Abstract
Despite the many recent advances in the field of epigenetics, application of this knowledge in environmental health risk assessment has been limited. In this paper, we identify opportunities for application of epigenetic data to support health risk assessment. We consider current applications and present a vision for the future.
Keywords: Epigenetics, Epigenomic modifications, Risk assessment, Environmental, Mechanisms, Adverse outcome pathway
1. Introduction
Approximately 90,000 chemicals are currently used in commerce with more introduced annually. Yet, the potential for chemical exposures to affect human health has been evaluated in only a small fraction (<2%) of those chemicals [1,2]. Environmental protection programs strive to understand and protect human health from the adverse consequences of chemical exposures under the authority of numerous statutes, including the Clean Air Act (1990), Safe Drinking Water Act (1974), and the Frank R. Lautenberg Chemical Safety for the 21st Century Act (2016). Human health risk assessments produce qualitative and, if possible, quantitative summaries of the health outcomes associated with chemical exposures which are used to support decision making [3].
A review of policies articulated by major risk assessment agencies suggests that detailed epigenomic risk assessment guidance has not been developed. Chemically associated epigenomic modifications, as potentially causal or contributory events to various diseases, however, are noted in several guidelines [4–8] and reports [9–17]. Additionally, agencies are developing testing guidance and repositories for standardized methods [5,17–20]. Importantly, agencies are confronting how to use epigenomics in practice, in chemical- and disease-specific assessments, e.g., di(2ethylhexyl) phthalate [21], endocrine disrupting chemicals [22], bisphenol A [23], as well as cancer and obesity [15,24]. The lack of extensive formal guidelines at this state of the science is not unusual. Detailed risk assessment guidelines tend to be developed to codify lessons learned in practice, rather than developed in advance of experience.
While the fundamentals of health risk assessment are well known, the application of data, methods, and models to support chemical evaluations is variable and related to the decision context. Although there is ongoing discussion regarding the benefits and validity of using epigenetic data in risk assessment, there is a growing body of evidence suggesting that epigenetic mechanisms have a critical role in determining adverse health outcomes. The epigenome functions to regulate gene expression through chemical modifications to DNA and its histone protein scaffolding, as well as through the use of noncoding RNAs. As a master regulator of gene expression, the epigenome is responsive to a range of environmental factors, including chemical exposure, diet, and stress. Within the context of an adverse outcome pathway (AOP), toxicant-induced changes in the epigenome could alter the dose–response relationship at one or more key element(s) and thus impact susceptibility, or result in inappropriate gene expression at a given time or space. In this manner epigenetic change can more directly affect an adverse outcome by modifying a key element. Thus, we believe the time is right to begin exploring how epigenetic data can be used to better inform risk assessment.
A variety of potential applications of epigenetic data in risk assessment are presented in Box 1 (adapted from Afshari et al., 2011 [25]). How data are used for risk assessment differs by the confidence in the overall body of evidence and the confidence necessary to support various decisions. We have outlined three illustrative risk assessment applications requiring increasing levels of data confidence and indicate how epigenetic data may be used in each case: 1) chemical screening and prioritization; 2) limited scope, often non-regulatory, risk assessments; and 3) major scope, potentially regulatory, risk assessments (further described in Refs. [26,27]).
Box 1. Potential applications of epigenetic data in risk assessment.
Identify chemicals with potential to cause diverse and long lasting effects
Elucidate mechanisms of action and affect weight of evidence conclusions
Categorize compounds by mechanistic class, including chemicals of unknown apical outcome
Rank by relative potency and epigenetic signature
Assist in identifying susceptible populations and life stages
Use as biomarkers of exposure and/or effect
Help identify cumulative risk factors
Better understand uncertainties of cross tissue, species and experimental paradigms
Discern potential lowest effect levels or dose–response relationships as surrogates for traditional apical outcomes
(adapted from Afshari et al., 2011 [25]).
2. Methods and considerations in the application of epigenetics data in risk assessment
Measuring epigenetic alterations is substantially easier than interpreting the resulting data in terms of public health impacts. Interpretation is particularly difficult when apical outcomes may occur years subsequent to the epigenetic modification [28–31]. Interpretation is further complicated by variations in endpoint expression based on multiple factors in addition to epigenetics that can contribute to disease, such as life stage when the exposure occurred, genetic profiles, lifestyles or conditions, or preexisting disease etc., [32–35].
Inclusion of epigenetic data in mechanistic characterizations can add confidence to an interpretation of observed alterations as being biologically significant and informative of adverse versus adaptive changes. Further, placing epigenetic data within the context of an adverse outcome pathway (AOP) or AOP network, which are chemically agnostic, enhances interpretation of mechanistic data, including data from well-studied, as well as less well-studied chemicals.
