Abstract
Autophagy is an evolutionarily conserved lysosome-mediated cellular degradation program. Accumulating evidence shows that autophagy is important to the maintenance of liver homeostasis. Autophagy involves recycling of cellular nutrients recycling as well as quality control of subcellular organelles. Autophagy deficiency in the liver causes various liver pathologies. Fatty liver disease (FLD) is characterized by the accumulation of lipids in hepatocytes and the dysfunction in energy metabolism. Autophagy is negatively affected by the pathogenesis of FLD and the activation of autophagy could ameliorate steatosis, which suggests a potential therapeutic approach to FLD. In this review, we will discuss autophagy and its relevance to liver diseases, especially FLD. In addition we will discuss recent findings on potential therapeutic applications of autophagy modulators for FLD.
Keywords: Autophagy, fatty liver disease, lipophagy, liver function, metabolism
1. Introduction
Autophagy (from the Greek, “auto” oneself, “phagy” to eat) is an evolutionarily conserved cellular degradation process that involves the delivery of cytoplasmic cargo (macromolecules or organelles) to the lysosome (Deter and De Duve 1967). It becomes an intensely studied area for its implications in fundamental cell biology as well as its important roles in the pathogenesis including tissue injury, microbial infection, cancer, neurodegeneration, and aging (Shintani and Klionsky 2004). Autophagy has been generally classified into three types including macroautophagy, microautophagy, and chaperone-mediated autophagy (Meijer and Codogno 2004). Macroautophagy is the most active form of autophagic process in which cytosolic materials are sequestered into autophagosomes and transported to the lysosome, where they would be degraded (Mizushima and Komatsu 2011). Microautophagy, which is mainly defined in the yeast, but is also observed in mammalian cells, mediates both invagination and vesicle scission into the lumen of lysosomes through direct engulfment of cytoplasmic cargo at a boundary membrane of the lysosome (Mijaljica et al. 2011; Farre et al. 2009). In chaperone mediated autophagy (CMA), specific proteins are targeted by chaperones, for example, heat shock-cognate protein of 70 kda (Hsc70), and directly shuttled across the lysosomal membrane for degradation in the lumen (Kaushik and Cuervo 2012).
In this review, we will focus on the signaling and roles of the macroautophagic process, referred to hereafter simply as autophagy, in fatty liver diseases (FLDs). Meanwhile, we will discuss potential therapeutic applications by targeting autophagy in FLDs.
2. Autophagy: the basic process
2.1 Autophagy pathways and regulations
Autophagy occurs when an organelle known as autophagosome is formed. The autophagosome is characterized as a double-membrane cisterna that envelops cytosolic material like proteins and organelles (Lamb et al. 2013). The origin of the autophagosomal membrane and how it’s formation is still not well understood. Various models have been proposed, including the de novo production and the derivation from endoplasmic reticulum (ER), the Golgi apparatus, the mitochondria, the endocytic system or the plasma membrane (Hamasaki et al. 2013; Lamb et al. 2013). The autophagosome then becomes the autolysosome after fusing with the lysosome, where the components from the cytosols are degraded (Kaur and Debnath 2015; Madrigal-Matute and Cuervo 2016).
Several key molecular pathways that regulate the autophagy have been elucidated in the past decade. To date, more than 35 different proteins, named Atg or “autophagy-related” proteins, have been characterized in yeast, including 15 core Atg proteins required for both selective and starvation-induced autophagy are highly conserved in mammal cells (Atg1-10, 12-14, 16, 18) (Nakatogawa et al. 2009; Mizushima et al. 2011). These proteins work together as the main regulators for autophagy. Generally, Atg proteins could be functionally classified into six clusters (Shibutani and Yoshimori 2014; Mizushima et al. 2011).
The Atg1/ULK complex is comprised of the autophagy-initiating UNC-5 like autophagy activating kinase 1 (ULK1), FIP200, Atg13L and Atg101 in mammals.
The class III phosphatidylinositol 3-kinase (PI3K) complex consists of VPS34, VPS15, Beclin-1, autophagy/beclin 1 regulator 1 (AMBRA1) and Atg14 (L)/Barkor in mammals.
The Atg2-Atg18/WIPI complex includes the PI3P-binding protein Atg18/WIPI and its binding partner Atg2.
The Atg12 conjugation system includes Atg12, Atg7, Atg10, Atg5 and Atg16L1/2 in mammals.
The Atg8/LC3 conjugation system includes LC3A/B/C, GABARAP, GABARAPL1/2/3, Atg4A-D, Atg7 and Atg3 in mammals.
Atg9 vesicles. Atg9 was detected on small vesicles and tubular structures in mammals.
During the biogenesis of autophagosome, the formation of Atg1/ULK complex is one of the earliest detectable events and is at the most upstream position of the recruitment of Atg proteins (Lamb et al. 2013; Shimizu et al. 2014). In the initial stage (vesicle nucleation), the Atg1/ULK complex activates the PI3K complex, which recruits several Atg proteins to the phagophore. This is followed by the stages of vesicle elongation and completion, which includes the recruitment of Atg12-Atg5-Atg16 complex to the autophagosome membrane and the lipidation of Atg8/LC3 through the conjugation to PE on the autophagosomal membrane. The mature autophagosome then migrates to and fuses with lysosomes to form autolysosome, in which the sequestered cytoplasmic material of the autophagosome is degraded (Liu and Levine 2015).
Other than the aforementioned core pathway of mammalian autophagy, certain subcellular systems, such as the secretory and endocytic pathway, and the cytoskeletal network, may also play important roles during autophagy (He and Klionsky 2009). Meanwhile, autophagy is inhibited or stimulated by multiple stimuli, including the change of nutritional status, hormonal factors, and other environmental differences like temperature, oxygen concentrations, and cell density (Levine and Kroemer 2008). These stimuli regulate autophagy via different mechanisms. The mammalian target of rapamycin (mTOR) is one of the major inhibitor. mTOR is a master regulator of cellular metabolism and promotes cell growth in response to environmental cues. During the autophagy process, mTOR complex 1 (mTORC1) inhibits ULK complex by phosphorylating Atg13 and ULK1/2, which results in autophagy suppression (Kim and Guan 2015). In addition, mTORC1 has also been suggested to inhibit ULK1 stability by inhibitory phosphorylation of AMBRA1, and to regulate autophagy at the transcriptional level by inhibiting transcription factor EB (TFEB) (Kim and Guan 2015). mTORC1 could be inactivated by an important energy sensing molecule, AMP-activated protein kinase (AMPK), via phosphorylating tuberous sclerosis 2 (TSC2) and the mTORC1 component Raptor, which subsequently activate the ULK1 complex (Kim and Guan 2015). Furthermore, AMPK could also phosphorylate and activate ULK1 directly (Kim et al. 2013a). AMPK and mTORC1 also regulate the class III PI3K complex, in which AMPK phosphorylates Beclin1, leading to the activation of the complex, whereas mTORC1 phosphorylates Atg14L, resulting in an inhibitory effect (Kim and Guan 2015). Other nutrient sensing pathways, such as the Ras/cAMP-dependent protein kinase A (PKA) signaling pathway, and the insulin/growth factor pathways are also involved in the regulation of autophagy (He and Klionsky 2009).
Autophagosome formation is also potently induced under various extra- and intra-cellular stresses, which is beneficial for organisms to rid themselves of damaging cytoplasmic components (He and Klionsky 2009; Levine and Kroemer 2008). These stress stimuli include ER stress, hypoxia, redox stress, pathogen-associated molecular patterns (PAMPs) and danger-associated molecular patterns (DAMPs), and mitochondrial damage (Kroemer et al. 2010). Under these stress conditions, autophagy is regulated through transcriptional reprogramming, activation of autophagy-inducing proteins, cell cycle modulation, and some other unclear events (Kroemer et al. 2010). For example, when the folding capacity is exceeded in the ER, the unfolded protein response (UPR) signaling triggers and mediates the transcriptional activation of the autophagy related proteins, such as LC3 and Atg5 (Ogata et al. 2006; Ding et al. 2007; Ding and Yin 2008; Kroemer et al. 2010).
Considering the critical role of autophagy in degradative process, its potential regulation mechanisms have been extensively studied. An increasing amount of evidence suggests that autophagy can be regulated at different levels, including transcriptional, post-transcriptional and post-translational level in order to maintain cellular homeostasis (Feng et al. 2015). At the transcriptional level, several transcription factors are involved in the autophagic progress, including the E2F family, FoxO family, p53, STAT1/3, TFEB, FXR, PPARα, GATA, and ATF4, et al (Feng et al. 2015; Seok et al. 2014; Lee et al. 2014). Several of them are also nutrient-sensing nuclear receptors, which may also explain the nutritional regulation of autophagy. TFEB, one target of mTORC1 as mentioned above, has been shown to regulate autophagy process by modulating expression of autophagy-related genes, such as UVRAG, WIPI, MAPLC3B, SQSTM1, VPS11, VPS18, and ATG9B, during starvation (Settembre et al. 2011). Under the condition of nutrient deprivation, forkhead box O 3a (FoxO3a) is phosphorylated by AMPK in the nucleus, which results in transcriptional repression of S-phase kinase-associated protein 2 (SKP2) (Shin et al. 2016). Coactivator associated arginine methyltransferase (CARM1) protein is increased after the decrease of SKP2 and subsequently increases the dimethylation of histone H3 Arg17 (Shin et al. 2016). Eventually the whole event activates autophagy-related and lysosomal genes through TFEB, which results in autophagy stimulation after nutrient starvation (Shin et al. 2016). Besides transcriptional regulation, autophagy is also regulated at the post-transcriptional or post-translational level. The most typical post-transcriptional regulation is the action of noncoding RNAs, especially microRNAs (miRNA), which target to the core machinery during autophagy process (Feng et al. 2015). Meanwhile, post-translational regulations, such as phosphorylation, ubiquitination, acetylation, epigenetic regulation, and protein-protein interaction, are also essential for the modulation of autophagy (Feng et al. 2015). Accumulating evidence suggests that autophagy modulation could be a potential therapeutic modality for different disease. Some of the aforementioned factors may be considered as therapeutic targets, which will be further discussed below.
2.2 Roles of autophagy in metabolic homeostasis
The liver is a gland that plays a critical role in metabolism in the human body (Abdel-Misih and Bloomston 2010). FLD is characterized by hepatic steatosis, inflammation and fibrosis. FLD is clinically classified into two broad entities, including alcoholic fatty liver disease (AFLD) and nonalcoholic fatty liver disease (NAFLD). The previous one is caused by excessive alcohol intake, and the later one is resulted from any non-alcohol etiologies (Reddy and Rao 2006). Disturbance of metabolic homeostasis seems to be one of the key events during the occurrence of FLD. Here we will discuss the roles of autophagy in hepatic metabolic homeostasis.
