Abstract
We present a method for using measurements of membrane voltage in individual neurons to estimate the parameters and states of the voltage-gated ion channels underlying the dynamics of the neuron’s behavior. Short injections of a complex time-varying current provide sufficient data to determine the reversal potentials, maximal conductances, and kinetic parameters of a diverse range of channels, representing tens of unknown parameters and many gating variables in a model of the neuron’s behavior. These estimates are used to predict the response of the model at times beyond the observation window. This method of data assimilation extends to the general problem of determining model parameters and unobserved state variables from a sparse set of observations, and may be applicable to networks of neurons. We describe an exact formulation of the tasks in nonlinear data assimilation when one has noisy data, errors in the models, and incomplete information about the state of the system when observations commence. This is a high dimensional integral along the path of the model state through the observation window. In this article, a stationary path approximation to this integral, using a variational method, is described and tested employing data generated using neuronal models comprising several common channels with Hodgkin–Huxley dynamics. These numerical experiments reveal a number of practical considerations in designing stimulus currents and in determining model consistency. The tools explored here are computationally efficient and have paths to parallelization that should allow large individual neuron and network problems to be addressed.
Keywords: Data assimilation, Neuronal dynamics, Ion channel properties
1 Introduction
Networks of neurons exhibit complex dynamical behaviors, including rhythmic bursting and patterned sequence generation (Stein et al. 1997; Laurent et al. 2001; Johnston and Wu 1995; Koch 1999). These dynamics derive from the intrinsic properties of individual neurons and from properties arising from excitatory and inhibitory connections within the network. The membrane voltage of a neuron depends on currents from a diverse collection of ion channels, many of which have nonlinear voltage-dependent dynamics (Johnston and Wu 1995). General forms for the dynamics of many of the major families of ion channels have been characterized (Brette et al. 2007; Graham 2002), but the kinetic parameters vary from isoform to isoform. Small differences in the complement and densities of the channel isoforms expressed by a neuron can lead to widely divergent physiological behavior. This sensitivity may be significant functionally but also complicates determining channel properties experimentally.
Patch-clamp recording techniques (Hamill et al. 1981) allow low-impedance intracellular injections of current while simultaneously measuring membrane potential. Depolarizing and hyperpolarizing currents can reveal much of a neuron’s response properties, but due to the large variety of channels expressed in most neurons, determining the contribution of specific channel types usually requires intracellular or extracellular pharmacological manipulation. The efficacy and specificity of the pharmacological agents is often a concern, and data typically have to be pooled from many neurons, possibly obscuring differences among individual cells. Furthermore, such pharmacological manipulations require numerous experimental steps and lengthy recording durations, which often has the practical implication of limiting analysis to exceptional recordings fortuitously maintained in a healthy condition for an extended period of time. Based on these results, biophysical Hodgkin–Huxley (HH) models of the neurons can be constructed, with the parameters fixed by experimental results or chosen based on simulation and iterative search (Brette et al. 2007).
This article describes a method for directly inferring the channel parameters and unobserved states of HH-type neuronal models using injections of complex current waveforms often less than a second in duration, allowing insight into a cell’s complement of ionic channels prior to or in lieu of any pharmacological manipulations. Beginning with a discussion of the overall problem of state and parameter estimation in nonlinear systems, we give a formally exact framework for assimilating data into models in order to determine the set of parameters and initial conditions likely to have given rise to the data (Evensen 2009). Based on a variational approximation to the exact formulation we construct an efficient algorithm for performing this task.
We test the utility of the method through “twin experiments,” where a biophysical neuronal model is used to generate data and then subsequently used to see if the parameters and underlying states can be accurately recovered through the estimation procedure in this article. We demonstrate that the procedure is robust to additive noise and model errors, and also we discuss what features of the stimulus current are necessary to obtain good estimates. For application to experimentally obtained data, where the underlying states are unknown, we describe tests of model consistency and validity. The methods we outline and explore here have wide applicability to equivalent questions in areas of inquiry well beyond neurobiology.
1.1 Ion channel models of neuron dynamics
Biophysical models of neuronal dynamics are based on the flux of ions through channels or different types and densities inserted into an impermeable membrane. Many of these channels open and close in a voltage-dependent manner, so the models consist of an equation of current conservation determining the voltage across the cell membrane V(t) as well as a set of state variables ai (t) describing the voltage-dependent permeability of each type of channel. These equations are described more fully in Sect. 2.2. More complex models may have multiple compartments with different densities and complements of channels, additional state variables for the concentration of intracellular signals like calcium, and synapses with channels gated by extra cellular ligands. Here, we focus our attention on single-compartment models of individual neurons to explore the methods and the numerical requirements of estimating states and parameters in a neurobiological context. To extend these methods, more complex settings may require richer models and additional data.
We consider the most common case, where the only measurement an experimenter makes of intracellular dynamics is the membrane voltage. (We note that optical proxies for intracellular concentrations and channel state continue to be developed.) A stimulus current Iapp(t) is applied through the recording electrode to change the polarity of the membrane and probe the responses of voltage-gated channels. Thus, a data set usually consists of the applied current and the observed voltage over some observation window t = [t0, tm = T ] with measurements made at times tn = {t0, t1, …, tm} within that window.
Working solely with observations of the membrane voltage and kinetic models of the channels, the goal is to estimate the parameters of the model, including maximal conductance, intracellular ion concentrations, and channel gating kinetics—as well as the unobserved gating dynamics ai (t)—throughout the observation window t = T. Knowledge of the quantities V(T ) and all of the ai (T ) along with estimates of all fixed parameters allow us to use the model to predict the response of the neuron for t > T given Iapp(t > T ). If these predictions are accurate for a broad range of biologically plausible stimuli, then the estimates of the model parameters provide a parsimonious, biophysically interpretable description of the neuron’s behavior. Furthermore, such results would motivate the hypothesis that the model neurons will respond accurately to a diverse set of stimuli when used in models of interesting functional networks.
2 Methods
2.1 General framework
In an experimental preparation (individual neuron or network of neurons), we have L observed quantities (i.e., membrane voltages) at times tn in the window [t0, tm = T ]. We label these observations yl (tn) = yl (n); l = 1, 2, …, L; n = 0, 1, …, m.
Using our ideas about biophysical processes associated with the preparation, we construct a model comprising a D-dimensional state vector including the voltage V(t) and the gating variables ai (t); i = 1, 2, …, D − 1. For brevity, we denote the state variables {V (t), ai (t)} as xa(tn) = xa(n); a = 1, 2, …, D. We also establish, as part of the model of the experiment, a connection between the state of the neuron x(n) and observed quantities. The observation function of the neuron state x(n), hl (x(n)); l = 1, 2, …, L, should accurately yield the observations yl (n), if the model is ‘good.’ In both the statement of the dynamical model of the neuron and the observation functions hl (x) there may be unknown parameters. The collection of parameters we denote as p.
The model comprises a rule in discrete time taking the state at time tn = n to the state at time n + 1: xa(n + 1) = fa(x(n), p) along with the observation functions hl (x(n)). The discrete time rule may be the specific formulation of a numerical solution algorithm for an underlying set of differential equations.
Using the observations yl (n) of a subset of the state variables we wish to estimate the full collection of state variables x(n) at times in the observation window, in particular at the end of the measurements t = T. With x(T ), the estimated parameters p, and knowledge of the stimulus for t > T we may use the model to predict the behavior for t > T. Our goal is to achieve yl (t) ≈ hl (x(t)) for t > T, as these are the observed quantities.
These requirements raise a number of practical and theoretical questions:
How many measurements L ≤ D are required? Typically for single neurons L = 1 (voltage), and given that most of the states of the system will remain unmeasurable, L ≪ D. Equivalently, given that observations are generally limited to V(t), is there an upper limit to the complexity of the models that can be completed from the data?
How often is it necessary to make measurements? Namely, given a fixed observation window [t0, …, tm = T ], how frequently should the measurable variables be sampled? How large can one choose the intervals between observations, tn+1 − tn?
Given that measurements are noisy and models are always wrong in some respect, how robust are the estimates of parameters and unobserved state variables to these errors?
What kinds of stimuli lead to adequate exploration of the state space of the model so that all the parameters and unobserved state variables can be estimated with a similar degree of confidence?
What metrics are most appropriate for testing the validity of a neuronal model, especially in light of the intrinsic variability of real neurons?
We do not suggest we have full answers to these questions. However, in this article and others in the references, we explore them within the context of various examples. The examples in this article are directed toward neurobiological instantiations of these questions.
In this article, the data are simulated from a model which has the form of a set of ordinary differential equations (Sect. 2.2). After choosing a realistic set of parameters and initial states, we generate a solution to the equations. We select a subset of the output, here the voltage alone, as observed quantities y(t) and, after adding noise, attempt to fit the model, with undetermined parameters, to the data. This process is called data assimilation following the name given in the extensive and well developed geophysical literature on this subject (Evensen 2009).
2.2 Neuron models
The first and simplest model we address consists of the dynamics of membrane voltage V(t) driven by two voltage-gated ion channels, Na and K, a ‘leak’ current and an external, applied current we call Iapp(t). The dynamics of the single compartment model comprise the usual equations for voltage
| (1) |
where the indicate maximal conductances and the reversal potentials, for each of the Na, K, and leak channels. IDC is a DC current, and Iapp(t) is an applied external current selected in an experiment. We refer to this as the NaKL HH model.
The gating variables ai (t) = {m(t), h(t), n(t)} are discussed in many textbooks and reviews (Johnston and Wu 1995; Brette et al. 2007; Graham 2002), and each satisfies a first order kinetic equation of the form
| (2) |
The kinetic terms ai0(V) and τi (V) are taken here in the form
| (3) |
The constants va, dva, … are selected to match the familiar forms for the kinetic coefficients usually given in terms of sigmoidal functions (1 ± exp((V − V1)/V2))−1. The tanh forms are numerically the same over the dynamic range of the neuron models but have better controlled derivatives when one goes out of that range during the required search procedures.
In the language used in our general formulation, the model state variables are the x(t) = {V(t), m(t), h(t), n(t)}, and the parameters are p = {C, gNa, ENa, gK, EK, …, dvat}. In a twin experiment, the data are generated by solving these HH equations for some initial condition x(0) = {V(0), m(0), h(0), n(0)} and some choice for the parameters, the DC current and Iapp(t), the stimulating current. The data presented to the model y(t) consist of the voltage alone V(t) alone along with additive noise. The measurement function h(x(t)) is here just the voltage V(t) produced by the model.
