Abstract
The ability of bacteria to recognize kin provides a means to form social groups. In turn these groups can lead to cooperative behaviors that surpass the ability of the individual. Kin recognition involves specific biochemical interactions between a receptor(s) and an identification molecule(s). To ensure that nonkin are excluded and kin are included, recognition specificity is critical and depends on the number of loci and polymorphisms involved. After recognition and biochemical perception, the common ensuing cooperative behaviors include biofilm formation, quorum responses, development and swarming motility. Although kin recognition is a fundamental mechanism through which cells might interact, microbiologists are only beginning to explore the topic. This review considers both molecular and theoretical aspects of bacterial kin recognition. Consideration is also given to bacterial diversity, genetic relatedness, kin selection theory, and mechanisms of recognition.
Keywords: kin recognition, kin selection, relatedness, greenbeard, bacteriocin
INTRODUCTION
All cells interact with other cells. These interactions begin with the birth of a cell, but after septation the extent of interactions varies widely. Some cells stay in continuous contact with neighbors, whereas others rarely interact. The ability of a cell or a multicellular individual to recognize its surrounding neighbors provides important clues about opportunities for exploitation, cooperation, or competition. One type of recognition is identification of others as like oneself. Recognition of kin allows the formation of groups of genetically related individuals in which combined functions exceed the capabilities of an individual. In turn, these advantages can increase the reproductive fitness of the individuals. The study of kin recognition originated with animal research (33, 50). Some animal species, using their five senses, memory, and brain cognition, along with local enviornmental clues, can identify others that are related to themselves. However, because of the complexity of kin recognition in animals, our understanding of it at the molecular level is lacking. In contrast, microbes offer an experimentally accessible path to identify the genetic determinants and molecular interactions responsible for kin recognition. In this review, I discuss the nascent field of kin recognition in bacteria from molecular and evolutionary perspectives.
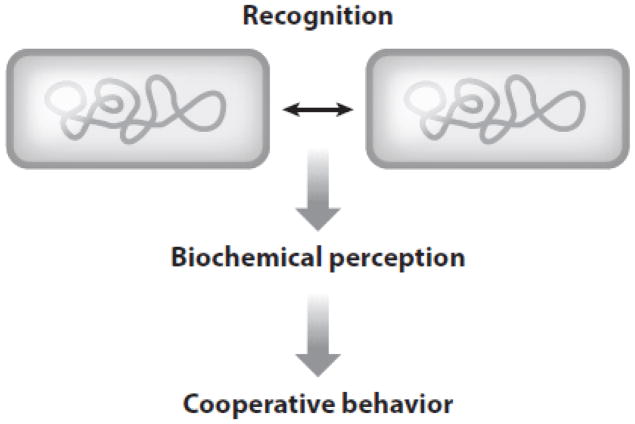
Bacterial kin recognition involves three steps (Figure 1). First, individuals recognize one another by receptor-ligand or receptor-receptor binding. Second, recognition leads to a signal or biochemical perception. Third, there is a behavioral response. Because brain cognition is typically involved in animal kin recognition, studies in these species are largely limited to observation of behavioral changes. That is, kin recognition is indirectly observed as the differential treatment of full siblings as compared to nonkin. In part because of these difficulties, studying kin recognition in animals can be problematic (44, 99). In contrast, bacterial kin recognition involves a molecular event(s) that can be directly observed—e.g., kin cells that adhere together. The end result of these interactions is a cooperative behavior that increases the fitness for the participating individuals.
Figure 1.
Bacterial kin recognition involves three steps.
TYPES OF RECOGNITION
In this review, I categorize kin recognition as either general or specific. General recognition involves low levels of specificity. For example, recognition may simply involve the presence or absence of a single receptor type. Thus recognition occurs when both cells contain the receptor, and discrimination occurs when one cell lacks the receptor. In contrast, specific recognition involves multiple recognition types. For instance, with polymorphic receptors that have different binding affinities, kin recognition depends not only on the presence of the receptor but also on its type (allele). As the diversity of receptor polymorphisms increases, the specificity of recognition correspondingly increases. Specific kin recognition can also occur when multiple loci are involved in recognition. Because bacterial kin recognition depends on interactions between molecules, e.g., proteins, the limits on specificity depend on chemical diversity. In contrast, recognition based on brain cognition potentially offers greater precision. For instance, a human might be able to identify a kin among a thousand or even a million different individuals. Although bacteria cannot discriminate between individuals (e.g., clonemates), such resolution is not necessarily relevant when cells are genetically identical. Instead, what might be critical is the ability of bacteria to differentiate between clonemates and conspecific strains (genetically distinct environmental isolates that belong to the same species) and other species. Examples of general and specific recognition, as well as greenbeard recognition, an evolutionary concept, are described below.
DIVERSITY IN NATURAL HABITATS
Describing his observations about microbiology, Baas Becking formulated the hypothesis that “everything is everywhere, but the environment selects” (25). Although not absolute, this statement captures salient features about microbial ecology. For instance, in similarly located tree canopies, the bacterial communities differ widely depending on the tree species (environment) (60). Additionally, microbes are easily dispersed by the movements of wind, water and animals. In a recent study, atmospheric clouds were shown to be a microbial mixing pot of diverse species at high concentrations (105 bacteria per milliliter) (58). Not surprisingly, hurricane winds further aerosolize particles, leading to even greater diversity and densities of microbes (26). These initial mixtures, once deposited by the winds, are then refined by environmental selection.
Bacteria often live in crowded and diverse habitats in which they interact with neighbors (1). In soils, densities reach 109 bacteria per gram (36), whereas in the human gut they can reach 1012 organisms per gram (53). Bacterial diversity also tends to be high in these environments. For example, the human gut contains ~103 species (and ~104 strains) (8, 53), and a gram of soil might contain 104 to 106 species (36, 75). Two groups also examined conspecific differences and found another ~50-fold increase in diversity at the species level (87, 88, 96, 97). These studies, involving Myxococcus xanthus and Bacillus subtilis, found not only genetic differences among isolates but also the formation of different social groups. These findings suggest that bacteria use recognition tools to form communities with increased fitness.
EVOLUTIONARY THEORY
Evolutionary biologists have played an important role in bringing the topic of microbial kin recognition to the forefront (34, 89, 95, 104). Attracted to simpler and faster models, some evolutionary biologists transitioned from more complex systems, e.g., insects and mammals, to microbes. Microbes not only allow evolutionary experiments to be conducted in the lab but also facilitate the understanding of behaviors and kin recognition at the molecular level. The influx of evolutionary biologists has also introduced a different language and set of questions to the field of microbiology.
Kin Selection and Cooperation
The underlying theme of kin recognition is that it provides a platform for individuals to identify others that are closely related and then form a cooperative group. At face value, the notion that individuals cooperate appears to be at odds with natural selection: Individuals compete for survival. However, the finding that cooperation is common in all kingdoms has inspired research on social evolutionary theory for the last 50 years. Central to this field is William Hamilton’s seminal work that provided a conceptual framework for social and cooperative behaviors (47, 48). Central to his work is Hamilton’s rule, which states, in accordance with Darwinian principles, that cooperative behaviors can occur when r > c/b, where r is the coefficient of relatedness between individuals, c is the fitness cost to the actor, and b is the fitness benefit to the recipient (72a, 104). Thus a cooperative act by an individual can be explained when r and b values are high relative to c. In a bacterial clonal population, clonemates have identical genomes, and thus r = 1. From a kin selection viewpoint, there is a reason (r = 1) for clonemates to cooperate. They are facilitating the propagation of the same genes. As the value of r decreases, the benefit to the recipient needs to correspondingly outweigh the cost of the deed to the actor for the cooperative behavior to evolve within a kin selection framework.