Hepatic autophagy occurs at the basal level and may be elevated under stressed conditions, thus contributing to the maintenance of normal hepatocyte functions and responding to pathogenic changes in the liver (Yin et al. 2008). Autophagy used to be considered as a bulk degradation process that is non-selective. However, recent evidence suggests that autophagy can be highly selective, especially during such processes as the removal of protein aggregates, damaged organelles, and intracellular pathogens (Okamoto 2014; Stolz et al. 2014). Studies have shown that autophagy not only can modulate cellular energy stores like lipids and carbohydrates, but also could target dysfunctional or superfluous organelles as well as toxic protein aggregates (Mizushima and Klionsky 2007). Based on the targets, several selective types of autophagy have been described, including aggrephagy (targets to aggregated proteins), mitophagy (targets to the damaged mitochondria), reticulophagy or ER-phagy (targets to the ER), pexophagy (targets to peroxisomes), lipophagy (targets to lipid droplets), ferritinophagy (targets to ferritin) and xenophagy (targets to intracellular microorganisms) (Stolz et al. 2014). Several autophagic adaptor proteins have been found to act as autophagy adaptors, such as p62/SQSTM1, NBR1 (neighbor of BRCA1 gene 1), NDP52 (nuclear domain 10 protein 52kDa), and OPTN (Optineurin), which bind to the cargo (usually ubiquitinated) and some key components of the autophagy machinery like the LC3 protein (Rogov et al. 2014; Johansen and Lamark 2011; Stolz et al. 2014). Failure to eliminate potentially dangerous substrates in the liver disturbs cellular homeostasis and subsequently results in hepatic diseases (Czaja et al. 2013). In mouse models with hepatic deletion of Atg7 or Atg5, protein aggregates and subcellular organelles are massively accumulated, leading to liver enlargement, severe liver injury, inflammation, fibrosis, cirrhosis, and tumorigenesis (Komatsu et al. 2005; Takamura et al. 2011; Ni et al. 2014).
In general, autophagy plays a role in hepatic metabolic homeostasis in three ways (Fig. 1), promoting nutrient recycle, removing abnormal organelles and toxic protein aggregates, and altering the level of metabolic factors (Christian et al. 2013; Kim and Lee 2014). The liver can actively participate in several parts of lipid metabolisms, such as fatty acids (FAs) uptake, de novo synthesis, and β-oxidation; cholesterol synthesis and biotransformation into bile acid; and lipoprotein uptake and secretion (Singh et al. 2009). The storage form of lipids is named lipid droplets (LDs). Accumulating evidence has revealed an important role for autophagy in LDs breakdown. Inhibition of autophagy by either 3-methyladenine (an autophagy inhibitor) or RNA interference against Atg5 or Atg7 increases triglyceride level and LD number and size in both cultured hepatocytes and mouse livers (Singh et al. 2009). Mice deficient in Atg5 in the liver also shows excessive hepatic lipid accumulation (Ni et al. 2014). In addition to the effects on lipid metabolism, the role of autophagy in hepatic carbohydrate metabolism has been shown to affect gluconeogenesis and glycogen storage (Kim and Lee 2014). In hepatic Atg7 deficient mice, the absence of amino acid release by autophagy affects gluconeogenesis during starvation (Ezaki et al. 2011). Atg7 is also necessary for statin-induced gluconeogenesis in the liver (Wang et al. 2015a). Nonetheless, in tissue-specific Atg5–deficient mice, another study suggested that Atg5-related autophagy did not influence gluconeogenesis, but may affect ketogenesis during starvation (Takagi et al. 2016). Other than gluconeogenesis, autophagy is also involved in glycogen breakdown in newborn hepatocytes of the rat and skeletal muscles of Drosophila melanogaster (Zirin et al. 2013; Kotoulas et al. 2006). However, a decrease or no change in hepatic glycogen content was observed in other studies with hepatic Atg7- or VPS34-deficient mice (Komatsu et al. 2005; Jaber et al. 2012; Ezaki et al. 2011). These contradictive studies imply differential roles of ATG genes in carbohydrate metabolism, which still need to be clarified for different circumstances.
Fig. 1. Roles of autophagy in metabolic homeostasis.
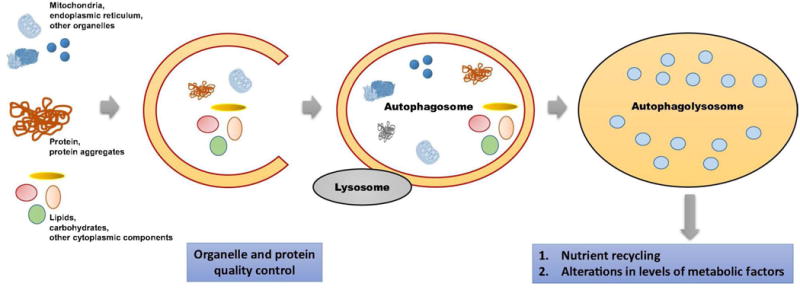
Autophagy plays a key role in hepatic metabolic homeostasis in three ways: nutrient recycle, clearance of abnormal organelles and toxic protein aggregates, and regulation of the level of metabolic factors.
Carbohydrates supply energy at the beginning of fasting in most organisms, followed with breakdown of proteins to supply substrates for gluconeogenesis (Mizushima and Klionsky 2007). When these stores are exhausted, cells start to employ autophagy to reuse existing macromolecules. During starvation, about 30% of hepatic proteins are decreased in wild-type mice in 24h of fasting, which becomes insignificant in conditional knockout mice of Atg7 (Mizushima and Klionsky 2007; Komatsu et al. 2005). In fact, protein turnover via autophagy is related to multiple hepatic pathophysiological conditions, including the clearance of misfolded mutant proteins in alpha-1-antitrypsin deficiency (Puri and Chandra 2014; Hidvegi et al. 2010; Yamamura et al. 2014), and in the clearance of the inclusion bodies (Strnad et al. 2008; Zatloukal et al. 2007; Harada et al. 2008).
In order to maintain nutrients homeostasis, autophagy has been suggested to assist the quality control of organelles, a process also known as organellophagy (Okamoto 2014). As mentioned above, organellophagy could be classified into different forms depending on the target organelles, including mitophagy, reticulophagy, and pexophagy etc. Mitophagy is important to remove defective and unhealthy mitochondria, which can boost cellular oxidative stress and may induce cell death via apoptosis. Recent studies have shown that the PINK1/Parkin pathway of mitophagy is important to mitochondrial homeostasis regulation (Eiyama and Okamoto 2015; Wei et al. 2015; Ding et al. 2010b; Zhang 2013). Another important organellophagy is reticulophagy, also called ER-phagy, which is a process that degrades defective or excessive ER by delivering them to the lysosomes. Xenobiotic substances, like drugs and chemicals, are mostly processed in the liver by a variety of enzymes. These enzymes are associated to hepatocyte ER, the proliferation of ER usually occurs in the liver after being exposed to xenobiotics (Komatsu et al. 2005; Yang et al. 2016). Autophagy is known to be part of the ER quality control (ERQC) system via both micro-or macro-ER-phagy, using p62 as an adaptor or requiring core autophagy related proteins, respectively (Schuck et al. 2014; Khaminets et al. 2015; Ding and Yin 2008; Lipatova and Segev 2014, 2015; Lipatova et al. 2013). Another common organellophagy is pexophagy, a type of autophagy that participates in the quality control of peroxisomes, which is important to lipid metabolism and hydrogen peroxide production and degradation (Nordgren et al. 2013). Several proteins that are involved in peroxisome biogenesis, e.g., peroxins (encoded by PEX genes), including Pex3, Pex5 and Pex14, are suggested to participate in macropexophagy (Sakai et al. 2006; Manjithaya et al. 2010; Neuhaus et al. 2014; Subramani 2015). An oncoprotein, hypoxia inducible factor 2α (HIF2α), has also been indicated to induce hepatocyte pexophagy (Walter et al. 2014). All these observations indicate the critical role of autophagy in cellular metabolic homeostasis.
3. Relevance of autophagy to liver diseases
The liver has the highest protein turn-over rate compared to other organs. In mouse livers, protein degradation rate increases from 1.5% of total liver protein per hour to 4.5% under the starving condition, and 40% of total protein would be degraded after a 48h starvation (Schworer et al. 1981; Mortimore et al. 1983). Previous researches suggested that autophagy assists the degradation of protein in the liver. In mice deficient in Atg5 or Atg7 in the liver, severe hepatomegaly is observed with an abundant amount of polyubiquitinated proteins and with an increased amount of defective mitochondria and peroxisomes (Rusten and Stenmark 2010; Takamura et al. 2011).
Autophagy is important to liver diseases. Recent studies have shown that autophagy is involved in the life cycle Hepatitis C and B (Virus HCV and HBV). HBV could stimulate autophagy by the X protein (HBx) and the Small Surface Protein (SHBs) (Tang et al. 2009). Inhibition of autophagosome formation could significantly inhibit HBV production, and the induction of autophagy markedly contributed to HBV production (Mao et al. 2011). Meanwhile, HBx could impair lysosomal acidification, possibly through interaction with V-ATPase, which results in a drop in lysosomal degradative capacity and the accumulation of immature lysosomes (Liu et al. 2014). HCV has also been reported to induce autophagy, but to suppress autophagic protein degradation by inhibiting the fusion of autophagosomes and lysosomes (Ait-Goughoulte et al. 2008; Sir et al. 2008). HCV and HBV can also induce intracellular events that trigger the mitochondrial dysfunction leading to mitophagy induction (Kim et al. 2013c).
Autophagy has been suggested to play various roles in tumorigenesis and tumor growth (Brech et al. 2009; White et al. 2011). Hepatic deficiency of Atg5 or Atg7 can lead to the development of hepatocellular adenomas with mitochondrial swelling, accumulation of p62 protein aggregates, oxidative stress and genomic in the tumor cells (Takamura et al. 2011; Ni et al. 2014). Multiple tumors including liver tumors are also observed in mice with Beclin haplo-insufficiency (Qu et al. 2003; Yue et al. 2003). In contrast, autophagy has been indicated to be an important surviving mechanism for cancer, and in a rat tumor model of N-diethylnitrosamine-induced hepatocarcinogenesis, the inhibition of autophagy by chloroquine at the tumor-forming stage could restrained tumor formation (Sun et al. 2013). Autophagy is also critical for the invasion and metastasis of hepatocellular carcinoma (HCC) (Li et al. 2013a; Peng et al. 2013).
Liver-specific deletion of Atg5 or Atg7 causes hepatocyte hypertrophy and liver injury in mice (Takamura et al. 2011; Komatsu et al. 2005; Ni et al. 2014). SQSTM1/p62 is significantly increased in the liver of mice with hepatic autophagy deficiency, which weakens the interaction between Kelch-like ECH-associated protein 1 (Keap1) and nuclear factor erythroid 2–related factor 2 (Nrf2). Nrf2 is thus dissociated from Keap1 and enters into the nucleus, resulting in the transcriptional activation of Nrf2 target molecules, including antioxidant proteins and detoxifying enzymes (Komatsu et al. 2010). Co-deletion of p62/SQSTM1 or Nrf2 in the liver ameliorates the liver injury from the autophagy-deficiency (Komatsu et al. 2007; Komatsu et al. 2010; Ni et al. 2014). However, Keap1 co-deletion further exacerbates the injury (Komatsu et al. 2010). Progressive accumulation of extracellular matrix (ECM) occurs in chronic liver injury due to the activation of hepatic myofibroblasts, which then causes hepatic fibrosis (Mederacke et al. 2013). Hepatic stellate cells (HSC) are suggested to be the dominant contributor to the myofibroblast pool in a variety of hepatic pathological conditions, including toxic, biliary, and fatty liver fibrosis (Mederacke et al. 2013). Autophagy is increased in HSC isolated from murine fibrotic livers, or from liver specimens of patients chronically infected with Hepatitis B Virus (Hernandez-Gea et al. 2012). Autophagy flux is also increased during HSC activation in vitro and pharmacological blockage of autophagy inhibited the HSC activation (Thoen et al. 2011). Additionally, HSCs-specific deletion of Atg7 in mice attenuated fibrosis after carbon tetrachloride or thioacetamide induced sustained liver injury caused (Hernandez-Gea et al. 2013). All of these pieces of evidence indicate a critical role of autophagy during hepatic fibrosis.