2.3 Data assimilation
The data presented to the model consist of noisy measurements made at times {t0, t1, …, tm = T }. We wish to determine the parameters p and the state of the neuron model x(tm) = x(m) = x(T ) at the end of the observation window. The parameters are independent of the external currents IDC and Iapp(t), while x(m) is dependent on them. If we know p, x(m) = x(T ) and the external currents for t > tm = T, we can predict the response of the model neuron.
The first step is to specify a cost function, which quantifies the ‘distance’ between the model output V(t) which is a function of x(0) and the p, and the observations y(t) to which we wish it to correspond. We choose a least squares distance
| (4) |
and we seek to minimize this cost function with respect to the x(0) and the p subject to the HH Eqs. (1) and (2). The assumption is that this will yield accurate estimates of p and values of x(tn) though the observation window. Unless there are some symmetries among the parameters leaving the solutions invariant, this is likely to be the case.
If we know x(0) then using the model equations, we may evaluate x(n); n = 1, 2, …, m. In the cases, we discuss in this article that may be quite a stable procedure. However, in a nonlinear problem which has chaotic trajectories, small errors in x(0) can lead to large errors in x(n > 0). This leads us to use the so-called direct method for performing the minimization indicated here: this treats all values of x(n) and all parameters as variables in the minimization process, subject to equality constraints enforcing the dynamics across each time interval tn+1−tn (Gill et al. 1998). The values of x(n) = x(tn) come directly from the optimization procedure, and one does not have to contend with growing errors in x(0) to reach the end of the observation period {t0, t1, …, tm} where accurate x(tm) are required for accurate forecasts. The details of this approach are discussed in Appendix II. Justification for using the least squares cost function is presented in Appendix I where the exact formulation of data assimilation tasks is outlined. Huys et al. (2006) provide methods for linear estimation and summarize earlier studies of the problems we address. That earlier study does not recognize the variational principle as a first approximation to the exact integral representation of data assimilation questions as it outlined in Appendix I.
2.3.1 Control term in cost function
When minimizing cost functions such as Eq. 4 we have shown there can be multiple local minima in C(x(0), p) associated with stability properties of the nonlinear equation (Creveling et al. 2008; Abarbanel et al. 2009). To regulate this unwanted phenomena, we argued that one can utilize an external control added to the differential equation for the measured state variable. In our case, we would add to the HH voltage equation a term u(t)(y(t) − V(t)); u(t) ≥ 0. The time dependence of u(t) is introduced because the magnitude of this ‘control’ required to accomplish V(t) ≈ y(t) is not uniform along the orbit of the model in its state space. The role of the control u(t) is to drive the solution of the model equation V(t) to the observations y(t). Indeed, as u(t) becomes large, V(t) ≈ y(t) with an error of order u(t)−1.
However, the control u(t) is not part of the biophysics of the HH model, and while we may use it during the estimation procedure, it should be absent when we have concluded the data assimilation task. Certainly, we make forecasts with u(t) = 0. To accomplish this, we argued that one should add to the cost function a term (a cost) for the control. This should vanish as the model and the data synchronize V(t) ≈ y(t).
Many such ‘costs’ can be explored, however, the simple term u(t)2 works well (Crevelingetal. 2008). This now changes the optimization problem seeking to estimate fixed parameters and unobserved state variables to the minimization of
| (5) |
subject to
| (6) |
along with unchanged kinetic equations for the gating variables. We have called this general procedure dynamical state and parameter estimation (Abarbanel et al. 2009).
3 Results
The optimization procedure was initially applied to data generated using the NaKL model (Sect. 2.2), which has four state variables and 26 fixed parameters (Table 1). The applied current waveform was taken from the output of a small chaotic system providing a spectrum of frequencies in a band approximating 50% of the frequencies in the neuron model voltage output. The time scale of the current ρI is defined relative to the time step for the integration of the model (here 0.01 ms). Initially no additional noise was added to the voltage, so any error is associated with numerical round off and the signal to noise ratio is large.
Table 1.
Parameters in NaKL HH model: Na, K, and leak currents
| Name | Value | Name | Value |
|---|---|---|---|
| C | 1.0μF/cm2 | vh | −60.0mV |
| gNa | 120.0mS/cm2 | dvh | −15.0mV |
| ENa | 50.0mV | th0 | 1.0ms |
| gK | 20.0mS/cm2 | th1 | 7.0ms |
| EK | −77.0mV | vht | −60.0mV |
| gL | 0.3mS/cm2 | dvht | −15.0mV |
| EL | −54.4mV | vn | −55.0mV |
| vm | −40.0mV | dvn | 30.0mV |
| dvm | 15.0mV | tn0 | 1.0ms |
| tm0 | 0.1ms | tn1 | 5.0ms |
| tm1 | 0.4ms | υnt | −55.0mV |
| vmt | −40.0mV | dvnt | 30.0mV |
| dvmt | 15.0mV | IDC | 7.3pA/cm2 |
The idea of using ρI is to give a rough measure of the time scale on which a selected stimulus waveform is sampled. Once we have chosen a waveform—in particular its dynamical range of amplitude variation so all depolarized and hyperpolarized dynamics are stimulated—then we need to select how much it is compressed—its frequency is raised— or expanded—its frequency is lowered—relative to the frequency of activity of the spiking response of the neuron to which the stimulus is presented. We have selected ρI as described so that when it is small, the frequency of variation of the stimulus is slow compared to the neuron response spiking, and vice versa.
The cost function in Eq. 5 was minimized while allowing the state variables and 18 of the parameters to be free. We selected some of the parameters to be equal, for example vm = vmt reducing the possible 26–18 because no change in the overall dynamical performance of the model neuron was changed by this. This made the example neurons used here a bit simpler without sacrificing any generality in the biophysics of the model. We also performed the same kind of twin experiments reported on here with all 26 parameters to be determined, and that worked just as well as the results we report.
The membrane capacitance and IDC were fixed, and some of the parameters were constrained to be equal. We used 9000 data points spaced at dtM = 0.01 ms, corresponding to 90 ms of data, and used observations at each time step. Table 2 shows the estimated values for each of the parameters in comparison to the values used to generate the data. ρI = 0.5 in these calculations, i.e., the step size was twice as big for the injected current as was the integration time of the model. The estimated values of the parameters are quite accurate.
Table 2.
Parameters in ‘Data’ and estimated parameters in NaKL HH model; ρI = 0.5
| Parameter | Data | Estimate | Units |
|---|---|---|---|
| gNa | 120 | 118.79 | mS/cm2 |
| ENa | 50.0 | 49.99 | mV |
| gK | 20 | 20.35 | mS/cm2 |
| EK | −77.0 | −76.97 | mV |
| gL | 0.3 | 0.2955 | mS/cm2 |
| EL | −54.4 | −54.11 | mV |
| vm = vmt | −40.0 | −40.08 | mV |
| dvm=dvmt | 15.0 | 14.90 | mV |
| tm0 | 0.1 | 0.1009 | ms |
| tm1 | 0.4 | 0.3982 | ms |
| vh=vht | −60.0 | −59.91 | mV |
| dvh=dvht | −15.0 | −14.88 | mV |
| th0 | 1.0 | 1.004 | ms |
| th1 | 7.0 | 7.047 | ms |
| vn=vnt | −55.0 | −54.91 | mV |
| dvn=dvnt | 30.0 | 29.28 | mV |
| tn0 | 1.0 | 1.012 | ms |
| tn1 | 5.0 | 4.99 | ms |
IDC and C were fixed in all calculations
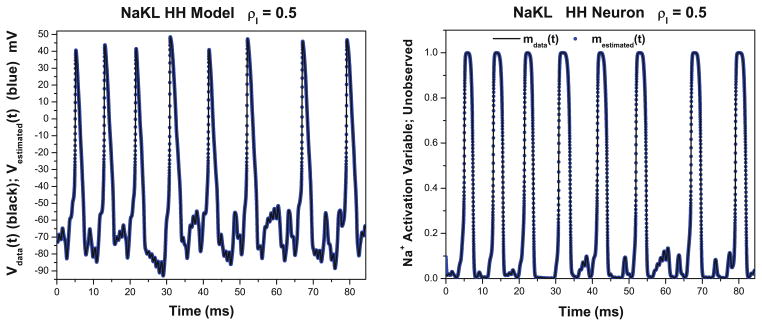
As shown in Fig. 1 Left, the estimated values for the membrane voltage are also very close to the true values. This is unsurprising, because these are the data to which the model output was compared, and the control term u(t) allows the estimation to be nearly perfect even if the dynamics of the estimated model have errors.
Fig. 1.
NaKL model. Left We display the membrane voltage Vdata(t) from ‘data’ generated in a twin experiment using the NaKL model as a data source. Also we display the estimated membrane voltage Vestimated(t) using the dynamical state and estimation approach. The frequency scale of the stimulus to the neuron response is ρI = 0.5. Right We display the Na+ activation gating variable mdata(t) from ‘data’ generated in a twin experiment using the NaKL model as a data source. Also we display the estimated membrane voltage Na+ activation gating variable mestimated(t) using the dynamical state and estimation approach. The frequency scale of the stimulus to the neuron response is ρI = 0.5. In a laboratory experiment, m(t) would be an unobserved state variable. In this twin experiment, the unobserved variables are known to us and comparison is possible
More importantly, the estimates of the unobserved state variables, which are not directly passed to the model during the optimization, also closely approximate the true values in the simulated data Fig. 1 Right. We display here only the Na+ activation variable m(t), but the accuracy shown here is also reflected in the estimates of h(t) and n(t). In effect, the model acts as a nonlinear filter using data on the observed variables through the estimation procedure to estimate the state variables that are unobserved. More specific interpretations are appropriate for the neurobiological setting of the example problem, but this description is of more general utility.
3.1 Testing model consistency
As noted earlier, an excellent estimation for observed state variables and an associated small C(x(0), p) could result from the action of the control variable u(t) introduced into the regulated data assimilation procedure, because large values of u(t)would drive the model output V (t) to the observed data y(t). u(t) is also determined by the numerical optimization task in Eqs. 5 and 6. This provides a useful consistency test, because we can examine the time course of the control variable to determine when the dynamics of the fitted model fail to match the data. We define a dimensionless ratio
| (7) |
which measures the importance of the control term u(t) in the dynamical equation for voltage compared to the usual HH term FV (V(t), m(t), h(t), n(t)). At the end of the estimation, procedure given an optimization problem in Eqs. 5 and 6 we use the values of V(t), ai (t), and u(t) over the observation interval to evaluate R(t).
