: where did surveillance fail? The American journal of gastroenterology. 2013;108:425–32. doi: 10.1038/ajg.2012.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Singal AG, Yopp AC, Gupta S, Skinner CS, Halm EA, Okolo E, et al. Failure rates in the hepatocellular carcinoma surveillance process. Cancer prevention research. 2012;5:1124–30. doi: 10.1158/1940-6207.CAPR-12-0046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Joshi K, Mendler M, Gish R, Loomba R, Kuo A, Patton H, et al. Hepatocellular carcinoma surveillance: a national survey of current practices in the USA. Dig Dis Sci. 2014;59:3073–7. doi: 10.1007/s10620-014-3256-6. [DOI] [PubMed] [Google Scholar]
- 9.El-Serag HB, Davila JA. Surveillance for hepatocellular carcinoma: in whom and how? Therapeutic advances in gastroenterology. 2011;4:5–10. doi: 10.1177/1756283X10385964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yu NC, Chaudhari V, Raman SS, Lassman C, Tong MJ, Busuttil RW, et al. CT and MRI improve detection of hepatocellular carcinoma, compared with ultrasound alone, in patients with cirrhosis. Clinical gastroenterology and hepatology : the official clinical practice journal of the American Gastroenterological Association. 2011;9:161–7. doi: 10.1016/j.cgh.2010.09.017. [DOI] [PubMed] [Google Scholar]
- 11.Sherman M, Bruix J, Porayko M, Tran T. Screening for hepatocellular carcinoma: the rationale for the American Association for the Study of Liver Diseases recommendations. Hepatology. 2012;56:793–6. doi: 10.1002/hep.25869. [DOI] [PubMed] [Google Scholar]
- 12.Gopal P, Yopp AC, Waljee AK, Chiang J, Nehra M, Kandunoori P, et al. Factors that affect accuracy of alpha-fetoprotein test in detection of hepatocellular carcinoma in patients with cirrhosis. Clin Gastroenterol Hepatol. 2014;12:870–7. doi: 10.1016/j.cgh.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sherman M. Surveillance for hepatocellular carcinoma. Semin Oncol. 2001;28:450–9. doi: 10.1016/s0093-7754(01)90138-1. [DOI] [PubMed] [Google Scholar]
- 14.Wang M, Devarajan K, Singal AG, Marrero JA, Dai J, Feng Z, et al. The Doylestown Algorithm: A Test to Improve the Performance of AFP in the Detection of Hepatocellular Carcinoma. Cancer Prev Res (Phila) 2016;9:172–9. doi: 10.1158/1940-6207.CAPR-15-0186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Block TM, Comunale MA, Lowman M, Steel LF, Romano PR, Fimmel C, et al. Use of targeted glycoproteomics to identify serum glycoproteins that correlate with liver cancer in woodchucks and humans. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:779–84. doi: 10.1073/pnas.0408928102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Comunale MA, Lowman M, Long RE, Krakover J, Philip R, Seeholzer S, et al. Proteomic analysis of serum associated fucosylated glycoproteins in the development of primary hepatocellular carcinoma. Journal of Proteome Research. 2006;5:308–15. doi: 10.1021/pr050328x. [DOI] [PubMed] [Google Scholar]
- 17.Comunale MA, Mattu TS, Lowman MA, Evans AA, London WT, Semmes OJ, et al. Comparative proteomic analysis of de-N-glycosylated serum from hepatitis B carriers reveals polypeptides that correlate with disease status. Proteomics. 2004;4:826–38. doi: 10.1002/pmic.200300625. [DOI] [PubMed] [Google Scholar]
- 18.Comunale MA, Rodemich-Betesh L, Hafner J, Wang M, Norton P, Di Bisceglie AM, et al. Linkage Specific Fucosylation of Alpha-1-Antitrypsin in Liver Cirrhosis and Cancer Patients: Implications for a Biomarker of Hepatocellular Carcinoma. PLoS ONE. 2010;5:e12419. doi: 10.1371/journal.pone.0012419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Comunale MA, Wang M, Hafner J, Krakover J, Rodemich L, Kopenhaver B, et al. Identification and development of fucosylated glycoproteins as biomarkers of primary hepatocellular carcinoma. Journal of Proteome Research. 