In addition to mechanistic context, confidence in the body of evidence will depend on the following:
The epigenome is highly complex and cannot be well represented by only considering the relationship between individual epigenetic modifications (e.g., DNA methylation) and exposure outcomes. Thus, research efforts should increasingly evaluate contributions from multiple epigenetic modifications and/or consider the integration of data from complimentary studies to assemble a more comprehensive representation of the epigenetic landscape at loci relevant to the outcomes of interest [36,37];
Significant variability in tissues and experimental paradigms can exist, so confidence in data interpretation from non-target tissue, in vitro cell lines or, to a lesser extent, from primary cell cultures will be increased by coherence with other evidence streams [38];
Species, strain, sex, inter-individual, and life-stage differences also can exist, so information on these factors is useful if available [36,39,40];
Confidence in epigenetic data can be strengthened by evidence for causality, including pharmacologically blocking a specific epigenetic modification, resulting in the elimination or amelioration of the adverse outcome, and comparison of disease states with and without the epigenetic modification of interest.
Notably, corroboration of evidence can be garnered from the broader knowledge of systems biology rather than just chemical-specific evidence. For example, chemically induced epigenetic changes may be shown to alter expression of a specific gene, but the role of that specific gene in health may be derived from general knowledge and thus, the role of epigenetics in health implied.
Considering the three major epigenetic mechanisms (i.e., chemical modification of DNA, chemical modification of the histone scaffold, and the differential expression of non-coding RNAs), data from the chemical modification of DNA and the differential expression of non-coding RNAs are most ready to incorporate into the risk assessment paradigm. Thus, there are good examples where chemical exposure causes changes in DNA methylation with subsequent alterations in gene expression that are associated with an adverse outcome (e.g., [31,32,41]). Similarly, chemical-induced changes in non-coding RNA expression have been identified that are both an adverse outcome biomarker (e.g., [42]), but also have been demonstrated to be involved in the mechanism of toxicity (e.g., [43]). In contrast, despite an abundant literature describing the roles of histone modifications in regulating gene expression (reviewed in Ref. [44]), the exploration of these modifications in the context of classical toxicology and epigenetics, has only begun recently. Thus their incorporation into the risk assessment paradigm is more challenging at this time, yet whenever possible, is warranted because of the importance of this mechanism in modifying risk. Further, rapid advances in this field are already showing promise with regards to both facile and cost-effective assessments (e.g., [45,46]).
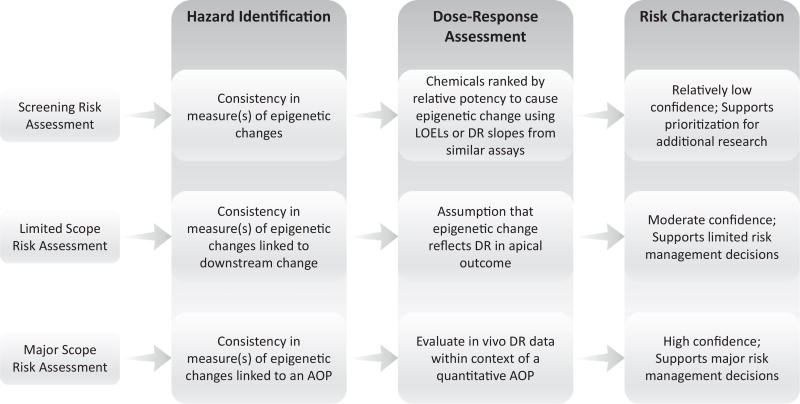
Three illustrative applications of epigenomic data in chemical evaluation follow, with Fig. 1 illustrating the application of evidence in hazard identification, dose–response assessment and risk characterization.
Fig. 1.
Potential application of epigenetic information to support risk screening, prioritization and assessment. LOEL = Lowest Observed Effect Level; DR = Dose Response; AOP = Adverse Outcome Pathway.
2.1. Screening assessments and prioritization
Screening assessments generally rely on relatively limited data to identify and prioritize chemicals for additional testing and research and, when necessary, for support in emergency response situations (e.g., Deepwater Horizon – Judson et al., 2010 [47]). Over time, screening assessments have begun to utilize higher throughput assays that can generate data more rapidly. This allows for much improved comparisons across chemicals due to similar assays being applied to groups of chemicals.
The crux for hazard identification generally is how to define adverse, which is particularly problematic for chemicals for which data are scarce. We propose that identifying the capability of a chemical to modify the epigenome is sufficient to assign it a higher priority for additional testing and research. In this context, it is unlikely that available epigenetic data alone would predict specific adverse health outcomes in humans.
Rather, epigenomic modification as observed in various assays could be indicative of the ability of a chemical to disrupt the epigenetic machinery (i.e., DNA methyltransferases, histone deacetylase, bromodomain-containing protein, etc.). Given the importance of these enzymes for cell fate and function, demonstrating disruption of any of these could be an indicator that important biological processes are likely to be altered. For example, BRD2 is a bromodomain-containing protein that is a constituent of multi-protein transcription factor complexes that play key roles in regulating gene expression [48]. BRD2 is considered to be an “epigenetic reader” because its bromodomains binds to acetylated histone protein, thus interpreting the epigenetic landscape and directing its parent transcriptional complexes to specific loci within the genome. By screening chemicals for their potential to alter the ability of the bromodomains within BRD2 to bind acetylated histones we could be able to identify those that should be investigated further (i.e., prioritized). In such case, inhibition of BRD2 binding to acetylated histones would be the molecular initiating event (MIE) that would lead to a divergent AOP network comprised of several potential adverse outcomes. The final determination of the specific adverse outcome(s) associated with that chemical would occur during subsequent testing.