3.1 AFLD
Typical pathological features of AFLD include steatosis, inflammation, fibrosis, and cirrhosis, which are considered to be related to oxidative stress resulted from acetaldehyde accumulation, increased NADH/NAD + ratio, or reactive oxidative species (ROS) generation (Lumeng and Crabb 2000; Lieber 2004; Zakhari and Li 2007). Oxidative stress may be involved in the functional and structural changes of mitochondria during the progress of AFLD, leading to the alteration of oxidative phosphorylation, increase of mitochondrial DNA damage, and changes of mitochondrial protein profiles (Bailey and Cunningham 2002; Demeilliers et al. 2002; Mansouri et al. 2001; Mansouri et al. 1999; Cahill et al. 2002; Cahill et al. 1999). The increase of oxygen free radicals, together with lipids accumulation caused by ethanol metabolism, leads to lipid peroxidation that can enhance oxidative damage in AFLD (Wang et al. 2015b). Autophagy induced by ethanol seems to be selective for damaged mitochondria and accumulated lipid droplets, but not long-lived proteins (Ding et al. 2010a). Stimulating or repressing autophagy could correspondingly ease or exacerbate hepatic steatosis in both acute and chronic AFLD (Ding et al. 2010a; Lin et al. 2013).
Effects of autophagy on AFLD can vary in different pathological stages. Hepatic autophagy is activated in vivo and in cultured primary hepatocytes after acute alcohol treatment (Ding et al. 2010a; Thomes et al. 2012). This activation requires ethanol metabolism, reactive oxygen species, the alteration of signaling pathway, and some other mechanisms. Oxidative stress is one stimulus for autophagy in AFLD. Acetaldehyde, a major ethanol metabolite and a pro-oxidant as well, has been indicated to induce autophagy (Thomes et al. 2013), and ethanol-induced autophagy can be suppressed by anti-oxidants, such as N-acetyl cysteine (NAC) also has been observed (Ding et al. 2010a; Wu et al. 2012). Inhibition of mTOR signaling and activation of AMPK have been indicated to participate in autophagy induced by ethanol under oxidative stress (Sid et al. 2013; Ding et al. 2011). In addition to this, ethanol treatment also activates FoxO3a, \which transcriptionally regulates several autophagy genes (Ni et al. 2013). Other autophagy stimulators, including ethanol-induced proteasome inhibition, ER stress, and metal elements like zinc, also participate in autophagy alteration after ethanol treatment (Ding and Yin 2008; Ding et al. 2007; Liuzzi and Yoo 2013).
Autophagy may be suppressed in chronic AFLD. In mice fed with Lieber-DeCarli diet for 4 weeks, autophagy was elevated at a lower dose of ethanol, accounting for 29% of the caloric need, but was inhibited at a higher ethanol dose (accounting for 36% of the caloric need) (Lin et al. 2013). Decrease in both the amount and the function of lysosomes was found in chronic alcohol treated rat livers, which may contribute to the autophagy suppression (Kharbanda et al. 1996; Kharbanda et al. 1997; Dolganiuc et al. 2012). However, the liver injury is worse when autophagy is suppressed while enhancement of autophagy improves the condition (Lin et al. 2013). A histological character of chronic AFLD is the development of Mallory-Denk body (MDB). MDB is ubiquitin and SQSTM1/p62 positive, and is accumulated more significantly in autophagy deficiency. MDB can be better cleaned using rapamycin, an mTOR inhibitor and autophagy activator, which further supported the connection between MDB formation and autophagy and the suppression of autophagy in chronic AFLD (Harada et al. 2008).
3.2 NALFD
NAFLD, which accounts for 75% of all chronic liver diseases, shares several similar pathological changes with AFLD, such as steatosis, inflammation, fibrosis and cirrhosis (Clark et al. 2003; Ruhl and Everhart 2013). However, it is more prevalent than AFLD, and can occur with obesity and diabetes (Tiniakos et al. 2010). There are no effective therapeutic approaches for NAFLD have been found. Studies have shown that autophagy can be involved in the pathogenesis of NAFLD. NAFLD can also affect autophagy. Severe hepatic steatosis was observed in mice with a hepatic deletion of either Atg5 or Atg7, and high fat diet seems to exacerbate the condition (Singh et al. 2009; Ni et al. 2014; Kim et al. 2013b). Hepatic autophagy can be inhibited in both genetic and dietary obesity models, which is partially resulted from a reduction in the expression of key autophagy molecules like Atg7 (Yang et al. 2010). ER stress is found in both autophagy deficiency MEF cells and in vivo Atg7 know-down mice, which could be improved by the restoration of Atg7 expression (Yang et al. 2010). Hepatic autophagy is also involved in the stimulation of fatty acid β-oxidation via thyroid hormone (TH), and suppression of autophagy dramatically decreased fatty acid β-oxidation in both cultured cells and mouse models (Sinha et al. 2012).
A number of signal pathways and regulators are found to regulate autophagy in NAFLD, including the insulin-mTOR signaling pathway (Liu et al. 2009). Steatosis can impair autophagy at different levels. SQSTM1/p62 accumulation is observed in the liver of ob/ob mice, and its aggregation is correlated with serum alanine aminotransferase activity and inflammatory activity by NAFLD score (Inami et al. 2011; Fukuo et al. 2014). Further studies have shown a decrease of lysosome proteolytic activity accompanied with the reduction of hepatic levels of cathepsin B and L in both ob/ob mice and NAFLD patients, which suggests that the deficiency of lysosomes may affect autophagic degradation and may contribute to SQSTM1/p62 accumulation (Inami et al. 2011; Fukuo et al. 2014). Additionally, fusion of isolated hepatic lysosomal and autophagosomes were different in mice fed with high fat diet or regular diet, indicating that an excessive amount of lipids can suppress autophagosome/lysosome fusion (Koga et al. 2010). Furthermore, high fat diets seem to suppress the expression of certain key ATG genes at the transcriptional level, which may attribute to the activity of transcriptional factors like FoxO1 (Liu et al. 2009; Xiong et al. 2012). Abnormal activation of proteases, such as calpain, may also be involved in the suppression of autophagy by reducing the protein level of certain key autophagy molecules, such as Atg5, Atg7 and Beclin 1 (Yang et al. 2010).
Autophagy can also affect liver injury during NAFLD. Autophagy can be a stress adaptation pathway that avoids cell death and suppresses, although it serves as an alternative cell-death pathway in specific cellular settings (Maiuri et al. 2007). In mice with a hepatic deletion of either Atg5 or Atg7, serum alanine aminotransferase activity is increased and inflammatory cells are accumulated in the liver (Ni et al. 2014; Komatsu et al. 2005), indicates that autophagy is critical to protect hepatocytes against cellular injury. Inhibiting autophagy in cultured hepatocytes by knocking down Atg5 sensitizes cells to death from superoxide-mediated oxidative stress, which may partly attribute to the activation of c-Jun N-terminal kinase (JNK) pathway (Wang et al. 2010). Another study have also shown autophagy could protect hepatocytes against the injury induced by lipopolysaccharide (LPS) in mice with an inducible hepatocyte-specific knockout of Atg7 (Lalazar et al. 2016). The observation may be resulted from a defect in Akt signaling in response to LPS, which set the condition in which the autophagy deficient hepatocytes were sensitized to tumor necrosis factor (TNF)-dependent liver damage (Amir et al. 2013; Lalazar et al. 2016). During the development of NAFLD, hepatic JNK and hepatocyte death receptor pathways are activated and autophagy activation can prevent apoptosis from TNF and Fas (Czaja 2016). These pieces of evidence suggest that suppression of autophagy may promote liver injury in NAFLD. However, due to limited evidence of the effects of autophagy on liver injury in in vivo NAFLD models, the precise roles of autophagy in the development of NAFLD and related mechanisms still requires further studies.
By affecting innate immunity, autophagy can modulate inflammatory response by sequestering microorganisms and regulating pathogen recognition receptors like the toll-like receptors (TLRs) (Czaja 2016). Macrophage autophagy can be impaired in primary bone marrow-derived macrophages (BMDM) and peritoneal macrophages from mice fed with high fat diet (Czaja 2016). Mice with Atg5 deficiency in macrophage, when treated with LPS after high fat diet feeding, developed systemic and hepatic inflammation. Abnormalities in macrophage polarization were observed in BMDM and Kupffer cells with an increase of proinflammatory M1 and a decrease of anti-inflammatory M2 polarization (Liu et al. 2015). This study showed a possible connection between macrophage autophagy and hepatic inflammation in obese mice, which provided a novel view of therapeutic methods for NALFD.
Liver fibrosis usually occurs in the liver with chronic injury, which is mostly attributed to the activation of hepatic myofibroblasts, hepatocytes apoptosis and/or necroptosis, and sustained hepatic inflammation (Mallat et al. 2014). The extent of fibrosis is the most important clinical determinant in patients with nonalcoholic steatohepatitis (NASH) (Czaja 2016). Loss of Atg5 in hepatocytes can induce liver fibrosis in mice (Ni et al. 2014), furthermore, mice with mutations in Atg5 in the myeloid lineage also show more susceptibility to liver fibrosis in chronic carbon tetrachloride treatment (Lodder et al. 2015). In contrast to the effects of autophagy on live fibrosis in hepatocytes and macrophages, autophagy plays a role during stellate cell activation and the development of liver fibrosis (Hernandez-Gea et al. 2012; Thoen et al. 2011). These studies provide a potential connection between autophagy and liver fibrosis. However, the degree of HSC autophagy in murine or human NAFLD is still unclear. Whether autophagy is a friend or foe in liver fibrosis in the pathogenesis of NAFLD still need to be clarified.
4. Therapeutic Effects of Autophagy Modulators on FLDs
The excessive accumulation of fatty acids is one of the main pathological features of fatty liver diseases. The reduction of hepatic triglyceride levels and the improvement of hepatic functions by autophagy activation suggest a potential therapeutic application of autophagy regulators for FLD (Singh et al. 2009; Ding et al. 2010a; Lin et al. 2013). Indeed, in both AFLD and NAFLD mouse models, clinically available chemicals that induce autophagy like rapamycin and carbamazepine, have shown the anticipated effects (Lin et al. 2013). As more key targets in the autophagy pathway have been identified, the number of novel therapeutic agents targeting autophagy have been increased as well (Cheng et al. 2013). In this part, we will discuss the therapeutic effects of autophagy modulators especially some new agents which have been indicated to affect FLD.
4.1 Autophagy Inhibitors
4.1.1 Mechanisms of action
The evolutionarily conserved function of autophagy in helping cell survival during metabolic stress has led to therapeutic resistance to cytotoxic therapy (Rubinsztein et al. 2012). Therefore, several studies have undertaken to determine therapeutic targets of autophagy and some of the inhibitors of autophagy are already in clinical trials. The major step in affecting autophagy initiation is the membrane nucleation, which is controlled by Beclin1, Vps34 and ULK complex. Several ULK1 inhibitors like MRT67307, MRT68921, SBI-0206965 have been tested in in vitro systems for their autophagy inhibition potency, no detailed information about their application to in vivo autophagy inhibition is available yet (Egan et al. 2015; Petherick et al. 2015). In addition, wortmannin, 3-methyl adenine (3-MA), and LY294002 [2-(4-morpholinyl)-8-phenyl-4H-1-benzopyran-4-one hydrochloride] are common inhibitors that interfere with the formation of autophagosomes by targeting the activity of Vps34 (Petiot et al. 2000; Pasquier 2016).