R(t) lies between zero and unity. When it is close to one, it indicates that the role of the control term in achieving a good estimation, or small cost function, is not important and the model is consistent with the data presented to it. Excursions of R(t) toward values near zero indicate increasing inconsistency of the model with the observed data. R(t) ≈ 1 does not demonstrate that the model is valid, just that it is consistent with the data presented to it. In the example of voltage data from the NaKL HH model presented to itself, R(t) deviates from unity by order 10−6 or less across the observation window (not shown).
The importance of the use of the control term u(t) in the numerical accuracy of the state and parameter estimations was assessed by performing the calculation that led to Table 2 with precisely the same model, the same data, the same stimulus, and the same algorithm for nonlinear estimation, but setting u(t) = 0.0 throughout. This result is presented in Table 3 where we see clearly that the overall quality of the estimation is degraded. This suggests that the use of the control variable u(t) during the data assimilation will prove a good strategy even when the impediments of chaotic behavior are absent.
Table 3.
Parameters in ‘Data’ and estimated parameters in NaKL HH model; ρI = 0.5
| Parameter | Data | Estimate | Units |
|---|---|---|---|
| gNa | 120 | 100.0 | mS/cm2 |
| ENa | 50.0 | 50.71 | mV |
| gK | 20 | 10.0 | mS/cm2 |
| EK | −77.0 | −115.0 | mV |
| gL | 0.3 | 0.262 | mS/cm2 |
| EL | −54.4 | −75 | mV |
| vm = vmt | −40.0 | −57.9 | mV |
| dvm=dvmt | 15.0 | 0.378 | mV |
| tm0 | 0.1 | 0.2368 | ms |
| tm1 | 0.4 | 7.34 × 10−14 | ms |
| vh=vht | −60.0 | −58.13 | mV |
| dvh=dvht | −15.0 | −0.47 | mV |
| th0 | 1.0 | 0.48 | ms |
| th1 | 7.0 | 3.43 | ms |
| vn=vnt | −55.0 | −63.06 | mV |
| dvn=dvnt | 30.0 | 55.0 | mV |
| tn0 | 1.0 | 1.00 | ms |
| tn1 | 5.0 | 0.285×10−12 | ms |
No coupling of observed data into the dynamical equations: u(t) = 0.0. In all other aspects, this is the same calculation as displayed in Table 2
3.2 Frequency content of the stimulus Iapp(t)
Intuitively, proper estimation of the parameters of the model will depend on providing a stimulus current that adequately explores regions of the state space where different subsets of the channels are opened or closed. If the frequency content of the stimulus is such that it fails to spend sufficient time in these regions the fit may be poor. To test this, the relative frequency of the stimulus current ρI was increased by selecting the same waveform for Iapp(t) and sampling it more frequently. In the initial experiment ρI was 0.5, which means it was relatively slow in comparison to the dynamics of the model. We found that ρI could be increased up to about 5.0 without compromising the parameter and state estimates. However, at ρI ≈ 10.0, these estimates became of low quality (Table 4).
Table 4.
Parameters in ‘Data’ and estimated parameters in NaKL HH model; ρI = 10.0
| Parameter | Data | Estimate | Units |
|---|---|---|---|
| gNa | 120.0 | 100.0 | mS/cm2 |
| ENa | 50.0 | 50.0 | mV |
| gK | 20.0 | 14.6 | mS/cm2 |
| EK | −77.0 | −115.0 | mV |
| gL | 0.3 | 1.23 × 10−13 | mS/cm2 |
| EL | −54.4 | −58.22 | mV |
| vm = vmt | −40.0 | −47.78 | mV |
| dvm=dvmt | 15.0 | 30.0 | mV |
| tm0 | 1.0 | 0.636 | ms |
| tm1 | 0.4 | 3.1 | ms |
| vh=vht | −60.0 | −83.72 | mV |
| dvh=dvht | −15.0 | −6.5 | mV |
| th0 | 1.0 | 1.602 | ms |
| th1 | 7.0 | 12.0 | ms |
| vn=vnt | −55.0 | −52.04 | mV |
| dvn=dvnt | 30.0 | 13.80 | mV |
| tn0 | 1.0 | 7.25 | ms |
| tn1 | 5.0 | 15.0 | ms |
In all other aspects, this is the same calculation whose results are reported in Table 2
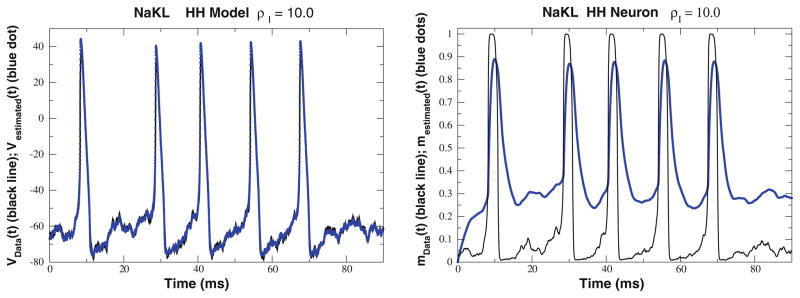
This experiment provides a useful demonstration of the utility of the consistency test. In Fig. 2 Left, the estimated membrane voltage is compared to the known Vdata(t) when ρI = 10.0. This plot alone is misleading, however, as seen when the unobserved state variables are compared to their estimated values as shown in Fig. 2 Right. Thus, by changing ρI from 0.5 to 10.0, we have lost the capability of the model to transfer information from the observations to an unobserved state variable.
Fig. 2.
NaKL Model. Left We display the membrane voltage Vdata(t) from ‘data’ generated in a twin experiment using the NaKL model as a data source. Also we display the estimated membrane voltage Vestimated(t) using the dynamical state and estimation approach. The frequency scale of the stimulus relative to the neuron response is ρI = 10.0. The apparent good fit results from the large magnitudes of the control u(t). Right We display the Na+ activation gating variable mdata(t) from ‘data’ generated in a twin experiment using the NaKL model as a data source. Also we display the estimated membrane voltage Na+ activation gating variable mestimated(t) using the dynamical state and estimation approach. The frequency scale of the stimulus to the neuron response is ρI = 10.0. In a laboratory experiment, m(t) would be an unobserved state variable
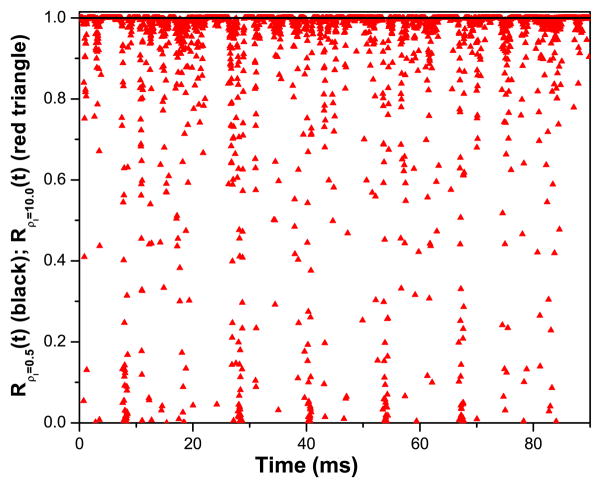
In an experiment on physical neurons, we cannot check how well unobserved variables are estimated by our approach, but we can still test whether the fitted model is consistent with the data. As seen in Fig. 3, R(t) deviates significantly from 1.0 when ρI = 10.0. As discussed above this indicates that with this higher frequency stimulus, our model is not consistent with the data, because the control term u(t) cannot be reduced to zero. This result also helps to emphasize that seemingly good fits of observed variables Vdata(t) likely have little biological significance if the control term is not reduced to zero.
Fig. 3.
The consistency check for the NaKL model in two twin experiments. They differ only in the frequency of the stimulus current Iapp(t) compared to that of the model. We display results for R(t) both for ρI = 0.5 (solid line) and ρI = 10.0 (triangles). When R(t) is near unity, the model is consistent with the data, including properties of the stimulus. When R(t) deviates significantly from one, the model is inconsistent with the data. By increasing the frequency content of the stimulus, we have reached a setting where, through its own RC filtering, the model has removed information from the stimulus. The message is that in experimental protocols, one must attend to the ratio of model and stimulus frequencies
3.3 Additive noise in the observations
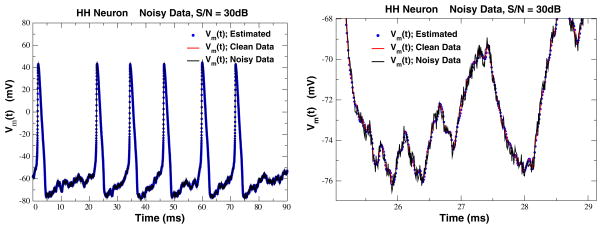
In the calculations presented thus far on the NaKL model, we have allowed only numerical roundoff error to contaminate the ‘observations’ y(t). To assess the role of noise in measurements, which will eventually degrade any state and parameter estimation procedure, we added noise drawn from a uniform distribution to y(t), reducing the signal to noise ratio to 30 dB. As shown in Fig. 4, the added noise does not compromise the data assimilation procedure. In fact, the estimated voltage is much closer to the original ‘clean’ data than to the noisy data presented to the model. This phenomenon has been seen in some of our earlier study (Abarbanel et al. 2011) and discussed there at more length. Here, we simply observe that for modestly noisy data the dynamical state and parameter estimation approach succeeds in resolving a clean signal uncontaminated by instrumental or environmental addition of noise. Earlier calculations suggested that until the signal to noise ratio decreased to about 20 dB, the procedure remained successful.
Fig. 4.
Left Estimated membrane voltage (dots) along with original clean data (grey line) and the noisy voltage data (solid line) presented to the NaKL model in the dynamical data assimilation approach. Here the signal to noise ratio is 30 dB, and, as above, ρI = 0.5. Right A blowup of a section of Fig. 4 showing how the estimation procedure has removed much of the noise added to the original clean data. The model acts here as a nonlinear noise filter. See Abarbanel et al. (2011) for a longer discussion of this effect
In neurobiological experiments making intracellular recordings, a noise floor in the range of 0.5mV is commonly achieved. In favorable recordings, activity results in signals of up to 100mV and certainly at least 40–60mV, so the signal to noise ratio required for accurate modeling is easily achieved. Smaller potentials representing input to a cell from other cells are often in the range of 4mV, however, so this may represent a signal to noise ratio problem when we extend these techniques to modeling interactions between neurons in a network.