Although Hamilton’s rule provides important insights into how cooperation evolves, the utility of Hamilton’s rule has limits from an experimental perspective. For instance, evolutionary theory has not provided a framework for how to measure r in bacteria that are not clonemates. With diploid organisms, r is frequently measured as the probability of two individuals sharing a gene relative to the whole population (39, 67). For instance, the r coefficient for two brothers is defined as 0.5, whereas two individuals randomly selected from a population would have an r coefficient of 0. Although the literature typically describes r in the context of genes, with diploids its description really pertains to alleles; for example, two brothers would share an identical gene set, but half of their alleles will differ. In bacteria there are not only allelic differences but also a wide variation in gene content between conspecific individuals (discussed below).
A particularly relevant idea for bacterial kin recognition that has emerged from the field of social evolution is the concept of a greenbeard gene. The idea originated with Hamilton (48) and was significantly embellished by Dawkins (23, 24) and others (39, 72a, 103). A greenbeard gene or locus has three features: (a) a recognizable trait, (b) the ability to recognize others that bear the same trait (gene/allele), and (c) the ability to confer a cooperative or helpful behavior on those individuals that bear the same trait. Although the greenbeard was originally conceived as a hypothetical gene, Haig (45, 46) made the idea concrete by suggesting that homotypic cell surface receptors could perform all of these functions. As described below, several recent reports describe greenbeard functions in bacteria, and this type of recognition may constitute a major form of kin recognition in these organisms. Because greenbeard involves a single locus, there is also the potential for nonkin to be recognized. For instance, if a single greenbeard locus was horizontally transferred to an unrelated bacterium, that recipient would now recognize the donor or siblings thereof. In this scenario, recognition is instead called greenbeard or kind recognition (89), because the individuals are related only at the greenbeard locus; the rest of their genomes are not related. Although nonkin greenbeard recognition is theoretically plausible, the extent to which it occurs in nature is unknown. As described above, greenbeard recognition can be either specific or general, depending on whether the locus is polymorphic.
Darwinian Selection
It is important to note that, from a theoretical perspective, kin recognition and kin selection not necessarily have to be invoked to explain cooperative behaviors; there are other theoretical explanations for how cooperation might evolve (67). In Darwinian theory the underlying principle is that individuals compete for limited resources and that the most fit prevail. This leads to the conclusion that individuals will tend not to assist their neighbors but instead will antagonize them. Bacteria are no exception—they compete. Perhaps the clearest example of this is the propensity of bacteria to antagonize or kill their neighbors. This behavior is frequently directed toward conspecific relatives (16, 55, 74). Because related strains share genes, traits, and niches, they are strong competitors for shared resources. This leads to a conundrum: Does a bacterium antagonize a close relative or cooperate with it? Before we consider this, it would first be useful to discuss bacterial relatedness.
BACTERIAL RELATEDNESS
The concept of species, and hence relatedness, in bacteria is different from relatedness in plants and animals, which is typically described in terms of sexual reproduction. Bacteria undergo asexual reproduction; they divide by binary fission. This results in genetically identical offspring. However, microbial communities are far from monocultures; populations rapidly diverge by mutation and horizontal gene transfer, as well as environmental conditions that continuously mix populations (64). For this reason, understanding bacterial relatedness is important in the context of kin recognition.
Individual bacterial strains are similar if they belong to the same species. Classically, bacterial species are defined as a group of wild strains that share phenotypes (77). This definition has been augmented by 16S rRNA classification, which defines bacterial strains as belonging to the same species when their 16S rRNA sequences are ≥98.7% identical. Strains whose 16S rRNA identity is <98.7% are generally viewed as distinct species. However, the highly conserved nature of 16S rRNA genes results in poor resolution between strains and species. An alternative criterion for relatedness classification is DNA-DNA hybridization (DDH) (77). A DDH value of 70% is a widely accepted standard to delineate species. Today, with the availability of genome sequences, DDH is being replaced with average nucleotide identity (ANI), which is a measure of homology between shared sequences. For species cutoff values, 70% DDH correlates to 95% ANI (43). However, two strains can share a high ANI value, e.g., 96%, yet their shared genome content might be only 70% (43).
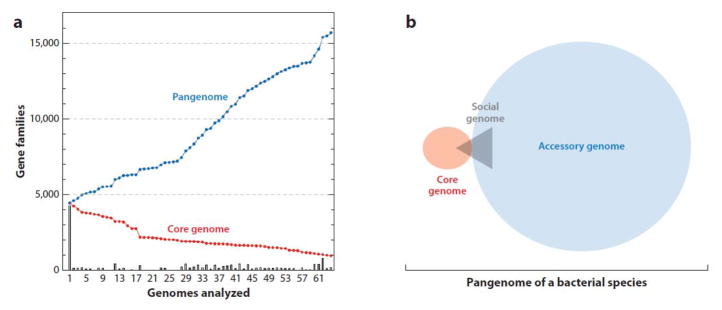
Ussery and colleagues carried out a global analysis of a species by comparing over 60 E. coli genomes (63). Genome size ranged from 4.57 to 5.93 Mbp, a nearly 30% difference between the smallest and largest genomes. The pangenome consisted of 15,741 gene families, and only 993 gene families were represented in every (core) genome (Figure 2). Since E. coli strains contain about 5,000 genes, each strain contains only a third of its species’s pangenome. From this analysis, one can conclude that a group of conspecific strains that share a similar gene content, a measure of relatedness, should be defined as a genovar (72). A genovar is a more precise indication of relatedness than serovar (which describes antigen relatedness) and in this review suggests r > 0.
Figure 2.
Core genomes and pan genomes among conspecific strains. (a) Plot of 61 completed Escherichia coli genomes showing a cumulative count of common (core; red) and total (pan; blue) gene families as a function of the number of analyzed genomes. The bars represent the number of unique (new) gene families as genomes are added along the x-axis. Genomes 62–64 are phylogenetically distant strains. Adapted with permission from Reference 63. (b) Schematic illustration comparing types of genes. The core genome consists of genes shared by conspecific isolates that are presumably important for the survival of the species. Accessory genes provide adaptation advantages for growth under different conditions or in different niches. Strains that share accessory gene sets belong to the same genovar. The social genome (gray) can allow kin recognition and cooperative behaviors; it includes genes that can belong to the core or accessory genome.
DISCRIMINATION AGAINST RELATED INDIVIDUALS
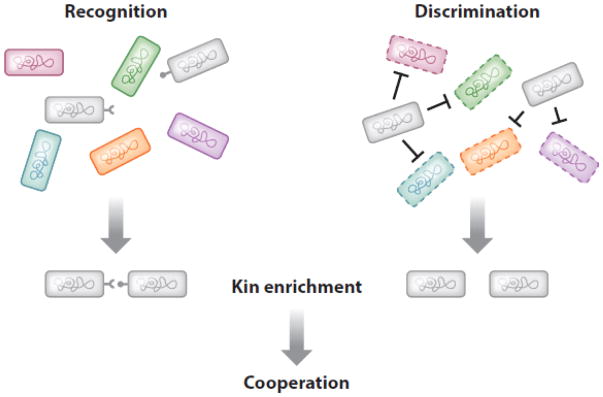
Although I have described what may be viewed as two very different paths—bacterial killing of related individuals and kin recognition—the ability to discriminate against (antagonize) others can lead to outcomes that are similar to those of kin recognition (Figure 3). That is, the removal of competing bacteria from a habitat creates a population that is more closely related. In this regard, bacteria are well known for their ability to produce a diverse array of toxins that kill or inhibit other bacteria (16, 74, 81). Bacteriocins are one common class of toxins that have a narrow range of activity that is directed toward related strains. For example, colicins, a well-known class of bacteriocins made by E. coli, target E. coli strains (16). Bacteriocins are released into the environment by either transport systems or cell lysis (16, 74). Once released, bacteriocins bind to specific cell surface receptors on target cells and then kill by a variety of mechanisms. Producer cells and clonemates are resistant to their own bacteriocin(s) because they make a cognate immunity protein(s). Because bacteriocin systems are relatively simple and provide an effective means for dominating an environmental niche, nearly all bacteria produce bacteriocins (57, 74, 107). For toxins to be effective, they need to be unique to help ensure that other cells in the population are not resistant. Because of this selective pressure, there is a diverse array of polymorphic toxins (55, 107). Toxin modules that are homologs to bacteriocins are also found associated with bacterial delivery systems. Such systems include type VI secretion (T6S), contact-dependent inhibition (CDI; discussed below), and others (49, 55, 81). As discussed below, polymorphic toxins have likely contributed to diversification of kin recognition systems (87a).
