Abstract
We have a mutually beneficial relationship with the trillions of microorganisms inhabiting our gastrointestinal tract. However, maintaining this relationship requires recognizing these organisms as affable and restraining inflammatory responses to these organisms when encountered in hostile settings. How and when the immune system develops tolerance to our gut microbial members is not well understood. Here we identify a specific pre-weaning interval in which gut microbial antigens are encountered by the immune system to induce antigen specific tolerance to gut bacteria. Intriguingly for some bacterial taxa, physiologic encounters with the immune system are restricted to this interval, despite abundance of these taxa in the gut lumen at later times outside this interval. Antigen specific tolerance to gut bacteria induced during this pre-weaning interval is stable and maintained even if these taxa are encountered later in life in an inflammatory setting. However, inhibiting microbial antigen encounter during this interval or extending these encounters beyond the normal interval, results in a failure to induce tolerance and robust antigen specific effector responses to gut bacteria upon reencounter in an inflammatory setting. Thus, we have identified a defined pre-weaning interval critical for developing tolerance to gut bacteria and maintaining the mutually beneficial relationship with our gut microbiota.
Introduction
The gastrointestinal tract (GI) is home to trillions of microorganisms (1). The relationship between the host and a healthy gut microbiota is mutually beneficial, with the microorganisms receiving an environment in which to reside and the host receiving benefits related to development, protection from pathogens, immunity, and metabolism (2–6). To maintain these mutual benefits, interactions between the host and the gut microbiota are orchestrated to avoid infection yet maintain tolerance and prevent inappropriate inflammatory responses. Indeed, long-lived effector responses to gut commensal antigens can occur when they are encountered in an inflammatory setting (7). Moreover loss of tolerance to the gut microbiota, as evidenced by systemic immune responses to gut commensals (8–12), is believed to underlie the pathogenesis of inflammatory bowel disease (IBD), a chronic inflammatory condition of the GI tract. Damage from inappropriate inflammatory responses is not limited to the host cells, but also induces dysbiosis of the gut microbiota, which has been associated with multiple disorders (13) and in turn promotes host inflammatory responses (14, 15). Accordingly, how immune tolerance to the microbiota is established, maintained, and becomes altered are central topics to understanding multiple facets of health and disease.
Immune tolerance to environmental antigens is largely mediated by peripheral Foxp3+ regulatory T cells (pTregs). Specific gut commensal bacteria taxa promote the induction of pTregs (16–18) via bacterial products(19, 20) or metabolites (21, 22). However, the bacterial antigens to which the majority of colonic pTregs respond originate from gut bacteria taxa that are distinct from those identified to promote pTreg induction (16–18, 23, 24). This suggests that tolerance promoting bacterial taxa provide an environment promoting the induction of pTregs directed toward other antigens from other gut bacteria, which may enforce homeostasis and limit inflammatory responses when members of the gut microbiota are re-encountered in other settings. Yet, how and when the process of tolerance induction to the larger community of the gut microbiota occurs is largely unknown.
Initial exposure to microbes via the GI tract early in life has been associated with reduced risk for inflammatory disorders. Observations from multiple studies have suggested the ‘hygiene hypothesis’, where a decreased susceptibility for immune mediated diseases later in life is associated with early life exposure to microbes and microbial antigens, which are largely encountered in the GI tract (25, 26). Further, mouse studies have shown that initial exposure to microbes via the GI tract pre-weaning, but not later in life, reduces susceptibility to colitis later in life (27). These outcomes may be related to encountering microbial antigens in the setting of tolerance promoting gut bacterial taxa (16–18), establishing immune tolerance to these antigens and suppressing inflammatory responses upon future encounters. However, the role for tolerance-inducing gut bacteria is not straightforward, as these species are most abundant in later childhood and adulthood, when microbe introduction via the GI tract is less effective at reducing the risk of disease. Thus, there exist additional early life and time-limited aspects to the induction of immune tolerance to the gut microbiota.
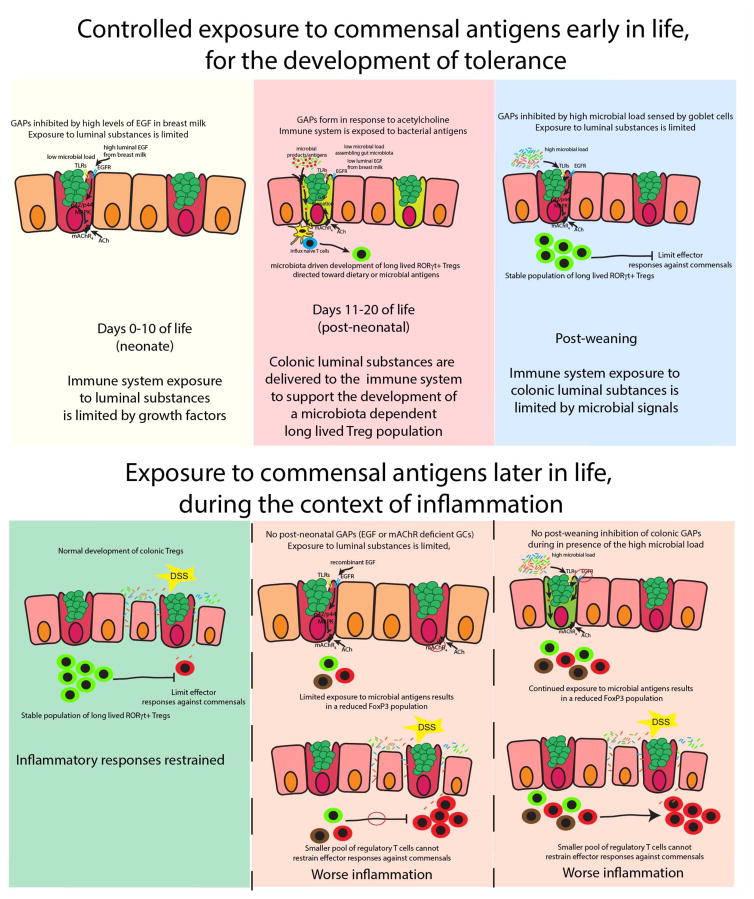
Here we identify distinct phases in early life in mice; the neonatal phase, days of life (DOL) 0–10, in which luminal antigens are not encountered by the small intestine (SI) or colonic immune system, the post-neonatal phase, DOL 11-weaning, in which luminal antigens are encountered almost exclusively by the colonic immune system, and the post-weaning phase, in which luminal antigens are encountered almost exclusively by the SI immune system. The inhibition of antigen delivery to the SI and colon in the neonatal phase and to the SI in the post-neonatal phase was mediated by high levels of luminal epidermal growth factor (EGF) acting on the EGFR on goblet cells (GCs) suppressing the formation of goblet cell associated antigen passages (GAPs). This pattern of antigen delivery allowed for the immune system’s encounter with gut microbial antigens and the development of pTregs directed toward members of the gut microbiota at this specific and crucial time in early-life. Inhibition of bacterial antigen encounter pre-weaning or shifting the timing of bacterial antigen encounter by the colonic immune system to the post-weaning period abrogated the development of pTregs directed toward members of the gut microbiota and resulted in worse colitis in response to epithelial injury and inflammatory responses to gut commensals later in life. These observations identify a specific period in early life that are critical for the induction of tolerance to the gut microbiota, which once established serves to limit inflammatory responses upon encounter of gut resident bacteria in inflammatory settings.