If data allow, relative rankings of chemicals based on epigenetic alterations, within similar assays, could be estimated by comparing some consistent measure of dose–response relationships across chemicals. This concept is supported by the work of Kuppusamy et al. (2015) [49] who investigated the relationship between in vivo DNA methylation and cancer bioassay dose–response for 8 environmental toxicants. Their results indicated similarity between modeled points of departure (PODs) for cancer incidence and DNA methylation. While this work is based on a relatively small number of chemicals, these findings open the door to using epigenetics data, possibly even in vitro epigenetics data, in risk assessment.
In emergency response situations where decisions must be made rapidly, data on epigenetic alterations may be suitable to support urgent decisions. The inclusion of epigenetic endpoints in high throughput screening of the toxicological properties of chemicals would provide an opportunity to broaden the spectrum of potential effects that are assessed under such assays and strengthen confidence in the outcome of screening-level assessments.
2.2. Limited scope assessments
Limited scope assessments generally require additional data and greater confidence in the results beyond that required for screening chemicals for prioritization purposes. Limited scope assessments are usually consistent with the scale of the problem being addressed (e.g., local or regional impact), the type of solutions being considered (e.g., voluntary compliance), and/or data availability. An important feature of this type of assessment is the consideration of mechanistic-level information. In the case of epigenetic data, that could range from linking epigenetic modification(s) to an up- or down-stream event, such as a change in signaling linked to a suspected key event in a putative adverse outcome pathway or a downregulation in target gene transcription, respectively, to their inclusion in a fairly robust description of chemical-induced changes of multiple key events in a complete adverse outcome pathway or network. In limited scope assessments, information from multiple data streams may be available, including in vitro epigenetic data and traditional short-term bioassay data. A variety of new types of short-term in vivo assays are becoming increasingly available and will facilitate these types of assessments [50]. Further, the overlay of epigenetic data on top of the short-term in vivo and in vitro assays may help in creating an understanding of the probability of the effects to be long-lasting. There are a range of assays that are used to quantify changes in different aspects of the epigenome (see examples, Table 1); however, each individual method has advantages and disadvantages. Ideally, either data from multiple epigenetic modifications (e.g. DNA methylation and histone modifications) or the net functional effect of epigenetic modifications on chromatin accessibility will be assessed together to get a more comprehensive perspective on epigenomic states and their relationship with exposures and health effects.
Table 1.
Examples of epigenetic techniques and applications.
| Epigenetic technique | Type of epigenetic modification assessed |
Purpose | Application | Advantages | Disadvantages | Protocol | Application example |
|---|---|---|---|---|---|---|---|
| Chromatin Immunoprecipitation (ChIP) | Histone modifications | Identify relationships between specific histone modifications and/or histone proteins and genomic loci | Targeted or genome-wide | Histone modifications are the key active mediators of gene expression Can be used with both qPCR and high-throughput sequencing | There are over 130 unique histone modifications Complex experimental procedure | McCullough et al., 2017 [51] | McCullough et al., 2016 [36] |
| Methylation Bead Array | DNA methylation | Identify CpG loci with a change in protection from bisulfite conversion | Genome-wide | Straightforward assessment of bisulfite-insensitive loci | Assess a subset of CpG loci Protection from bisulfite conversion is not specific for 5-mC | Bibikova et al., 2011 [52] | Nwanaji-Enwerem et al., 2016 [53] |
| Reduced Representation Bisulfite Sequencing (RRBS) | Identify CpG loci with a change in protection from bisulfite conversion | Genome-wide sampling of gene regulatory regions | Relatively selective for assessment of bisulfite-insensitive loci within gene regulatory regions | Does not survey all CpG loci Protection from bisulfite conversion is not specific for 5-mC | Meissner et al., 2005 [54] | Meissner et al., 2008 [55] | |
| Methylation-sensitive Quantitative PCR | Quantify changes in sensitivity to bisulfite conversion at specific loci | Targeted | Quantitative and high-resolution | Low throughput Protection from bisulfite conversion is not specific for 5-mC | Yang et al., 2004 [56] | Koturbash et al., 2011 [40] | |
| Formaldehyde-Assisted Isolation of Regulatory Elements (FAIRE) | Chromatin accessibility | Identify changes in accessibility of genomic DNA | Targeted or genome-wide | Data represent the net effects of epigenetic modifications | The contributions of specific epigenetic modifications cannot be assessed | Simon et al., 2012 [57] | Pattenden et al., 2016 [45] |
| Assay for Transposase-Accessible Chromatin Sequencing (ATAC-Seq) | Identify changes in accessibility of genomic DNA | Genome-wide | Data represent the net effects of epigenetic modifications | The contributions of specific epigenetic modifications cannot be assessed | Buenrostro et al., 2015 [58] | Vallaster et al., 2017 [59] | |
