Abstract
We report the synthesis of a 5-formyl-2′-deoxyuridine (5fU) phosphoramidite and the preparation of oligonucleotides comprising all known, naturally observed eukaryotic thymidine modifications. Biophysical characterization of the synthetic oligonucleotides indicates that 5fU, but not the other T-derivatives, can alter DNA structures.
Natural chemical modifications to DNA nucleobases have the potential to influence biological events. Previous reports on cytosine (C) modifications have suggested regulatory roles for 5-hydroxymethylcytosine (5hmC) and 5-formylcytosine (5fC) in the active demethylation of 5-methylcytosine (5mC), and also via protein recruitment during cell differentiation and development in mammals.1 The less studied thymine (T) modifications (Scheme 1A) exist in the genome of eukaryotic cells and may also have biological functions. 5-Hydroxymethyluridine (5hmU) can be generated from T by the action of ten-eleven translocation (TET) enzymes in mouse embryonic stem cells and can influence the recruitment of proteins.2 5hmU, 5-formyluridine (5fU) and base J (ß-glucosylated 5hmU) co-exist in the DNA of eukaryotic parasite genomes, where base J is proposed to act as an epigenetic mark affecting Pol II transcription and gene expression.3 Although originally reported as a product of oxidative DNA damage,4 recent studies suggest that 5fU may also arise via enzymatic oxidization of 5hmU.5 5fU has been reported to disrupt the sequence-specific interactions between DNA and the transcription factor NFκB,6 suggesting a role in modulating protein–DNA interactions. It also induces mutations and serves as a substrate for DNA glycosylases in the formation of abasic sites,7,8 thereby contributing to genomic instability.
Scheme 1.
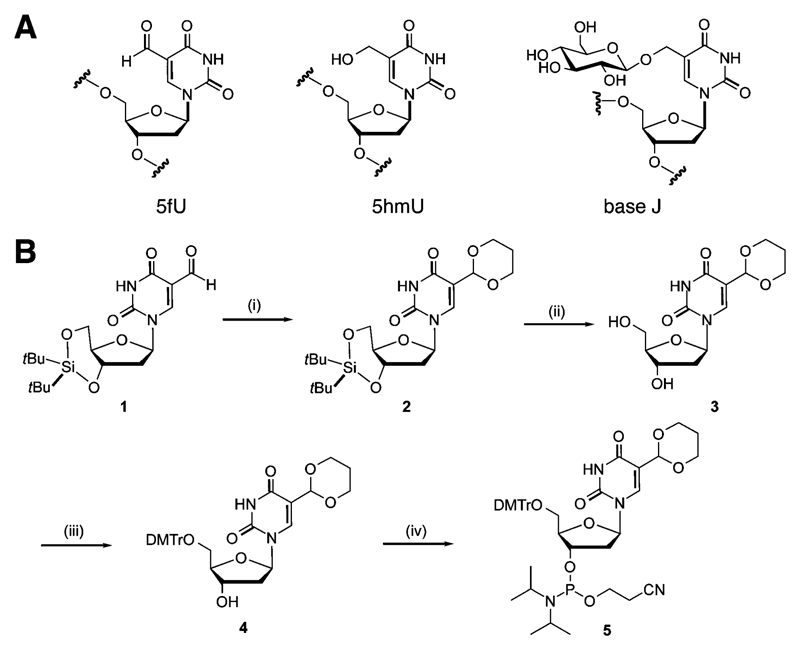
Chemical structure of eukaryotic thymidine modifications and synthesis of the 5-formyl-2′-deoxyuridine phosphoramidite. (A) Structure of 5hmU, 5fU and base J. (B) Synthesis of the 5fU phosphoramidite: (i) HO(CH2)3OH, TiCl4, (EtO)3CH, CH2Cl2, 0 °C, 1 h, 80%; (ii) HF·pyridine, THF, 0° to rt, 1 h, 99%; (iii) DMTrCl, Et3N, pyridine, rt, 4 h, 92%; (iv) ClPN(iPr)2(OCH2CH2CN), Et3N, 0 °C to rt, 3 h, 57%.