Figure 3.
Two paths that lead to kin enrichment. The recognition model is based on binding affinities between related cells. The discrimination model is based on antagonism (bacteriocin production). Colors indicate distinct genovars/strains; dashed borders represent cells inhibited by blue cells.
Discrimination Against Clonemates
Considering the increased competition among genetically similar bacteria, as noted above, it is perhaps not surprising that discrimination is not restricted to distant species and relatives but also includes self. A number of examples have emerged in which bacteria kill their clonemates. Paenibacillus dendritiformis produces a bacteriocin called sibling lethal factor that functions in territoriality behavior, where approaching bacterial swarms repel each other or form demarcation lines between colony swarms (9, 10). Bacillus subtilis and Streptococcus pneumoniae similarly produce bacteriocins that kill clonemates in response to starvation and stress, respectively (17, 18, 32, 42). Although clonemate competition is not readily explained by kin selection theory, it may be more prevalent in bacterial populations than originally thought, but this has not yet been confirmed.
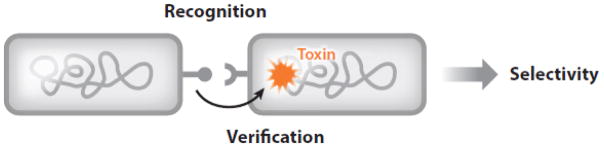
Precision of recognition can be increased when recognition and discrimination functions are combined (Figure 4). Examples of such recognize and verify systems are given below.
Figure 4.
The recognize-and-verify model for kin recognition. The first stage involves receptor-ligand binding. During the second stage (verification), a polymorphic toxin (orange) is delivered to the recognized cell. Clonemates will express a cognate immunity factor and survive, whereas a cell that is not a clonemate will lack immunity and die.
BACTERIAL KIN RECOGNITION
Myxobacteria and Outer Membrane Exchange
The roots of bacterial sociobiology research can be traced back to myxobacteria (30). These terrestrial microbes exhibit complex social behaviors that culminate in the production of multicellular fruiting bodies. Because these bacteria form multicellular aggregates from cells in their environment, they use forms of kin recognition to identify partnering siblings (100). In support of this, fruiting bodies derived from natural sources tend to consist of clonemates and closely related strains (59, 86). Although the mechanism for clonemate selection is poorly understood, early studies showed that myxobacteria isolates antagonize one another from joining a fruit, apparently through bacteriocin activity (86). Other mechanisms may also contribute to clonemate selection.
One of the best understood kin recognition systems in bacteria is called outer membrane exchange (OME). OME is a process by which myxobacteria transiently fuse their outer membranes and exchange their outer membrane proteins and lipids (Figure 5a) (13). OME is initiated when two cells make contact on a solid surface (101). The ensuing exchange is robust, as partnering cells share private goods in essentially equal amounts (68). Because a substantial amount of outer membrane material is exchanged, OME can repair damaged cells (93). For instance, if one cell lacks an outer membrane protein essential for motility, it can be functionally rescued for that defect by a partnering cell that contains the corresponding protein (Figure 5a,c). Similarly, if one cell contains defective lipopolysaccharide (LPS), another cell with wild-type LPS can repair the defective cell by OME (92). Based on these and other findings, we hypothesize that one role of OME is to help create a population that is more homogenous by sharing outer membrane components. In turn the whole population benefits because it is more fit to conduct multicellular functions (93).
Figure 5.
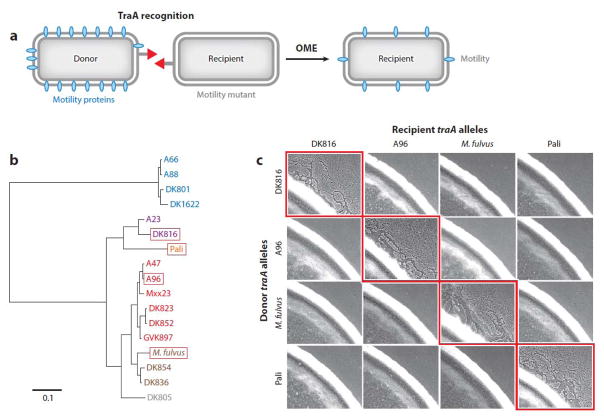
Kin recognition and outer membrane exchange (OME) in myxobacteria. A) A model of OME. Two cells have compatible TraA proteins (red) for recognition, which leads to membrane fusion and bi-directional cell component exchange. Here, the ‘recipient’ lacks a particular outer membrane motility protein and can move only once it receives that protein (blue) from the donor. (b) A phylogenetic tree showing the relatedness of traA alleles from different isolates. The tree contains six functionally distinct recognition groups that are color coded; those in red boxes pertain to panel c, which shows phenotypic assays demonstrating kin (allele)-specific recognition by TraA. Colony edges contain 1:1 mixtures of isogenic donor and recipient strains that contain the indicated traA alleles. All strains contain mutations that block motility; however, motility in the recipients can be restored by OME (see panel a). OME occurs only between identical traA alleles (red boxes). M. fulvus = Myxococcus fulvus. Panels b and c adapted with permission from Reference 70.
OME requires two cell surface proteins, called TraA and TraB, that must be present on both partnering cells (69). TraA/B forms the machinery to catalyze the fusion of the outer membranes, although the mechanistic details are not well understood. Importantly, the TraA cell surface receptor is the recognition determinant (70). For recognition to occur, the partnering cells must have identical or very similar receptors, providing for a homotypic interaction. Among environmental isolates, traA is polymorphic (Figure 5b). In studies from a limited set of M. xanthus isolates, six distinct TraA recognition groups were identified (Figure 5b) (70). Based on an expanded study consisting of a bioinformatic and functional analysis of 100 traA alleles from a diverse pool of myxobacteria, we propose that this collection represents ~50 distinct TraA recognition groups (P. Cao, R. Awal, R. Muller, D. Wall, unpublished manuscript). By extending this dataset, we hypothesize that in nature there are hundreds of different TraA recognition groups and that these polymorphisms determine recognition specificity. Thus by simply swapping traA alleles recognition can be reprogrammed in M. xanthus cells (Figure 5C) (70). Recently, we reengineered TraA specificity by site-directed mutagenesis (P. Cao & D. Wall, unpublished manuscript). In some cases single–amino acid substitutions changed recognition specificity, whereas in other cases alleles were created with unique specificities. Thus TraA is a kin recognition determinate in which simple polymorphisms can lead to completely altered specificities.
The polymorphic nature of TraA recognition raises Crozier’s paradox (21, 38, 52), which states that genetic variation should erode because larger cooperative groups are more fit than small groups. So for TraA, how can diversity be maintained if the sharing of private goods by OME leads to beneficial outcomes? That is, members of a large population will share more frequently and will incur more benefit than those in small groups. In turn, traA alleles that represent small groups should be driven to extinction (52). We think the answer to this question lies in the nature of the goods that are exchanged. Recently, we discovered that, along with beneficial goods, harmful toxins (bacteriocins) are also exchanged (28). Thus indiscriminate pairing of individuals leads to lethal outcomes. Stated another way, OME represents a dual model involving the recognition and verification of partner identity (Figure 4). In this scheme, to guard against lethal OME events, TraA polymorphisms are selected for and maintained to provide recognition specificity that helps ensure that OME only occurs between clonemates.