2009;8:595–602. doi: 10.1021/pr800752c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Comunale MA, Wang M, Rodemich-Betesh L, Hafner J, Lamontagne A, Klein A, et al. Novel changes in glycosylation of serum Apo-J in patients with hepatocellular carcinoma. Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2011;20:1222–9. doi: 10.1158/1055-9965.EPI-10-1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hann HW, Wang M, Hafner J, Long RE, Kim SH, Ahn M, et al. Analysis of GP73 in patients with HCC as a function of anti-cancer treatment. Cancer biomarkers : section A of Disease markers. 2010;7:269–73. doi: 10.3233/CBM-2010-0190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marrero JA, Romano PR, Nikolaeva O, Steel L, Mehta A, Fimmel CJ, et al. GP73, a resident Golgi glycoprotein, is a novel serum marker for hepatocellular carcinoma. J Hepatol. 2005;43:1007–12. doi: 10.1016/j.jhep.2005.05.028. [DOI] [PubMed] [Google Scholar]
- 23.Wang M, Long RE, Comunale MA, Junaidi O, Marrero J, Di Bisceglie AM, et al. Novel fucosylated biomarkers for the early detection of hepatocellular carcinoma. Cancer Epidemiol Biomarkers Prev. 2009;18:1914–21. doi: 10.1158/1055-9965.EPI-08-0980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang M, Comunale MA, Herrera H, Betesh L, Kono Y, Mehta A. Identification of IgM as a contaminant in lectin-FLISA assays for HCC detection. Biochem Biophys Res Commun. 2016;476:140–5. doi: 10.1016/j.bbrc.2016.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Block TM, Comunale MA, Lowman M, Steel LF, Romano PR, Fimmel C, et al. Use of targeted glycoproteomics to identify serum glycoproteins that correlate with liver cancer in woodchucks and humans. Proc Natl Acad Sci U S A. 2005;102:779–84. doi: 10.1073/pnas.0408928102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harrell F. Regression Modeling Strategies. Springer-Verlag; NY: 2001. [Google Scholar]
- 27.Hosmer D, Lemeshow S, Sturdivant R. Applied Logistic Regression. 3rd. Wiley Online Library: Wiley Online Library; 2013. [Google Scholar]
- 28.Hilbe JM. Logistic Regression Models. Chapman & Hall/CRC Texts in Statistical Science: Chapman and Hall/CRC. 2009 [Google Scholar]
- 29.Japkowicz N, Shah M. Evaluating Learning Algorithms. Cambridge University Press; 2014. [Google Scholar]
- 30.Carpenter J, Bithell J. Bootstrap confidence intervals: when, which, what? A practical guide for medical statisticians. Stat Med. 2000;19:1141–64. doi: 10.1002/(sici)1097-0258(20000515)19:9<1141::aid-sim479>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 31.Doherty M, McManus CA, Duke R, Rudd PM. High-throughput quantitative N-glycan analysis of glycoproteins. Methods in molecular biology. 2012;899:293–313. doi: 10.1007/978-1-61779-921-1_19. [DOI] [PubMed] [Google Scholar]
- 32.Saldova R, Fan Y, Fitzpatrick JM, Watson RW, Rudd PM. Core fucosylation and alpha2-3 sialylation in serum N-glycome is significantly increased in prostate cancer comparing to benign prostate hyperplasia. Glycobiology. 2011;21:195–205. doi: 10.1093/glycob/cwq147. [DOI] [PubMed] [Google Scholar]
- 33.Rudd PM, Guile GR, Kuster B, Harvey DJ, Opdenakker G, Dwek RA. Oligosaccharide sequencing technology. Nature. 1997;388:205–7. doi: 10.1038/40677. [DOI] [PubMed] [Google Scholar]
- 34.Guile GR, Rudd PM, Wing DR, Prime SB, Dwek RA. A rapid high-resolution high-performance liquid chromatographic method for separating glycan mixtures and analyzing oligosaccharide profiles. Anal Biochem. 1996;240:210–26. doi: 10.1006/abio.1996.0351. [DOI] [PubMed] [Google Scholar]
- 35.Rudd PM, Mattu TS, Zitzmann N, Mehta A, Colominas C, Hart E, et al. Glycoproteins: rapid sequencing technology for N-linked and GPI anchor glycans. Biotechnol Genet Eng Rev. 1999;16:1–21. doi: 10.1080/02648725.1999.10647969. [DOI] [PubMed] [Google Scholar]