The introduction of an extra chemical group can modulate hydrophobic and hydrogen-bonding interactions resulting in potential changes in the structure, stability and flexibility of the DNA double helix, which may in turn influence DNA recognition and biological functions.9 We recently reported a crystal structure of a 5fC-containing DNA dodecamer showing that 5fC alters the geometry of the grooves and base pairs associated with the modified base, leading to local helical underwinding of a DNA duplex.10 The findings for 5fC suggest that formyl substitution can profoundly alter the properties of DNA, prompting us to explore the consequences of 5fU modifications on DNA. Previous studies showed that single alteration of T to 5fU does not significantly affect the B-DNA conformation of a DNA double helix with the effect on DNA stability varied by sequences and conditions,8,11,12 which calls for a further study in more sequence contexts as well as higher 5fU density. Herein we report a robust synthetic route to a 5fU phosphoramidite, which allows the synthesis of oligonucleotides bearing multiple 5fUs together with all known T-modifications. We also report biophysical studies to assess the effect of 5fU, as compared to other T modifications, on the stability and structure of DNA. We found that 5fU can substantially affect both the structure and the stability of double-stranded DNA and can promote the formation of alternative secondary structures.
We developed a route to the 5fU phosphoramidite (Scheme 1B) that allows efficient synthesis of oligodeoxynucleotides (ODNs) with multiple 5fU modifications in addition to other T-modifications. A common strategy to introduce 5fU to ODNs utilizes a phosphoramidite with a protected 1,2-diol group, which is then converted to the formyl group by sodium periodate oxidation following oligonucleotide backbone synthesis.13 However, this strategy is inefficient for preparing ODNs with multiple formyl groups14 and is not compatible with other T modifications, notably base J, due to uncontrolled oxidation of the glucose residue. We thus utilized a cyclic acetal group, previously used for 5fC,14 to protect the 5fU formyl group during solid phase oligonucleotide synthesis. This strategy allowed the generation of the formyl group during the deprotection step under mild acidic conditions without affecting other T modifications. Briefly, phosphoramidite 5 was prepared in 4 steps from di-tert-butylsilyl protected fdU 1, which was synthesised from 5-iodo-dU according to literature procedures (Scheme 1B).15 Activation of the formyl group using titanium tetrachloride as a Lewis acid and treatment with 1,3-dihydroxypropane gave the acetal 2 in good yield (80%). Silyl deprotection, introduction of a 4,4′-dimethoxytrityl group and standard phosphitylation gave the 5fU phosphoramidite building block 5, which was subsequently used to prepare oligonucleotides by standard phosphoramidite solid phase synthesis.
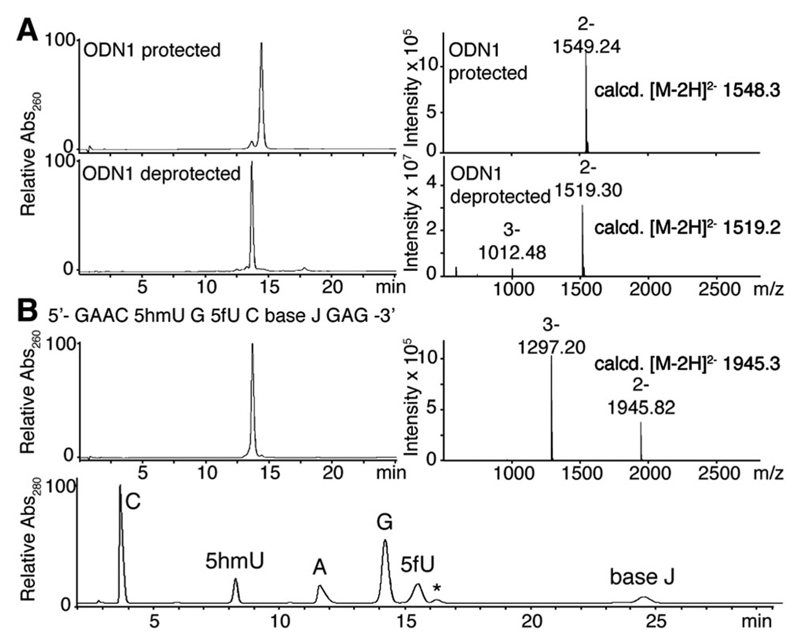
The cyclic acetal group introduced in ODN1 (Table 1) was readily removed in 2% (v/v) aqueous acetic acid without any notable side reactions (Fig. 1A). The deprotection reaction also proceeded in the presence of multiple 5fU modifications on a single strand to give ODN2 and 3 (Table 1 and Fig. S1, ESI†). Incorporation of 5fU was confirmed by LC-MS analysis and mononucleoside composition analysis followed by enzymatic digestion of ODNs (Fig. 1A and Fig. S1f, ESI†). By combining phosphoramidite 5 with the phoshoramidites of base J and 5hmU prepared using reported procedures,16,17 we were able to synthesize an ODN bearing all eukaryotic T modifications (5hmU, 5fU and base J), proving the wide applicability and robustness of our synthetic strategy (Fig. 1B).