The properties of TraA match the definition of a greenbeard locus (23, 24). Consistent with this, we found that a traA allele from an M. fulvus isolate was phylogenetically related and functionally compatible with some M. xanthus traA alleles (Figure 5b) (70). At face value, TraA recognition can thus occur between distinct species, a characteristic of greenbeard (kind) recognition (89). However, there are caveats to this conclusion. First, M. fulvus and M. xanthus are closely related, and it is unclear whether these isolates actually represent distinct species. Second, although TraA recognition and OME occur, the cells involved do not necessarily remain viable (28, 70), because OME involves a verification step with a toxin at a different locus. In conclusion, TraA has greenbeard qualities; however, a successful OME outcome also depends on genetic relatedness at other loci in the genome, a requirement for kin recognition.
Adhesins and Contact-Dependent Inhibition
The specific adhesion of sibling cells represents a direct form of bacterial kin recognition. Some adhesins recognize the same adhesin on adjacent cells, and thus kin recognition occurs through homotypic receptors. Examples of homotypic receptors include members of the autotransporter family, a subclass of the type V secretion system, in which a single polypeptide facilitates its own transport across the outer membrane (22). TibA and Ag43 are members of the autotransporter family and share homology in their adhesin domains, which contain repeat sequences (51, 85). Crystal structures have shown that self-associated Ag43 proteins bind by a Velcro-like handshake (51). Expression of these adhesins in E. coli results in self-recognition manifested as autoaggregation or cell clumping. Ag43 expression also results in self-associated chains of cells, suggesting that adhesins are concentrated at cell poles (94). The ability of TibA/Ag43 to recognize clonemates allows those bacteria to form social groups in the form of biofilms or aggregates that can protect cells from environmental insults.
Some adhesins are multifunctional. A case in point is CDI, a system that is widely distributed in alpha-, beta- and gammaproteobacteria (5). The CDI system is also a member of the type V secretion family and belongs to the two-partner secretion subclass. The CDI system uses large and extended adhesins (CdiA) on the cell surface. Related cells are recognized when they attach to cell surface receptors. The C terminus of CdiA proteins is highly polymorphic and contains different types of toxin modules (5). Thus upon binding to related cells, toxins are delivered to the recipient. If the recipient does not contain the cognate CdiI immunity protein, because it is not a clonemate, then its growth is blocked. CDI is thus another example of a recognize-and-verify system that increases kin selectivity (Figure 4).
Importantly, the function of CDI is not simply to antagonize related strains. CDI also functions to assemble multicellular biofilm communities (3, 37). CdiA recognition occurs by binding to heterologous cell surface receptors as well as through homotypic interactions (79). Such CdiA-CdiA interactions illustrate a greenbeard recognition mechanism. By recognizing and verifying other cells, the CDI system helps to ensure that biofilms consist of clonemates.
Other interesting aspects of the CDI system involve its distribution and receptor recognition. A genomic analysis determined that cdi loci are found in 90 of the 576 sequenced E. coli genomes (78). These polymorphic loci are thus part of the accessory genome and are typically associated with genomic islands. The best-characterized CDI system is from E. coli EC93, in which CdiA was shown to bind BamA as the cell surface receptor (6). BamA is an essential β-barrel protein involved in outer membrane protein folding (6). Although BamA homologs are universally found in gram-negative bacteria, the specificity of CdiA to binding to BamA is in extracellular loops 6 and 7, whose sequence conservation are species restricted to E. coli (80). Surprisingly, these surface-exposed loops are identical in hundreds of E. coli isolates. This finding is unexpected, as cell surface sequences are under selective pressure to diversify and these loop sequences are not essential for BamA function (71, 80). Although it is unknown why loops 6 and 7 are invariant in E. coli but are polymorphic in other species (80), their utilization allows CdiA-EC93 to selectively recognize fellow E. coli strains for CDI functions.
Proteus Territoriality
Proteus mirabilis exhibits a behavior known as territoriality (7, 11, 40). The behavior is seen on swarm agar plates on which different motile isolates frequently form demarcations called Dienes lines when the swarm colonies meet. In contrast, two sibling colonies will merge their swarms in the absence of Dienes lines. The Dienes lines are thus thought, in part, to reflect a molecular identification system(s) for self/nonself recognition to ensure that territories are inhabited by fellow clonemates. Interestingly, the selectivity of Proteus discrimination is high, because in one study Dienes typing of 204 isolates resulted in 98 distinct compatibility groups (83). Discrimination, in part, is correlated with the production of bacteriocins (40, 84). Thus isolates that are sensitive to a particular bacteriocin are killed, and this outcome seems to contribute toward Dienes line formation. However, not all Dienes lines are attributed to bacteriocins. In these cases, boundary formation seems to depend on cell-cell recognition. To identify genetic determinants involved in recognition, Gibbs and coworkers (41, 102) conducted forward screens. They found three gene clusters that govern self-recognition in P. mirabilis. One gene cluster encodes homologs of the T6S system (tss). The T6S system is a transport apparatus that delivers effector proteins to other cells (81). Frequently the effectors are toxins and can be homologous to bacteriocins. A second gene cluster is called idr and encodes putative cytotoxins delivered by tss. Both idr and tss are involved in growth inhibition, and similar results were independently reported (2). The role of the third gene cluster, ids, is less clear. The ids gene cluster contains two genes that are polymorphic, suggesting a role in recognition (41). The ids genes are not required for competition with foreign strains but are required for nonlethal interactions with the parental strain (102). Biochemically, the polymorphic proteins IdsD and IdsE interact in an allele-specific manner through their transmembrane domains (15). Additional studies are needed to elucidate the molecular mechanism by which IdsD-IdsE binding leads to behavioral changes manifested as territoriality. Although T6S systems are typically associated with aggressive behaviors, they may also be involved in nonantagonistic interactions (81), and perhaps Ids is one example of this exception.
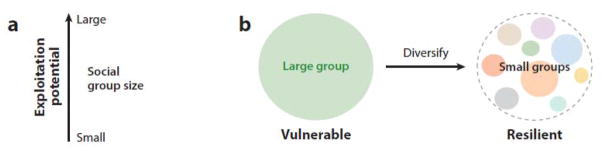
As microbiologists turn more attention toward how bacteria interact, we have an increasing number of examples of territorial behaviors, as judged by demarcation lines, between conspecific isolates (65, 87, 87a, 97). As seen with P. mirabilis, the resolution of discrimination is high even among isolates that originate from the same location and that are highly related (87, 97). These findings suggest that kin recognition and discrimination systems are widespread in the bacterial kingdom. Moreover, these findings suggest that the diversification of social groups within ecotypes may contribute to the stability of the overall population (Figure 6).
Figure 6.
(a) Larger kin groups are less stable. A single individual cannot be socially exploited, whereas a large social group can. Exploitation mechanisms include social cheats and the propagation of a phage infection in a social group. (b) A model suggesting that large social groups are under environmental and exploitation pressures that tend to lead to diversification. In contrast, a collection of smaller groups provides overall population resiliency for two reasons. First, if an exploitation phenotype develops, it is limited to one social group. Second, different groups (genovars; see accessory genes, Figure 2) expand the genetic diversity, which in turn increases the potential, of the entire population, to adapt when faced with changing environmental conditions and competitors. The relative size of each group is dynamic and will temporally fluctuate depending on the conditions. For ecological examples of niche diversity and diversification, see References 73, 87, 87a, 97 and 97a.