Results
Luminal antigen encounter by the gut immune system occurs in phases in early life
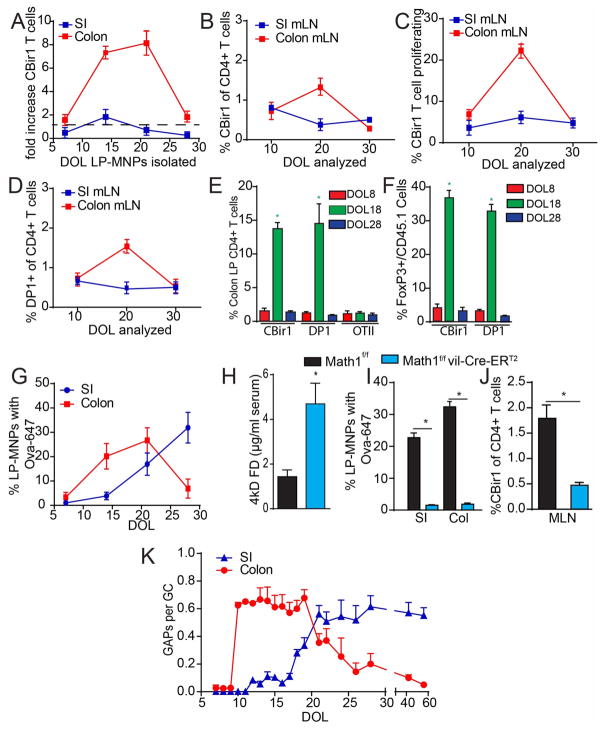
While multiple studies have documented that gut bacteria can affect the outcome of T cell responses, fewer have examined how T cells encounter and respond to gut commensal bacterial antigens. Results from the few studies examining this topic indicate that T cell responses to gut bacteria are dominated by a relatively small number of taxa that are closely adherent to the epithelium, such as segmented filamentous bacteria (SFB) or Helicobacter, while in contrast antigens from other bacteria, such as Bacteriodes vulgatus and the Lachnospiraceae bacterium A4, do not elicit responses from antigen specific T cells in the absence of inflammation (7, 24, 28, 29). A limitation of these studies is that they have examined adult mice or gnotobiotic mice, which may not reflect the physiologic interactions of the developing immune system with the microbiota in early life. To overcome this limitation, we isolated lamina propria (LP) CD11c+ MHCII+ populations, referred to as mononuclear phagocytes (MNPs), from the SI and colon of conventionally housed mice during early life and evaluated their ability to stimulate CBir1 CD4+ TCR transgenic T cells specific for a flagellin epitope produced by the Lachnospiraceae bacterium, A4, and COE1(12). We observed that SI LP-MNPs were relatively ineffective at inducing CBir1 T cell expansion at all days examined (Fig. 1A). In contrast colonic LP-MNPs isolated on DOL14 and DOL21, but not earlier on DOL7 or later on DOL28, were able to induce strong proliferation of CBir1 T cells suggesting these LP-MNPs were loaded with the CBir1 flagellin epitope (Fig. 1A). Adoptive transfer of CFSE labeled CBir1 T cells recapitulated the temporal and regional pattern of T cell stimulation in the gut in vivo, with effective accumulation and proliferation of CBir1 T cells in the colon draining MLN around DOL21 but not earlier on DOL10 or later on DOL30, and not in the SI draining MLN on any day examined (Fig. 1B and C, Fig. S1). Likewise, we observed that adoptively transferred DP1 CD4+ TCR transgenic T cells, specific for an antigen produced by B. vulgatus, localized to the colon draining MLN around DOL20 but not earlier on DOL10 or later on DOL30 and not to the SI draining MLN on any day examined (Fig. 1D). CBir1 and DP1 TCR transgenic T cells transferred on DOL18, but not earlier on DOL8 or later on DOL28, and not ovalbumin (Ova) specific OTII TCR transgenic T cells in the absence of Ova, expanded in the colon LP, and expressed Foxp3 seven days later (Fig. 1E and F, Fig. S1). This signifies that antigen specific Foxp3 pTreg responses are made towards multiple members of the commensal microbiota selectively during this pre-weaning interval. The lack of responses in the SI is likely due to the preferential localization of these bacterial taxa in the colon and the lack of responses in the colon around DOL10 may be accounted for by the low levels of these bacteria in the lumen during this time of life (Fig. S2). However, the lack of responses in the colon around DOL30 were paradoxical as the flagellin peptide recognized by CBir1 T cells and the B. vulgatus bacteria recognized by DP1 T cells became more abundant in the colonic luminal contents around DOL30 (Fig. S2), suggesting that LP-MNPs became unable to acquire these luminal microbial antigens.
Figure 1. Bacterial antigen encounter occurs during a specific pre-weaning interval, is dependent upon GCs, and correlates with the presence of colonic GAPs.
A) Fold increase in CBir1 T cells after ex vivo culture with LP-MNPs isolated from the SI or colon on DOL7, 14, 21, or 28. B) Percent of (CD45.1+) CBir1 T cells of total CD4+ T cells or C) percent of proliferating CBir1 T cells of the total CD45.1+ CBir1 T cells within the SI or colon draining MLNs three days following transfer into recipient CD45.2 mice on DOL 7,17, or 27. D) Percent of (CD45.1+) DP1 T cells of CD4+ T cells within the MLNs three days following transfer. E) Percent of CD45.1 cells of the total CD4+CD3+ T cell population or F) percentage of Foxp3+ cells of the CD45.1+ cells in the colon LP seven days following transfer of CD45.1+ CBir1, DP1 or OTII T cells into recipient mice not receiving ovalbumin; DOL represents day of transfer. G) Percentage of LP-MPNs staining for luminally administered Ova-647 on DOL7, 14, 21, or 28. H) 4kD FITC dextran in serum following gavage in DOL18 GC knockout and littermate control mice. I) Percentage of LP-MNPs staining for luminally administered Ova-647 in DOL18 GC knockout and littermate control mice J) Percent of CD45.1+ CBir1 cells of CD4+ T cells within the MLNs three days following transfer into DOL18 GC knockout and littermate control mice. M) Ratio of GAPs per GC in the SI and colon from DOL7-60. *= p<0.001, ns = not significant, n = 4 mice per group. Experiments in B, C, G, H, and K were repeated two independent times.
To overcome the inability to assure microbial antigen presence at all time points in early life, we evaluated the ability of SI and colon LP-MNPs to acquire a surrogate antigen Ova. Fluorescent Ova acquisition by colonic LP-MNPs occurred in a pattern similar to that of CBir1 antigen (Fig. 1G and Fig. S1). In contrast SI LP-MNPs acquired fluorescent Ova around the time of weaning, DOL21, with limited capacity at earlier time points (Fig. 1G). Thus, the inability to stimulate gut bacteria antigen T cell responses prior to DOL10 might reflect both the limited presence of the antigen and the inability of LP-MNPs to acquire luminal antigens, while in contrast the inability to stimulate gut bacteria antigen T cell response in the colon post-weaning is reflective of the inability of colonic LP-MNPs to acquire antigen. Further the lack of T cell responses to bacterial antigen in the SI is not due to the inability of SI LP-MNPs to acquire luminal antigens post-weaning, but likely is reflective of the low levels of this bacterial antigen in the SI lumen (Fig. S2).