Table 1.
Sequence and melting temperature of ODNs
| X | Y | Tm (°C)a,b,c (ΔTm based on T control) |
|---|---|---|
| ODN1: 5′-ATCGCAXGTA-3′ | ||
| ODN4: 3′-TAGCGYACAT-5′ | ||
| T | T | 43.5 ± 1.1 |
| 5fU | 5fU | 45.2 ± 0.1 (1.7) |
| ODN2: 5′-CGXACXAGXACG-3′ (self-complementary) | ||
| T | — | 51.6 ± 0.4 |
| 5fU | — | 52.5 ± 0.4 (0.9) |
| U | — | 50.1 ± 0.4 (−1.5) |
| 5hmU | — | 47.8 ± 0.1 (−3.8) |
| ODN3: 5′-GAACXGXCXGAG-3′ | ||
| ODN5: 3′-CTTGACAGACTC-5′ | ||
| T | — | 49.8 ± 0.3 |
| 5fU | — | 49.4 ± 0.1 (−0.4) |
| U | — | 51.1 ± 0.2 (1.2) |
| 5hmU | — | 46.7 ± 0.1 (−3.1) |
| base J | — | 47.9 ± 0.2 (−1.9) |
Mean ± sd values of three measurements are shown.
Tm was measured at 5 μM of each ODN in PBS.
See the ESI for melting curves.
Fig. 1.
(A) LC trace of ODN1 before (top) and after (bottom) deprotection together with the corresponding ESI-MS (m/z) spectra of the largest peak. The small peak at 14.3 min in the top LC trace was characterized as the deprotected ODN1 by MS. (B) LC trace of the ODN with three different T modifications (top, left) with ESI-MS (m/z) spectra at the largest peak (top, right) and a LC trace of the ODN enzymatically digested into mononucleosides (bottom). The peak marked * is derived from the digestion buffer.
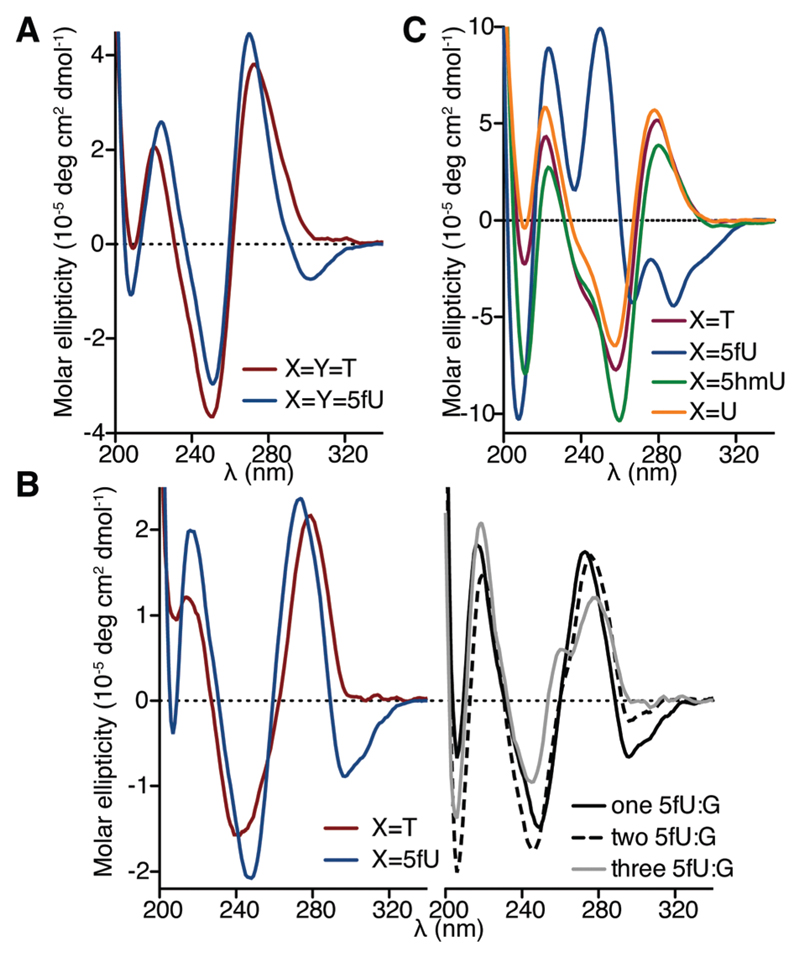
We then prepared ODNs comprising different densities of each T modification, ranging from one to three modifications in a single strand, to three in both strands of the DNA duplex (Table 1), and evaluated the effects of the modifications on the DNA duplexes using biophysical methods. We first assessed the thermodynamic stability of modified ODN duplexes by thermal melting UV analysis (Table 1 and Table S1, ESI†). 5fU did not significantly affect the thermal stability of the ODN1–ODN4 duplex, the self-complementary ODN2 duplex, and the ODN3–ODN5 duplex. On the other hand, multiple 5hmU modifications were found to destabilize duplexes, which may be due to the hydroxyl group substitution disrupting water-mediated hydrogen bonding in the major groove. The CD spectrum of the ODN1–ODN4 duplex containing just one modification (X = T, dU, 5hmU or 5fU) on one strand showed patterns characteristic of