Recognition from Afar—Diffusible Signals
Kin recognition does not require direct cell-cell contact; recognition can be perceived through diffusible signals. Recognition by diffusible signals provides the advantage that it allows individuals to decipher the composition of kin in a local population. A limitation is that the identity of adjacent cells is not necessarily known. Quorum sensing is an example of how many bacterial species recognize their kin and their population densities in local environments. In turn, quorum signaling, which is mediated by diffusible autoinducers, is used to regulate diverse cooperative behaviors, including bioluminescence, competence, virulence, biofilm formation, and motility, among others (35, 82). Quorum-sensing studies in gram-negative bacteria have found that acylated homoserine lactones (acyl-HSLs) represent a major class of signaling molecule, whereas gram-positive bacteria typically carry out this function with oligopeptides (66, 82). The specificity of recognition is derived from receptors that selectively bind their cognate signaling molecules. In the case of acyl-HSLs, their chemical structures vary depending on substitutions, saturation, and overall length of the acyl chains (35, 90). Quorum sensing in gram-negative bacteria can result in general kin recognition at the species level or can be more specific. For example, some Vibrio cholera and Pseudomonas aeruginosa strains make two and three types of signaling molecules, respectively, which adds complexity and specificity to the recognition process and to the regulatory control of gene expression (82, 98).
Gram-positive bacteria do contain polymorphic quorum-sensing molecules and cognate receptors (66, 91, 105). For instance, Staphylococcus aureus uses quorum sensing to control the expression of the agr virulence regulon. In S. aureus there are four known kin recognition groups that are determined by allelic variation in the peptide signal derived from AgrD and the cognate coevolved receptor, AgrC (66). The interaction between the signal and AgrC is highly specific, as a single amino acid substitution can change specificity. Interestingly, an AgrD variant that does not function as a cognate signal with a particular AgrC variant will block the expression of the agr regulon in that strain (56, 66). This antagonism prevents an established infection from being invaded by another S. aureus strain. In evolutionary terms, social antagonism may contribute to the diversification and hence selectivity found in kin recognition systems (31). Similar polymorphic recognition systems are found in B. subtilis and S. pneumoniae quorum control of natural competence and in Enterococcus spp for pheromone-controlled conjugation (4, 19, 29, 54, 88).
In other paradigms, the ligand and receptor that mediate kin recognition may have additional functions. One example is Rhizobium spp. that associate with plants where the roots form protected nodules, excluding other microorganisms. Here Rhizobium cells differentiate into bacteroids that fix nitrogen for the plant. In this mutualistic interaction, the plant in turn provides bacteroids with nutrients that the bacterium uses to synthesize rhizopines, which are metabolites that are unique to Rhizobium spp. (27, 76, 106). Rhizobium spp. located outside of nodules and that are not differentiated can selectively take up and use rhizopines as a carbon/energy source. In essence this is a form of greenbeard recognition in which rhizopines represent the specificity ligand that binds catabolite enzymes (receptor) and results in their utilization (benefit) (106).
In an interesting twist on kin recognition, Mougous and colleagues propose that P. aeruginosa recognizes danger in its environment by the lysis of kin cells (61, 62). In this scheme, sensor kinases/response regulators (RetS/GacS/A) detect a diffusible signal that originates from the lysis of siblings. In turn, this activates a response to the danger by the induction of a regulon that includes the T6S system, a weapon used to attack hostile bacteria (a likely source of the danger). This response is one of many examples whereby different bacterial groups induce bacteriocin/toxin production in response to competition or stress as an apparent counterattack measure (20).
PERSPECTIVE
Kin recognition in bacteria is a frontier in science. Currently there are few labs that are making a concerted effort to study this topic. However, there is a growing appreciation that bacteria recognize their neighbors and this leads to important social functions, such as multicellular development, biofilm formation, and quorum sensing (57, 82, 100). Given the vast and unknown diversity of bacteria, there are likely many examples where kin recognition is involved in unforeseen microbial processes, whether in the human body or the environment.
An advantage of studying kin recognition and kin selection in bacteria is the ability to understand each step at the molecular level (Figure 1). Experimental methods in bacteria allow hypotheses to be tested and bacterial systems to be reengineered. In addition, with the advent of next-generation sequencing and other sensitive molecular techniques, particular ecological, evolutionary, and mechanistic questions can be explored in natural populations.
FUTURE ISSUES
Experimental
How are bacterial populations spacially organized and how do they interact in natural habitats?
With the large diversity of bacterial species, what other types of cooperative behaviors and kin recognition systems have evolved? Do some recognition systems rely on multiple loci, instead of a single polymorphic receptor, to confer specificity of recognition?
When kin recogntion governs cooperative behavoirs how is selectivity of recognition maintained when Cozier’s paradox says it should erode?
Can cooperative behaviors and fitness gains be precisely defined and quantified?
Definitions
Can evolutionary theory provide a matrix or a protocol to measure relatedness in bacteria beyond clonemates?
What constitutes a kin group within a bacterial species?
Acknowledgments
I thank Joan Strassmann for helpful comments. This work was supported by the National Institutes of Health grant GM101449.
Footnotes
DISCLOSURE STATEMENT
The author is not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Achtman M, Wagner M. Microbial diversity and the genetic nature of microbial species. Nat Rev Microbiol. 2008;6:431–40. doi: 10.1038/nrmicro1872. [DOI] [PubMed] [Google Scholar]
- 2.Alteri CJ, Himpsl SD, Pickens SR, Lindner JR, Zora JS, et al. Multicellular bacteria deploy the type VI secretion system to preemptively strike neighboring cells. PLOS Pathog. 2013;9:e1003608. doi: 10.1371/journal.ppat.1003608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anderson MS, Garcia EC, Cotter PA. Kind discrimination and competitive exclusion mediated by contact-dependent growth inhibition systems shape biofilm community structure. PLOS Pathog. 2014;10:e1004076. doi: 10.1371/journal.ppat.1004076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ansaldi M, Marolt D, Stebe T, Mandic-Mulec I, Dubnau D. Specific activation of the Bacillus quorum-sensing systems by isoprenylated pheromone variants. Mol Microbiol. 2002;44:1561–73. doi: 10.1046/j.1365-2958.2002.02977.x. [DOI] [PubMed] [Google Scholar]