3.4 Additional currents: Ih
Most neurons express a number of voltage-gated channels in addition to the sodium and potassium channels directly responsible for action potential generation (Graham 2002). These additional channels contribute to bursting, firing rate adaptation, and other behaviors. Determining which channels are expressed in a population of neurons typically requires molecular or pharmacological data, but the data assimilation procedure we describe here has the potential to infer which channels contribute to a neuron’s response properties. As a first step, we expanded the NaKL model to include the Ih current, which has slower kinetics than the other channels in the model, and is activated by hyperpolarization. Similar results should obtain for other fast-gated channels. We do not discuss voltage-gated Ca2+ currents or Ca2+ activated K currents. These require a model of intracellular Ca2+ concentration which has slow kinetics causing the differential equations to become stiff. The importance of these currents in neurobiological characterization is undeniable, and clearly one future step is to address the building and testing of model with their participation.
The inclusion of an additional current in the model allows us to address several questions that represent our first attempts to evaluate if the data assimilation procedure can indeed infer the existence of specific channel types:
If one presents a stimulus that does not activate Ih strongly, but produces spikes, can we still evaluate the biophysical characteristics of Ih (or other currents acting at hyperpolarized voltages) ?
If data from a more complex model including Ih (NaKLh) are used to estimate states and parameters in a simpler (NaKL) model, are the parameter estimates still accurate?
If data from a model that lacks an Ih or other current (NaKL) are presented to amore complex model (NaKLh), will the dynamical data assimilation procedure inform us that Ih is absent?
The Ih current was represented by an additional term in the HH voltage equation,
| (8) |
as well as an additional equation for the dynamics of the Ih gating variable
| (9) |
3.5 NaKLh ‘twin experiments’
Data were simulated from the NaKLh model using the parameter values in Table 1 and in Table 5. We now have five state variables {V(t), m(t), h(t), n(t), hc(t)} and 26 unknown parameters.
Table 5.
Parameters in NaKLh HH model: Na, K, leak, and Ih currents
| Name | Value | Name | Value |
|---|---|---|---|
| gh | 1.21mS/cm2 | thc0 | 0.1 ms |
| Eh | −40.0mV | thc1 | 193.5 ms |
| vhc | −75.0mV | vhct | −80 mV |
| dvhc | −11.0mV | dvhct | 21.0 mV |
We used two different stimulation currents, with the same ρI and shape as earlier, but with different amplitudes. The ‘strong’ stimulus was 1.85 times the amplitude of the ‘weak’ stimulus. The NaKLh model was then fit separately to the data from each of these simulations. As shown in Table 6 the estimates for all the parameters in the NaKL model are accurate using either stimulus, but only the data for the strong stimulus gives good estimates of the parameters for the Ih current. This suggests that the weaker stimulus did not adequately explore the dynamics associated with the Ih current. As shown in Fig. 5, the estimated voltage and state variables are good fits for all the currents when the stimulus is strong. When the stimulus is weak, the estimated voltage and state variables for the NaKL model are still good, but the estimated state associated with the Ih current is a poor fit (Fig. 6).
Table 6.
Parameters in a NaKLh HH Model where the maximal conductance of the Ih current from two different amplitudes of the stimulus current
| Parameter | Data | Estimate
|
Units | |
|---|---|---|---|---|
| Strong | Weak | |||
| gNa | 120 | 120.27 | 119.88 | mS/cm2 |
| ENa | 55.0 | 54.98 | 54.993 | mV |
| gK | 20 | 20.148 | 20.148 | mS/cm2 |
| EK | −77.0 | −77.013 | −77.019 | mV |
| gL | 0.3 | 0.30675 | 0.29851 | mS/cm2 |
| EL | −54.4 | −54.706 | −54.121 | mV |
| vm = vmt | −34.0 | −33.983 | −33.994 | mV |
| dvm=dvmt | 34.0 | 33.996 | 33.985 | mV |
| tm0 | 0.01 | 0.01023 | 0.01013 | ms |
| tm1 | 0.5 | 0.50022 | 0.50045 | ms |
| vh=vht | −60.0 | −59.991 | −59.961 | mV |
| dvh=dvht | −19.0 | −18.982 | −18.973 | mV |
| th0 | 0.2 | 0.19885 | 0.19993 | ms |
| th1 | 8.5 | 8.5163 | 8.5078 | ms |
| vn=vnt | −65.0 | −64.834 | −64.94 | mV |
| dvn=dvnt | 45.0 | 44.966 | 45.147 | mV |
| tn0 | 0.8 | 0.80137 | 0.80155 | ms |
| tn1 | 5.0 | 5.0123 | 4.9932 | ms |
| gh | 1.21 | 1.2274 | 1.000 | mS/cm2 |
| Eh | −40.0 | −39.359 | −32.745 | mV |
| vhc | −75.0 | −76.119 | −60.996 | mV |
| dvhc | −11.0 | −11.122 | −7.6066 | mV |
| thc0 | 0.1 | 0.09632 | 0.1224 | ms |
| thc1 | 193.5 | 190.26 | 67.750 | ms |
| vhct | −80 | −81.063 | −59.644 | mV |
| dvhct | 21.0 | 20.781 | 18.279 | mV |
The “strong” stimulus is 1.85 times in amplitude of the “weak” stimulus. Only the “strong” stimulus hyperpolarizes the neuron sufficiently to activate the Ih current. The “weak” stimulus only weakly activates Ih and the reduced information on the activity of Ih leads to reduced accuracy in estimating the parameters of Ih
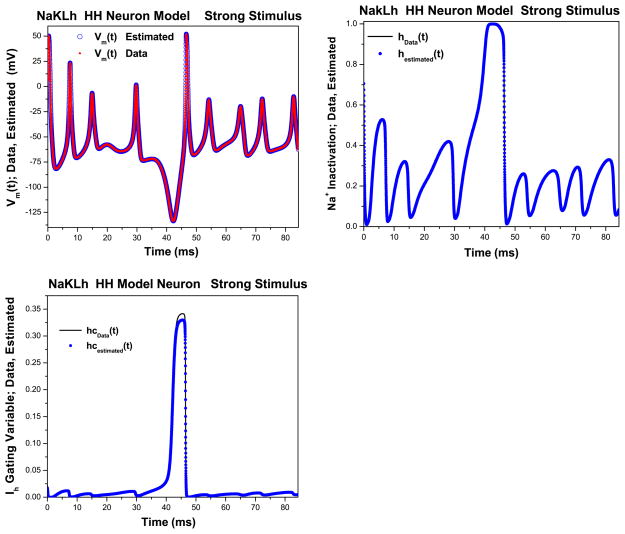
Fig. 5.
NaKLh twin experiments: strong stimulus. Left Estimated membrane voltage (circles) along with original data (triangles) as presented to the NaKLh model. The stimulus current was taken as usual from a chaotic system. This result uses the ‘strong’ stimulus which excites both polarized state variables and hyperpolarized state variables. Right Estimated Na+ inactivation gating variable (circles) along with original data (solid line) as presented to the NaKLh model. The stimulus current was taken as usual from a chaotic system. This result uses the ‘strong’ stimulus which excites both polarized state variables and hyperpolarized state variables; as above. Bottom Estimated Ih activation gating variable (circles) along with original data (solid line) as presented to the NaKLh model. The stimulus current was taken as usual from a chaotic system. This result uses the ‘strong’ stimulus which excites both polarized state variables and hyperpolarized state variables. ρI = 0.5
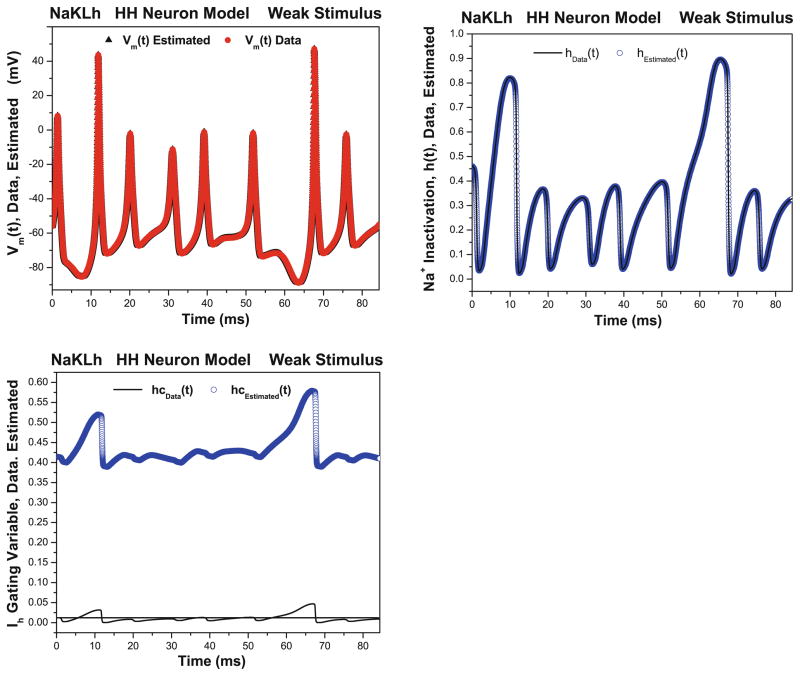
Fig. 6.
NaKLh twin experiments: weak stimulus Left Estimated membrane voltage (triangles) along with original data (circles) as presented to the NaKLh model. The stimulus current was taken as usual from a chaotic system. This result uses the ‘weak’ stimulus which excites polarized state variables. The ‘weak’ stimulus provides sufficient excitation to the Na and K channels to allow accurate estimation of their properties. Right Estimated Na+ inactivation gating variable (circles) along with original data (solid line) as presented to the NaKLh model. The stimulus current was taken as usual from a chaotic system. This result uses the ‘weak’ stimulus which excites polarized state variables. The ‘weak’ stimulus provides sufficient excitation to the Na and K channels to allow accurate estimation of their properties. Bottom Estimated Ih activation gating variable (circles) along with original data (solid line) as presented to the NaKLh model. The stimulus current was taken as usual from a chaotic system. This result uses the ‘weak’ stimulus which excites polarized state variables state variables. The Ih current is not well stimulated by this ‘weak’ Iapp(t). ρI = 0.5.