B-form DNA, suggesting that a single substitution by any T modification does not significantly affect the duplex structure (Fig. S2A, ESI†). This result agrees with a previous study, where crystal structures of short duplexes with a single 5fU residue demonstrated a B-DNA structure.12,18 Interestingly, changes in the DNA structure were observed when increasing the density of 5fU. When 5fU was introduced to both ODN1 and ODN4 (X = Y = 5fU), the CD spectrum of ODN1–ODN4 duplex displayed an absorbance band with negative ellipticity in the near-UV region (Fig. 2A). The CD spectrum of 5fU-containing the ODN3–ODN5 duplex also displayed a similar absorption pattern (Fig. 2B, left), which was not observed in the corresponding spectra of the counter-parts comprising T, U, 5hmU or base J (Fig. 2B, left and Fig. S2B, ESI†). The CD spectrum of the self-complementary ODN2, bearing three 5fU modifications per strand, displayed a broad negative ellipticity band in the near-UV region (λ > 260 nm), two positive ellipticity bands around 250 nm and 223 nm, and a sharp negative ellipticity band around 210 nm (Fig. 2C). These features in the CD spectra are distinct from the characteristic B-DNA observed for its T, U, and 5hmU counterparts (Fig. 2C) and suggest a significant structural alteration as a result of formylated nucleobases. A negative ellipticity band in the near-UV region was also observed previously for 5fC-containing dodecamers,10 which adopt a non-B form DNA structure. Although there has been no theoretical premise to correlate this CD spectra signature with specific DNA structural changes so far, the similarity in CD signatures suggests that the presence of 5fU also leads to a non B-DNA double helix. This negative band was not observed at pH 3.5 and decreased at pH 8.6 (Fig. S2C, ESI†), implying that the contribution of 5fU to the DNA structure of ODN2 is pH-dependent.
Fig. 2.
CD spectra of ODNs at pH 7.2. (A) ODN1–ODN4 duplexes. (B) ODN3–ODN5 duplex (left) and 5fU-containing ODN3–ODN5 duplex with one (annealed to 5′-CTCAGGCAGTTC-3′), two (annealed to 5′-CTCG GACGGTTC-3′), and three (annealed to 5′-CTCGGGCGGTTC-3′) 5fU:G pairs (right). (C) Self-complementary ODN2 duplex.