- 5.Aoki SK, Diner EJ, de Roodenbeke CT, Burgess BR, Poole SJ, et al. A widespread family of polymorphic contact-dependent toxin delivery systems in bacteria. Nature. 2010;468:439–42. doi: 10.1038/nature09490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aoki SK, Malinverni JC, Jacoby K, Thomas B, Pamma R, et al. Contact-dependent growth inhibition requires the essential outer membrane protein BamA (YaeT) as the receptor and the inner membrane transport protein AcrB. Mol Microbiol. 2008;70:323–40. doi: 10.1111/j.1365-2958.2008.06404.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Armbruster CE, Mobley HL. Merging mythology and morphology: The multifaceted lifestyle of Proteus mirabilis. Nat Rev Microbiol. 2012;10:743–54. doi: 10.1038/nrmicro2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Backhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI. Host-bacterial mutualism in the human intestine. Science. 2005;307:1915–20. doi: 10.1126/science.1104816. [DOI] [PubMed] [Google Scholar]
- 9.Be’er A, Ariel G, Kalisman O, Helman Y, Sirota-Madi A, et al. Lethal protein produced in response to competition between sibling bacterial colonies. PNAS. 2010;107:6258–63. doi: 10.1073/pnas.1001062107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Be’er A, Florin EL, Fisher CR, Swinney HL, Payne SM. Surviving bacterial sibling rivalry: Inducible and reversible phenotypic switching in Paenibacillus dendritiformis. mBio. 2011;2:e00069–11. doi: 10.1128/mBio.00069-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Budding AE, Ingham CJ, Bitter W, Vandenbroucke-Grauls CM, Schneeberger PM. The Dienes phenomenon: competition and territoriality in swarming Proteus mirabilis. J Bacteriol. 2009;191:3892–900. doi: 10.1128/JB.00975-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deleted in proof
- 13.Cao P, Dey A, Vassallo CN, Wall D. How myxobacteria cooperate. J Mol Biol. 2015;427:3709–21. doi: 10.1016/j.jmb.2015.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deleted in proof
- 15.Cardarelli L, Saak C, Gibbs KA. Two proteins form a heteromeric bacterial self-recognition complex in which variable subdomains determine allele-restricted binding. mBio. 2015;6:e00251. doi: 10.1128/mBio.00251-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cascales E, Buchanan SK, Duche D, Kleanthous C, Lloubes R, et al. Colicin biology. Microbiol Mol Biol Rev. 2007;71:158–229. doi: 10.1128/MMBR.00036-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Claverys JP, Havarstein LS. Cannibalism and fratricide: mechanisms and raisons d’etre. Nat Rev Microbiol. 2007;5:219–29. doi: 10.1038/nrmicro1613. [DOI] [PubMed] [Google Scholar]
- 18.Claverys JP, Martin B, Havarstein LS. Competence-induced fratricide in streptococci. Mol Microbiol. 2007;64:1423–33. doi: 10.1111/j.1365-2958.2007.05757.x. [DOI] [PubMed] [Google Scholar]
- 19.Cook LC, Federle MJ. Peptide pheromone signaling in Streptococcus and Enterococcus. FEMS Microbiol Rev. 2014;38:473–92. doi: 10.1111/1574-6976.12046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cornforth DM, Foster KR. Competition sensing: the social side of bacterial stress responses. Nat Rev Microbiol. 2013;11:285–93. doi: 10.1038/nrmicro2977. [DOI] [PubMed] [Google Scholar]
- 21.Crozier RE. Genetic clonal recognition abilities in marine invertebrates must be maintained by selection for something else. Evolution. 1986;40:1100–1. doi: 10.1111/j.1558-5646.1986.tb00578.x. [DOI] [PubMed] [Google Scholar]
- 22.Dautin N, Bernstein HD. Protein secretion in gram-negative bacteria via the autotransporter pathway. Annu Rev Microbiol. 2007;61:89–112. doi: 10.1146/annurev.micro.61.080706.093233. [DOI] [PubMed] [Google Scholar]
- 23.Dawkins R. The Selfish Gene. Oxford, UK: Oxford Univ. Press; 1976. [Google Scholar]
- 24.Dawkins R. The Extended Phenotype: The Gene as the Unit of Selection. Oxford, UK: W. H. Freeman; 1982. [Google Scholar]
- 25.de Wit R, Bouvier T. ‘Everything is everywhere, but, the environment selects’; what did Baas Becking and Beijerinck really say? Environ Microbiol. 2006;8:755–58. doi: 10.1111/j.1462-2920.2006.01017.x. [DOI] [PubMed] [Google Scholar]
- 26.DeLeon-Rodriguez N, Lathem TL, Rodriguez RL, Barazesh JM, Anderson BE, et al. Microbiome of the upper troposphere: species composition and prevalence, effects of tropical storms, and atmospheric implications. PNAS. 2013;110:2575–80. doi: 10.1073/pnas.1212089110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Denison RF. Legume sanctions and the evolution of symbiotic cooperation by rhizobia. Am Nat. 2000;156:567–76. doi: 10.1086/316994. [DOI] [PubMed] [Google Scholar]
- 28.Dey A, Vassallo CN, Conklin AC, Pathak DT, Troselj V, Wall D. Sibling rivalry in Myxococcus xanthus is mediated by kin recognition and a polyploid prophage. J Bacteriol. 2016;198:994–1004. doi: 10.1128/JB.00964-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dunny GM, Antiporta MH, Hirt H. Peptide pheromone-induced transfer of plasmid pCF10 in Enterococcus faecalis: probing the genetic and molecular basis for specificity of the pheromone response. Peptides. 2001;22:1529–39. doi: 10.1016/s0196-9781(01)00489-2. [DOI] [PubMed] [Google Scholar]
- 30.Dworkin M. The myxobacteria: new directions in studies of procaryotic develoment. CRC Crit Rev Microbiol. 1972;1:435–52. [Google Scholar]
- 31.Eldar A. Social conflict drives the evolutionary divergence of quorum sensing. PNAS. 2011;108:13635–40. doi: 10.1073/pnas.1102923108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ellermeier CD, Hobbs EC, Gonzalez-Pastor JE, Losick R. A three-protein signaling pathway governing immunity to a bacterial cannibalism toxin. Cell. 2006;124:549–59. doi: 10.1016/j.cell.2005.11.041. [DOI] [PubMed] [Google Scholar]
- 33.Fletcher DJC, Michener CD. Kin Recognition in Animals. Chichester, UK: John Wiley; 1987. [Google Scholar]
- 34.Foster KR, Parkinson K, Thompson CR. What can microbial genetics teach sociobiology? Trends Genet. 2007;23:74–80. doi: 10.1016/j.tig.2006.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fuqua C, Parsek MR, Greenberg EP. Regulation of gene expression by cell-to-cell communication: acyl-homoserine lactone quorum sensing. Annu Rev Genet. 2001;35:439–68. doi: 10.1146/annurev.genet.35.102401.090913. [DOI] [PubMed] [Google Scholar]
- 36.Gans J, Wolinsky M, Dunbar J. Computational improvements reveal great bacterial diversity and high metal toxicity in soil. Science. 2005;309:1387–90. doi: 10.1126/science.1112665. [DOI] [PubMed] [Google Scholar]
- 37.Garcia EC, Anderson MS, Hagar JA, Cotter PA. Burkholderia BcpA mediates biofilm formation independently of interbacterial contact-dependent growth inhibition. Mol Microbiol. 2013;89:1213–25. doi: 10.1111/mmi.12339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gardner A, West SA. Social evolution: the decline and fall of genetic kin recognition. Curr Biol. 2007;17:R810–12. doi: 10.1016/j.cub.2007.07.030. [DOI] [PubMed] [Google Scholar]
- 39.Gardner A, West SA. Greenbeards. Evolution. 2010;64:25–38. doi: 10.1111/j.1558-5646.2009.00842.x. [DOI] [PubMed] [Google Scholar]
- 40.Gibbs KA, Greenberg EP. Territoriality in Proteus: advertisement and aggression. Chem Rev. 2011;111:188–94. doi: 10.1021/cr100051v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gibbs KA, Urbanowski ML, Greenberg EP. Genetic determinants of self identity and social recognition in bacteria. Science. 2008;321:256–59. doi: 10.1126/science.1160033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gonzalez-Pastor JE, Hobbs EC, Losick R. Cannibalism by sporulating bacteria. Science. 2003;301:510–13. doi: 10.1126/science.1086462. [DOI] [PubMed] [Google Scholar]