| Non-coding RNA-Seq/PCR | Non-coding RNA | Identify changes in non-coding RNA expression/secretion | Targeted or genome-wide | Can be used with both qPCR and high-throughput sequencing Soluble circulating biomarker of injury for inaccessible tissues | Normalization is challenging in biofluids. | Kroh et al., 2010 [60] | Bollati et al., 2013 [61] |
Establishment of a dose–response relationship also will strengthen the ability to conduct a limited scope assessment. Epigenetic modifications, particularly from primary cell culture or non-human in vivo data, could serve as a surrogate for estimating human dose–response. For dose–response assessment, either the lowest observed or modeled (e.g., using benchmark dose methods [62]), effects levels could be combined with uncertainty factors; or, ideally, dose–response relationships in the range of environmental exposures that are linked to internal dosimetry through pharmacokinetic modeling. Processes downstream from the epigenetic modification tend to modulate the apical outcome and thus an assessment based on epigenetic changes can be viewed as a conservative estimate (i.e., public health protective). However, in the human population, it should be recognized that other factors such as lifestyle or co-exposures to other chemicals may exacerbate effects observed in the laboratory. Importantly, large amounts of data from efforts like the NIH Epigenome Roadmap Consortium (http://www.roadmapepigenomics.org) are becoming available and will facilitate epigenetic data-informed risk assessment.
2.3. Major scope assessments
Major assessments are those intended to address problems of national significance (e.g., particulate air pollution). Because of the economic cost of pollution control to the nation (often ≥ $100M/year), these assessments have high data/high confidence requirements in order to demonstrate that pollution controls will benefit public health by an equal or larger amount [63]. Consequently, these assessments are generally driven by epidemiological, clinical, and in vivo toxicology data.
For these assessments, epigenetic data are likely to serve several purposes: 1) provide insight into chemical exposure effects that are beyond the sensitivity or scope of detection of other methods (e.g. changes to the epigenome that alter long-term and/or multi-/trans-generational disease risk and/or susceptibility to subsequent chemical/non-chemical exposures); 2) provide a combination of highly reliable in vivo data and in vitro-derived mechanistic data; 3) contribute to the weight-of-evidence that an effect is causally related to outcome via a well described mechanism; 4) identify sensitive sub-populations and life-stages; 5) add to dose–response information; and 6) increase confidence in the overall scientific evidence used for decision-making.
3. Conclusions
In this paper, we illustrate pragmatic paths to the consideration of epigenomic data in the toxicological evaluation of chemicals. There appears to be a scientific consensus that: 1) epigenomic modifications can have a role in or be a driver for a wide variety of diseases, such as cancer, obesity, diabetes, and cardiovascular diseases; 2) chemicals may alter the epigenome or disrupt mechanisms of epigenomic regulation; 3) epigenomic events are part of a complex, multifactorial determinants of disease (including environment and genetics) that can be adaptive, adverse or neutral; 4) understanding the mechanistic links from epigenomic modifications to adverse effects (i.e., AOP) is a key component of interpreting the significance of the event(s) to public health; 5) early life stages maybe particularly susceptible to epigenomic modifications and have the potential to cause later-in-life disease as a result of early-life exposures or alter intergenerational disease susceptibility or incidence. In contrast, there is some disagreement in the scientific community as to whether conduct of quantitative epigenomic risk assessment is premature due to incomplete knowledge. We propose, however, that epigenomic modifications that differ significantly from background and lead to pathogenic alterations in gene expression or cell signaling may be suitable for use in hazard identification and dose-response assessment, including the derivation of a point of departure.
Footnotes
Disclaimer: The contents of this article have been reviewed by the Environmental Protection Agency and approved for publication, and do not necessarily represent Agency policy, nor does mention of trade names or commercial products constitute endorsement or recommendations for use.
Conflict of interest
The authors declare no competing interests.
References
- 1.NRC (National Research Council) Toxicity testing in the 21st century: a vision and a strategy. Washington, D.C: National Academy Press; 2007. [Accessed 22 April 2017]. Available at: https://www.nap.edu/catalog/11970/toxicity-testing-in-the-21st-century-a-vision-and-a. [Google Scholar]
- 2.Hayes AW, Kruger CL. Hayes’ principles and methods of toxicology. 6. CRC Press; Oct 10, 2014. p. 1711. [Google Scholar]
- 3.NRC (National Research Council) Science and decisions. Washington, DC: Advancing Risk Assessment; 2009. [Accessed 22 April 2017]. Available at: http://www.nap.edu/catalog/12209.html. [Google Scholar]
- 4.USEPA (U.S. Environmental Protection Agency) Guidelines for carcinogen risk assessment. Washington, DC: Risk Assessment Forum; 2005. EPA/630/P-03/001F March 2005. [Google Scholar]
- 5.ECHA (European Chemical Agency) [Accessed 25 July 2017];Guidance on information requirements and chemical safety assessment. 2011 Chapter R.7a: Endpoint specific guidance. Available at, https://echa.europa.eu/documents/10162/13632/information_requirements_r7a_en.pdf/e4a2a18f-a2bd-4a04-ac6d-0ea425b2567f.