- 36.Rudd PMCC, Royle L, Murphy N, Hart E, Merry AH, Hebestreit HF, Dwek RA. A high-performance liquid chromatography based strategy for rapid, sensitive sequencing of N-linked oligosaccharide modifications to proteins in sodium dodecyl sulphate polyacrylamide electrophoresis gel bands. Proteomics. 2001;1:285–289. doi: 10.1002/1615-9861(200102)1:2<285::AID-PROT285>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 37.Bones J, Mittermayr S, O’Donoghue N, Guttman A, Rudd PM. Ultra performance liquid chromatographic profiling of serum N-glycans for fast and efficient identification of cancer associated alterations in glycosylation. Analytical Chemistry. 2010;82:10208–15. doi: 10.1021/ac102860w. [DOI] [PubMed] [Google Scholar]
- 38.Peracaula R, Sarrats A, Rudd PM. Liver proteins as sensor of human malignancies and inflammation. Proteomics Clinical applications. 2010;4:426–31. doi: 10.1002/prca.200900170. [DOI] [PubMed] [Google Scholar]
- 39.Tharmalingam T, Adamczyk B, Doherty MA, Royle L, Rudd PM. Strategies for the profiling, characterisation and detailed structural analysis of N-linked oligosaccharides. Glycoconjugate journal. 2013;30:137–46. doi: 10.1007/s10719-012-9443-9. [DOI] [PubMed] [Google Scholar]
- 40.Comunale MA, Lowman M, Long RE, Krakover J, Philip R, Seeholzer S, et al. Proteomic analysis of serum associated fucosylated glycoproteins in the development of primary hepatocellular carcinoma. J Proteome Res. 2006;5:308–15. doi: 10.1021/pr050328x. [DOI] [PubMed] [Google Scholar]
- 41.Mehta AS, Long RE, Comunale MA, Wang M, Rodemich L, Krakover J, et al. Increased levels of galactose-deficient anti-Gal immunoglobulin G in the sera of hepatitis C virus-infected individuals with fibrosis and cirrhosis. J Virol. 2008;82:1259–70. doi: 10.1128/JVI.01600-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Norton PA, Comunale MA, Krakover J, Rodemich L, Pirog N, D’Amelio A, et al. N-linked glycosylation of the liver cancer biomarker GP73. J Cell Biochem. 2008;104:136–49. doi: 10.1002/jcb.21610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mehta A, Norton P, Liang H, Comunale MA, Wang M, Rodemich-Betesh L, et al. Increased Levels of Tetra-antennary N-Linked Glycan but Not Core Fucosylation Are Associated with Hepatocellular Carcinoma Tissue. Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2012;21:925–933. doi: 10.1158/1055-9965.EPI-11-1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Comunale MA, Wang M, Anbarasan N, Betesh L, Karabudak A, Moritz E, et al. Total serum glycan analysis is superior to lectin-FLISA for the early detection of hepatocellular carcinoma. Proteomics Clinical applications. 2013;7:690–700. doi: 10.1002/prca.201200125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Comunale MA, Wang M, Hafner J, Krakover J, Rodemich L, Kopenhaver B, et al. Identification and development of fucosylated glycoproteins as biomarkers of primary hepatocellular carcinoma. J Proteome Res. 2009;8:595–602. doi: 10.1021/pr800752c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang M, Long RE, Comunale MA, Junaidi O, Marrero J, Di Bisceglie AM, et al. Novel fucosylated biomarkers for the early detection of hepatocellular carcinoma. Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2009;18:1914–21. doi: 10.1158/1055-9965.EPI-08-0980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pompach P, Ashline DJ, Brnakova Z, Benicky J, Sanda M, Goldman R. Protein and site specificity of fucosylation in liver-secreted glycoproteins. Journal of Proteome Research. 2014;13:5561–9. doi: 10.1021/pr5005482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mehta A, Comunale MA, Rawat S, Casciano JC, Lamontagne J, Herrera H, et al. Intrinsic hepatocyte dedifferentiation is accompanied by upregulation of mesenchymal markers, protein sialylation and core alpha 1,6 linked fucosylation. Sci Rep. 2016;6:27965. doi: 10.1038/srep27965. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