Several processes have been implicated in luminal antigen capture by LP-MNPs, however these processes have not been investigated in the pre-weaning intestine. Consistent with observations in the adult intestine, villous M-cells, or M-cells in the non-follicle bearing epithelium, were absent from the colon pre-weaning (Fig. S3). CD11c+ APCs can extend trans-epithelial dendrites (TEDs) into the lumen of the SI for the purpose of sampling luminal antigens (30–32), however this has not been observed in the colon of adult mice (33, 34) where the majority of antigen is loading onto LP-MNPs prior to DOL 21 (Fig. 1A and G). In vivo two-photon imaging of a DOL21 CD11cYFP reporter mice did not reveal dendrites protruding past the intestinal epithelium in either the SI or the colon, though we could readily see dendrites in the LP probing the epithelium (Fig. S4). Studies evaluating TED formation in adult mice removed luminal contents and mucus prior to imaging (30–32, 35, 36), a process, which induces TED formation in the SI, but not the colon of adult mice (34). We observed that even after removal of the luminal contents and mucus layer, TEDs did not form in the SI or colon of DOL18 mice (Fig. S4). Goblet cells (GCs) can form goblet cell associated antigen passages (GAPs) and deliver luminal substances to LP-MNPs in the SI of adult mice. Mouse atonal homologue 1 (Math1) is a transcription factor required for GC development (37), and inducible deletion of Math1 in intestinal epithelial cells results in GC deletion (38–40). We observed that deletion of GCs in DOL18 Math1f/fvil-Cre-ERT2 mice resulted in a ~3 fold increase in intestinal permeability as evaluated by 4kD FITC dextran presence in the serum following gavage (Fig. 1H). Surprisingly, LP-MNPs from the SI or colon of DOL18 mice lacking GCs could not capture luminal fluorescent Ova (Fig. 1I and Fig. S1), yet had a normal number of LP-MNPs and could capture fluorescent Ova when administered systemically (Fig. S5). Moreover, CBir1T cells transferred into DOL 18 mice lacking GCs failed to expand in vivo (Fig. 1J), despite the ability of colonic LP-MNPs from mice lacking GCs to expand CBir1 T cells similar to controls when antigen was added to ex vivo cultures (Fig. S5). Thus, the presence of increased intestinal permeability did not correlate with the ability of LP-MNPs to capture luminal antigen and to stimulate immune responses in early life, and suggested that GCs and GAPs are required for LP-MNPs to acquire luminal antigens in the pre-weaning period. Therefore, we evaluated the regional pattern and timing of GAP formation during early life. GAPs were not seen in the colon or small intestine prior to DOL10 (Fig. 1K). Around DOL10, GAPs were present in the colon (Fig. S6) at a rate of 0.6 GAPs per goblet cells, and remained at that level until DOL21, the time of weaning, when GAP formation decreased to become rare in the colon at DOL24 (Fig 1K). At DOL18 GAPs became consistently present in the SI across multiple experiments and became prevalent at 0.6 GAPs per GC on DOL21 and remained at that level throughout adulthood (Fig. 1K). Thus, GCs were required for the delivery of antigens to the LP-MNPs in early life, and the presence of GAPs, but not TED formation, increased intestinal permeability, or villous M cells correlated with the regional pattern and timing of antigen delivery during early life. We defined three phases of luminal antigen delivery in the gut; the neonatal phase (DOL0-10) in which antigens are not delivered to the SI or colonic immune system, the post-neonatal phase (DOL11-weaning) in which antigens are predominantly delivered to the colonic immune system, and the post-weaning phase when antigens are delivered predominantly to the SI immune system.
The gut microbiota and breast milk control luminal antigen delivery in early life
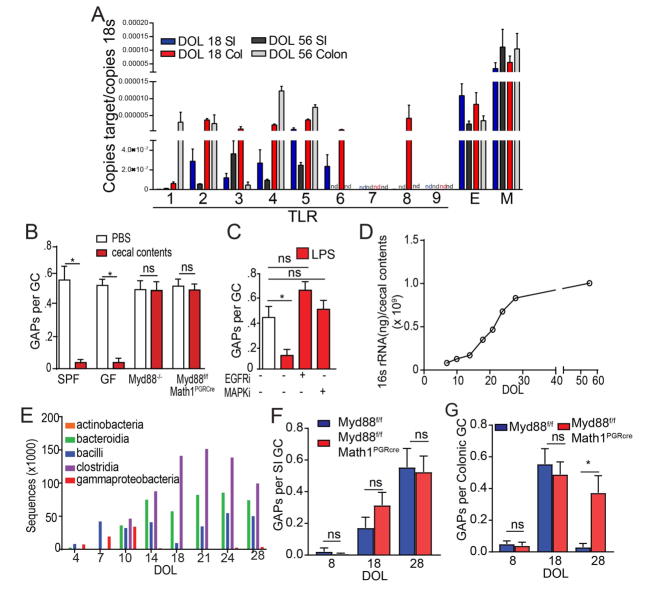
GCs in the colon of adult mice are largely unable to form GAPs due to Myd88 dependent GC intrinsic sensing of the abundant colonic microbes and microbial products (39). GC microbial sensing activates the epidermal growth factor receptor (EGFR) and p42/p44 mitogen activated protein kinase (MAPK), inhibiting GC responses to acetylcholine (ACh), the stimulus inducing spontaneous GAP formation (39, 41). Like intestinal GCs from adults, GCs from pre-weaning DOL18 mice expressed TLRs, Myd88, and EGFR (Fig. 2A) (39), suggesting that they would respond similarly to microbial products. Indeed, colonic GAPs on DOL18 could be inhibited by heat killed cecal contents from adult SPF housed mice and this inhibition was dependent upon Myd88 in Math1 expressing epithelial lineages, which is largely restricted to GCs in the colon (Fig. 2B). Intraluminal LPS inhibited GAP formation in the DOL18 colon, and this inhibition was dependent upon activation of EGFR and MAPK (Fig. 2C), confirming the pathways downstream of microbial sensing seen in GCs in the adult colon applies to pre-weaning colonic GCs as well. The gut microbiota undergoes dramatic changes in quantity and diversity from birth to weaning(42–45). The cecal bacterial load increased little during the neonatal phase, but then logarithmically increased during the post-neonatal phase and post-weaning before plateauing during adulthood (Fig. 2D). This quantitative increase in bacterial load correlated with qualitative changes as seen by 16s deep sequencing (Fig. 2E), with a switch from gammaproteobacteria to clostridia, bacilli, and bacteroidia in early life. Indeed, when Myd88 was deleted from GCs, GAPs formed spontaneously in the post-weaning colon, but the density of colonic GAPs in earlier phases of life, and in the small intestine remained unchanged (Fig. 2F and G), indicating that Myd88 dependent signals suppress GAP formation specifically in the colon, in the post-weaning period. Therefore, while both pre-weaning and post-weaning colonic GCs can respond to microbial signals to inhibit GAP formation, the gut microbiota is permissive for the spontaneous formation of colonic GAPs pre-weaning.
Figure 2. The microbiota inhibits colonic GAPs and antigen delivery post-weaning.
A) Expression of TLRs 1–9 (labeled by the number), Myd88 (M), and EGFR (E) on FACS-sorted GCs from the SI (blue) or colon (red) of DOL18 mice. B) Ratio of GAPs per GCs in the colon of DOL18 specific pathogen free (SPF) housed mice, germ-free (GF) housed mice, SPF housed Myd88−/− mice or SPF housed mice lacking Myd88 in GCs, with or without luminal heat-killed cecal contents from a DOL56 SPF housed mice. C) Ratio of GAPs per GC in the colon of DOL18 mice following luminal LPS with or without inhibition of EGFR (EGFRi) or p42/p44 MAPK (MAPKi) activation. D) Quantification of 16s rRNA in the cecal contents from DOL7-60 E) Number of 16s rRNA sequences grouped by bacteria class across the first 28 days of life. Ratio of GAPs per GC in the F) SI and G) colon of Myd88f/fMath1PGRCre mice lacking Myd88 in GCs, or cre negative littermates on DOL8, 18, or 28. nd = not detected, * = p<0.05, ns = not significant, n = 4 mice per group. Experiments in A were repeated three independent times, B, C, F, and G were repeated two independent times.