We then assessed the effect of 5fU on duplexes with T:G mismatches, since 5fU can potentially pair with G via wobble base pair formation through its tautomer.19,20 The negative absorbance band in the CD spectra of 5fU-containing the ODN3–ODN5 duplex became less apparent by increasing numbers of G opposite to 5fU (Fig. 2B, right). This observation suggests that 5fU affects the DNA duplex structure when stably paired with its complementary nucleobase A and has less impact when paired with G or partially unpaired. Additionally, the decrease in the thermal stability of 5fU-modified ODN3 on introducing 5fU:G pairs was larger than that of the non-modified counterparts under near physiological conditions (Fig. S3, ESI†), indicating that 5fU further destabilizes DNA duplexes bearing T:G mismatches in spite of its capacity to base pair with G. The observed destabilization of DNA in 5fU:G contexts and the structural alteration of DNA in 5fU:A contexts may have relevance to the mutagenic properties of 5fU.7
Modified Ts are enriched at the GGGTTA telomeric repeats of Trypanosomatid genomes.21 The GGGTTA repeats potentially fold into G-quadruplexes, single-stranded DNA structures comprising stacked guanine tetrads and loops in between.22 We therefore studied the effect of 5fU on ODNs with the GGGTTA telomeric repeat (ODN6) either paired with its C-rich complementary sequence (ODN7) or single-stranded (Table 2 and Table S1, ESI†). UV thermal melting studies showed that the introduction of 5fU in this specific context decreases the melting temperature of the duplex (Table 2).
Table 2.
Tm of ODNs in the GGGTTA/TAACCC context
| X | Y | Tma (ΔTm based on T control) |
|---|---|---|
| ODN6: 5′-AGGGXTAGGGXTAGGGXTAGGGT-3′ | ||
| ODN7: 3′-TCCCAAYCCCAAYCCCAAYCCCA-5′ | ||
| T | T | 68.8 ± 0.1b [62.6 ± 0.2]c |
| 5fU | 5fU | 63.3 ± 1.3 (−5.5)b [55.6 ± 0.6 (−7.0)]c |
| ODN6 (G-quadruplex) | ||
| T | — | 63.2 ± 0.2d |
| 5fU | — | 67.8 ± 0.3 (4.6)d |
Mean ± sd values for three measurements.
Tm values were obtained by UV260 absorbance at 5 μM of each ODN in PBS.
Tm values were obtained by UV260 absorbance in 10 mM phosphate buffer (pH 7.4) with 70 mM KCl.
Tm values were obtained by UV295 absorbance at 10 μM of ODN6 in 10 mM phosphate buffer (pH 7.4) with 70 mM KCl.
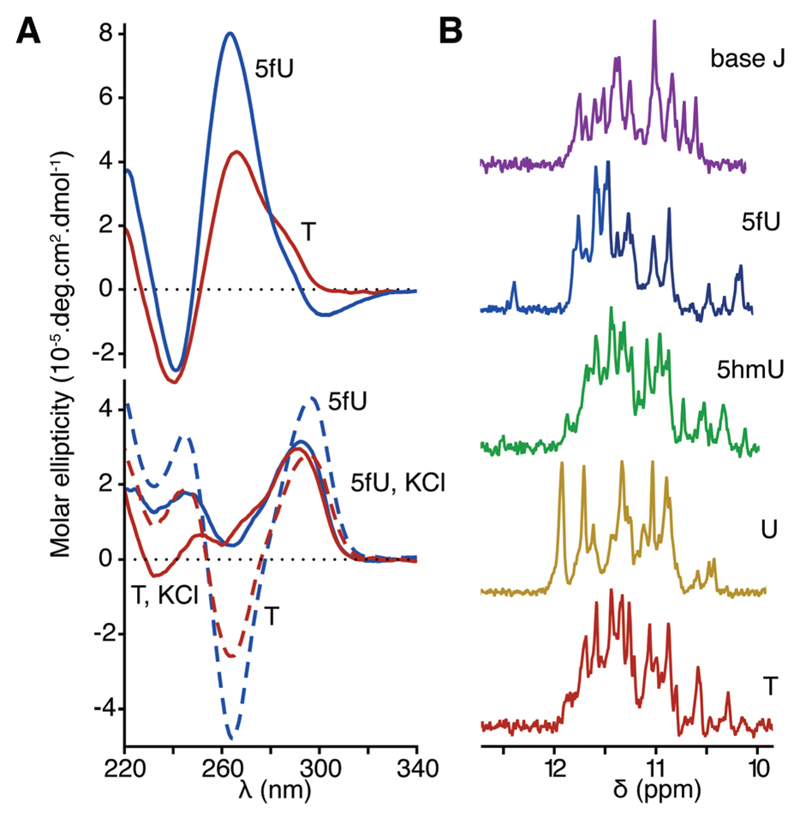
Upon the introduction of three 5fU modifications on both strands of the ODN6–ODN7 duplex, we observed a negative band in the near-UV region of the corresponding CD spectrum consistent with previous observations for other ODNs (Fig. 3A, top). When annealed in the absence of its complementary sequence, regardless of the presence of modifications, ODN6 displayed CD spectra characteristic of antiparallel quadruplexes (a positive maximum at 295 nm and a negative minimum at 260 nm) and hybrid quadruplexes (a positive maximum