- 6.IARC (International Agency for Research on Cancer) [Accessed 25 July 2017];Preamble to the IARC monographs (amended 2006, updated 2015) 2015 Available at, http://monographs.iarc.fr/ENG/Preamble/index.php.
- 7.ECHA (European Chemical Agency) [Accessed 25 July 2017];CLH report proposal for harmonised classification and labelling based on regulation (EC) No 1272/2008 (CLP regulation), annex VI, Part 2, dibenzo[b,def] chrysene. 2016 Available at: https://echa.europa.eu/documents/10162/13626/clh_report_dibenzo_bdef_chrysene_en.pdf/f8dcbcb6-d242-4357-8a72-894d159fb123.
- 8.ECHA (European Chemical Agency) [Accessed 25 July 2017];Guidance on the biocidal products regulation, volume III assessment & evaluation (parts B+C) 2017 Available at: Version 2.1. https://echa.europa.eu/documents/10162/23036412/biocides_guidance_human_health_ra_iii_part_bc_en.pdf/30d53d7d-9723-7db4-357a-ca68739f5094.
- 9.Greally JM. [Accessed 25 July 2017];OECD review: endocrine disruptors and the epigenome. 2011 Available at: www.oecd.org/chemicalsafety/testing/48435503.pdf.
- 10.Herceg Z, Lambert MP, van Veldhoven K, Demetriou C, Vineis P, Smith MT, Straif K, Wild CP. Towards incorporating epigenetic mechanisms into carcinogen identification and evaluation. Carcinogenesis. 2013 Sep;34(9):1955–1967. doi: 10.1093/carcin/bgt212. https://doi.org/10.1093/carcin/bgt212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.USEPA (U.S. Environmental Protection Agency) Next generation risk assessment: incorporation of recent advances in molecular, computational, and systems biology (final report) Washington, DC: U.S: EPA; 2014. [accessed July 2017]. EPA/600/R-14/004. Available at: http://cfpub.epa.gov/ncea/risk/recordisplay.cfm?deid=286690. [Google Scholar]
- 12.Khoury MJ. [Accessed 25 July 2017];Epigenetics and public health: why we should pay attention by. 2014 Available at: https://blogs.cdc.gov/genomics/2014/10/09/epigenetics/
- 13.Chappell G, Pogribny IP, Guyton KZ, Rusyn I. Epigenetic alterations induced by genotoxic occupational and environmental human chemical carcinogens: a systematic literature review. Mutat Res Rev Mutat Res. 2016 Apr-Jun;768:27–45. doi: 10.1016/j.mrrev.2016.03.004. https://doi.org/10.1016/j.mrrev.2016.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.EFSA (European Food Safety Authority) Bahadori T, Bell D, Ceccatelli S, Corvi R, Hogstrand C, Munn S, Nilsson E, Spurgeon D, Vom Brocke J, Wray-Cahen D, Wright M, Binaglia M, Dorne JL, Georgiadis N, Germini A, Kass G, Robinson T, Rossi A, Schoonjans R, Terron A, Noteborn H. EFSA scientific colloquium 22-epigenetics and risk assessment: where do we stand? EFSA Support Publ 2016. 2016;13(12):28. https://doi.org/10.2903/sp.efsa.2016.EN-1129. EN-1129. [Google Scholar]
- 15.Smith MT, Guyton KZ, Gibbons CF, Fritz JM, Portier CJ, Rusyn I, DeMarini DM, Caldwell JC, Kavlock RJ, Lambert PF, Hecht SS, Bucher JR, Stewart BW, Baan RA, Cogliano VJ, Straif K. Key characteristics of carcinogens as a basis for organizing data on mechanisms of carcinogenesis. Environ Health Perspect. 2016 Jun;124(6):713–721. doi: 10.1289/ehp.1509912. https://doi.org/10.1289/ehp.1509912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.JSTA (Japanese Science and Technology Agency) [Accessed 25 July 2017];What’s epigenetic? 2017 Available at: http://www.epigenetics.jst.go.jp/english/epigenetics.html.
- 17.Parfett CL, Desaulniers DA. Tox21 approach to altered epigenetic landscapes: assessing epigenetic toxicity pathways leading to altered gene expression and oncogenic transformation. In Vitro Int J Mol Sci. 2017 Jun 1;18(6):E1179. doi: 10.3390/ijms18061179. https://doi.org/10.3390/ijms18061179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.OECD (Organisation for Economic Co-operation and Development) Guidance document of standardised test guidelines for evaluation chemical for endocrine disruption. 2012:22. Series on Testing and Assessment No. 150. ENV/JM/MONO(2012. [Google Scholar]
- 19.OECD (Organisation for Economic Co-operation and Development) Fundamental and guiding principles for (Q)SAR analysis of chemical carcinogens with mechanistic considerations series on testing. 2015:46. ENV/JM/MONO(2015) [Google Scholar]
- 20.NIH (National Institutes of Health) [Accessed 25 July 2017];Epigenomics laboratory methods. 2017 Available at: https://commonfund.nih.gov/epigenomics/resources/methods.