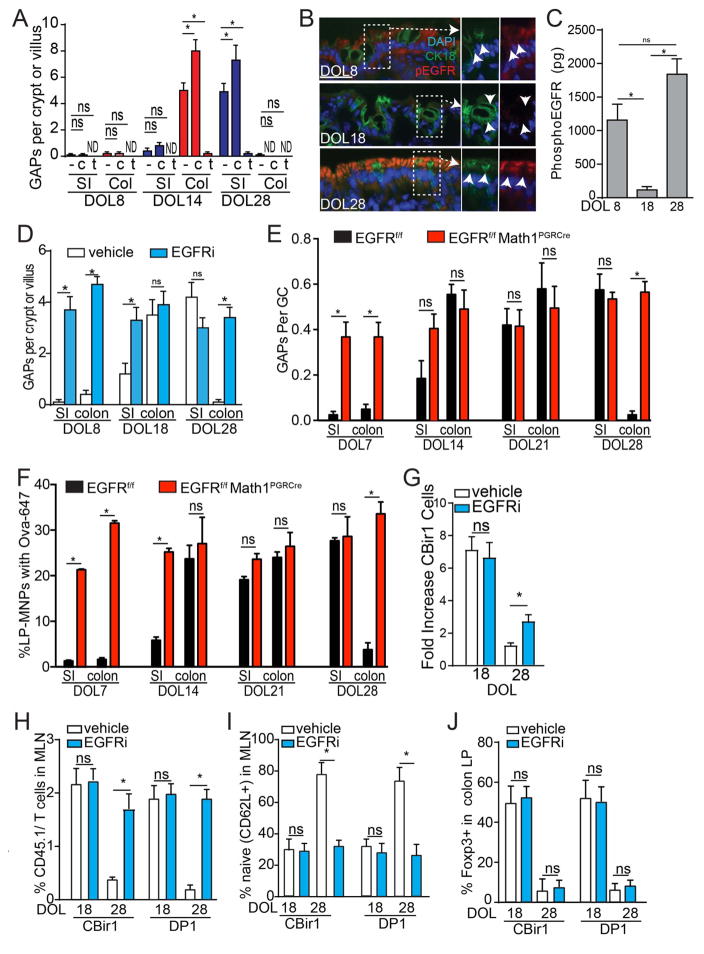
The lack of GAPs in the neonatal phase of mice lacking Myd88 in GCs could be due to the lack of ACh, the stimulus spontaneously inducing GAPs or due to lack of GC responsiveness to ACh due to other pathways activating the EGFR in GCs. We evaluated the ability of GCs to respond to ACh analogue carbamylcholine (CCh) to form GAPs. SI and colonic GCs in DOL8 mice, the neonatal phase, did not form GAPs in response to CCh (Fig. 3A). SI GCs in DOL 14 mice, the post-neonatal phase, were relatively unresponsive to CCh, while colonic GCs were responsive to CCh to form GAPs (Fig. 3A). Moreover, spontaneously forming colonic GAPs on DOL 14 were suppressed by tropicamide, an antagonist of muscarinic ACh receptor 4 (mAChR4; Fig. 3A), the ACh receptor on GCs inducing GAPs in the adult SI (39). In contrast, GCs in the post-weaning phase responded similar to what has been reported for adults (39) with SI GCs increasing GAPs in response to CCh and spontaneously forming GAPs via mAChR4 signals, and colonic GCs being unresponsive to CCh to form GAPs (Fig. 3A). This suppression post-weaning was relieved by deletion of Myd88 in GCs, which allowed GAPs to be form spontaneously in a mAChR4 dependent manner and to be further induced by CCh (Fig. S7). This indicates that the pattern of GC responsiveness to ACh to form GAPs changes throughout early life, prompting us to explore if other ligands/pathways activate EGFR and p42/p44 MAPK at these times of life. Consistent with EGFR activation inhibiting GAPs in the pre-weaning colon, phosphorylation of the EGFR in GCs inversely correlated with the presence of colonic GAPs pre-weaning (compare Fig. 3B–C with Fig. 1K) and inhibition of EGFR phosphorylation with tryphostin AG1483 (EGFRi), or deletion of EGFR in GCs, allowed GAP formation in the SI and colon during all phases of early life, independent of changes to the number of GCs (Fig. 3D–E, Fig. S6).
Figure 3. EGFR activation in GCs inhibits GAP formation and luminal antigen delivery throughout life.
A) GAPs per crypt or villius cross section in the SI (blue) or colon (red) of SPF housed mice in the presence of tropicamide (t), or carbamycholine (CCh) treatment on DOL8, 18, or 28. B) Immunofluorescent staining of phosphorylated EGFR (red; pEGFR) and cytokeratin 18 (green; CK18) in colon sections from DOL8, DOL18, and DOL28 mice; DAPI nuclear stain (blue). C) amount of phosphorylated EGFR in the colon epithelium colon of DOL8,18, or 28 mice measured by ELISA. D) GAPs per crypt (colon) or villus (SI) cross section in DOL8, 18, or 28 SPF housed mice in the presence or absence of EGFR inhibition (EGFRi). E) Ratio of GAPs per GC and F) percentage of LP-MNPs cells staining with luminal ova-647 in the SI and colon of DOL7, 14, 21, or 28 mice lacking EGFR in GCs or Cre negative littermate controls. G–J) Mice were treated with vehicle or inhibition of EGFR activation (EGFRi) on DOL14 and 16 or 24 and 26 and LP-MNPs isolated or TCR transgenic T cells adoptively transferred on DOL18 or 28 respectively. G) Fold increase in CBir1 transgenic T cells after ex vivo culture with LP-MNPs from colon isolated on DOL18 or DOL28. H) Percent of CD45.1+ CBir1 or DP1 cells of total CD4+ T cells in the MLNs three days following transfer into recipient mice on DOL18 or DOL 28. I) Percent of naïve (CD62L+) CBir1 or DP1 cells in the MLNs three days following transfer into recipient mice on DOL18 or DOL28. J) Percent of Foxp3+ CBir1 or DP1 cells in the colon LP seven days following transfer into recipient mice on DOL18 or DOL 28. * = p<0.05, ns = not significant, n = 4 mice per group. Experiments in A–D, and H were repeated three independent times, E–G, and I–J were repeated two independent times.
Intraluminal fluorescent Ova-647 was captured by SI and colonic LP-MNPs in all phases of life in mice lacking EGFR in GCs (Fig. 3F), indicating that EGFR activation in GCs was controlling the regional and temporal pattern of antigen delivery in early life. Inhibition of EGFR activation allowed colonic LP-MNPs from post-weaning mice to capture microbial antigen to stimulate CBir1 T cells ex-vivo (Fig. 3G). The presence of GAPs correlated with MNP recruitment to the colonic epithelium and the acquisition of luminal ova (Fig. S5). However, these GAP manipulations did not affect the overall number of MNPs in the colonic LP, the ability of MNPs to capture systemic antigens, or their ability to stimulate bacterial antigen specific T cell proliferation when antigen was added to ex vivo cultures (Fig. S5). This indicates that GAP manipulation specifically affected luminal antigen delivery to MNPs but not their presence within the colonic LP or their antigen presentation capacity. Additionally, inhibition of EGFR activation allowed adoptively transferred CBir1 or DP1 cells to expand and become activated in the MLN of post-weaning mice while cells transferred into post-weaning vehicle treated mice, lacking colonic GAPs, remained naïve (Fig. 3H and I). However, unlike microbial antigen specific T cells encountering antigen in the post-neonatal phase of life, DOL18, CBir1and DP1 T cells encountering microbial antigen post-weaning, DOL28, had little Foxp3 expression seven days post transfer (Fig. 3J), indicating that the colon LP of post-neonatal mice is uniquely favorable for the induction of regulatory responses.