at 295 nm and a shoulder at 260 nm) in the absence or the presence of KCl respectively (Fig. 3A, bottom).23 While the introduction of 5fU modifications did not significantly affect the CD spectra of the G-quadruplex formed by ODN6, it increased the thermal stability of the G-quadruplex by 4.6 °C (Table 2). This increased thermal stability of single-stranded G-quadruplex motifs may contribute to destabilizing the DNA duplex formed by the telomeric sequence in favour of a G-quadruplex structure. We used 1D 1H NMR spectroscopy to further investigate the structure of G-quadruplexes formed by ODN6 containing different T modifications. While imino proton signals characteristic of G-quadruplexes (10–12 ppm) were observed regardless of the presence of any modifications, their patterns varied in each T modified ODN6 (Fig. 3B). Telomeric DNA is known to fold into a mixture of G-quadruplex conformers,22 and the observed variations suggest that the introduction of modified Ts in the loops affects the equilibrium between the different conformers. The 1H NMR spectrum of the 5fU-modified ODN6 displayed additional peaks at 12.5 ppm as well as some upfield-shifted peaks. These peaks broadened at elevated temperatures (Fig. S4, ESI†), suggesting that imino protons corresponding to these peaks are not involved in the formation of the G-tetrads. This result suggests that 5fU introduces extra H-bond interactions, which may result in stabilizing the G-quadruplex motif. Taken together these observations suggest that the 5fU may have a specific function associated with the telomere structure.
Fig. 3.
CD spectrum and NMR spectra of ODNs in the GGGTTA/TTACCC context. (A) ODN6–ODN7 duplex in PBS (top) and single stranded ODN6 in the presence (75 mM, solid line) or absence (dashed line) of KCl at pH 7.4 (bottom). Spectra of 5fU modified ODNs are in blue and T controls are in red. (B) 1H NMR spectra (500 MHz) of ODN6 in the presence of 100 mM KCl at pH 7.4.
In conclusion, the synthesis of DNA oligonucleotides comprising various eukaryotic T modifications has enabled us to reveal that the formyl group of 5fU has a distinct ability to influence the structure and the stability of DNA, though the degree of its contribution to DNA structures was dependent on its local density. This, taken together with the previously reported 5fC mediated DNA structure alteration, suggests that formylationmay generally regulate the DNA structure.
Supplementary Material
† Electronic supplementary information (ESI) available: Experimental protocols, supplementary figures, and characterization of synthetic compounds. See DOI: 10.1039/c6cc08670e
Acknowledgments
This work was supported by the following grants: the Wellcome Trust (099232/Z/12/Z), Cancer Research UK (C14303/A17197), ERC Advanced grant (PM), Herchel Smith Funding and A*STAR (ZL).