In an experimental setting, however, only the quality of the estimation of the voltage trace (Fig. 6) and the consistency test would be available. Neither of these indicate that the weaker stimulus fails to excite one of the currents in the model, though the latter may well indicate an inconsistency of the model with the data. Thus, an additional test for model validity is needed, which we discuss in the following section.
3.6 Model validation using state and parameter estimates
An important piece of a model investigation we have not covered in our discussion until now is the ability of the model neuron with parameters and state variables estimated in an observation window to predict the response of that neuron to new stimuli. If this is successful for a class of stimuli that one might expect to be given to this neuron when it is placed into a network, we can expect that it will respond properly in the network. We examined this issue in the context of our two stimuli: weak and strong, knowing the former was not giving sufficient stimulation of the Ih current.
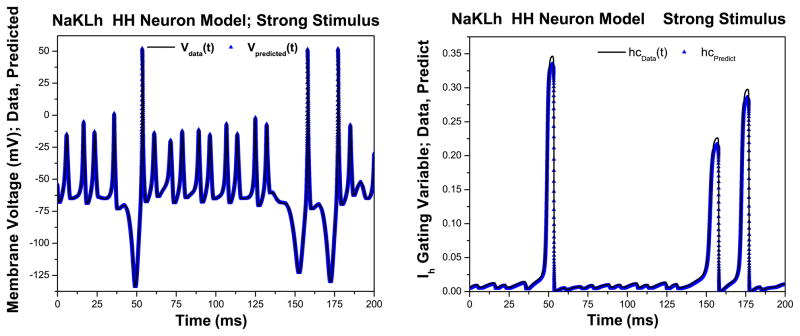
To validate the models’ consistency to weak and strong stimuli, we tested their ability to predict responses to the other stimulus. Using the parameters estimated with data in the observation window, we employed the estimated state variables {V(T ), m(T ), h(T ), n(T ), hc(T )} to predict the behavior of the model neuron for t > T. When the model fit to the strong stimulus was tested on the weak stimulus, we found that the model was able to predict the response of the neuron to the weak stimulus with a high degree of accuracy (Fig. 7 Left). In this twin experiment, we are also able to compare the predicted values for the state variables against their true values (Fig. 7 Right), which display a similar degree of accuracy.
Fig. 7.
Left Prediction of the membrane voltage for times greater than the observation window with state variables and parameters estimated with the strong stimulus. Right Prediction of the Ih gating variable hc(t) for times greater than the observation window with state variables and parameters estimated with the strong stimulus. In both cases, the values of the current’s parameters and state variables are accurately estimated
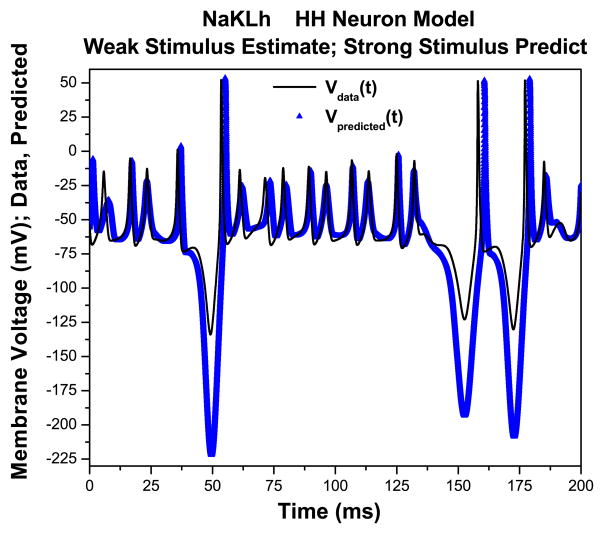
In contrast, the model fit to the weak stimulus was unable to predict the response to the strong stimulus (Fig. 8). Both the phase and amplitude of the predictions is degraded, especially at times when the stimulus was strongly hyperpolarizing. Thus, because the weak stimulus did not give sufficient information about Ih, the predictions are inadequate. Corresponding to twin experiments, predictive experiments in a laboratory setting are a useful way to test whether a candidate stimulus in conjunction with a neuron model is capable of adequately probing the model in question. These results also emphasize the importance of using model predictions to cross-validate them against different kinds of stimuli.
Fig. 8.
Using the parameters and state variables estimated using the ‘weak’ stimulus, we predict the response of the NaKLh neuron to the ‘strong’ stimulus
3.7 Robustness to model errors
3.7.1 NAKLh data presented to NAKLh model
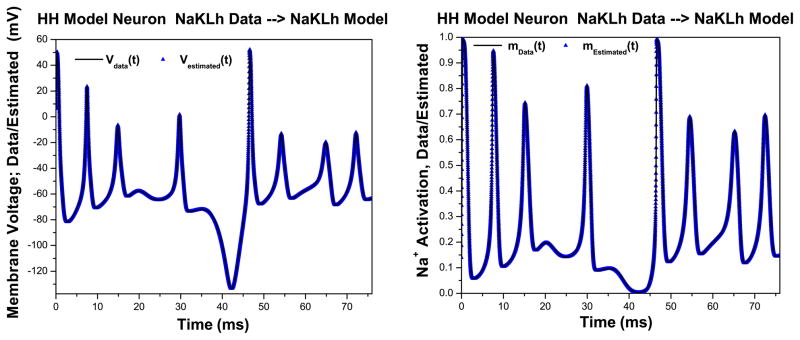
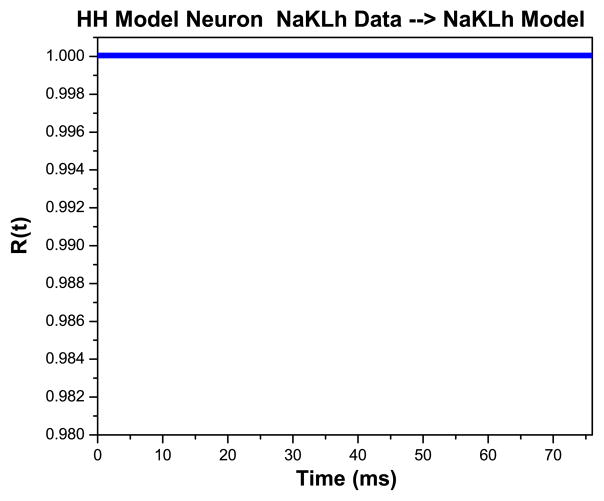
We want to contrast what happens when we present data from the NaKLh model first to itself, a now-familiar twin experiment, to what happens when we present NaKLh voltage data to the NaKL model. The latter presents the data to a wrong model; namely one missing Ih. First, we take the NaKLh model with a strong stimulus and ρI = 0.5, our two good stimulus settings, and generate ‘data’ without added noise. We present this NaKLh model output voltage along with the same stimulus, and we are able to accurately estimate the parameters, as reported, and as we show in Fig. 9 Left, also the voltage response. The is the minimum we would expect to qualify as an interesting model for the source of our data. We confirm this quality of the model, in particular its consistency with the data, by evaluating R(t)as shown in Fig. 10. As additional confirmation, we examine the accuracy of our estimation of an unobserved state variable m(t) for the NaKLh model. This is shown in Fig. 9 Right.
Fig. 9.
Left Voltage estimated with a NaKLh HH model presented with data created from the same model—a twin experiment. The stimulus used is our ‘strong’ stimulus and has ρI = 0.5 which we determined earlier allows accurate estimations. Right Estimation of the Na+ activation variable m(t) when NaKLh voltage data is presented to a NaKLh model. The high quality of this estimation is consistent with the R(t) values in Fig. 10
Fig. 10.
R(t) for the situation represented in Fig. 9. The value of R(t) does not deviate from unity, indicating the model is consistent with the data
3.7.2 NAKLh data presented to NAKL model
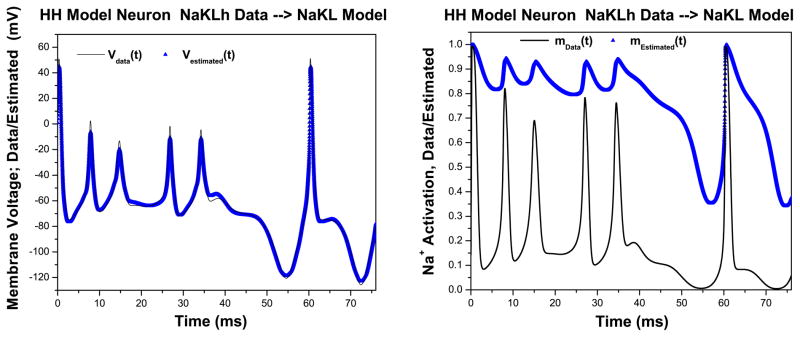
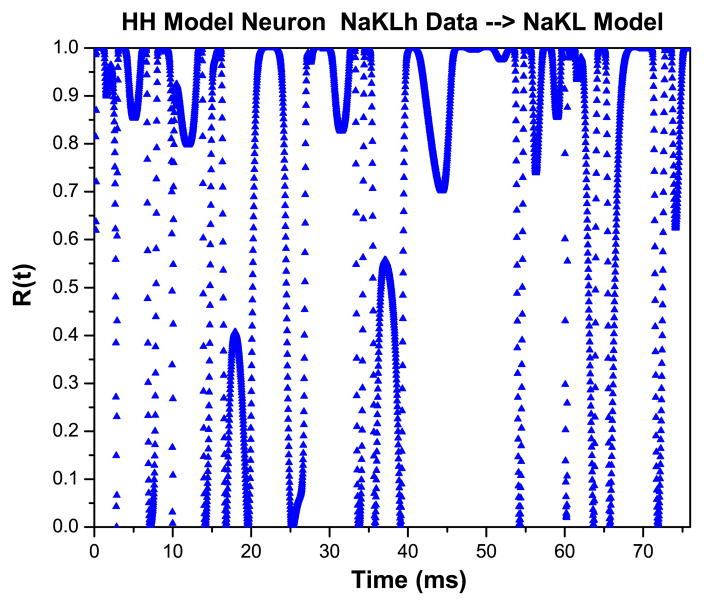
We contrast these results with those resulting when we present the wrong data to our NAKL model. Again we generated data using our NaKLh model with a strong stimulus and ρI = 0.5 and presented these data to our NaKL model, which we recall does not have an Ih current. In Fig. 11 Left, we show that a rather accurate, perhaps impressive ‘fit’ to the data is achieved by the dynamical data assimilation procedure. However, when we evaluate R(t) for this configuration, we see the result in Fig. 12 which indicates the apparent success of the estimate comes from large values of the control u(t) because R(t) makes significant excursions to values less than one.