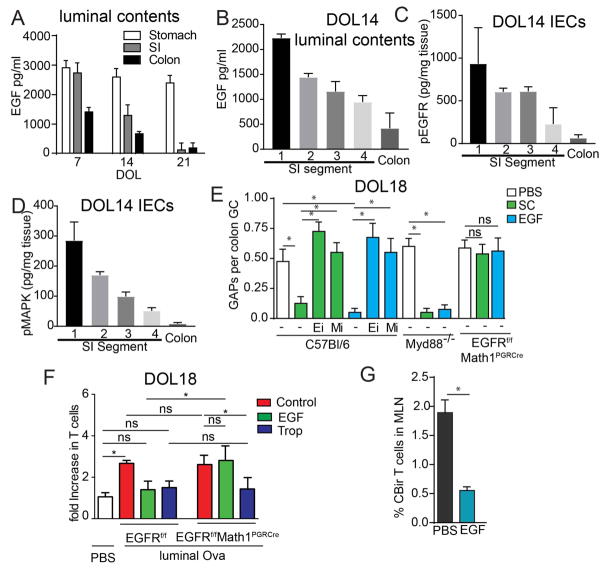
Because pathways independent of Myd88 were activating EGFR in GCs to suppress GAPs, we evaluated the pattern of EGFR ligands in the pre-weaning gut. EGFR ligands, including EGF, amphiregulin, and heparin-binding EGF like growth factor, are abundant in the breast milk after parturition and decrease throughout lactation until weaning (46–50). Of these, EGF is the most abundant in breast milk, being ~5 fold higher than the others (46). Furthermore, ingested breast milk EGF reaches the offspring’s GI tract in a biologically active form (49, 51, 52). We observed that EGF was present in luminal contents throughout the GI tract of mice prior to weaning in a concentration pattern inversely correlating with the presence of GAPs (Fig. 4A compare with Fig. 1K). The concentration of EGF decreased both throughout the GI tract, and over time, and correlated with the activation of the EGFR and p42/p44 MAPK in the epithelium (Fig 4A–D), indicating that the proximal to distal gradient and temporal decrement of luminal EGF could allow GAP formation to occur in a distal to proximal and temporal pattern as we observed in the gut during early life. Luminal EGF inhibits GAPs(39), and accordingly the stomach contents from DOL10 mice, which contains high levels of EGF, inhibited GAPs in the colon of DOL18 mice in a manner dependent upon EGFR and p42/p44 MAPK activation (Fig. 4E). Stomach contents also inhibited colonic GAP formation in DOL18 Myd88−/− mice (Fig. 4E) confirming the inhibitory ligands present in the stomach contents of neonatal mice were not microbial ligands acting via Myd88. Indeed, luminal recombinant EGF inhibited GAPs and luminal antigen loading of LP-MNPs in post-neonatal colon, in a EGFR and MAPK dependent, but Myd88 independent manner (Fig. 4E–F and Fig. S7). Moreover, CBir1 cells failed to expand in the colon draining MLN three days post transfer in post-neonatal mice given intracolonic recombinant EGF (Fig. 4G). These data indicate that breast milk EGF and potentially other EGFR ligands control the temporal and regional luminal antigen exposure of the gut immune system pre-weaning.
Figure 4. Luminal EGF inhibits GAPs and antigen delivery prior to weaning.
A) Concentration of EGF in the luminal contents of the stomach, SI, or colon in on DOL7, 14, or 21. B) Concentration of EGF in the luminal contents, C) amount of phosphorylated EGFR in the epithelium or D) amount of phosphorylated p42/p44 MAPK in the epithelium of SI segments or colon of DOL14 mice measured by ELISA; SI segments, 1=0–6 cm, 2=6–12 cm, 3=12–18cm, 4=18–24cm measured from the pylorus. E) Ratio of GAPs per GC following incubation with stomach contents (SC) from DOL10 mice, or recombinant EGF (EGF) in the presence or absence of EGFR inhibition (EGFRi) or p42/p44MAPK inhibition (MAPKi). F) Fold increase in Ova specific OTI T cells after culture with colonic LP-MNPs isolated from DOL18 mice lacking EGFR in GCs or Cre negative littermate controls given luminal Ova or PBS and treated with intracolonic EGF, i.p. tropicamide (Trop), or untreated (Control). G) Percentage of CBir1 T cells of total CD4+ T cells in the MLNs three days following cell transfer into DOL18 in mice that received intracolonic EGF (1μg) or PBS daily from DOL10-21. * = p <0.05, ns = not significant, n = 4 mice per group. Experiments in A–D, F, and G were repeated two independent times. E was repeated three independent times.
Altering the timing of exposure to bacterial antigens results in loss of tolerance to gut bacteria, worsened colitis, and inflammatory responses to gut bacteria following epithelial injury
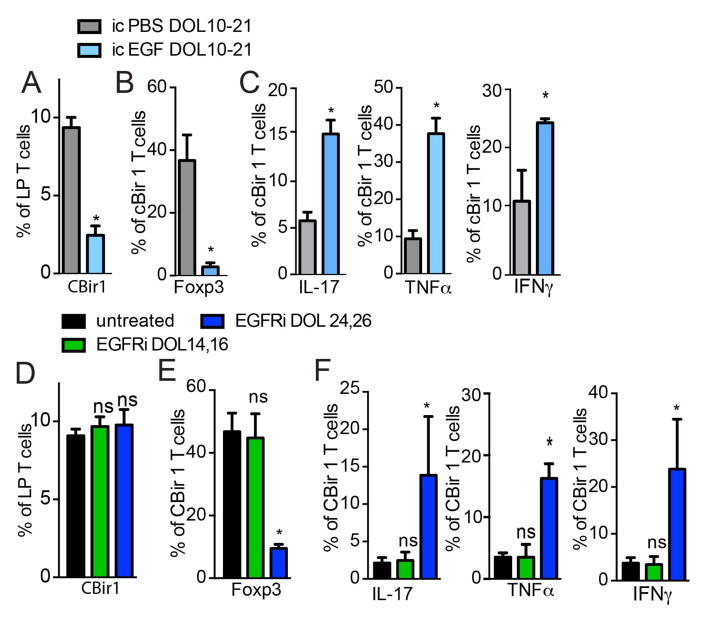
We evaluated whether GAP dysregulation or disruption affects the development of microbial specific Tregs. CBir1 T cells transferred into mice given intracolonic EGF on DOL10-21, to inhibit colonic GAPs, had significantly less expansion and expressed significantly less Foxp3 on DOL30 (Fig. 5A and B and Fig. S8). However, a significantly larger proportion of the remainig CBir1 T cell population expressed inflammatory cytokines, when compared to PBS treated control mice (Fig. 5C). Conversely, inhibition of EGFR on DOL14 and 16, during the post-neonatal phase, or on DOL24 and 26, post-weaning, did not affect the expansion of transferred CBir1 T cells (Fig. 5D). However, unlike control treated mice or mice given EGFR inhibitors on DOL14 and 16 during the post-neonatal phase, CBir1 T cells in mice given EGFR inhibitors on DOL24 and 26 post-weaning, to allow encounters with microbial antigen outside of the normal window, expressed significantly less Foxp3 and a significantly greater proportion of the population expressed inflammatory cytokines (Fig. 5E and F). Inhibition of EGFR during the post-neonatal phase on DOL14 and 16, when colonic GAPs are already present, resulted in no significant difference in the expression of Foxp3+ or inflammatory cytokines by transferred CBir1 T cells when compared with controls (Fig. 5E and F). This supports that the effects of EGFR inhibition post-weaning on CBir1 T cells is a result of the induction of colonic GAPs and extension of the window of microbial antigen encounter.
Figure 5. Inhibiting or altering the timing of microbial antigen encounter results in inflammatory T cell responses against gut bacteria.
A) percentage of CD45.1+ CBir1 T cells of total colon LP CD4+ T cells or B) percentage of Foxp3+ or C) IL-17+, TNFα+, or IFNγ+ CBir1 T cells among all CBir1 T cells in the colon LP following transfer on DOL16 and analysis on DOL30 in mice treated with intracolonic (ic) vehicle (PBS), or EGF on DOL10-21. D) percentage of CD45.1+ CBir1 T cells of total colon LP CD4+ T cells or E) percentage of Foxp3+ or F) IL-17+, TNFα+, or IFNγ+ CBir1 T cells among all CBir1 T cells in the colon LP following transfer on DOL16 and analysis on DOL30 in mice treated with inhibition of EGFR activation (EGFRi) on DOL14 and 16, or on DOL24 and 26. * = p<0.05, ns = not significant, n = 4 mice per group.