- 21.Agency for Toxic Substances and Disease Registry (ATSDR) Toxicological profile for Di(2-ethylhexyl) phthalate (DEHP) Atlanta, GA: U.S. Department of Health and Human Services, Public Health Service; 2002. [PubMed] [Google Scholar]
- 22.WHO (World Health Organization) Bergman Åke, Heindel Jerrold J, Jobling Susan, Kidd Karen A, Thomas Zoeller R., editors. [Accessed 25 July 2017];State of the science of endocrine disrupting chemicals. 2012 Available at: www.who.int/iris/bitstream/10665/78101/1/9789241505031_eng.pdf.
- 23.ECHA (European Chemical Agency) [Accessed 25 July 2017];SVHC support document-4,4”-isopropylidenediphenol (bisphenol A) 2016 Available at: https://echa.europa.eu/documents/10162/b10d6a00-8e47-9b14-4f61-c779a8dc8450.
- 24.WHO (World Health Organization) Consideration of the evidence on childhood obesity for the commission on ending childhood obesity: report of the ad hoc working group on science and evidence for ending childhood obesity. Geneva, Switzerland: [Google Scholar]
- 25.Afshari CA, Hamadeh HK, Bushel PR. The evolution of bioinformatics in toxicology: advancing toxicogenomics. Toxicol Sci. 2011 Mar;120(Suppl 1):S225–S237. doi: 10.1093/toxsci/kfq373. https://doi.org/10.1093/toxsci/kfq373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.USEPA (U.S. Environmental Protection Agency) Next generation risk assessment: incorporation of recent advances in molecular, computational, and systems biology (final report) Washington, DC: U.S. Environmental Protection Agency; 2014. 2014. EPA/600/R-14/004. [Google Scholar]
- 27.Thomas RS, Philbert MA, Auerbach SS, Wetmore BA, Devito MJ, Cote I, Rowlands JC, Whelan MP, Hays SM, Andersen ME, Meek ME, Reiter LW, Lambert JC, Clewell HJ, 3rd, Stephens ML, Zhao QJ, Wesselkamper SC, Flowers L, Carney EW, Pastoor TP, Petersen DD, Yauk CL, Nong A. Incorporating new technologies into toxicity testing and risk assessment: moving from 21st century vision to a data-driven framework. Toxicol Sci. 2013 Nov;136(1):4–18. doi: 10.1093/toxsci/kft178. https://doi.org/10.1093/toxsci/kft178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pembrey M, Saffery R, Bygren LO. Human transgenerational responses to early-life experience: potential impact on development, health and biomedical research. J Med Genet. 2014;51:563–572. doi: 10.1136/jmedgenet-2014-102577. https://doi.org/10.1136/jmedgenet-2014-102577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Boekelheide K, Blumberg B, Chapin RE, Cote I, Graziano JH, Janesick A, Lane R, Lillycrop K, Myatt L, States JC, Thayer KA, Waalkes MP, Rogers JM. Predicting later-life outcomes of early-life exposures. Environ Health Perspect. 2012 Oct;120(10):1353–1361. doi: 10.1289/ehp.1204934. https://doi.org/10.1289/ehp.1204934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Greathouse KL, Bredfeldt T, Everitt JI, Lin K, Berry T, Kannan K, Mittelstadt ML, Ho S-M, Walker CL. Environmental estrogens differential engage the histone methyltransferase EZH2 to increase risk of uterine tumorigenesis. Mol Cancer. 2012 Apr;10(4):546–557. doi: 10.1158/1541-7786.MCR-11-0605. https://doi.org/10.1158/1541-7786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smith AH, Marshall G, Liaw J, Yuan Y, Ferreccio C, Steinmaus C. Mortality in young adults following in utero and childhood exposure to arsenic in drinking water. Environ Health Perspect. 2012 Nov;120(11):1527–1531. doi: 10.1289/ehp.1104867. https://doi.org/10.1289/ehp.1104867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ray PD, Yosim A, Fry RC. Incorporating epigenetic data into the risk assessment process for the toxic metals arsenic, cadmium, chromium, lead, and mercury: strategies and challenges. Front Genet. 2014 Jul 16;5:201. doi: 10.3389/fgene.2014.00201. https://doi.org/10.3389/fgene.2014.00201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tobi EW, Slieker RC, Stein AD, Suchiman HE, Slagboom PE, van Zwet EW, Heijmans BT, Lumey LH. Early gestation as the critical time-window for changes in the prenatal environment to affect the adult human blood methylome. Int J Epidemiol. 2015 Aug;44(4):1211–1223. doi: 10.1093/ije/dyv043. https://doi.org/10.1093/ije/dyv043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zeise L, Bois FY, Chiu WA, Hattis D, Rusyn I, Guyton KZ. Addressing human variability in next-generation human health risk assessments of environmental chemicals. Environ Health Perspect. 