Notes and references
- 1.Wu H, Zhang Y. Cell. 2014;156:45–68. doi: 10.1016/j.cell.2013.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pfaffeneder T, Spada F, Wagner M, Brandmayr C, Laube SK, Eisen D, Truss M, Steinbacher J, Hackner B, Kotljarova O, Schuermann D, et al. Nat Chem Biol. 2014;10:574–581. doi: 10.1038/nchembio.1532. [DOI] [PubMed] [Google Scholar]
- 3.van Luenen HG, Farris C, Jan S, Genest PA, Tripathi P, Velds A, Kerkhoven RM, Nieuwland M, Haydock A, Ramasamy G, Vainio S, et al. Cell. 2012;150:909–921. doi: 10.1016/j.cell.2012.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bjelland S, Eide L, Time RW, Stote R, Eftedal I, Volden G, Seeberg E. Biochemistry. 1995;34:14758–14764. doi: 10.1021/bi00045a017. [DOI] [PubMed] [Google Scholar]
- 5.Bullard W, Lopes da Rosa-Spiegler J, Liu S, Wang Y, Sabatini R. J Biol Chem. 2014;289:20273–20282. doi: 10.1074/jbc.M114.579821. [DOI] [PMC free article] [PubMed] [Google Scholar]; Pais JE, Dai N, Tamanaha E, Vaisvila R, Fomenkov AI, Bitinaite J, Sun Z, Guan S, Correa IR, Jr, Noren CJ, Cheng X, et al. Proc Natl Acad Sci U S A. 2015;112:4316–4321. doi: 10.1073/pnas.1417939112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kittaka A, Takayama H, Kurihara M, Horii C, Tanaka H, Miyasaka T, Inoue J. Nucleosides, Nucleotides Nucleic Acids. 2001;20:669–672. doi: 10.1081/NCN-100002347. [DOI] [PubMed] [Google Scholar]
- 7.Anensen H, Provan F, Lian AT, Reinertsen SH, Ueno Y, Matsuda A, Seeberg E, Bjelland S. Mutat Res. 2001;476:99–107. doi: 10.1016/s0027-5107(01)00086-0. [DOI] [PubMed] [Google Scholar]
- 8.Liu P, Burdzy A, Sowers LC. DNA Repair. 2003;2:199–210. doi: 10.1016/s1568-7864(02)00198-2. [DOI] [PubMed] [Google Scholar]
- 9.Liu MY, DeNizio JE, Schutsky EK, Kohli RM. Curr Opin Chem Biol. 2016;33:67–73. doi: 10.1016/j.cbpa.2016.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Raiber EA, Murat P, Chirgadze DY, Beraldi D, Luisi BF, Balasubramanian S. Nat Struct Mol Biol. 2015;22:44–49. doi: 10.1038/nsmb.2936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ono A, Okamoto T, Inada M, Nara H, Matsuda A. Chem Pharm Bull. 1994;42:2231–2237. doi: 10.1248/cpb.42.2231. [DOI] [PubMed] [Google Scholar]
- 12.Tsunoda M, Kondo J, Karino N, Ueno Y, Matsuda A, Takenaka A. Biophys Chem. 2002;95:227–233. doi: 10.1016/s0301-4622(01)00259-9. [DOI] [PubMed] [Google Scholar]
- 13.Sugiyama H, Matsuda S, Kino K, Zhang Q-M, Yonei S, Saito I. Tetrahedron Lett. 1996;37:9067–9070. [Google Scholar]
- 14.Schroder AS, Steinbacher J, Steigenberger B, Gnerlich FA, Schiesser S, Pfaffeneder T, Carell T. Angew Chem, Int Ed. 2014;53:315–318. doi: 10.1002/anie.201308469. [DOI] [PubMed] [Google Scholar]
- 15.Dai Q, Song CX, Pan T, He C. J Org Chem. 2011;76:4182–4188. doi: 10.1021/jo200566d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de Kort M, de Visser PC, Kurzeck J, Meeuwenoord NJ, van der Marel GA, Ruger W, van Boom JH. Eur J Org Chem. 2001:2075–2082. [Google Scholar]
- 17.Sowers LC, Beardsley GP. J Org Chem. 1993;58:1664–1665. doi: 10.1021/jo00059a011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tsunoda M, Sakaue T, Naito S, Sunami T, Karino N, Ueno Y, Matsuda A, Takenaka A. Nucleic Acids Res Suppl. 2001:279–280. doi: 10.1093/nass/1.1.279. [DOI] [PubMed] [Google Scholar]
- 19.Yoshida M, Makino K, Morita H, Terato H, Ohyama Y, Ide H. Nucleic Acids Res. 1997;25:1570–1577. doi: 10.1093/nar/25.8.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Masaoka A, Terato H, Kobayashi M, Ohyama Y, Ide H. J Biol Chem. 2001;276:16501–16510. doi: 10.1074/jbc.M008598200. [DOI] [PubMed] [Google Scholar]
- 21.Genest PA, Ter Riet B, Cijsouw T, van Luenen HG, Borst P. Nucleic Acids Res. 2007;35:2116–2124. doi: 10.1093/nar/gkm050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Phan AT. FEBS J. 2010;277:1107–1117. doi: 10.1111/j.1742-4658.2009.07464.x. [DOI] [PubMed] [Google Scholar]
- 23.Kypr J, Kejnovska I, Renciuk D, Vorlickova M. Nucleic Acids Res. 2009;37:1713–1725. doi: 10.1093/nar/gkp026. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.