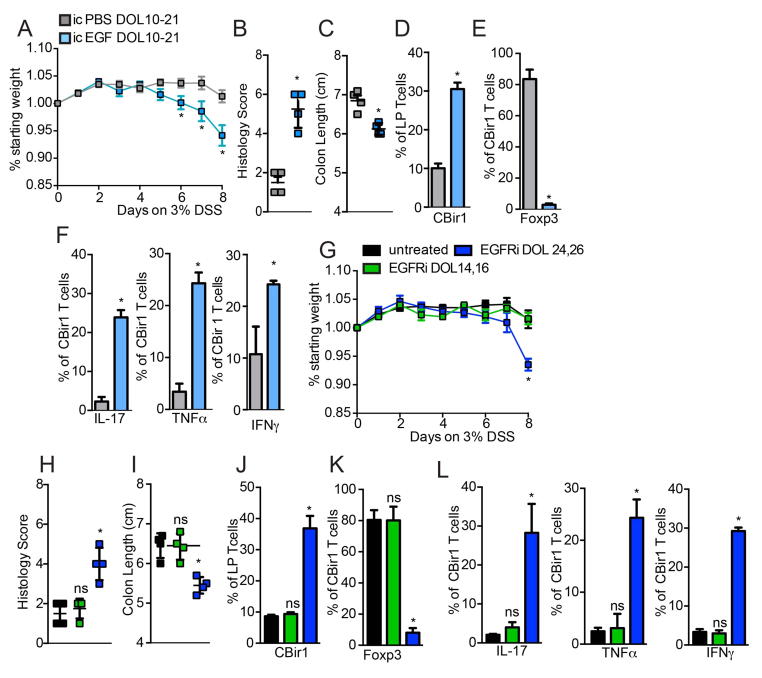
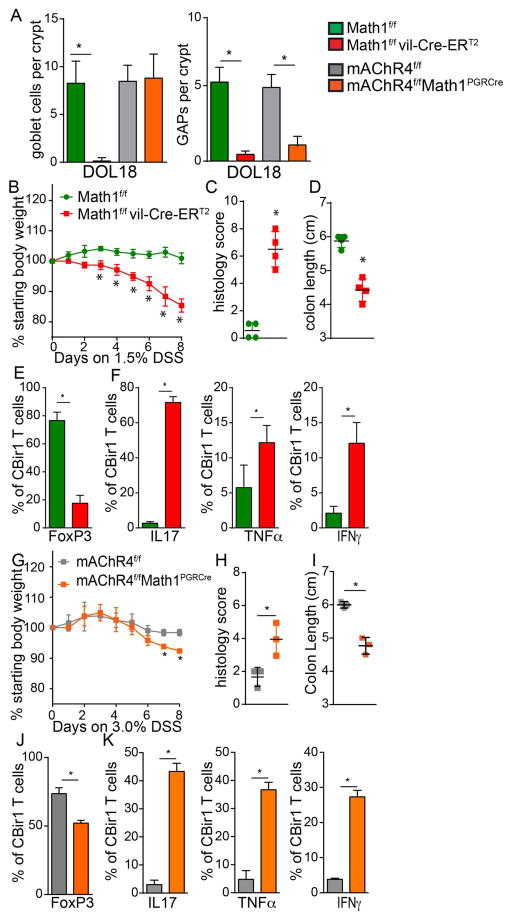
Altering the timing of microbial antigen encounter resulted in inflammatory cytokine production in the MLN one week later, but no overt pathology in the colon (Fig S9). We reasoned that the lack of pathology might in part be due to the relatively low level of microbial antigen encountered by the immune system post-weaning, as we observed little or no responses by bacterial antigen specific T cells adoptively transferred into mice post-weaning (Fig. 1B–D). Therefore, mice given intracolonic EGF on DOL10-21 to inhibit microbial antigen encounter and pTreg development, mice given EGFR inhibitors post-weaning on DOL 24 and 26 to alter the timing of microbial antigen encounter, and control mice were given 3% DSS in drinking water on DOL30-38 to disrupt the colonic epithelial barrier and allow exposure of the immune system to luminal bacteria. Mice treated with EGF on DOL10-21, or EGFR inhibition on DOL 24 and 26, lost significantly more weight (Fig. 6A and G) and developed significantly worse disease as evidenced by histology demonstrating increased edema, increased ulceration, increased cellular infiltration, and shortened colons (Fig. 6B, C, H, and I and Fig. S10). In untreated mice, mice receiving intrarectal PBS on DOL10-21, and mice receiving EGFR inhibitors in the post-neonatal phase on DOL14 and 16 when colonic GAPs are normally present, transferred CBir1 T cells did not expand further in the colon LP (compare Fig. 1E with Fig. 6D and J), and continued expressing Foxp3 but not inflammatory cytokines after DSS treatment (Fig. 6E, F, K and L). However, in mice where microbial antigen encounter was inhibited by EGF, or inappropriately induced post-weaning by EGFR inhibition on DOL24 and 26, DSS treatment resulted in significant expansion of CBir1 T cells in colon LP (Fig. 6D and J). The transferred CBir1 T cells had significantly reduced Foxp3 expression and significantly increased inflammatory cytokines (Fig. 6E, F, K, and L) indicative of effector T cell expansion during inflammation in response commensal bacteria. Similarly, mice lacking GCs and GAPs beginning on DOL 12, or mice with deletion of mAChR4 on GCs between DOL 10 and 21, which have GCs but are unable to form GAPs during this interval (Fig 7A), experienced worse DSS induced colitis (Fig. 7B–D and G–I). Similar to mice treated with EGF on DOL 10–21 to inhibit GAPs during the post-neonatal phase, CBir1 T cells adoptively transferred on DOL16 into mice lacking GCs or GAPs had significantly reduced Foxp3 expression and significantly increased inflammatory cytokines following disruption of the epithelial barrier with DSS (compare Fig. 6F and G with Fig. 7G, H, N and O). These data demonstrate that altering GAPs and the timing of gut bacterial antigen encounter in early life is a critical component to establish durable tolerance and limit inflammatory responses upon encounter later in life.
Figure 6. Inhibiting or altering the timing of microbial antigen encounter results in inflammatory T cell responses against gut bacteria and worsened colitis upon epithelial damage.
A–F) Mice were given intracolonic PBS or EGF on DOL10-21 or G–L) inhibition of EGFR activation (EGFRi) on DOL 14 and 16 or 24 and 26, adoptively transferred CBir1 T cells on DOL 16, given DSS in drinking water from DOL30-38. A) and G) Weight loss B) and H) histology score C) and I) colon length, D) and J) percentage of CBir1 T cells in the colon LP and percentage of E) and K) Foxp3+ and F) and L) percentage of IL-17+, TNFα+, or IFNγ+ CBir1 T cells following 8 days of DSS treatment. * = p<0.05, ns = not significant, n = 4 mice per group.
Figure 7. Deletion of GCs or GAPs during early life results in inflammatory T cell responses against gut bacteria and worsened colitis later in life.
A) Enumeration of GCs and GAPs per colonic crypt in DOL18 GC deficient mice or mice with mAChR4 deleted from GCs and their littermate controls. A–F) GCs were deleted beginning on DOL12 or G–K) mAChR4 was deleted from GCs between DOL10-21, CBir1 T cells adoptively transferred on DOL16, and mice placed on DSS from DOL30-38. B and G) Weight loss C) and H) histology score D) and I) colon length, E) and J) percentage of CBir1 T cells expressing Foxp3, and F and K) percentage of CBir1 T cells expressing IL17, TNFα, or IFNγ in the colon LP following 8 days of DSS treatment. * = p<0.05, ns = not significant, n = 4 mice per group A–F, n=3 mice per group G–K.
Discussion
Here we identify a specific pre-weaning interval in which bacterial antigens are encountered by the colonic immune system. Surprisingly, in the course of normal development the immune system’s encounter with some bacterial antigens is largely restricted to this interval in the post-neonatal phase of life. However, a limitation of this study is that these events, and their time-limited nature, do not pertain to all gut bacterial antigens, as some bacteria in the fully developed gut microbiota are not present during this interval, and gut bacteria closely adhering to the epithelium have been observed to generate antigen specific T cell responses outside of this interval in later life (24, 28, 29). Blocking the encounter of the immune system with bacterial products during this window resulted in failure to develop pTregs specific for antigens from these bacteria that was not compensated for by later encounters, indicating that the timing of these events is critical for developing tolerance to some gut bacteria. Further, initial encounter of these bacterial antigens outside of this interval or extending encounters beyond this interval resulted in the induction of effector T cell responses to gut bacteria. Thus, the encounter of antigens from some gut bacterial taxa during a specific pre-weaning interval is critical for the development of tolerance to members of the gut microbiota.