In the later stage of the autophagic process, chemicals that inhibit lysosome acidification can block the formation of autolysomes and therefore the autophagic degradation by suppressing the function of lysosome (Rubinsztein et al. 2012). Pepstatin A and Cathepsin proteases inhibitor E64d are the well-known acid protease inhibitors that can effectively suppress autophagy (Li et al. 2013b). In addition, inhibitors like bafilomycin A1 can inhibit V-ATPases by blocking the proton transport and then inhibit the autophagy process (Rubinsztein et al. 2012). Furthermore, a number of preclinical reports now have shown that the two lysosomotropic agents known as chloroquine and hydrochloroquine, effectively inhibit autophagy pathway by increasing pH inside the lysosome (Homewood et al. 1972; Pasquier 2016; Slater 1993; O’Neill et al. 1998).
4.1.2 Compounds inhibiting autophagy in liver diseases
Modulation of autophagy can be a novel therapeutic target for the treatment of HCC (Amaravadi et al. 2011). In combination of a number of cancer drugs with autophagy inducers as for example rapamycin, sorafenib (a tyrosine kinase inhibitor), NPC-16, cannabinoids have been used to target cancer cells. Autophagy was found to be upregulated in metastatic HCC (Peng et al. 2013). Use of inhibitors of autophagy like chloroquine and hydrochloroquine in combination of therapeutic drugs may be a better option for treating metastatic HCC.
There is evidence showing that autophagy suppression might be harmful in fatty liver diseases (Gracia-Sancho et al. 2014; Ding et al. 2010a; Lin et al. 2013). However, a PI3K-Akt pathway inhibitor, wortmannin, was indicated to have a dual effect on AFLD in mice (Zeng et al. 2012). In this earlier study, low dose of wortmannin significantly reduced the hepatic triglyceride level in an acute ethanol-induced fatty liver mouse model, but aggravated fatty liver at a higher dose treatment. The contradictory findings may be resulted from the effects of the compounds other than autophagy inhibition, but the exact mechanisms still need to be further explored.
4.2 Autophagy Activators
Discovering new therapeutic interventions to treat lipid disorders is of great interest and the discovery of autophagy as a regulator of lipid metabolism has opened up new avenues for targeting modulators of this pathway. The primary rationales on developing autophagy inducers for the treatment of fatty liver comes from the observation that genetic defect in autophagy gene cause accumulation of fat in hepatocytes (Singh et al. 2009). Meanwhile, autophagy gene therapy via adenovirus-associated viral delivery in liver tissue resulted in significant reduction in hepatic steatosis in Ob/Ob mice model (Yang et al. 2010). Moreover, pharmacological enhancement of autophagy with inducers such as rapamycin and carbamazepine reduced hepatic and blood triglycerides, blood glucose levels and markers of hepatic damage in high fat fed rodent model (Lin et al. 2013). All of these observations suggest that upregulation of autophagy could be clinically beneficial.
Autophagy inducers can be divided into two categories - non-pharmaceutical interventions and pharmacological activators. Non-pharmaceutical interventions includes the long-term caloric restriction and regular exercise that strongly upregulates autophagy and improve the overall health (Cui et al. 2013). Nutritional supplements such as consumption of coffee and vitamin D, may also influence health through autophagy induction. A previous study showed that caffeine could reduce hepatic steatosis by stimulating autophagy in mice with non-alcoholic FLD (Sinha et al. 2014). Pharmacological activators can be those approved by US Food and Drug Administration (FDA) or those identified through high throughput chemical screening, which are less characterized and not suitable for immediate clinical use.
4.2.1 Mechanisms of action
Various strategies have been utilized for inducing autophagy. Both mTOR-dependent and-independent pathways are considered.
4.2.1.1 mTOR-dependent autophagy inducers
This group of inducers includes rapamycin and rapalogs, such as CCI-779, RAD001 and AP23573. They are basically lipophilic macrolide antibiotics that inhibit mTOR1 Complex 1 (Noda and Ohsumi 1998). Recently, two selective ATP-competitive small molecules mTOR inhibitors, PP242 and Torin 1 have been identified, which directly inhibit both mTORC1 and mTORC2 (Thoreen et al. 2009).Moreover, a new class of stronger inducer of autophagy, PI-103 and NVP-BEZ235 (imidazo[4,5-c]quinoline derivative) have also been reported, which dually inhibits mTORC and class I PI3K-AKT signaling (Degtyarev et al. 2008; Maira et al. 2008).
4.2.1.2 mTOR-independent autophagy inducers
Metformin, which is widely used for diabetes treatment, is one of the most popular inducers in this category. Metformin activates AMPK, which in turn increases autophagy through mTOR inhibition and direct ULK1 activation (Alers et al. 2012). Some other reagents, including carbamazepine, lithium, and sodium valproate, are another category of mTOR-independent autophagy inducers. They reduce intracellular levels of inositol and inositol-1, 4, 5-triphosphate (Ins (1, 4, 5) P3) and positively regulate autophagy (Sarkar et al. 2005; Williams et al. 2008). Carbamazepine has been shown to have beneficial effects in mouse models of high fat diet induced steatosis, and of α1-antitrypsin deficiency (Lin et al. 2013; Hidvegi et al. 2010). The third group of inducers include FDA approved drugs and pharmacological probes, such as rilmenidine, clonidin, and verampil, which induce autophagy by targeting the pathways involved in cyclic AMP (Williams et al. 2008; Rose et al. 2010). Other examined inducers of autophagy including BH3 mimetics, resveratrol, trehalose, spermidine, EGFR agonists and L-NAME (Rubinsztein et al. 2012). Among these inducers, BH3 mimetics is well known to disrupt the inhibitory interaction between the BH3 domain of beclin-1 and BCL-2, stimulating the beclin-1 dependent allosteric activation of the pro-autophagic lipid kinase PIK3C3 (Malik et al. 2011).
4.2.2 Compounds targeting autophagy for liver diseases
Currently there is no approved pharmacological therapies for FLD (Musso et al. 2012). However, a number of novel agents specifically targeting non-alcoholic FLD pathogenesis have entered into clinical trials. Some mTORC1 and mTORC2 Inhibitors, including rapamycin, AZD3147, Z1001, rottlerin and XL388, are in preclinical stage of drug development (Musso et al. 2016). AMPK activators such as monascin, ankaflavin, quercetin, berberin, curcumin and oltipraz are also in developmental stage for the NAFLD treatment. Among the AMPK activators, oltipraz is currently at phase IIa state of development (Musso et al. 2016). Better understanding of the autophagy role in the pathogenesis of fatty liver (both alcoholic and non-alcoholic) disease and simultaneous preclinical or clinical studies of the current or emerging pharmacological inducers is necessary to develop and identify the promising molecules for applications in FLDs.
5. Future Directions
Although recent studies have elucidated the molecular mechanism of autophagy and the role of autophagy in FLDs, most of recent studies are focusing on potential roles of autophagy in hepatocytes. Only a few studies have explored the effects of autophagy in hepatic non-parenchymal cells on the pathogenesis of FLDs. Accumulating evidences have shown that autophagy in non-parenchymal cells is critical to the pathogenesis of liver diseases, like liver fibrosis (Thoen et al. 2011; Thoen et al. 2012; Lodder et al. 2015). Further understanding of the roles of autophagy in hepatic non-parenchymal cells during the pathogenesis of FLDs would be important. Cautions have to be exercised that autophagy may play different roles in hepatocytes and hepatic non-parenchymal cells, which results in opposite effects on FLDs (Czaja 2016). Agents that can target to autophagy in tissue-specific way would be most useful for the treatment of FLD in future.
6. Conclusion
A basal level of autophagy is essential for the maintenance of normal liver function. Accumulating evidence from various FLD models suggests FLDs can affect the autophagic process, and conversely autophagy can affect the pathogenesis of FLDs. Early studies have opened up the avenue for potential therapeutic applications of autophagy regulators in FLDs. Indeed, various modulators of autophagy have been suggested to be beneficial to ameliorate the fatty livers. The connection between autophagy and FLDs at different stages, and potential therapeutic applications of autophagy modulators are summarized in Fig. 2. We hope that this summary will be helpful in the further exploration of the role of autophagy in FLDs, and the use of autophagy modulators to alleviate FLDs.
Fig. 2.

Relationship of autophagy and fatty liver diseases at different stages, and potential therapeutic applications of autophagy modulators in treating fatty liver diseases.
Acknowledgments
The authors are partially supported by NIH research grants (R21-AA021450 and R01-AA021751).
Abbreviations
- 3-MA
3-methyl adenine
- AFLD
Alcoholic fatty liver disease
- AMPK
AMP-activated protein kinase
- Atg
Autophagy-related proteins
- CMA
Chaperone-mediated autophagy
- DAMPs
Danger-associated molecular patterns
- ECM
Extracellular matrix
- ER
Endoplasmic reticulum
- ERQC
Endoplasmic reticulum quality control
- FAs
Fatty acids
- FLD
Fatty liver disease
- HCC
Hepatocellular carcinoma
- HSC
Hepatic stellate cells
- LDs
Lipid droplets
- MDB
Mallory-Denk body
- mTOR
Mammalian target of rapamycin
- NAC
N-acetyl cysteine
- NAFLD
Nonalcoholic fatty liver disease
- PAMPs
Pathogen-associated molecular patterns DAMPs, Danger-associated molecular patterns
- PI3K
Class III phosphatidylinositol 3-kinase
- PKA
Ras/cAMP-dependent protein kinase A
- TFEB
Transcription factor EB
- ULK1
UNC-5 like autophagy activating kinase 1
- UPR
Unfolded protein response
Footnotes
The authors declare there are no conflicts of interest.