2013 Jan;121(1):23–31. doi: 10.1289/ehp.1205687. https://doi.org/10.1289/ehp.1205687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Krewski D, Westphal M, Andersen ME, Paoli GM, Chiu WA, Al-Zoughool M, Croteau MC, Burgoon LD, Cote I. A framework for the next generation of risk science. Environ Health Perspect. 2014 Aug;122(8):796–805. doi: 10.1289/ehp.1307260. https://doi.org/10.1289/ehp.1307260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McCullough SD, Bowers EC, On DM, Morgan DS, Dailey LA, Hines RN, Devlin RB, Diaz-Sanchez D. Baseline chromatin modification levels may predict interindividual variability in ozone-induced gene expression. Toxicol Sci. 2016 Mar;150(1):216–224. doi: 10.1093/toxsci/kfv324. https://doi.org/10.1093/toxsci/kfv324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bowers EC, McCullough SD. Linking the epigenome with exposure effects and susceptibility: the epigenetic seed and soil model. Toxicol Sci. 2017 Feb;155(2):302–314. doi: 10.1093/toxsci/kfw215. https://doi.org/10.1093/toxsci/kfw215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chappell G, Kobets T, O’Brien B, Tretyakova N, Sangaraju D, Kosyk O, Sexton KG, Bodnar W, Pogribny IP, Rusyn I. Epigenetic events determine tissue-specific toxicity of inhalational exposure to the genotoxic chemical 1,3-butadiene in male C57BL/6J mice. Toxicol Sci. 2014 Dec;142(2):375–384. doi: 10.1093/toxsci/kfu191. https://doi.org/10.1093/toxsci/kfu191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Waxman DJ, O’Connor C. Mol Endocrinol. 2006;20(11):2613–2629. doi: 10.1210/me.2006-0007. https://doi.org/10.1210/me.2006-0007. [DOI] [PubMed] [Google Scholar]
- 40.Koturbash I, Scherhag A, Sorrentino J, Sexton K, Bodnar W, Swenberg JA, Beland FA, Pardo-Manuel Devillena F, Rusyn I, Pogribny IP. Epigenetic mechanisms of mouse interstrain variability in genotoxicity of the environmental toxicant 1,3-butadiene. Toxicol Sci. 2011 Aug;122(2):448–456. doi: 10.1093/toxsci/kfr133. https://doi.org/10.1093/toxsci/kfr133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tang W-Y, Newbold R, Mardilovich K, Jefferson W, Cheng RYS, Medvedovic M, Ho S-M. Persistent hypomethylation in the promoter of nucleosomal binding protein 1 (Nsbp1) correlates with overexpression of Nspb1 in mouse uteri neonatally exposed to diethylstilbestrol or genistein. Endocrinology. 2008;149(12):5922–5931. doi: 10.1210/en.2008-0682. https://doi.org/10.1210/en.2008-0682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang X, Salminen WF, Shi Q, Greenhaw J, Gill PS, Bhattacharyya S, Beger RD, Mendrick DL, Mattes WB, James LP. Potential of extracellular microRNAs as biomarkers of acetaminophen toxicity in children. Toxicol Appl Pharmacol. 2015;284:180–187. doi: 10.1016/j.taap.2015.02.013. https://doi.org/10.1016/j.taap.2015.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Goeman F, Strano S, Blandino G. MicroRNAs as key effectors in the p53 network. Int Rev Cell Mol Biol. 2017;333:51–90. doi: 10.1016/bs.ircmb.2017.04.003. https://doi.org/10.1016/bs.ircmb.2017.04.003. [DOI] [PubMed] [Google Scholar]
- 44.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. https://doi.org/10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 45.Pattenden SG, Simon JM, Wali A, Jayakody CN, Troutman J, McFadden AW, Wooten J, Wood CC, Frye SV, Janzen WP, David IJ. High-throughput small molecular screen identifies inhibitors of aberrant chromatin accessibility. Proc Natl Acad Sci USA. 2016;113:3018–3023. doi: 10.1073/pnas.1521827113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang Q, Trevino LS, Wong RL, Medvedovic M, Chen J, Ho SM, Shen J, Foulds C, Coarfa C, O’Malley BW, Shilatifard A, Walker CL. Reprogramming of the epigenome by MLL1 links early-live environmental exposures to prostate cancer risk. Mol Endocrinol. 2016;30(8):856–871. doi: 10.1210/me.2015-1310. https://doi.org/10.1210/me.2015-1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Judson RS, Martin MT, Reif DM, Houck KA, Knudsen TB, Rotroff DM, Xia M, Sakamuru S, Huang R, Shunn P, Austin CP, Kavlock RJ, Dix DJ. Environ Sci Technol. 2010;44:5979–5985. doi: 10.1021/es102150z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Denis GV, McComb ME, Faller DV, Sinha A, Romesser PB, Costello CE. Identification of transcription complexes that contain the double bromodomain protein Brd2 and chromatin remodeling machines. J Proteome Res. 2006;5(3):502. doi: 10.1021/pr050430u. https://doi.org/10.1021/pr050430u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kuppusamy SP, Kaiser JP, Wesselkamper SC. Epigenetic regulation in environmental chemical carcinogenesis and its applicability in human health risk assessment. Int J Toxicol. 2015 Sep-Oct;34(5):384–392. doi: 10.1177/1091581815599350. https://doi.org/10.1177/1091581815599350. [DOI] [PubMed] [Google Scholar]