The microbiota drives the development of a long-lived population of pTregs in the pre-weaning colon, and that these pTregs can control effector responses (53–55). Superimposing our findings on these studies suggests that these pTregs can be specific microbial antigens encountered in the post-neonatal phase, and that these encounters serve to develop a long-lived pTreg population with the ability to control inflammatory responses. While a limitation of our study is that it is restricted to mice, it is notable that individuals with Crohn’s disease make systemic immune responses to the CBir1 antigen, and we observed that pTregs specific for CBir1 antigen were largely induced during the post-neonatal phase of life in mice. This indicates that the events we identified in pre-weaning mice have relevance to humans and combined with observations that antibiotic use in the first year of life is associated with an increased incidence of asthma, allergy and inflammatory bowel disease (56–58), strongly suggests that altering microbial antigen encounters during specific intervals in pre-weaning children increases the risk of disease.
Effective acquisition of luminal antigens by LP-MNPs was abrogated in the absence of GCs and correlated with the density of GAPs in all situations, and was not correlated with paracellular leak, villous M-cells, or the extension of TEDs by LP-MNPs. While it is not possible to entirely exclude contributions from these other pathways, and a limitation is that the pharmacologic and genetic manipulations may have other effects beyond altering GAPs, multiple strategies manipulating GAPs correlated with antigen acquisition by LP-MNPs strongly implicating that GAPs are major contributors to antigen acquisition in the pre-weaning intestine. The stratified mucus layer, which separates bacteria from the epithelium, develops in the first week of life (59–61). However, this mucus layer was not sufficient to prevent encounters of the colonic immune system with gut bacterial antigens. This suggests that bacterial antigens, as opposed to whole bacteria, are delivered via GAPs, or alternatively encounters with live bacteria occur in the proximal colon where the mucus layer is more permeable(62). The need to restrict the encounter of gut bacterial antigens with the immune system to a specific interval is not well understood. The need to limit encounters in the neonatal phase might be related to the limited diversity of the gut bacterial population and limited T cell repertoire (63, 64). This might preclude developing a diverse and stable responding T cell population as well as Th2 skewing from T cells derived from fetal T cell precursors (65, 66), resulting in unbalanced responses to gut bacterial antigens. The need to limit exposure to gut bacterial antigens after weaning, when the immune system is fully competent may be related to the complex environment in which these antigens are encountered, as we observed that inappropriate exposure to gut bacterial antigens post-weaning does not result in the induction of pTregs, and previous observations indicate these encounters result in inflammatory responses (34, 41). In total, these observations indicate that timing of exposure is a critical component to the development of tolerance to some gut bacterial antigens, and highlights the unique and time-limited events occurring in the pre-weaning GI tract.
The gut microbiota contributes to multiple facets of health, and dysbioisis of the gut microbiota is implicated to contribute to multiple diseases. Interactions of the gut microbiota with the immune system shape each other resulting in co-development of these two organs, and accordingly these interactions are important to establish a stable relationship that is able to withstand transient perturbations by either member. In fact, the unstable relationship between the immune system and the gut microbiota, characterized by reciprocal inappropriate inflammatory responses and dysbiosis, is believed to underlie the pathogenesis of IBD. Here we demonstrate that the encounters of bacterial antigens with the immune system that establish this relationship are much more complex than previously appreciated and extend beyond the mere presence of specific bacterial taxa in the lumen and responding T cells in the immune compartment. For antigens from some bacterial taxa, these encounters initially occur, and are largely limited to, a specific interval in early life, which is critical for developing tolerance to these bacteria and fostering a stable relationship with our gut microbiota.
Materials and Methods
Mice
All mice were ten generations or greater on the C57BL/6 background, with the exception of mAChr4f/f mice, which were six or more generations on the C57BL/6 background. C57BL/6 mice, OTII T cell receptor transgenic mice(67), and CD11cYFP transgenic mice(68), Math1f/f mice(37), Myd88f/f mice (69), Math1PGRCre mice(70) were purchased from The Jackson Laboratory (Bar Harbor, ME) and bred and maintained in house. EGFRf/f mice(71) were a gift from Dr. David Threadgill (University of North Carolina, Chapel Hill, North Carolina). Myd88−/− mice(72), a gift from Dr. Akira (Osaka University, Osaka, Japan), were bred onto the C57BL/6 background by the Speed Congenics Facility of the Rheumatic Diseases Core Center, and maintained in house. mAChR4f/f mice (73) were a kind gift from Jurgen Wess (National Institute Health, Bethesda, MD). CBir1 TCR transgenic mice and DP1 transgenic mice have been previously described (12, 23). Transgenic mice bearing a tamoxifen-dependent Cre recombinase expressed under the control of the villin promoter (vil-Cre-ERT2) mice (74) were a gift from Sylvie Robine (Institut Curie, Paris, France). To deplete Myd88, EGFR, or mAChR4 in GCs, mice were bred to generate Myd88f/fMath1PGRCre mice, EGFRf/fMath1PGRCre mice, and mAChR4f/fMath1PGRCre mice. These mice and Cre negative littermate controls were injected i.p. with mifepristone (10mg/kg) every day starting four days prior to use in experiments. Generation of Math1fl/fl ERT2ViCre mice and inducible deletion of GCs by treatment with tamoxifen has been previously described(39). Mice used in these experiments were bred in house and weaned at DOL 21. Cohoused littermates were used as experimental groups and controls to minimize differences in the gut microbiota. For pharmacologic manipulations, littermates of the same sex were assigned a cage which contained animals of all treatment groups. For genetic manipulations, cages of weaned mice contained littermates of the same sex and both genotypes. Animal procedures and protocols were carried out in accordance with the institutional review board at Washington University School of Medicine.
Statistical Analysis
Data analysis using a student’s t test or one-way ANOVA with a Dunnett or Tukey test to correct for multiple comparisons was performed using GraphPad Prism (GraphPad Software Inc., San Diego, CA). Biologic variables measured were normally distributed. Test for significance were two-sided. Alpha of p value <0.05 was considered significant.
Supplementary Material
Figure 8.
Summary Schematic
Acknowledgments
The authors would like to thank Dr. Mark J. Miller for insightful critiques and assistance with 2P imaging and analysis. The 2P imaging was performed at the In Vivo Imaging Core at Washington University School of Medicine. The Washington University Digestive Diseases Research Center Core (DDRCC) Gnotobiotic Facility, supported by NIH grant P30 DK052574, provided the germfree mice. The High Speed Cell Sorter Core at the Alvin J. Siteman Cancer Center at Washington University School of Medicine and Bar nes-Jewish Hospital in St. Louis, MO, supported in part by NCI Cancer Center Support Grant P30 CA91842, provided flow cytometric cell sorting services. The Speed Congenics Facility of the Rheumatic Diseases Core Center, supported by NIH grant P30AR048335 bred the Myd88−/− mice onto the C57BL/6 background.
Funding: Supported by grants: AI131342-RDN and PIT, DK097317-RDN, AI1126260 – SPH and RDN, Children’s Discovery Institute Grant MD-II-2015-481–RDN and PIT, P30 DK052574, and DK109006-KAK.
Footnotes
Author Contributions: K.A.K. performed animal colitis models, T cell transfers, ELISAs, cell isolation, in vitro studies, microarray analysis, flow cytometry, and data analysis. K.A.K, J.K.G, K.G.M, and P.E.C. performed immunofluorescence and data analysis. K.A.K., and K.G.M. performed two-photon imaging experiments and data analysis. K.A.K, S.M., and D.K. performed RTPCR and data analysis. P.I.T, C.H, S.P.H and C.O.E. provided input on experimental design and data analysis. K.A.K, and R.D.N conceived of the study, directed the experiment design, analyzed the data, and wrote the initial manuscript draft. All authors reviewed and discussed the manuscript.
Competing interests: The authors declare no competing interests.