References
- Abdel-Misih SR, Bloomston M. Liver anatomy. Surg Clin North Am. 2010;90(4):643–653. doi: 10.1016/j.suc.2010.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ait-Goughoulte M, Kanda T, Meyer K, Ryerse JS, Ray RB, Ray R. Hepatitis C virus genotype 1a growth and induction of autophagy. J Virol. 2008;82(5):2241–2249. doi: 10.1128/JVI.02093-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alers S, Loffler AS, Wesselborg S, Stork B. Role of AMPK-mTOR-Ulk1/2 in the regulation of autophagy: cross talk, shortcuts, and feedbacks. Mol Cell Biol. 2012;32(1):2–11. doi: 10.1128/MCB.06159-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amaravadi RK, Lippincott-Schwartz J, Yin XM, Weiss WA, Takebe N, Timmer W, DiPaola RS, Lotze MT, White E. Principles and current strategies for targeting autophagy for cancer treatment. Clin Cancer Res. 2011;17(4):654–666. doi: 10.1158/1078-0432.CCR-10-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amir M, Zhao E, Fontana L, Rosenberg H, Tanaka K, Gao G, Czaja MJ. Inhibition of hepatocyte autophagy increases tumor necrosis factor-dependent liver injury by promoting caspase-8 activation. Cell Death Differ. 2013;20(7):878–887. doi: 10.1038/cdd.2013.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey SM, Cunningham CC. Contribution of mitochondria to oxidative stress associated with alcoholic liver disease. Free Radic Biol Med. 2002;32(1):11–16. doi: 10.1016/s0891-5849(01)00769-9. [DOI] [PubMed] [Google Scholar]
- Brech A, Ahlquist T, Lothe RA, Stenmark H. Autophagy in tumour suppression and promotion. Mol Oncol. 2009;3(4):366–375. doi: 10.1016/j.molonc.2009.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahill A, Cunningham CC, Adachi M, Ishii H, Bailey SM, Fromenty B, Davies A. Effects of alcohol and oxidative stress on liver pathology: the role of the mitochondrion. Alcohol Clin Exp Res. 2002;26(6):907–915. [PMC free article] [PubMed] [Google Scholar]
- Cahill A, Stabley GJ, Wang X, Hoek JB. Chronic ethanol consumption causes alterations in the structural integrity of mitochondrial DNA in aged rats. Hepatology. 1999;30(4):881–888. doi: 10.1002/hep.510300434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y, Ren X, Hait WN, Yang JM. Therapeutic targeting of autophagy in disease: biology and pharmacology. Pharmacol Rev. 2013;65(4):1162–1197. doi: 10.1124/pr.112.007120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christian P, Sacco J, Adeli K. Autophagy: Emerging roles in lipid homeostasis and metabolic control. Biochim Biophys Acta. 2013;1831(4):819–824. doi: 10.1016/j.bbalip.2012.12.009. [DOI] [PubMed] [Google Scholar]
- Clark JM, Brancati FL, Diehl AM. The prevalence and etiology of elevated aminotransferase levels in the United States. Am J Gastroenterol. 2003;98(5):960–967. doi: 10.1111/j.1572-0241.2003.07486.x. [DOI] [PubMed] [Google Scholar]
- Cui M, Yu H, Wang J, Gao J, Li J. Chronic caloric restriction and exercise improve metabolic conditions of dietary-induced obese mice in autophagy correlated manner without involving AMPK. J Diabetes Res. 2013;2013:852754. doi: 10.1155/2013/852754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czaja MJ. Function of Autophagy in Nonalcoholic Fatty Liver Disease. Dig Dis Sci. 2016;61(5):1304–1313. doi: 10.1007/s10620-015-4025-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czaja MJ, Ding WX, Donohue TM, Jr, Friedman SL, Kim JS, Komatsu M, Lemasters JJ, Lemoine A, Lin JD, Ou JH, Perlmutter DH, Randall G, Ray RB, Tsung A, Yin XM. Functions of autophagy in normal and diseased liver. Autophagy. 2013;9(8):1131–1158. doi: 10.4161/auto.25063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degtyarev M, De Maziere A, Orr C, Lin J, Lee BB, Tien JY, Prior WW, van Dijk S, Wu H, Gray DC, Davis DP, Stern HM, Murray LJ, Hoeflich KP, Klumperman J, Friedman LS, Lin K. Akt inhibition promotes autophagy and sensitizes PTEN-null tumors to lysosomotropic agents. J Cell Biol. 2008;183(1):101–116. doi: 10.1083/jcb.200801099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demeilliers C, Maisonneuve C, Grodet A, Mansouri A, Nguyen R, Tinel M, Letteron P, Degott C, Feldmann G, Pessayre D, Fromenty B. Impaired adaptive resynthesis and prolonged depletion of hepatic mitochondrial DNA after repeated alcohol binges in mice. Gastroenterology. 2002;123(4):1278–1290. doi: 10.1053/gast.2002.35952. [DOI] [PubMed] [Google Scholar]
- Deter RL, De Duve C. Influence of glucagon, an inducer of cellular autophagy, on some physical properties of rat liver lysosomes. J Cell Biol. 1967;33(2):437–449. doi: 10.1083/jcb.33.2.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding WX, Li M, Chen X, Ni HM, Lin CW, Gao W, Lu B, Stolz DB, Clemens DL, Yin XM. Autophagy reduces acute ethanol-induced hepatotoxicity and steatosis in mice. Gastroenterology. 2010a;139(5):1740–1752. doi: 10.1053/j.gastro.2010.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding WX, Li M, Yin XM. Selective taste of ethanol-induced autophagy for mitochondria and lipid droplets. Autophagy. 2011;7(2):248–249. doi: 10.4161/auto.7.2.14347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding WX, Ni HM, Gao W, Yoshimori T, Stolz DB, Ron D, Yin XM. Linking of autophagy to ubiquitin-proteasome system is important for the regulation of endoplasmic reticulum stress and cell viability. Am J Pathol. 2007;171(2):513–524. doi: 10.2353/ajpath.2007.070188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding WX, Ni HM, Li M, Liao Y, Chen X, Stolz DB, Dorn GW, 2nd, Yin XM. Nix is critical to two distinct phases of mitophagy, reactive oxygen species-mediated autophagy induction and Parkin-ubiquitin-p62-mediated mitochondrial priming. J Biol Chem. 2010b;285(36):27879–27890. doi: 10.1074/jbc.M110.119537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding WX, Yin XM. Sorting, recognition and activation of the misfolded protein degradation pathways through macroautophagy and the proteasome. Autophagy. 2008;4(2):141–150. doi: 10.4161/auto.5190. [DOI] [PubMed] [Google Scholar]
- Dolganiuc A, Thomes PG, Ding WX, Lemasters JJ, Donohue TM., Jr Autophagy in alcohol-induced liver diseases. Alcohol Clin Exp Res. 2012;36(8):1301–1308. doi: 10.1111/j.1530-0277.2012.01742.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan DF, Chun MG, Vamos M, Zou H, Rong J, Miller CJ, Lou HJ, Raveendra-Panickar D, Yang CC, Sheffler DJ, Teriete P, Asara JM, Turk BE, Cosford ND, Shaw RJ. Small Molecule Inhibition of the Autophagy Kinase ULK1 and Identification of ULK1 Substrates. Mol Cell. 2015;59(2):285–297. doi: 10.1016/j.molcel.2015.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eiyama A, Okamoto K. PINK1/Parkin-mediated mitophagy in mammalian cells. Curr Opin Cell Biol. 2015;33:95–101. doi: 10.1016/j.ceb.2015.01.002. [DOI] [PubMed] [Google Scholar]
- Ezaki J, Matsumoto N, Takeda-Ezaki M, Komatsu M, Takahashi K, Hiraoka Y, Taka H, Fujimura T, Takehana K, Yoshida M, Iwata J, Tanida I, Furuya N, Zheng DM, Tada N, Tanaka K, Kominami E, Ueno T. Liver autophagy contributes to the maintenance of blood glucose and amino acid levels. Autophagy. 2011;7(7):727–736. doi: 10.4161/auto.7.7.15371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farre JC, Krick R, Subramani S, Thumm M. Turnover of organelles by autophagy in yeast. Curr Opin Cell Biol. 2009;21(4):522–530. doi: 10.1016/j.ceb.2009.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y, Yao Z, Klionsky DJ. How to control self-digestion: transcriptional, post-transcriptional, and post-translational regulation of autophagy. Trends Cell Biol. 2015;25(6):354–363. doi: 10.1016/j.tcb.2015.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuo Y, Yamashina S, Sonoue H, Arakawa A, Nakadera E, Aoyama T, Uchiyama A, Kon K, Ikejima K, Watanabe S. Abnormality of autophagic function and cathepsin expression in the liver from patients with non-alcoholic fatty liver disease. Hepatol Res. 2014;44(9):1026–1036. doi: 10.1111/hepr.12282. [DOI] [PubMed] [Google Scholar]
- Gracia-Sancho J, Guixe-Muntet S, Hide D, Bosch J. Modulation of autophagy for the treatment of liver diseases. Expert Opin Investig Drugs. 2014;23(7):965–977. doi: 10.1517/13543784.2014.912274. [DOI] [PubMed] [Google Scholar]
- Hamasaki M, Shibutani ST, Yoshimori T. Up-to-date membrane biogenesis in the autophagosome formation. Curr Opin Cell Biol. 2013;25(4):455–460. doi: 10.1016/j.ceb.2013.03.004. [DOI] [PubMed] [Google Scholar]
- Harada M, Hanada S, Toivola DM, Ghori N, Omary MB. Autophagy activation by rapamycin eliminates mouse Mallory-Denk bodies and blocks their proteasome inhibitor-mediated formation. Hepatology. 2008;47(6):2026–2035. doi: 10.1002/hep.22294. [DOI] [PubMed] [Google Scholar]
- He C, Klionsky DJ. Regulation mechanisms and signaling pathways of autophagy. Annu Rev Genet. 2009;43:67–93. doi: 10.1146/annurev-genet-102808-114910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez-Gea V, Ghiassi-Nejad Z, Rozenfeld R, Gordon R, Fiel MI, Yue Z, Czaja MJ, Friedman SL. Autophagy releases lipid that promotes fibrogenesis by activated hepatic stellate cells in mice and in human tissues. Gastroenterology. 2012;142(4):938–946. doi: 10.1053/j.gastro.2011.12.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez-Gea V, Hilscher M, Rozenfeld R, Lim MP, Nieto N, Werner S, Devi LA, Friedman SL. Endoplasmic reticulum stress induces fibrogenic activity in hepatic stellate cells through autophagy. J Hepatol. 2013;59(1):98–104. doi: 10.1016/j.jhep.2013.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hidvegi T, Ewing M, Hale P, Dippold C, Beckett C, Kemp C, Maurice N, Mukherjee A, Goldbach C, Watkins S, Michalopoulos G, Perlmutter DH. An autophagy-enhancing drug promotes degradation of mutant alpha1-antitrypsin Z and reduces hepatic fibrosis. Science. 2010;329(5988):229–232. doi: 10.1126/science.1190354. [DOI] [PubMed] [Google Scholar]
- Homewood CA, Warhurst DC, Peters W, Baggaley VC. Lysosomes, pH and the anti-malarial action of chloroquine. Nature. 1972;235(5332):50–52. doi: 10.1038/235050a0. [DOI] [PubMed] [Google Scholar]
- Inami Y, Yamashina S, Izumi K, Ueno T, Tanida I, Ikejima K, Watanabe S. Hepatic steatosis inhibits autophagic proteolysis via impairment of autophagosomal acidification and cathepsin expression. Biochem Biophys Res Commun. 2011;412(4):618–625. doi: 10.1016/j.bbrc.2011.08.012. [DOI] [PubMed] [Google Scholar]