- 50.Kamstra JH, Alestrom J, Kooter JM, Legler J. Zebrafish as a model to study the role of DNA methylation in environmental toxicology. Environ Sci Pollut Res. 2015;22:16262–16276. doi: 10.1007/s11356-014-3466-7. https://doi.org/10.1007/s11356-014-3466-7. [DOI] [PubMed] [Google Scholar]
- 51.McCullough SD, On DM, Bowers EC. Using chromatin immunoprecipitation in toxicology: a step-by-step guide to increasing efficiency, reducing variability, and expanding applications. Curr Protoc Toxicol. 2017;72:3.14.1–3.14.28. doi: 10.1002/cptx.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bibikova M, Barnes B, Chan Tsan, Ho V, Klotzle B, Le JM, Delano D, Zhang L, Schroth GP, Gunderson KL, Fan J-B, Shen R. High density DNA methylation array with single CpG site resolution. Genomics. 2011;98:288–295. doi: 10.1016/j.ygeno.2011.07.007. [DOI] [PubMed] [Google Scholar]
- 53.Nwanaji-Enwerem JC, Colicino E, Trevisi L, Kloog I, Just AC, Shen J, Brennan K, Dereix A, Hou L, Vokonas P, Schwartz J, Baccarelli AA. Long-term ambient exposures and blood DNA methylation age: findings from the VA normative aging study. Environ Epigenet. 2016;2 doi: 10.1093/eep/dvw006. [epub] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Meissner A, Gnirke A, Bell GW, Ramsahoye B, Lander ES, Jaenisch R. Reduced representation bisulfite sequencing for comprehensive high-resolution DNA methylation analysis. Nucleic Acids Res. 2005;33:5868–5877. doi: 10.1093/nar/gki901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Meissner A, Mikkelsen TS, Gu H, Wernig M, Hanna J, Sivachenko A, Zhang X, Bernsteain BE, Nusbaum C, Jaffe DB, Gnirke A, Jaenisch R, Lander ES. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454:766–771. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yang AS, Estecio MRH, Doshi K, Kondo Y, Tajara EH, Issa J-PJ. A simple method for estimating global DNA methylation using bisulfite PCR of repetitive DNA elements. Nucleic Acids Res. 2004;32:e38. doi: 10.1093/nar/gnh032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Simon JM, Giresi PG, Davis IJ, Lieb JD. Using formaldehyde-assisted isolation of regulatory elements (FAIRE) to isolate active regulatory DNA. Nat Protoc. 2012;7:256–267. doi: 10.1038/nprot.2011.444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Buenrostro J, Wu B, Chang H, Greenleaf W. ATAC-Seq: a method for assaying chromatin accessibility genome-wide. Curr Protoc Mol Biol. 2015;109:21.29.1–21.29.9. doi: 10.1002/0471142727.mb2129s109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vallaster MP, Kukreja S, Bing XY, Ngolab J, Zhao-Shea R, Gardner PD, Tapper AR, Rando OJ. Paternal nicotine exposure alters hepatic xenobiotic metabolism in offspring. eLife. 2017;6:e24771. doi: 10.7554/eLife.24771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kroh EM, Parkin RK, Mitchell PS, Tewari M. Analysis of circulating microRNA biomarkers in plasma serum using quantitative reverse transcription-PCR (qRT-PCR) Methods. 2010;50:298–301. doi: 10.1016/j.ymeth.2010.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bollati V, Angelici L, Rizzo G, Pergoli L, Rota F, Hoxha M, Nordio F, Bonzini M, Tarantini L, Cantone L, Pesatori AC, Apostoli P, Baccarelli AA, Bertazzi PA. Microvesicle-associated microRNA expression is altered upon particulate matter exposure in healthy workers and in A549 cells. J Appl Toxicol. 2013;35:59–67. doi: 10.1002/jat.2987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.USEPA (U.S. Environmental Protection Agency) Benchmark dose technical guidance. Washington, DC: Risk Assessment Forum; 2012. EPA/100/R-12/001. [Google Scholar]
- 63.EO (Executive Order) 12866. Regulatory Planning and Review. 58 FR 51735. 1993 Sept. http://govinfo.library.unt.edu/npr/library/direct/orders/2646.html.