Fig. 11.
Left Estimating the membrane voltage in the NaKL model when it is presented with voltage output from an NaKLh model. The accuracy of the ‘fit’ to the voltage data is misleading as the voltage data is incorrect for this model. Right Estimation of the Na+ activation variable m(t) when NAKLh voltage data is presented to a NaKL model. The low quality of this estimation is consistent with the R(t) value in Fig. 12
Fig. 12.
R(t) for the situation represented in Fig. 9. The value of R(t) deviates significantly from unity, indicating the model is inconsistent with the data
We see features of this ‘wrong data’ once again, as we are able in a twin experiment, in the quite inaccurate estimation of the Na+ activation variable m(t) in Fig. 11 Right.
3.8 NaKL data driving a NAKLh model
In building a model for an experimentally observed neuron, we face the problem, once we have selected an HH framework for the model, of which currents to include in the model. Though it is hardly elegant, one rather straightforward idea is to include in the model ‘all’ the currents one can plausibly argue should enter the biophysics of that neuron. All such currents will have the form (Brette et al. 2007; Graham 2002)
| (10) |
and the kinetics of the gating variables will be selected as well. One would expect that if this particular channel is absent in the data from the experimental preparation, then gcurrent will be zero.
To examine whether the data assimilation methods, we have developed are able to identify currents in a model which are absent in the data, and thus prune a larger model down to those required by the data, we presented data from an NaKL model to a larger model having an Ih current as well. The NaKL data was generated by the NaKLh model where gh = 0 but constants for the Ih kinetics were specified although the Ih contribution to the voltage equation was absent. Table 7 displays the results for this situation. We see that all the information about the Na+, K+, and leak currents was accurately reproduced, the absence of the Ih current was recorded as a very small gh, namely gh = 1.9 × 10−9 mS/cm2, and estimates for Eh and the parameters in the kinetics which are wrong, but irrelevant as the current is absent. In other words, the idea that we could build a ‘large’ model of all neurons and use the data to prune off currents that are absent is plausible and supported by this calculation. Whether this optimistic viewpoint will persist as we confront laboratory data with our methods is yet to be seen. The independence of the prediction of Ih parameters from the prediction for the other currents gives some, albeit only preliminary, reason to hope such an approach might work.
Table 7.
Parameters in a NaKLh HH model where the maximal conductance of the Ih current was set to zero; so only the NaKL model was active (The NaKL parameters are slightly different from our earlier NaKL model)
| Name | Data | Estimate | Units |
|---|---|---|---|
| gNa | 120 | 120.4258 | mS/cm2 |
| ENa | 55.0 | 54.92950 | mV |
| gK | 20 | 20.05503 | mS/cm2 |
| EK | −77.0 | −77.01366 | mV |
| gL | 0.3 | 0.2990211 | mS/cm2 |
| EL | −54.4 | −54.24781 | mV |
| vm = vmt | −34.0 | 33.97316 | mV |
| dvm=dvmt | 34.0 | 34.00498 | mV |
| tm0 | 0.01 | 0.009585590 | ms |
| tm1 | 0.5 | 0.5008398 | ms |
| vh=vht | −60.0 | −59.98665 | mV |
| dvh=dvht | −19.0 | −18.97535 | mV |
| th0 | 0.2 | 0.1998151 | ms |
| th1 | 8.5 | 8.525130 | ms |
| vn=vnt | −65.0 | −64.94673 | mV |
| dvn=dvnt | 45.0 | 45.01288 | mV |
| tn0 | 0.8 | 0.7997343 | ms |
| tn1 | 5.0 | 5.016256 | ms |
| gh | 0 | 1.907150×10−9 | mS/cm2 |
| Eh | −40.0 | −60.57007 | mV |
| vhc | −75.0 | −6.88913 | mV |
| dvhc | −11.0 | 4.203612 | mV |
| thc0 | 0.1 | 3.589662 | ms |
| thc1 | 193.5 | 179.5280 | ms |
| vhct | −80 | −44.07163 | mV |
| dvhct | 21.0 | 55.30240 | mV |
This data was presented to a NaKLh model with free parameters, the dynamical estimation procedure yielded a maximal conductance for Ih of 1.907150 × 10−9 mS/cm2. The other Ih parameters are badly estimated, but they are not relevant as the Ih current was absent in the data, though present as a possibility in the NaKLh model
4 Discussion
We have touched upon a number of items of significance in the estimation of the parameters and unobserved state variables of HH neuron models. The context of our discussion is an exact formula (Appendix I) for the evaluation of the answers to questions arising when one wishes to estimate model parameters and unobserved model states when the model is presented with observed data. That exact formula is a path integral through model states encountered while observations are made, and in this article we have focused on the saddle path approximation to that integral representation of answers to data assimilation questions. More direct approaches to the evaluation of the desired path integral are in preparation (Kostuk et al. 2011).
In the framework of this approximation, we encounter a numerical optimization problem which asks that we maximize the transfer of information to a model from observations subject to the satisfaction of a biophysical model of the neural activities being observed. To regulate the impediments encountered in nonlinear estimation problems (Creveling et al. 2008; Abarbanel et al. 2009) we introduce a ‘control’ to drive the model output to the observations while assuring that, if the model is consistent with the data, the control is set to zero within the optimization routine.
Our methods have been focused on single compartment models of HH neurons. One has Na, K, and leak channels in the model, (NaKL model) and the second has an additional Ih current (NaKLh model). We described a series of ‘twin experiments’ wherein data is generated by the model, then, with or without noise added, one variable, the observable membrane voltage V(t) is presented to the model with free parameters and we use our saddle path variational principle (numerical optimization problem) to estimate the parameters and the unobserved (gating variables) state variables of the model.
The outcome of the estimation procedure is a single path satisfying the optimization task through the observation window. The path integral also allows the evaluation of errors estimates for each parameter and state variable, though we do not address this here.
In the solution of the numerical optimization problem, posed in Eqs. 5 and 6, we use the ‘direct method’ (Gill et al. 1998) where all state variables, all parameters, and, here, all control variables are varied in the optimization procedure. Most of the computational effort is centered, in our evaluations, in the numerical linear algebra required in the programs. We have shown (Toth 2010) that using linear algebra libraries optimized for parallel computing, we can achieve substantial speedup of our results.
When employing the regularized search method, called dynamical state and parameter estimation (Abarbanel et al. 2009), we may form a dimensionless estimate (Eq. 7) of the importance of the control u(t) that indicates whether the model is consistent with the data presented to it.
In the context of the two models we explored: NaKL and NaKLh, we addressed the following questions with associated results:
-
when presenting NaKL data to the NaKL model, is the use of the control variable important ? We found that the identical numerical optimization calculation to find the fixed parameters and unobserved state variables of the neuron model with u(t) ≠ 0 and u(t) = 0 yielded much better estimations for all quantities when u(t) ≠ 0 during the optimization procedure. By dint of the procedure itself u(t) = 0 results at the end of the optimization. This is good as the term in the equations of motion, Eq. 6, with u(t) is not part of the biophysics of the neuron but only a regularization tool.
The same outcome is seen when the model is enhanced with other currents; here only the addition of Ih was explored.
-
each twin experiment requires the selection of characteristics of the stimulating current, called Iapp(t). We showed that if the frequencies in Iapp(t) are too high compared to the basic frequency for the membrane voltage activity, the response of the neuron was washed out though its RC low pass filtering capability. We quantified this through the ratio of the neuron sampling time and the characteristic time of the stimulus, called ρI here. When ρI is in the range 0.5–1.0 we stimulate the neuron in a manner that does not get removed by the neuron membrane time constants.
This will play a significant role in the design of experimental protocols for using our estimation procedure.
we explored the role of additive noise in the measurements presented to the model. As in our earlier discussions (Abarbanel et al. 2011), the model through the dynamical state and parameter estimation regularized optimization method, acts as a nonlinear filter for the noise. In this article, we only examined an example for NaKL models with a signal to noise ratio of 30 dB.
in the context of NaKL and NaKLh models we established that when the stimulating current is strong enough during the observation window to effectively excite currents acting at polarized (Na and K) levels and hyperpolarized levels for the neuron, we are able to effectively predict beyond the observation window using the estimated parameters and estimated state variables at the end of observations. This also suggests stimulation protocols in experiments which result in quantitatively effective estimations.
in presenting the ‘wrong’ data from an NaKLh model to an NaKL model, we were able to detect the inconsistency of the model with the data, thus, indicating that the model required improvement. Our method, and probably any data assimilation method, when it fails in this manner does not suggest how to repair the model utilized. However, the inconsistency of the model with presented data is likely to be quite valuable information for model development.
-
one key to the success of the methods we explore in this article is the presentation of a sufficient length of data in time to show the model the full variations of the response (here V(t)) waveform. In our examples, we used about 60–90ms of simulated data. This may, in the examples investigated here, actually have been more data than was required. However, we did not investigate how short a data set would have sufficed for accurate estimates of parameters and unobserved states.
The accurate results we have presented, however, suggest that one may be able to utilize rather short (order of 100 ms) voltage signals for the exploration of neural dynamics in complicated nervous systems.
One of our results has stronger implications than these. We asked whether when presenting membrane voltage data from an NaKL model to an NaKLh model, we could detect in the latter, the absence of the Ih current. This was accomplished by noting that the maximal conductance gh in the HH form of Ih:
| (11) |
should be zero when NaKL data is presented to an NaKLh model with this current. Indeed, we found this to be the case, within numerical resolution gh ≈ 2×10−9 when other maximal conductances were order 0.1–100 in the same estimation procedure.
This suggests one may be able to devise a ‘grand’ model for a large class of neurons which contains a substantial collection of currents, voltage gated and ligand gated (in networks), as well as intracellular processes such as Ca2+ dynamics, and utilize the data, though DSPE, to prune those ingredients in the large model that are not required by the data. How one selects the components of the grand model is not dictated by the arguments presented in this article but calls for insightful biophysical reasoning about the classes of neurons of interest.