- Jaber N, Dou Z, Chen JS, Catanzaro J, Jiang YP, Ballou LM, Selinger E, Ouyang X, Lin RZ, Zhang J, Zong WX. Class III PI3K Vps34 plays an essential role in autophagy and in heart and liver function. Proc Natl Acad Sci U S A. 2012;109(6):2003–2008. doi: 10.1073/pnas.1112848109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansen T, Lamark T. Selective autophagy mediated by autophagic adapter proteins. Autophagy. 2011;7(3):279–296. doi: 10.4161/auto.7.3.14487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur J, Debnath J. Autophagy at the crossroads of catabolism and anabolism. Nat Rev Mol Cell Biol. 2015;16(8):461–472. doi: 10.1038/nrm4024. [DOI] [PubMed] [Google Scholar]
- Kaushik S, Cuervo AM. Chaperone-mediated autophagy: a unique way to enter the lysosome world. Trends Cell Biol. 2012;22(8):407–417. doi: 10.1016/j.tcb.2012.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaminets A, Heinrich T, Mari M, Grumati P, Huebner AK, Akutsu M, Liebmann L, Stolz A, Nietzsche S, Koch N, Mauthe M, Katona I, Qualmann B, Weis J, Reggiori F, Kurth I, Hubner CA, Dikic I. Regulation of endoplasmic reticulum turnover by selective autophagy. Nature. 2015;522(7556):354–358. doi: 10.1038/nature14498. [DOI] [PubMed] [Google Scholar]
- Kharbanda KK, McVicker DL, Zetterman RK, Donohue TM., Jr Ethanol consumption alters trafficking of lysosomal enzymes and affects the processing of procathepsin L in rat liver. Biochim Biophys Acta. 1996;1291(1):45–52. doi: 10.1016/0304-4165(96)00043-8. [DOI] [PubMed] [Google Scholar]
- Kharbanda KK, McVicker DL, Zetterman RK, MacDonald RG, Donohue TM., Jr Flow cytometric analysis of vesicular pH in rat hepatocytes after ethanol administration. Hepatology. 1997;26(4):929–934. doi: 10.1002/hep.510260419. [DOI] [PubMed] [Google Scholar]
- Kim J, Kim YC, Fang C, Russell RC, Kim JH, Fan W, Liu R, Zhong Q, Guan KL. Differential regulation of distinct Vps34 complexes by AMPK in nutrient stress and autophagy. Cell. 2013a;152(1–2):290–303. doi: 10.1016/j.cell.2012.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KH, Jeong YT, Oh H, Kim SH, Cho JM, Kim YN, Kim SS, Kim DH, Hur KY, Kim HK, Ko T, Han J, Kim HL, Kim J, Back SH, Komatsu M, Chen H, Chan DC, Konishi M, Itoh N, Choi CS, Lee MS. Autophagy deficiency leads to protection from obesity and insulin resistance by inducing Fgf21 as a mitokine. Nat Med. 2013b;19(1):83–92. doi: 10.1038/nm.3014. [DOI] [PubMed] [Google Scholar]
- Kim KH, Lee MS. Autophagy–a key player in cellular and body metabolism. Nat Rev Endocrinol. 2014;10(6):322–337. doi: 10.1038/nrendo.2014.35. [DOI] [PubMed] [Google Scholar]
- Kim SJ, Khan M, Quan J, Till A, Subramani S, Siddiqui A. Hepatitis B virus disrupts mitochondrial dynamics: induces fission and mitophagy to attenuate apoptosis. PLoS Pathog. 2013c;9(12):e1003722. doi: 10.1371/journal.ppat.1003722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YC, Guan KL. mTOR: a pharmacologic target for autophagy regulation. J Clin Invest. 2015;125(1):25–32. doi: 10.1172/JCI73939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koga H, Kaushik S, Cuervo AM. Altered lipid content inhibits autophagic vesicular fusion. FASEB J. 2010;24(8):3052–3065. doi: 10.1096/fj.09-144519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu M, Kurokawa H, Waguri S, Taguchi K, Kobayashi A, Ichimura Y, Sou YS, Ueno I, Sakamoto A, Tong KI, Kim M, Nishito Y, Iemura S, Natsume T, Ueno T, Kominami E, Motohashi H, Tanaka K, Yamamoto M. The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nat Cell Biol. 2010;12(3):213–223. doi: 10.1038/ncb2021. [DOI] [PubMed] [Google Scholar]
- Komatsu M, Waguri S, Koike M, Sou YS, Ueno T, Hara T, Mizushima N, Iwata J, Ezaki J, Murata S, Hamazaki J, Nishito Y, Iemura S, Natsume T, Yanagawa T, Uwayama J, Warabi E, Yoshida H, Ishii T, Kobayashi A, Yamamoto M, Yue Z, Uchiyama Y, Kominami E, Tanaka K. Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell. 2007;131(6):1149–1163. doi: 10.1016/j.cell.2007.10.035. [DOI] [PubMed] [Google Scholar]
- Komatsu M, Waguri S, Ueno T, Iwata J, Murata S, Tanida I, Ezaki J, Mizushima N, Ohsumi Y, Uchiyama Y, Kominami E, Tanaka K, Chiba T. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J Cell Biol. 2005;169(3):425–434. doi: 10.1083/jcb.200412022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotoulas OB, Kalamidas SA, Kondomerkos DJ. Glycogen autophagy in glucose homeostasis. Pathol Res Pract. 2006;202(9):631–638. doi: 10.1016/j.prp.2006.04.001. [DOI] [PubMed] [Google Scholar]
- Kroemer G, Marino G, Levine B. Autophagy and the integrated stress response. Mol Cell. 2010;40(2):280–293. doi: 10.1016/j.molcel.2010.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalazar G, Ilyas G, Malik SA, Liu K, Zhao E, Amir M, Lin Y, Tanaka KE, Czaja MJ. Autophagy confers resistance to lipopolysaccharide-induced mouse hepatocyte injury. Am J Physiol Gastrointest Liver Physiol. 2016;311(3):G377–386. doi: 10.1152/ajpgi.00124.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb CA, Yoshimori T, Tooze SA. The autophagosome: origins unknown, biogenesis complex. Nat Rev Mol Cell Biol. 2013;14(12):759–774. doi: 10.1038/nrm3696. [DOI] [PubMed] [Google Scholar]
- Lee JM, Wagner M, Xiao R, Kim KH, Feng D, Lazar MA, Moore DD. Nutrient-sensing nuclear receptors coordinate autophagy. Nature. 2014;516(7529):112–115. doi: 10.1038/nature13961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132(1):27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Yang B, Zhou Q, Wu Y, Shang D, Guo Y, Song Z, Zheng Q, Xiong J. Autophagy promotes hepatocellular carcinoma cell invasion through activation of epithelial-mesenchymal transition. Carcinogenesis. 2013a;34(6):1343–1351. doi: 10.1093/carcin/bgt063. [DOI] [PubMed] [Google Scholar]
- Li M, Khambu B, Zhang H, Kang JH, Chen X, Chen D, Vollmer L, Liu PQ, Vogt A, Yin XM. Suppression of lysosome function induces autophagy via a feedback down-regulation of MTOR complex 1 (MTORC1) activity. J Biol Chem. 2013b;288(50):35769–35780. doi: 10.1074/jbc.M113.511212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieber CS. Alcoholic fatty liver: its pathogenesis and mechanism of progression to inflammation and fibrosis. Alcohol. 2004;34(1):9–19. doi: 10.1016/j.alcohol.2004.07.008. [DOI] [PubMed] [Google Scholar]
- Lin CW, Zhang H, Li M, Xiong X, Chen X, Chen X, Dong XC, Yin XM. Pharmacological promotion of autophagy alleviates steatosis and injury in alcoholic and non-alcoholic fatty liver conditions in mice. J Hepatol. 2013;58(5):993–999. doi: 10.1016/j.jhep.2013.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipatova Z, Segev N. Ypt/Rab GTPases regulate two intersections of the secretory and the endosomal/lysosomal pathways. Cell Logist. 2014;4(3):e954870. doi: 10.4161/21592780.2014.954870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipatova Z, Segev N. A Role for Macro-ER-Phagy in ER Quality Control. PLoS Genet. 2015;11(7):e1005390. doi: 10.1371/journal.pgen.1005390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipatova Z, Shah AH, Kim JJ, Mulholland JW, Segev N. Regulation of ER-phagy by a Ypt/Rab GTPase module. Mol Biol Cell. 2013;24(19):3133–3144. doi: 10.1091/mbc.E13-05-0269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Fang M, Hu Y, Huang B, Li N, Chang C, Huang R, Xu X, Yang Z, Chen Z, Liu W. Hepatitis B virus X protein inhibits autophagic degradation by impairing lysosomal maturation. Autophagy. 2014;10(3):416–430. doi: 10.4161/auto.27286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu HY, Han J, Cao SY, Hong T, Zhuo D, Shi J, Liu Z, Cao W. Hepatic autophagy is suppressed in the presence of insulin resistance and hyperinsulinemia: inhibition of FoxO1-dependent expression of key autophagy genes by insulin. J Biol Chem. 2009;284(45):31484–31492. doi: 10.1074/jbc.M109.033936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K, Zhao E, Ilyas G, Lalazar G, Lin Y, Haseeb M, Tanaka KE, Czaja MJ. Impaired macrophage autophagy increases the immune response in obese mice by promoting proinflammatory macrophage polarization. Autophagy. 2015;11(2):271–284. doi: 10.1080/15548627.2015.1009787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Levine B. Autosis and autophagic cell death: the dark side of autophagy. Cell Death Differ. 2015;22(3):367–376. doi: 10.1038/cdd.2014.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liuzzi JP, Yoo C. Role of zinc in the regulation of autophagy during ethanol exposure in human hepatoma cells. Biol Trace Elem Res. 2013;156(1–3):350–356. doi: 10.1007/s12011-013-9816-3. [DOI] [PubMed] [Google Scholar]
- Lodder J, Denaes T, Chobert MN, Wan J, El-Benna J, Pawlotsky JM, Lotersztajn S, Teixeira-Clerc F. Macrophage autophagy protects against liver fibrosis in mice. Autophagy. 2015;11(8):1280–1292. doi: 10.1080/15548627.2015.1058473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lumeng L, Crabb DW. Alcoholic liver disease. Curr Opin Gastroenterol. 2000;16(3):208–218. doi: 10.1097/00001574-200005000-00003. [DOI] [PubMed] [Google Scholar]
- Madrigal-Matute J, Cuervo AM. Regulation of Liver Metabolism by Autophagy. Gastroenterology. 2016;150(2):328–339. doi: 10.1053/j.gastro.2015.09.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maira SM, Stauffer F, Brueggen J, Furet P, Schnell C, Fritsch C, Brachmann S, Chene P, De Pover A, Schoemaker K, Fabbro D, Gabriel D, Simonen M, Murphy L, Finan P, Sellers W, Garcia-Echeverria C. Identification and characterization of NVP-BEZ235, a new orally available dual phosphatidylinositol 3-kinase/mammalian target of rapamycin inhibitor with potent in vivo antitumor activity. Mol Cancer Ther. 2008;7(7):1851–1863. doi: 10.1158/1535-7163.MCT-08-0017. [DOI] [PubMed] [Google Scholar]
- Maiuri MC, Zalckvar E, Kimchi A, Kroemer G. Self-eating and self-killing: crosstalk between autophagy and apoptosis. Nature Reviews Molecular Cell Biology. 2007;8(9):741–752. doi: 10.1038/nrm2239. [DOI] [PubMed] [Google Scholar]
- Malik SA, Orhon I, Morselli E, Criollo A, Shen S, Marino G, BenYounes A, Benit P, Rustin P, Maiuri MC, Kroemer G. BH3 mimetics activate multiple pro-autophagic pathways. Oncogene. 2011;30(37):3918–3929. doi: 10.1038/onc.2011.104. [DOI] [PubMed] [Google Scholar]