- 43.Goris J, Konstantinidis KT, Klappenbach JA, Coenye T, Vandamme P, Tiedje JM. DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int J Syst Evol Microbiol. 2007;57:81–91. doi: 10.1099/ijs.0.64483-0. [DOI] [PubMed] [Google Scholar]
- 44.Grafen A. Do animals really recognize kin? Anim Behav. 1990;39:42–54. [Google Scholar]
- 45.Haig D. Gestational drive and the green-bearded placenta. PNAS. 1996;93:6547–51. doi: 10.1073/pnas.93.13.6547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Haig D. The social gene. In: Krebs JR, Davies NB, editors. Behavioural Ecology: An Evolutionary Approach. Cambridge, UK: Wiley-Blackwell; 1997. pp. 284–304. [Google Scholar]
- 47.Hamilton WD. The genetical evolution of social behaviour. I. J Theor Biol. 1964;7:1–16. doi: 10.1016/0022-5193(64)90038-4. [DOI] [PubMed] [Google Scholar]
- 48.Hamilton WD. The genetical evolution of social behaviour. II. J Theor Biol. 1964;7:17–52. doi: 10.1016/0022-5193(64)90039-6. [DOI] [PubMed] [Google Scholar]
- 49.Hayes CS, Koskiniemi S, Ruhe ZC, Poole SJ, Low DA. Mechanisms and biological roles of contact-dependent growth inhibition systems. Cold Spring Harb Perspect Med. 2014;4:a010025. doi: 10.1101/cshperspect.a010025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hepper PG. Kin recognition: functions and mechanisms; a review. Biol Rev Camb Philos Soc. 1986;61:63–93. doi: 10.1111/j.1469-185x.1986.tb00427.x. [DOI] [PubMed] [Google Scholar]
- 51.Heras B, Totsika M, Peters KM, Paxman JJ, Gee CL, et al. The antigen 43 structure reveals a molecular Velcro-like mechanism of autotransporter-mediated bacterial clumping. PNAS. 2014;111:457–62. doi: 10.1073/pnas.1311592111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Holman L, van Zweden JS, Linksvayer TA, d’Ettorre P. Crozier’s paradox revisited: maintenance of genetic recognition systems by disassortative mating. BMC Evol Biol. 2013;13:211. doi: 10.1186/1471-2148-13-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hooper LV, Gordon JI. Commensal host-bacterial relationships in the gut. Science. 2001;292:1115–18. doi: 10.1126/science.1058709. [DOI] [PubMed] [Google Scholar]
- 54.Iannelli F, Oggioni MR, Pozzi G. Sensor domain of histidine kinase ComD confers competence pherotype specificity in Streptoccoccus pneumoniae. FEMS Microbiol Lett. 2005;252:321–26. doi: 10.1016/j.femsle.2005.09.008. [DOI] [PubMed] [Google Scholar]
- 55.Jamet A, Nassif X. New players in the toxin field: polymorphic toxin systems in bacteria. mBio. 2015;6:e00285–15. doi: 10.1128/mBio.00285-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ji G, Beavis R, Novick RP. Bacterial interference caused by autoinducing peptide variants. Science. 1997;276:2027–30. doi: 10.1126/science.276.5321.2027. [DOI] [PubMed] [Google Scholar]
- 57.Klaenhammer TR. Bacteriocins of lactic acid bacteria. Biochimie. 1988;70:337–49. doi: 10.1016/0300-9084(88)90206-4. [DOI] [PubMed] [Google Scholar]
- 58.Kourtev PS, Hill KA, Shepson PB, Konopka A. Atmospheric cloud water contains a diverse bacterial community. Atmos Environ. 2011;45:5399–405. [Google Scholar]
- 59.Kraemer SA, Velicer GJ. Endemic social diversity within natural kin groups of a cooperative bacterium. PNAS. 2011;108(Suppl 2):10823–30. doi: 10.1073/pnas.1100307108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lambais MR, Crowley DE, Cury JC, Bull RC, Rodrigues RR. Bacterial diversity in tree canopies of the Atlantic forest. Science. 2006;312:1917. doi: 10.1126/science.1124696. [DOI] [PubMed] [Google Scholar]
- 61.LeRoux M, Kirkpatrick RL, Montauti EI, Tran BQ, Peterson SB, et al. Kin cell lysis is a danger signal that activates antibacterial pathways of Pseudomonas aeruginosa. eLife. 2015;4:e05701. doi: 10.7554/eLife.05701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.LeRoux M, Peterson SB, Mougous JD. Bacterial danger sensing. J Mol Biol. 2015;427:3744–53. doi: 10.1016/j.jmb.2015.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lukjancenko O, Wassenaar TM, Ussery DW. Comparison of 61 sequenced Escherichia coli genomes. Microb Ecol. 2010;60:708–20. doi: 10.1007/s00248-010-9717-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Luo C, Walk ST, Gordon DM, Feldgarden M, Tiedje JM, Konstantinidis KT. Genome sequencing of environmental Escherichia coli expands understanding of the ecology and speciation of the model bacterial species. PNAS. 2011;108:7200–5. doi: 10.1073/pnas.1015622108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Munson EL, Pfaller MA, Doern GV. Modification of dienes mutual inhibition test for epidemiological characterization of Pseudomonas aeruginosa isolates. J Clin Microbiol. 2002;40:4285–88. doi: 10.1128/JCM.40.11.4285-4288.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Novick RP, Geisinger E. Quorum sensing in staphylococci. Annu Rev Genet. 2008;42:541–64. doi: 10.1146/annurev.genet.42.110807.091640. [DOI] [PubMed] [Google Scholar]
- 67.Nowak MA. Five rules for the evolution of cooperation. Science. 2006;314:1560–63. doi: 10.1126/science.1133755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nudleman E, Wall D, Kaiser D. Cell-to-cell transfer of bacterial outer membrane lipoproteins. Science. 2005;309:125–27. doi: 10.1126/science.1112440. [DOI] [PubMed] [Google Scholar]
- 69.Pathak DT, Wei X, Bucuvalas A, Haft DH, Gerloff DL, Wall D. Cell contact-dependent outer membrane exchange in myxobacteria: genetic determinants and mechanism. PLOS Genet. 2012;8:e1002626. doi: 10.1371/journal.pgen.1002626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pathak DT, Wei X, Dey A, Wall D. Molecular recognition by a polymorphic cell surface receptor governs cooperative behaviors in bacteria. PLOS Genet. 2013;9:e1003891. doi: 10.1371/journal.pgen.1003891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Petersen L, Bollback JP, Dimmic M, Hubisz M, Nielsen R. Genes under positive selection in Escherichia coli. Genome Res. 2007;17:1336–43. doi: 10.1101/gr.6254707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Porwollik S, Boyd EF, Choy C, Cheng P, Florea L, et al. Characterization of Salmonella enterica subspecies I genovars by use of microarrays. J Bacteriol. 2004;186:5883–98. doi: 10.1128/JB.186.17.5883-5898.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72a.Queller DC. Expanded social fitness and Hamilton’s rule for kin, kith and kind. PNAS. 2011;108:10792–99. doi: 10.1073/pnas.1100298108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rendueles O, Zee PC, Dinkelacker I, Amherd M, Wielgoss S, Velicer GJ. Rapid and widespread de novo evolution of kin discrimination. PNAS. 2015;112:9076–81. doi: 10.1073/pnas.1502251112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Riley MA, Wertz JE. Bacteriocins: Evolution, ecology, and application. Annu Rev Microbiol. 2002;56:117–37. doi: 10.1146/annurev.micro.56.012302.161024. [DOI] [PubMed] [Google Scholar]
- 75.Roesch LF, Fulthorpe RR, Riva A, Casella G, Hadwin AK, et al. Pyrosequencing enumerates and contrasts soil microbial diversity. ISME J. 2007;1:283–90. doi: 10.1038/ismej.2007.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rossbach S, Rasul G, Schneider M, Eardly B, de Bruijn FJ. Structural and functional conservation of the rhizopine catabolism (moc) locus is limited to selected Rhizobium meliloti strains and unrelated to their geographical origin. Mol Plant Microbe Interact. 1995;8:549–59. doi: 10.1094/mpmi-8-0549. [DOI] [PubMed] [Google Scholar]