Although not discussed here in detail: (1) one can use a well established neuron model as a sensor of its environment and by treating the time dependent stimulating current Iapp(tn) as a set of m+1 parameters, estimate these numbers from the voltage response data. This entails the estimation in the class of problems discussed here of many thousands of unknown parameters, and it is accomplished by our methods with high accuracy (C. Knowlton, personal communication). (2) Through the use of well designed presynaptic voltages, one can determine the critical parameters of a dynamical synapse (neurotransmitter docking and undocking time constants, synaptic maximal conductivity,…) using the methods discussed in this article.
There are two final items we wish to address. First, the introduction of the control u(n) into the dynamical equations and the numerical optimization procedure might induce a bias in the estimation of the biophysical parameters in the HH voltage equation or the kinetics of the gating variables. While we have no mathematical proof this is not so, we have the following observations which suggest it is not so:
The controls u(n) are parameters just as a maximal conductance or a reversal potential is a parameter. Namely there is no dynamical equation, .
In the numerical optimization process, each of the parameters at each time point is treated independently, that is with no correlations between pairs of them: u(12) is independent of u(341) is independent of ENa…, etc. So the presence of the control parameters in addition to the biophysical parameters should have no more effect on any of them than an optimization addressing just the biophysical parameters. Nonetheless, there might be correlations, and consequently bias in the estimations, if within the numerical optimization routine there is a correlation we do not know about that is induced by the software algorithm.
We have two suggestions why there is no bias: (1) we have done the optimization starting at quite different initial guesses and quite different bounds over which to search. Each time we end up with results more or less precisely what we reported in the table—sometimes the estimates are a bit more accurate, sometimes a bit less accurate. The values of u(n) as one proceeds through the iterations of the optimization algorithm are different for each of these runs, and there appears to be no difference in the outcomes. (2) At the end of the iterations of the numerical algorithms, u(n) is essentially zero (≈10−13) and while the fluctuations around this tiny value may be correlated, again through the inner working of the algorithm, the numerical size, since the fluctuations of u(n) are also of order 10−13, is tiny.
Second, the algorithms we present and use in this article are quite general, as the discussion and derivation suggest. The use of them for studying neurobiological questions is one of many potential applications. The limits on their practical use come from three main sources we know about: (1) as the number of degrees of the models and observed systems rises, practical numerical algorithms and procedures may fail to produce accurate answers in acceptable times. As IPOPT (Wächter and Biegler 2006), one of the open-source numerical optimization programs we use extensively, uses so much of its computing time in linear algebra operations and as that is nicely parallelizable, this concern may be simply a matter of waiting for computing to catch up with the scale of interesting scientific problems. (2) If the required number of measurements is not available, the quality of predictions will degrade. We do not have a metric for that degradation, but it can be quite severe as instabilities on the synchronization manifold will not be totally cured. (3) There is no guarantee that the class of model chosen to represent the data is correct in any sense. This goes under the discussion of model errors, and we do not know an algorithm to discover and cure model errors. The calculation we presented when NaKL data is presented to a model with more degrees of freedom (here NaKLh) and prunes away the currents not needed, is the first example of using any method to remove incorrect model choices. In some cases, there may not be such a method and model errors may never be found until more physics is known or better guesses are made.
This article should be seen as an initial exploration of the use of the general data assimilation formulation. The direct evaluation of the path integral using Monte Carlo methods will be addressed using the same examples here in a companion paper (Kostuk et al. 2011).
Acknowledgments
Support from the US Department of Energy (Grant DE-SC0002349), the National Science Foundation (Grants IOS-0905076, IOS-0905030 and PHY-0961153), and the National Institutes of Health (Grant 1 F32 DC008752 and 2 R01MH59831) are gratefully acknowledged. Partial support from the NSF sponsored Center for Theoretical Biological Physics is also appreciated. The underlying theory for this article was developed when one of us (HDIA) was on leave from UCSD at the Bernstein Center for Computational Neuroscience in Munich.
Appendix I: General formulation: noisy data, model errors, uncertain x(0)
In the general setting for data assimilation problems, we have noisy data, errors in the model from missing dynamics, errors in the model due to resolution in the numerical implementation, resolution model errors due to noisy environmental effects, and we have incomplete knowledge of the state x(t0) = x(0) when measurements begin. In this, probably typical, circumstance we are limited to questions about probability densities in the space of states x, and we wish to answer questions about expectation values of functions of the system states defined over the time interval of observations [t0, tm = T ]. Starting with deterministic differential equations for the states
| (12) |
we would be led to a Fokker-Planck equation for the probability density function (pdf) P(x(t)) which is linear in the pdf, and has the formal solution
| (13) |
with a transfer function K(x(m); x(0)). We will write this in the form
| (14) |
and we will now give a formula for the ‘action’ A0(X) defined on the path of the system state X = {x(m), x(m − 1), …, x(0)} through the observation window {t0, t1, …, tm}. The expectation values of functions on the path G(X) are given by the integral over the path
| (15) |
In the questions we ask in this article, we have a conditional pdf as we wish to know the probability of being at x(m) (within dx(m)) conditioned on the (m + 1) L-dimensional observations Y(m) = {y(m), y(m − 1), …, y(0)} up until time tm. In this discussion, we choose the measurement function to be the unit operation: hl (x)) = xl.
We need two ingredients: (a) identities following from the definition of conditional probabilities, and (b) an understanding that the dynamics moving the model system forward in time is Markov: the state x(tn+1) = x(n + 1) depends only on the state at time tn x(n) and the fixed parameters p. We write the latter in discrete time as the explicit statement
| (16) |
when we have a deterministic (infinite resolution, no noise) dynamical rule. Of course, we will relax this as we proceed.
If the dynamics is not Markov, it usually signals that we have projected the underlying dynamics described by sets of differential equations into a smaller space, and we can rectify this by enlarging the state space of the model. In practice, we have no knowledge of the ‘correct’ model, so we proceed assuming the Markov property.
We are interested then in an expression for the conditional pdf P(x(m)|Y(m)), the probability density for the state at time tm conditioned on measurements at times {t0, …, tm}. Using the definition of a conditional probability, we may write
| (17) |
We recognize the quantity in the curly brackets to be the exponential of the conditional mutual information between the D-dimensional state at time tm and the L-dimensional observation at time tm conditioned on the previous observations Y(m − 1) (Fano 1961):
| (18) |
This set of relations among conditional pdfs is often called Bayes’ rule.
Since we have assumed the dynamics taking x(n) → x(n +1) to be Markov, we may use the Chapman-Kolmogorov relation for such processes
| (19) |
where the transition probability for x(n) → x(n + 1) is P(x(n + 1)|x(n)). Together with the identity on conditional pdf’s this gives us a recursion relation connecting P(x(m)|Y(m)) to P(x(m − 1)|Y(m − 1)):
| (20) |
In the deterministic dynamics setting P(x(n+1)|x(n)) = δD(x(n + 1) − f(x(n), p))). In the presence of model error, this sharp distribution is broadened.
We may now move from time tm−1 to time tm−2 and back to t0 to write (Abarbanel 2009)
| (21) |
defining the action
| (22) |
Just as a note, this exact formula, simplifies if the dynamics is deterministic because the integrals over x(m), x(m − 1), …, x(1)) can all be performed.
This expression for P(x(m)|Y(m)) and the related formula for E[G(X)|Y(m)], the conditional expectation value for a function G(X) on the path X, are useful only when we make approximations to the action. We address them in a moment.
There are two well established ways to approach the evaluation of the integral (15) when, as happens in rare instances, one can evaluate it exactly. (1) Use a Monte Carlo method (Quinn and Abarbanel 2010) to sample the high dimensional space of paths X to estimate the distribution exp[–A0(X|Y)], and (2) use a saddle path approximation which leads to a standard perturbation theory (Zinn-Justin 2002). In the latter approach, there are resummation methods (Zinn-Justin 2002) to approximate subsets of the full answer.
The optimization problem we used to estimate parameters and states for neuron (or other) models given data Y(m) comes as the first approximation to the saddle path evaluation of the integral in (m+1)D dimensional path space. This consists of expanding the action about a stationary point S, for fixed Y (not displayed),
| (23) |
The full integral also gives the corrections to the variational approximation discussed above. In the optimization method, we sketched earlier, we seek the extremum S of the action using one or another numerical optimization routine. This is the method we will utilize in our further considerations in this article. We return to the use of Monte Carlo methods in a subsequent paper. Now we turn to approximations to the action that allow us to build a practical strategy for estimating properties of neuron models from time course data.
Approximating the Action
Two more or less standard approximations to the action consist of first assuming that the noise in each measurement yl (n) is independent of the noise in measurements at different times. The conditional mutual information then becomes
| (24) |
If the noise is taken as additive, yl = xl + noise, then, ignoring factors of y that cancel in any expectation value, we have
| (25) |
If, further, the noise is Gaussian, then up to constants canceling in any expectation value,
| (26) |
where Rm is an L × L covariance matrix. If this is diagonal with equal terms along the diagonal, then
| (27) |
This term of the action is now seen as our earlier cost function which gives a magnitude to the synchronization error between the data yl (n) and the corresponding model output xl (n) summed over the data set.
Looking now at the model error term, if we assign the error to the resolution associated with discretization of space and time as well as environmental fluctuations, perhaps at other spatial scales, we can represent P(x(n + 1)|x(n)) = δD(x(n + 1) − f(x(n), p))) by
| (28) |
When R f → ∞, we return to the delta function of deterministic dynamics. The resolution in state space of the model is proportional to . While numerous other approximations are possible, some of which may be tested within the path integral representation (Abarbanel 2011). With these ‘standard’ approximations
| (29) |
Appendix II: Numerical procedures for nonlinear optimization
The stationary path approximation to our path integral in the case when the dynamics is deterministic is approximated by the standard numerical optimization problem: Minimize the objective or cost function
| (30) |
subject to the equality constraints imposed by the model dynamical Eq. 6 along with kinetic equations for the gating variables.
We write this in the form
| (31) |
which includes the control term u(t)(y(t) − x1(t)) = u(t) (y(t) − V(t)) in the function Fa(x(t), p).