- Mallat A, Lodder J, Teixeira-Clerc F, Moreau R, Codogno P, Lotersztajn S. Autophagy: a multifaceted partner in liver fibrosis. Biomed Res Int. 2014;2014:869390. doi: 10.1155/2014/869390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manjithaya R, Nazarko TY, Farre JC, Subramani S. Molecular mechanism and physiological role of pexophagy. FEBS Lett. 2010;584(7):1367–1373. doi: 10.1016/j.febslet.2010.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansouri A, Demeilliers C, Amsellem S, Pessayre D, Fromenty B. Acute ethanol administration oxidatively damages and depletes mitochondrial dna in mouse liver, brain, heart, and skeletal muscles: protective effects of antioxidants. J Pharmacol Exp Ther. 2001;298(2):737–743. [PubMed] [Google Scholar]
- Mansouri A, Gaou I, De Kerguenec C, Amsellem S, Haouzi D, Berson A, Moreau A, Feldmann G, Letteron P, Pessayre D, Fromenty B. An alcoholic binge causes massive degradation of hepatic mitochondrial DNA in mice. Gastroenterology. 1999;117(1):181–190. doi: 10.1016/s0016-5085(99)70566-4. [DOI] [PubMed] [Google Scholar]
- Mao Y, Da L, Tang H, Yang J, Lei Y, Tiollais P, Li T, Zhao M. Hepatitis B virus X protein reduces starvation-induced cell death through activation of autophagy and inhibition of mitochondrial apoptotic pathway. Biochem Biophys Res Commun. 2011;415(1):68–74. doi: 10.1016/j.bbrc.2011.10.013. [DOI] [PubMed] [Google Scholar]
- Mederacke I, Hsu CC, Troeger JS, Huebener P, Mu X, Dapito DH, Pradere JP, Schwabe RF. Fate tracing reveals hepatic stellate cells as dominant contributors to liver fibrosis independent of its aetiology. Nat Commun. 2013;4:2823. doi: 10.1038/ncomms3823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijer AJ, Codogno P. Regulation and role of autophagy in mammalian cells. Int J Biochem Cell Biol. 2004;36(12):2445–2462. doi: 10.1016/j.biocel.2004.02.002. [DOI] [PubMed] [Google Scholar]
- Mijaljica D, Prescott M, Devenish RJ. Microautophagy in mammalian cells: revisiting a 40-year-old conundrum. Autophagy. 2011;7(7):673–682. doi: 10.4161/auto.7.7.14733. [DOI] [PubMed] [Google Scholar]
- Mizushima N, Klionsky DJ. Protein turnover via autophagy: implications for metabolism. Annu Rev Nutr. 2007;27:19–40. doi: 10.1146/annurev.nutr.27.061406.093749. [DOI] [PubMed] [Google Scholar]
- Mizushima N, Komatsu M. Autophagy: renovation of cells and tissues. Cell. 2011;147(4):728–741. doi: 10.1016/j.cell.2011.10.026. [DOI] [PubMed] [Google Scholar]
- Mizushima N, Yoshimori T, Ohsumi Y. The role of Atg proteins in autophagosome formation. Annu Rev Cell Dev Biol. 2011;27:107–132. doi: 10.1146/annurev-cellbio-092910-154005. [DOI] [PubMed] [Google Scholar]
- Mortimore GE, Hutson NJ, Surmacz CA. Quantitative correlation between proteolysis and macro- and microautophagy in mouse hepatocytes during starvation and refeeding. Proc Natl Acad Sci U S A. 1983;80(8):2179–2183. doi: 10.1073/pnas.80.8.2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musso G, Cassader M, Gambino R. Non-alcoholic steatohepatitis: emerging molecular targets and therapeutic strategies. Nat Rev Drug Discov. 2016;15(4):249–274. doi: 10.1038/nrd.2015.3. [DOI] [PubMed] [Google Scholar]
- Musso G, Cassader M, Rosina F, Gambino R. Impact of current treatments on liver disease, glucose metabolism and cardiovascular risk in non-alcoholic fatty liver disease (NAFLD): a systematic review and meta-analysis of randomised trials. Diabetologia. 2012;55(4):885–904. doi: 10.1007/s00125-011-2446-4. [DOI] [PubMed] [Google Scholar]
- Nakatogawa H, Suzuki K, Kamada Y, Ohsumi Y. Dynamics and diversity in autophagy mechanisms: lessons from yeast. Nat Rev Mol Cell Biol. 2009;10(7):458–467. doi: 10.1038/nrm2708. [DOI] [PubMed] [Google Scholar]
- Neuhaus A, Kooshapur H, Wolf J, Meyer NH, Madl T, Saidowsky J, Hambruch E, Lazam A, Jung M, Sattler M, Schliebs W, Erdmann R. A novel Pex14 protein-interacting site of human Pex5 is critical for matrix protein import into peroxisomes. J Biol Chem. 2014;289(1):437–448. doi: 10.1074/jbc.M113.499707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni HM, Du K, You M, Ding WX. Critical role of FoxO3a in alcohol-induced autophagy and hepatotoxicity. Am J Pathol. 2013;183(6):1815–1825. doi: 10.1016/j.ajpath.2013.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni HM, Woolbright BL, Williams J, Copple B, Cui W, Luyendyk JP, Jaeschke H, Ding WX. Nrf2 promotes the development of fibrosis and tumorigenesis in mice with defective hepatic autophagy. J Hepatol. 2014;61(3):617–625. doi: 10.1016/j.jhep.2014.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda T, Ohsumi Y. Tor, a phosphatidylinositol kinase homologue, controls autophagy in yeast. J Biol Chem. 1998;273(7):3963–3966. doi: 10.1074/jbc.273.7.3963. [DOI] [PubMed] [Google Scholar]
- Nordgren M, Wang B, Apanasets O, Fransen M. Peroxisome degradation in mammals: mechanisms of action, recent advances, and perspectives. Front Physiol. 2013;4:145. doi: 10.3389/fphys.2013.00145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill PM, Bray PG, Hawley SR, Ward SA, Park BK. 4-Aminoquinolines–past, present, and future: a chemical perspective. Pharmacol Ther. 1998;77(1):29–58. doi: 10.1016/s0163-7258(97)00084-3. [DOI] [PubMed] [Google Scholar]
- Ogata M, Hino S, Saito A, Morikawa K, Kondo S, Kanemoto S, Murakami T, Taniguchi M, Tanii I, Yoshinaga K, Shiosaka S, Hammarback JA, Urano F, Imaizumi K. Autophagy is activated for cell survival after endoplasmic reticulum stress. Mol Cell Biol. 2006;26(24):9220–9231. doi: 10.1128/MCB.01453-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto K. Organellophagy: eliminating cellular building blocks via selective autophagy. J Cell Biol. 2014;205(4):435–445. doi: 10.1083/jcb.201402054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasquier B. Autophagy inhibitors. Cell Mol Life Sci. 2016;73(5):985–1001. doi: 10.1007/s00018-015-2104-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng YF, Shi YH, Ding ZB, Ke AW, Gu CY, Hui B, Zhou J, Qiu SJ, Dai Z, Fan J. Autophagy inhibition suppresses pulmonary metastasis of HCC in mice via impairing anoikis resistance and colonization of HCC cells. Autophagy. 2013;9(12):2056–2068. doi: 10.4161/auto.26398. [DOI] [PubMed] [Google Scholar]
- Petherick KJ, Conway OJ, Mpamhanga C, Osborne SA, Kamal A, Saxty B, Ganley IG. Pharmacological inhibition of ULK1 kinase blocks mammalian target of rapamycin (mTOR)-dependent autophagy. J Biol Chem. 2015;290(48):28726. doi: 10.1074/jbc.A114.627778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petiot A, Ogier-Denis E, Blommaart EF, Meijer AJ, Codogno P. Distinct classes of phosphatidylinositol 3′-kinases are involved in signaling pathways that control macroautophagy in HT-29 cells. J Biol Chem. 2000;275(2):992–998. doi: 10.1074/jbc.275.2.992. [DOI] [PubMed] [Google Scholar]
- Puri P, Chandra A. Autophagy modulation as a potential therapeutic target for liver diseases. J Clin Exp Hepatol. 2014;4(1):51–59. doi: 10.1016/j.jceh.2014.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu X, Yu J, Bhagat G, Furuya N, Hibshoosh H, Troxel A, Rosen J, Eskelinen EL, Mizushima N, Ohsumi Y, Cattoretti G, Levine B. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J Clin Invest. 2003;112(12):1809–1820. doi: 10.1172/JCI20039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy JK, Rao MS. Lipid metabolism and liver inflammation. II. Fatty liver disease and fatty acid oxidation. Am J Physiol Gastrointest Liver Physiol. 2006;290(5):G852–858. doi: 10.1152/ajpgi.00521.2005. [DOI] [PubMed] [Google Scholar]
- Rogov V, Dotsch V, Johansen T, Kirkin V. Interactions between autophagy receptors and ubiquitin-like proteins form the molecular basis for selective autophagy. Mol Cell. 2014;53(2):167–178. doi: 10.1016/j.molcel.2013.12.014. [DOI] [PubMed] [Google Scholar]
- Rose C, Menzies FM, Renna M, Acevedo-Arozena A, Corrochano S, Sadiq O, Brown SD, Rubinsztein DC. Rilmenidine attenuates toxicity of polyglutamine expansions in a mouse model of Huntington’s disease. Hum Mol Genet. 2010;19(11):2144–2153. doi: 10.1093/hmg/ddq093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinsztein DC, Codogno P, Levine B. Autophagy modulation as a potential therapeutic target for diverse diseases. Nat Rev Drug Discov. 2012;11(9):709–730. doi: 10.1038/nrd3802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruhl CE, Everhart JE. The association of low serum alanine aminotransferase activity with mortality in the US population. Am J Epidemiol. 2013;178(12):1702–1711. doi: 10.1093/aje/kwt209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusten TE, Stenmark H. p62, an autophagy hero or culprit? Nat Cell Biol. 2010;12(3):207–209. doi: 10.1038/ncb0310-207. [DOI] [PubMed] [Google Scholar]
- Sakai Y, Oku M, van der Klei IJ, Kiel JA. Pexophagy: autophagic degradation of peroxisomes. Biochim Biophys Acta. 2006;1763(12):1767–1775. doi: 10.1016/j.bbamcr.2006.08.023. [DOI] [PubMed] [Google Scholar]
- Sarkar S, Floto RA, Berger Z, Imarisio S, Cordenier A, Pasco M, Cook LJ, Rubinsztein DC. Lithium induces autophagy by inhibiting inositol monophosphatase. J Cell Biol. 2005;170(7):1101–1111. doi: 10.1083/jcb.200504035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuck S, Gallagher CM, Walter P. ER-phagy mediates selective degradation of endoplasmic reticulum independently of the core autophagy machinery. J Cell Sci. 2014;127(Pt 18):4078–4088. doi: 10.1242/jcs.154716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schworer CM, Shiffer KA, Mortimore GE. Quantitative relationship between autophagy and proteolysis during graded amino acid deprivation in perfused rat liver. J Biol Chem. 1981;256(14):7652–7658. [PubMed] [Google Scholar]
- Seok S, Fu T, Choi SE, Li Y, Zhu R, Kumar S, Sun X, Yoon G, Kang Y, Zhong W, Ma J, Kemper B, Kemper JK. Transcriptional regulation of autophagy by an FXR-CREB axis. Nature. 2014;516(7529):108–111. doi: 10.1038/nature13949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Settembre C, Di Malta C, Polito VA, Garcia Arencibia M, Vetrini F, Erdin S, Erdin SU, Huynh T, Medina D, Colella P, Sardiello M, Rubinsztein DC, Ballabio A. TFEB links autophagy to lysosomal biogenesis. Science. 2011;332(6036):1429–1433. doi: 10.1126/science.1204592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibutani ST, Yoshimori T. A current perspective of autophagosome biogenesis. Cell Res. 2014;24(1):58–68. doi: 10.1038/cr.2013.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu S, Honda S, Arakawa S, Yamaguchi H. Alternative macroautophagy and mitophagy. Int J Biochem Cell Biol. 2014;50:64–66. doi: 10.1016/j.biocel.2014.02.016. [DOI] [PubMed] [Google Scholar]