- 77.Rossello-Mora R, Amann R. Past and future species definitions for Bacteria and Archaea. Syst Appl Microbiol. 2015;38:209–16. doi: 10.1016/j.syapm.2015.02.001. [DOI] [PubMed] [Google Scholar]
- 78.Ruhe ZC, Low DA, Hayes CS. Bacterial contact-dependent growth inhibition. Trends Microbiol. 2013;21:230–37. doi: 10.1016/j.tim.2013.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ruhe ZC, Townsley L, Wallace AB, King A, Van der Woude MW, et al. CdiA promotes receptor-independent intercellular adhesion. Mol Microbiol. 2015;98:175–92. doi: 10.1111/mmi.13114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ruhe ZC, Wallace AB, Low DA, Hayes CS. Receptor polymorphism restricts contact-dependent growth inhibition to members of the same species. mBio. 2013;4:e00480–13. doi: 10.1128/mBio.00480-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Russell AB, Peterson SB, Mougous JD. Type VI secretion system effectors: poisons with a purpose. Nat Rev Microbiol. 2014;12:137–48. doi: 10.1038/nrmicro3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Rutherford ST, Bassler BL. Bacterial quorum sensing: its role in virulence and possibilities for its control. Cold Spring Harb Perspect Med. 2012;2:a012427. doi: 10.1101/cshperspect.a012427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Senior BW. The Dienes phenomenon: Identification of the determinants of compatibility. J Gen Microbiol. 1977;102:235–44. doi: 10.1099/00221287-102-2-235. [DOI] [PubMed] [Google Scholar]
- 84.Senior BW, Larsson P. A highly discriminatory multi-typing scheme for Proteus mirabilis and Proteus vulgaris. J Med Microbiol. 1983;16:193–202. doi: 10.1099/00222615-16-2-193. [DOI] [PubMed] [Google Scholar]
- 85.Sherlock O, Vejborg RM, Klemm P. The TibA adhesin/invasin from enterotoxigenic Escherichia coli is self recognizing and induces bacterial aggregation and biofilm formation. Infect Immun. 2005;73:1954–63. doi: 10.1128/IAI.73.4.1954-1963.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Smith DR, Dworkin M. Territorial interactions between two Myxococcus species. J Bacteriol. 1994;176:1201–5. doi: 10.1128/jb.176.4.1201-1205.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Stefanic P, Kraigher B, Lyons NA, Kolter R, Mandic-Mulec I. Kin discrimination between sympatric Bacillus subtilis isolates. PNAS. 2015;112:14042–47. doi: 10.1073/pnas.1512671112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87a.Lyons NA, Kraighter B, Stefanic P, Mandic-Mulec I, Kolter R. A combinatorial kin discrimination system in Bacillus subtilis. Curr Biol. 2016;26:733–42. doi: 10.1016/j.cub.2016.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Stefanic P, Mandic-Mulec I. Social interactions and distribution of Bacillus subtilis pherotypes at microscale. J Bacteriol. 2009;191:1756–64. doi: 10.1128/JB.01290-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Strassmann JE, Gilbert OM, Queller DC. Kin discrimination and cooperation in microbes. Annu Rev Microbiol. 2011;65:349–67. doi: 10.1146/annurev.micro.112408.134109. [DOI] [PubMed] [Google Scholar]
- 90.Thiel V, Kunze B, Verma P, Wagner-Dobler I, Schulz S. New structural variants of homoserine lactones in bacteria. Chembiochem. 2009;10:1861–68. doi: 10.1002/cbic.200900126. [DOI] [PubMed] [Google Scholar]
- 91.Thoendel M, Kavanaugh JS, Flack CE, Horswill AR. Peptide signaling in the staphylococci. Chem Rev. 2011;111:117–51. doi: 10.1021/cr100370n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Vassallo C, Pathak DT, Cao P, Zuckerman DM, Hoiczyk E, Wall D. Cell rejuvenation and social behaviors promoted by LPS exchange in myxobacteria. PNAS. 2015;112:E2939–46. doi: 10.1073/pnas.1503553112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Vassallo C, Wall D. Tissue repair in myxobacteria: A cooperative strategy to heal cellular damage. BioEssays. 2016;38:306–315. doi: 10.1002/bies.201500132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Vejborg RM, Klemm P. Cellular chain formation in Escherichia coli biofilms. Microbiology. 2009;155:1407–17. doi: 10.1099/mic.0.026419-0. [DOI] [PubMed] [Google Scholar]
- 95.Velicer GJ, Vos M. Sociobiology of the myxobacteria. Annu Rev Microbiol. 2009;63:599–623. doi: 10.1146/annurev.micro.091208.073158. [DOI] [PubMed] [Google Scholar]
- 96.Vos M, Velicer GJ. Genetic population structure of the soil bacterium Myxococcus xanthus at the centimeter scale. Appl Environ Microbiol. 2006;72:3615–25. doi: 10.1128/AEM.72.5.3615-3625.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Vos M, Velicer GJ. Social conflict in centimeter- and global-scale populations of the bacterium Myxococcus xanthus. Curr Biol. 2009;19:1763–67. doi: 10.1016/j.cub.2009.08.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97a.Wielgoss S, Didelot X, Chaudhuri RR, Liu X, Weedall GD, Velicer GJ, Vos M. A barrier to homologous recombination between sympatric strains of the cooperative soil bacterium Myxococcus xanthus. ISME J. 2016 doi: 10.1038/ismej.2016.34. doi:10.1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wagner-Dobler I, Thiel V, Eberl L, Allgaier M, Bodor A, et al. Discovery of complex mixtures of novel long-chain quorum sensing signals in free-living and host-associated marine alphaproteobacteria. Chembiochem. 2005;6:2195–206. doi: 10.1002/cbic.200500189. [DOI] [PubMed] [Google Scholar]
- 99.Waldman B, Frumhoff PC, Sherman PW. Problems of kin recognition. Trends Ecol Evol. 1988;3:8–13. doi: 10.1016/0169-5347(88)90075-4. [DOI] [PubMed] [Google Scholar]
- 100.Wall D. Molecular recognition in myxobacterial outer membrane exchange: functional, social and evolutionary implications. Mol Microbiol. 2014;91:209–20. doi: 10.1111/mmi.12450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wei X, Pathak DT, Wall D. Heterologous protein transfer within structured myxobacteria biofilms. Mol Microbiol. 2011;81:315–26. doi: 10.1111/j.1365-2958.2011.07710.x. [DOI] [PubMed] [Google Scholar]
- 102.Wenren LM, Sullivan NL, Cardarelli L, Septer AN, Gibbs KA. Two independent pathways for self-recognition in Proteus mirabilis are linked by type VI-dependent export. mBio. 2013;4:e00374–13. doi: 10.1128/mBio.00374-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.West SA, Gardner A. Altruism, spite, and greenbeards. Science. 2010;327:1341–44. doi: 10.1126/science.1178332. [DOI] [PubMed] [Google Scholar]
- 104.West SA, Griffin AS, Gardner A, Diggle SP. Social evolution theory for microorganisms. Nat Rev Microbiol. 2006;4:597–607. doi: 10.1038/nrmicro1461. [DOI] [PubMed] [Google Scholar]
- 105.Wright JS, 3rd, Traber KE, Corrigan R, Benson SA, Musser JM, Novick RP. The agr radiation: An early event in the evolution of staphylococci. J Bacteriol. 2005;187:5585–94. doi: 10.1128/JB.187.16.5585-5594.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Zee PC, Bever JD. Joint evolution of kin recognition and cooperation in spatially structured rhizobium populations. PLOS ONE. 2014;9:e95141. doi: 10.1371/journal.pone.0095141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Zhang D, de Souza RF, Anantharaman V, Iyer LM, Aravind L. Polymorphic toxin systems: Comprehensive characterization of trafficking modes, processing, mechanisms of action, immunity and ecology using comparative genomics. Biol Dir. 2012;7:18. doi: 10.1186/1745-6150-7-18. [DOI] [PMC free article] [PubMed] [Google Scholar]