There are many methods for solving this optimization problem (Kirk 2004; Gill et al. 2005; Wächter and Biegler 2006; Gill et al. 1981). We adopt the ‘direct method’ in which the D state variables x(tn) = x(n) at each time {t0, t1, …, tm} the NP parameters, and the (m + 1) control variables u(tn) = u(n) are varied to minimize the cost function. This transform what could be seen as a problem in D initial conditions x(0), NP parameters p, and m +1 ‘other parameters’ u(n) into the larger problem in (D +1)(m +1)+ NP dimensional space. The apparent disadvantage of the direct method which acts in this larger space is significantly offset by the sparseness of the problem Jacobian in the larger space and, equally importantly, in that it determines as outputs the parameters, the controls and the state variables at each time point in the observation window. If we determined only the parameters and the x(0), then prediction for t > tm = T would require us to the dynamical equations integrate forward from approximate initial conditions with approximate parameters. In the case where the dynamical system has chaotic solutions, this would result in enhanced errors at t = T making prediction certainly inaccurate and uncertain.
As a first step, the dynamical system equality constraints must be transformed to discrete time, commensurate with the discrete time data y(tn). Many options exist for this discretization. Here we choose Simpson’s Rule, with functional midpoints estimated by Hermitian cubic interpolation. This Hermite-Simpson (Gill et al. 2005) choice gives accuracy of order Δt4 for discrete time steps Δt in the integration routine. Methods with similar accuracy will suffice just as well, but lower order methods must be used carefully to insure that the data sampling rate is both accurate enough and fast enough to capture the variation of the underlying dynamics of the system. These integrated equations become the equality constraints in our optimization:
Simpson Integration:
with Hermite Polynomial Interpolation:
where tn2 = (tn+1 − tn)/2.
A variety of optimization software and algorithms are available to solve this ‘direct method’ optimization problem. SNOPT (Gill et al. 2005) and IPOPT (Wächter and Biegler 2006) were chosen since these are both widely available, are designed for nonlinear problems with sparse Jacobian structure, and can handle large problems. Depending on the problem and the data set, a few thousand data points (or more) are necessary to explore the state space of the model in order to produce accurate solutions. The dynamical criterion is that the waveform of the observed dynamics be explored accurately as it moves along either the transient to the system attractor or along the attractor itself. Avery short observation window might produce only a small piece of the variation in the waveform and appear essentially as a straight line which cannot be distinguished from other straight lines and does not identify the underlying dynamics.
For problems of the size described here, two to three voltage spikes or more are needed to explore the model state space, and this results in tens of thousands of constraints and unknown variables. Because of the discretized temporal structure of the problem, however, the Jacobian of the constraints is sparse, and both SNOPT and IPOPT can take advantage of this sparsity. All the results discussed in this article have used IPOPT.
As described by Toth (2010), the problem is set up using symbolic differentiation and code generation techniques with the Python programming language. For each model, for instance the NaKL or NaKLh models, with dynamics described by a set of first order ordinary differential equations, a separate instance of the optimization problem is necessary. This instance is implemented via C++ code generation in Python as follows:
Model dynamics (differential equations) input to a text file. In this file the dynamical equations are given in ascii format along with the names of the state variables, the parameters, the controls, the data file, and the file containing the applied (stimulus) current.
-
Python script imports this text file and then performs the following steps:
Discretizes it in accord with the selected discretization rule;
Uses symbolic differentiation with SymPy1 to generate Jacobian and Hessian matrices;
Generates C++ files that set up the optimization problem in IPOPT.
The user next compiles the C++ code with generated Make file that links to IPOPT libraries; this produces an executable ‘problemname_cpp’ file.
-
Execution of the compiled code requires a second user-generated text file that specifies:
Problem size
Integration time step, or inverse of sampling rate.
Bounds for all states and parameters.
Initial optimization starting point for all states and parameters.
Names of data files which include the observed data.
-
Completion of the optimization problem gives the following files:
Estimates of all states, measured and unmeasured, at all time points in the observation window, as well as the values for all control u(tn) terms for tn = {t0, t1, …, tm}.
Estimates of all parameters
Calculation of the R(t) value
An output file with the saddle path through the observation window. This is a (m+1)D+NP dimensional vector: X = {xa(m), xa(m − 1), …, xa(0), p1, p2, …, pNP} for possible later use in the full calculation of the path integral.
From x(Tm) = x(T ), the parameters p and the dynamical equations with u(n) = 0, we can forecast the behavior of the dynamical system for t > T.
In our use of IPOPT in problems such as the individual neuron models discussed here, we estimate from timing analyses that about 70% of the computation time is used in linear algebra calculations. This suggests, and we have verified this on CPUs with a small number of cores, that a nearly linear speedup in computation can be achieved with the use of linear algebra libraries developed for parallel computation. If this extends to parallel machines with a large number of cores, the methods discussed in this article can be extended in a straightforward manner to quite large problems.
Footnotes
Contributor Information
Bryan A. Toth, Department of Physics, University of California, 9500 Gilman, Drive, San Diego, La Jolla, CA 92093-0402, USA
Mark Kostuk, Department of Physics, University of California, 9500 Gilman, Drive, San Diego, La Jolla, CA 92093-0402, USA.
C. Daniel Meliza, Department of Organismal Biology and Anatomy, University of Chicago, 1027 E 57th Street, Chicago, IL 60637, USA.
Daniel Margoliash, Department of Organismal Biology and Anatomy, University of Chicago, 1027 E 57th Street, Chicago, IL 60637, USA.
Henry D. I. Abarbanel, Department of Physics, University of California, 9500 Gilman, Drive, San Diego, La Jolla, CA 92093-0402, USA. Marine Physical Laboratory (Scripps Institution of Oceanography), Center for Theoretical Biological Physics, University of California, 9500 Gilman Drive, San Diego, USA
References
- Abarbanel HD. Effective actions for statistical data assimilation. Phys Lett A. 2009;373(44):4044–4048. [Google Scholar]
- Abarbanel HD. Self consistent model errors. QJ Roy Meteor Soc. 2011 submitted. [Google Scholar]
- Abarbanel HDI, Creveling DR, Farsian R, Kostuk M. Dynamical state and parameter estimation. SIAM J Appl Dyn Syst. 2009;8(4):1341–1381. [Google Scholar]
- Abarbanel HDI, Bryant P, Gill PE, Kostuk M, Rofeh J, Singer Z, Toth B, Wong E. Dynamical parameter and state estimation in neuronmodels, Chap 8. In: Ding M, Glanzman DL, editors. The Dynamic Brain. Oxford University Press; 2011. pp. 139–180. [Google Scholar]
- Brette R, Rudolph M, Carnevale T, Hines M, Beeman D, Bower J, Diesmann M, Morrison A, Goodman P, Harris F, Zirpe M, Natschläger T, Pecevski D, Ermentrout B, Djurfeldt M, Lansner A, Rochel O, Vieville T, Muller E, Davison A, El Boustani S, Destexhe A. Simulation of networks of spiking neurons: a review of tools and strategies. J Comp Neurosci. 2007;23:349–398. doi: 10.1007/s10827-007-0038-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creveling DR, Gill PE, Abarbanel HD. State and parameter estimation in nonlinear systems as an optimal tracking problem. Phys Lett A. 2008;372(15):2640–2644. [Google Scholar]
- Evensen G. Data assimilation: the ensemble Kalman filter. Springer; Berlin: 2009. p. 2. [Google Scholar]
- Fano R. Transmission of information: a statistical theory of communications. The MIT Press; Cambridge: 1961. [Google Scholar]
- Gill P, Barclay A, Rosen JB. Sqp methods and their application to numerical optimal control. In: Bulirsch R, Bittner L, Schmidt WH, editors. Heier KVariational calculus, optimal control and applications, international series of numerical mathematics. Vol. 124. Birkhauser; Basel, Boston and Berlin: 1998. pp. 207–222. [Google Scholar]
- Gill P, Murray W, Saunders M. Snopt: an sqp algorithm for large-scale constrained optimization. SIAM Rev. 2005;47(1):99–131. [Google Scholar]
- Gill PE, Murray W, Wright MH. Practical optimization. Academic Press; London: 1981. [Google Scholar]
- Graham L. Modelling neuronal biophysics. In: Arbib MA, editor. The handbook for brain theory and neural networks. Vol. 2. MIT Press; Cambridge: 2002. pp. 164–170. [Google Scholar]
- Hamill OP, Marty A, Neher E, Sakmann B, Sigworth FJ. Improved patch-clamp techniques for high-resolution current recording from cells and cell-free membrane patches. Pflugers Arch. 1981;391(2):85–100. doi: 10.1007/BF00656997. [DOI] [PubMed] [Google Scholar]
- Huys QJM, Ahrens MB, Paninski L. Efficient estimation of detailed single-neuron models. J Neurophysiol. 2006;96(2):872–890. doi: 10.1152/jn.00079.2006. [DOI] [PubMed] [Google Scholar]
- Johnston D, Wu SMS. Foundations of cellular neurophysiology. MIT Press; Cambridge: 1995. [Google Scholar]
- Kirk DE. Optimal control theory: an introduction. Dover Publications; Mineola: 2004. [Google Scholar]
- Koch C. Biophysics of computation: information processing in single neurons. Oxford University Press; New York: 1999. [Google Scholar]
- Kostuk M, Toth B, Meliza CD, Abarbanel HDI, Margoliash D. Dynamical estimation of neuron and network properties II: Monte carlo methods. Biol Cybern. 2011 doi: 10.1007/s00422-012-0487-5. (in preparation) [DOI] [PubMed] [Google Scholar]
- Laurent G, Stopfer M, Friedrich RW, Rabinovich MI, Volkovskii A, Abarbanel HDI. Odor encoding as an active dynamical process: experiments, computation, and theory. Annu Rev Neurosci. 2001;24:293–297. doi: 10.1146/annurev.neuro.24.1.263. [DOI] [PubMed] [Google Scholar]
- Quinn JC, Abarbanel HD. State and parameter estimation using monte carlo evaluation of path integrals. Q J Roy Meteor Soc. 2010;136(652):1855–1867. [Google Scholar]
- Stein PSG, Grillner S, Selverston AI, Stuart DG, editors. Neurons, Networks, and Motor Behavior. MIT Press; Cambridge: 1997. [Google Scholar]
- Toth B. Dynamical estimation of neuron and network properties. SIAG/OPT Views-and-News. 2010;21(1):1–8. [Google Scholar]
- Wächter A, Biegler LT. On the implementation of an interiorpoint filter line-search algorithm for large-scale nonlinear programming. Math Prog. 2006;106(1):25–57. [Google Scholar]
- Zinn-Justin J. Quantum field theory and critical phenomena. Oxford University Press; Oxford: 2002. p. 4. [Google Scholar]