- Shin HJ, Kim H, Oh S, Lee JG, Kee M, Ko HJ, Kweon MN, Won KJ, Baek SH. AMPK-SKP2-CARM1 signalling cascade in transcriptional regulation of autophagy. Nature. 2016;534(7608):553–557. doi: 10.1038/nature18014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shintani T, Klionsky DJ. Autophagy in health and disease: a double-edged sword. Science. 2004;306(5698):990–995. doi: 10.1126/science.1099993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sid B, Verrax J, Calderon PB. Role of AMPK activation in oxidative cell damage: Implications for alcohol-induced liver disease. Biochem Pharmacol. 2013;86(2):200–209. doi: 10.1016/j.bcp.2013.05.007. [DOI] [PubMed] [Google Scholar]
- Singh R, Kaushik S, Wang Y, Xiang Y, Novak I, Komatsu M, Tanaka K, Cuervo AM, Czaja MJ. Autophagy regulates lipid metabolism. Nature. 2009;458(7242):1131–1135. doi: 10.1038/nature07976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha RA, Farah BL, Singh BK, Siddique MM, Li Y, Wu Y, Ilkayeva OR, Gooding J, Ching J, Zhou J, Martinez L, Xie S, Bay BH, Summers SA, Newgard CB, Yen PM. Caffeine stimulates hepatic lipid metabolism by the autophagy-lysosomal pathway in mice. Hepatology. 2014;59(4):1366–1380. doi: 10.1002/hep.26667. [DOI] [PubMed] [Google Scholar]
- Sinha RA, You SH, Zhou J, Siddique MM, Bay BH, Zhu X, Privalsky ML, Cheng SY, Stevens RD, Summers SA, Newgard CB, Lazar MA, Yen PM. Thyroid hormone stimulates hepatic lipid catabolism via activation of autophagy. J Clin Invest. 2012;122(7):2428–2438. doi: 10.1172/JCI60580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sir D, Chen WL, Choi J, Wakita T, Yen TS, Ou JH. Induction of incomplete autophagic response by hepatitis C virus via the unfolded protein response. Hepatology. 2008;48(4):1054–1061. doi: 10.1002/hep.22464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slater AF. Chloroquine: mechanism of drug action and resistance in Plasmodium falciparum. Pharmacol Ther. 1993;57(2–3):203–235. doi: 10.1016/0163-7258(93)90056-j. [DOI] [PubMed] [Google Scholar]
- Stolz A, Ernst A, Dikic I. Cargo recognition and trafficking in selective autophagy. Nat Cell Biol. 2014;16(6):495–501. doi: 10.1038/ncb2979. [DOI] [PubMed] [Google Scholar]
- Strnad P, Zatloukal K, Stumptner C, Kulaksiz H, Denk H. Mallory-Denk-bodies: lessons from keratin-containing hepatic inclusion bodies. Biochim Biophys Acta. 2008;1782(12):764–774. doi: 10.1016/j.bbadis.2008.08.008. [DOI] [PubMed] [Google Scholar]
- Subramani S. A mammalian pexophagy target. Nat Cell Biol. 2015;17(11):1371–1373. doi: 10.1038/ncb3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun K, Guo XL, Zhao QD, Jing YY, Kou XR, Xie XQ, Zhou Y, Cai N, Gao L, Zhao X, Zhang SS, Song JR, Li D, Deng WJ, Li R, Wu MC, Wei LX. Paradoxical role of autophagy in the dysplastic and tumor-forming stages of hepatocarcinoma development in rats. Cell Death Dis. 2013;4:e501. doi: 10.1038/cddis.2013.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagi A, Kume S, Kondo M, Nakazawa J, Chin-Kanasaki M, Araki H, Araki S, Koya D, Haneda M, Chano T, Matsusaka T, Nagao K, Adachi Y, Chan L, Maegawa H, Uzu T. Mammalian autophagy is essential for hepatic and renal ketogenesis during starvation. Sci Rep. 2016;6:18944. doi: 10.1038/srep18944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takamura A, Komatsu M, Hara T, Sakamoto A, Kishi C, Waguri S, Eishi Y, Hino O, Tanaka K, Mizushima N. Autophagy-deficient mice develop multiple liver tumors. Genes Dev. 2011;25(8):795–800. doi: 10.1101/gad.2016211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H, Da L, Mao Y, Li Y, Li D, Xu Z, Li F, Wang Y, Tiollais P, Li T, Zhao M. Hepatitis B virus X protein sensitizes cells to starvation-induced autophagy via up-regulation of beclin 1 expression. Hepatology. 2009;49(1):60–71. doi: 10.1002/hep.22581. [DOI] [PubMed] [Google Scholar]
- Thoen LF, Guimaraes EL, Dolle L, Mannaerts I, Najimi M, Sokal E, van Grunsven LA. A role for autophagy during hepatic stellate cell activation. J Hepatol. 2011;55(6):1353–1360. doi: 10.1016/j.jhep.2011.07.010. [DOI] [PubMed] [Google Scholar]
- Thoen LF, Guimaraes EL, Grunsven LA. Autophagy: a new player in hepatic stellate cell activation. Autophagy. 2012;8(1):126–128. doi: 10.4161/auto.8.1.18105. [DOI] [PubMed] [Google Scholar]
- Thomes PG, Ehlers RA, Trambly CS, Clemens DL, Fox HS, Tuma DJ, Donohue TM. Multilevel regulation of autophagosome content by ethanol oxidation in HepG2 cells. Autophagy. 2013;9(1):63–73. doi: 10.4161/auto.22490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomes PG, Trambly CS, Thiele GM, Duryee MJ, Fox HS, Haorah J, Donohue TM., Jr Proteasome activity and autophagosome content in liver are reciprocally regulated by ethanol treatment. Biochem Biophys Res Commun. 2012;417(1):262–267. doi: 10.1016/j.bbrc.2011.11.097. [DOI] [PubMed] [Google Scholar]
- Thoreen CC, Kang SA, Chang JW, Liu Q, Zhang J, Gao Y, Reichling LJ, Sim T, Sabatini DM, Gray NS. An ATP-competitive mammalian target of rapamycin inhibitor reveals rapamycin-resistant functions of mTORC1. J Biol Chem. 2009;284(12):8023–8032. doi: 10.1074/jbc.M900301200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiniakos DG, Vos MB, Brunt EM. Nonalcoholic fatty liver disease: pathology and pathogenesis. Annu Rev Pathol. 2010;5:145–171. doi: 10.1146/annurev-pathol-121808-102132. [DOI] [PubMed] [Google Scholar]
- Walter KM, Schonenberger MJ, Trotzmuller M, Horn M, Elsasser HP, Moser AB, Lucas MS, Schwarz T, Gerber PA, Faust PL, Moch H, Kofeler HC, Krek W, Kovacs WJ. Hif-2alpha promotes degradation of mammalian peroxisomes by selective autophagy. Cell Metab. 2014;20(5):882–897. doi: 10.1016/j.cmet.2014.09.017. [DOI] [PubMed] [Google Scholar]
- Wang HJ, Park JY, Kwon O, Choe EY, Kim CH, Hur KY, Lee MS, Yun M, Cha BS, Kim YB, Lee H, Kang ES. Chronic HMGCR/HMG-CoA reductase inhibitor treatment contributes to dysglycemia by upregulating hepatic gluconeogenesis through autophagy induction. Autophagy. 2015a;11(11):2089–2101. doi: 10.1080/15548627.2015.1091139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Khambu B, Zhang H, Yin XM. Autophagy in alcoholic liver disease, self-eating triggered by drinking. Clin Res Hepatol Gastroenterol. 2015b;39(Suppl 1):S2–6. doi: 10.1016/j.clinre.2015.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Singh R, Xiang Y, Czaja MJ. Macroautophagy and chaperone-mediated autophagy are required for hepatocyte resistance to oxidant stress. Hepatology. 2010;52(1):266–277. doi: 10.1002/hep.23645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei H, Liu L, Chen Q. Selective removal of mitochondria via mitophagy: distinct pathways for different mitochondrial stresses. Biochim Biophys Acta. 2015;1853(10 Pt B):2784–2790. doi: 10.1016/j.bbamcr.2015.03.013. [DOI] [PubMed] [Google Scholar]
- White EJ, Martin V, Liu JL, Klein SR, Piya S, Gomez-Manzano C, Fueyo J, Jiang H. Autophagy regulation in cancer development and therapy. Am J Cancer Res. 2011;1(3):362–372. [PMC free article] [PubMed] [Google Scholar]
- Williams A, Sarkar S, Cuddon P, Ttofi EK, Saiki S, Siddiqi FH, Jahreiss L, Fleming A, Pask D, Goldsmith P, O’Kane CJ, Floto RA, Rubinsztein DC. Novel targets for Huntington’s disease in an mTOR-independent autophagy pathway. Nat Chem Biol. 2008;4(5):295–305. doi: 10.1038/nchembio.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D, Wang X, Zhou R, Yang L, Cederbaum AI. Alcohol steatosis and cytotoxicity: the role of cytochrome P4502E1 and autophagy. Free Radic Biol Med. 2012;53(6):1346–1357. doi: 10.1016/j.freeradbiomed.2012.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong X, Tao R, DePinho RA, Dong XC. The autophagy-related gene 14 (Atg14) is regulated by forkhead box O transcription factors and circadian rhythms and plays a critical role in hepatic autophagy and lipid metabolism. J Biol Chem. 2012;287(46):39107–39114. doi: 10.1074/jbc.M112.412569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamura T, Ohsaki Y, Suzuki M, Shinohara Y, Tatematsu T, Cheng J, Okada M, Ohmiya N, Hirooka Y, Goto H, Fujimoto T. Inhibition of Niemann-Pick-type C1-like1 by ezetimibe activates autophagy in human hepatocytes and reduces mutant alpha1-antitrypsin Z deposition. Hepatology. 2014;59(4):1591–1599. doi: 10.1002/hep.26930. [DOI] [PubMed] [Google Scholar]
- Yang H, Ni HM, Guo F, Ding Y, Shi YH, Lahiri P, Frohlich LF, Rulicke T, Smole C, Schmidt VC, Zatloukal K, Cui Y, Komatsu M, Fan J, Ding WX. Sequestosome 1/p62 Protein Is Associated with Autophagic Removal of Excess Hepatic Endoplasmic Reticulum in Mice. J Biol Chem. 2016;291(36):18663–18674. doi: 10.1074/jbc.M116.739821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Li P, Fu S, Calay ES, Hotamisligil GS. Defective hepatic autophagy in obesity promotes ER stress and causes insulin resistance. Cell Metab. 2010;11(6):467–478. doi: 10.1016/j.cmet.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin XM, Ding WX, Gao W. Autophagy in the liver. Hepatology. 2008;47(5):1773–1785. doi: 10.1002/hep.22146. [DOI] [PubMed] [Google Scholar]
- Yue Z, Jin S, Yang C, Levine AJ, Heintz N. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc Natl Acad Sci U S A. 2003;100(25):15077–15082. doi: 10.1073/pnas.2436255100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakhari S, Li TK. Determinants of alcohol use and abuse: Impact of quantity and frequency patterns on liver disease. Hepatology. 2007;46(6):2032–2039. doi: 10.1002/hep.22010. [DOI] [PubMed] [Google Scholar]
- Zatloukal K, French SW, Stumptner C, Strnad P, Harada M, Toivola DM, Cadrin M, Omary MB. From Mallory to Mallory-Denk bodies: what, how and why? Exp Cell Res. 2007;313(10):2033–2049. doi: 10.1016/j.yexcr.2007.04.024. [DOI] [PubMed] [Google Scholar]
- Zeng T, Zhang CL, Song FY, Zhao XL, Yu LH, Zhu ZP, Xie KQ. PI3K/Akt pathway activation was involved in acute ethanol-induced fatty liver in mice. Toxicology. 2012;296(1–3):56–66. doi: 10.1016/j.tox.2012.03.005. [DOI] [PubMed] [Google Scholar]
- Zhang J. Autophagy and Mitophagy in Cellular Damage Control. Redox Biol. 2013;1(1):19–23. doi: 10.1016/j.redox.2012.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zirin J, Nieuwenhuis J, Perrimon N. Role of autophagy in glycogen breakdown and its relevance to chloroquine myopathy. PLoS Biol. 2013;11(11):e1001708. doi: 10.1371/journal.pbio.1001708. [DOI] [PMC free article] [PubMed] [Google Scholar]