Data and Materials Availability: Data and materials are available upon request from the corresponding author.
References
- 1.Turnbaugh PJ, et al. The human microbiome project. Nature. 2007;449:804–810. doi: 10.1038/nature06244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ng KM, et al. Microbiota-liberated host sugars facilitate post-antibiotic expansion of enteric pathogens. Nature. 2013;502:96–99. doi: 10.1038/nature12503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim YG, et al. Neonatal acquisition of Clostridia species protects against colonization by bacterial pathogens. Science. 2017;356:315–319. doi: 10.1126/science.aag2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Subramanian S, et al. Persistent gut microbiota immaturity in malnourished Bangladeshi children. Nature. 2014;510:417–421. doi: 10.1038/nature13421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Charbonneau MR, et al. Sialylated Milk Oligosaccharides Promote Microbiota-Dependent Growth in Models of Infant Undernutrition. Cell. 2016 doi: 10.1016/j.cell.2016.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mazmanian SK, Liu CH, Tzianabos AO, Kasper DL. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell. 2005;122:107–118. doi: 10.1016/j.cell.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 7.Hand TW, et al. Acute gastrointestinal infection induces long-lived microbiota-specific T cell responses. Science. 2012;337:1553–1556. doi: 10.1126/science.1220961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lodes MJ, et al. Bacterial flagellin is a dominant antigen in Crohn disease. J Clin Invest. 2004;113:1296–1306. doi: 10.1172/JCI20295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Quinton JF, et al. Anti-Saccharomyces cerevisiae mannan antibodies combined with antineutrophil cytoplasmic autoantibodies in inflammatory bowel disease: prevalence and diagnostic role. Gut. 1998;42:788–791. doi: 10.1136/gut.42.6.788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Targan SR, Karp LC. Defects in mucosal immunity leading to ulcerative colitis. Immunol Rev. 2005;206:296–305. doi: 10.1111/j.0105-2896.2005.00286.x. [DOI] [PubMed] [Google Scholar]
- 11.Cohavy O, et al. Colonic bacteria express an ulcerative colitis pANCA-related protein epitope. Infect Immun. 2000;68:1542–1548. doi: 10.1128/iai.68.3.1542-1548.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cong Y, Feng T, Fujihashi K, Schoeb TR, Elson CO. A dominant, coordinated T regulatory cell-IgA response to the intestinal microbiota. Proc Natl Acad Sci U S A. 2009;106:19256–19261. doi: 10.1073/pnas.0812681106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gilbert JA, et al. Microbiome-wide association studies link dynamic microbial consortia to disease. Nature. 2016;535:94–103. doi: 10.1038/nature18850. [DOI] [PubMed] [Google Scholar]
- 14.Nagao-Kitamoto H, et al. Functional Characterization of Inflammatory Bowel Disease-Associated Gut Dysbiosis in Gnotobiotic Mice. Cell Mol Gastroenterol Hepatol. 2016;2:468–481. doi: 10.1016/j.jcmgh.2016.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Elinav E, et al. NLRP6 inflammasome regulates colonic microbial ecology and risk for colitis. Cell. 2011;145:745–757. doi: 10.1016/j.cell.2011.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Round JL, Mazmanian SK. Inducible Foxp3+ regulatory T-cell development by a commensal bacterium of the intestinal microbiota. Proc Natl Acad Sci U S A. 2010;107:12204–12209. doi: 10.1073/pnas.0909122107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Atarashi K, et al. T induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature. 2013 doi: 10.1038/nature12331. [DOI] [PubMed] [Google Scholar]
- 18.Atarashi K, et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science. 2011;331:337–341. doi: 10.1126/science.1198469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mazmanian SK, Round JL, Kasper DL. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature. 2008;453:620–625. doi: 10.1038/nature07008. [DOI] [PubMed] [Google Scholar]
- 20.Chu H, et al. Gene-microbiota interactions contribute to the pathogenesis of inflammatory bowel disease. Science. 2016;352:1116–1120. doi: 10.1126/science.aad9948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Furusawa Y, et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature. 2013;504:446–450. doi: 10.1038/nature12721. [DOI] [PubMed] [Google Scholar]
- 22.Arpaia N, et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature. 2013;504:451–455. doi: 10.1038/nature12726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lathrop SK, et al. Peripheral education of the immune system by colonic commensal microbiota. Nature. 2011;478:250–254. doi: 10.1038/nature10434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chai JN, et al. Helicobacter species are potent drivers of colonic T cell responses in homeostasis and inflammation. Science Immunology Pending publication. 2017 doi: 10.1126/sciimmunol.aal5068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.von Mutius E. Allergies, infections and the hygiene hypothesis--the epidemiological evidence. Immunobiology. 2007;212:433–439. doi: 10.1016/j.imbio.2007.03.002. [DOI] [PubMed] [Google Scholar]
- 26.Strachan DP. Hay fever, hygiene, and household size. BMJ. 1989;299:1259–1260. doi: 10.1136/bmj.299.6710.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Olszak T, et al. Microbial exposure during early life has persistent effects on natural killer T cell function. Science. 2012;336:489–493. doi: 10.1126/science.1219328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gaboriau-Routhiau V, et al. The key role of segmented filamentous bacteria in the coordinated maturation of gut helper T cell responses. Immunity. 2009;31:677–689. doi: 10.1016/j.immuni.2009.08.020. [DOI] [PubMed] [Google Scholar]
- 29.Ivanov II, et al. Induction of Intestinal Th17 Cells by Segmented Filamentous Bacteria. Cell. 2009;139:485–498. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Niess JH, et al. CX3CR1-mediated dendritic cell access to the intestinal lumen and bacterial clearance. Science. 2005;307:254–258. doi: 10.1126/science.1102901. [DOI] [PubMed] [Google Scholar]
- 31.Chieppa M, Rescigno M, Huang AY, Germain RN. Dynamic imaging of dendritic cell extension into the small bowel lumen in response to epithelial cell TLR engagement. J Exp Med. 2006;203:2841–2852. doi: 10.1084/jem.20061884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Farache J, et al. Luminal bacteria recruit CD103+ dendritic cells into the intestinal epithelium to sample bacterial antigens for presentation. Immunity. 2013;38:581–595. doi: 10.1016/j.immuni.2013.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hapfelmeier S, et al. Microbe sampling by mucosal dendritic cells is a discrete, MyD88-independent stepin invG S. Typhimurium colitis. Journal of Experimental Medicine. 2008;205:437–450. doi: 10.1084/jem.20070633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Knoop KA, et al. Antibiotics promote the sampling of luminal antigens and bacteria via colonic goblet cell associated antigen passages. Gut microbes. 2017:1–12. doi: 10.1080/19490976.2017.1299846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vallon-Eberhard A, Landsman L, Yogev N, Verrier B, Jung S. Transepithelial pathogen uptake into the small intestinal lamina propria. Journal of immunology (Baltimore, Md: 1950) 2006;176:2465–2469. doi: 10.4049/jimmunol.176.4.2465. [DOI] [PubMed] [Google Scholar]
- 36.Kim KW, et al. In vivo structure/function and expression analysis of the CX3C chemokine fractalkine. Blood. 2011;118:e156–167. doi: 10.1182/blood-2011-04-348946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shroyer NF, et al. Intestine-specific ablation of mouse atonal homolog 1 (Math1) reveals a role in cellular homeostasis. Gastroenterology. 2007;132:2478–2488. doi: 10.1053/j.gastro.2007.03.047. [DOI] [PubMed] [Google Scholar]
- 38.McDole JR, et al. Goblet cells deliver luminal antigen to CD103+ dendritic cells in the small intestine. Nature. 2012;483:345–349. doi: 10.1038/nature10863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Knoop KA, McDonald KG, McCrate S, McDole JR, Newberry RD. Microbial sensing by goblet cells controls immune surveillance of luminal antigens in the colon. Mucosal Immunology. 2015;8:198–210. doi: 10.1038/mi.2014.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Peignon G, et al. Complex interplay between beta-catenin signalling and Notch effectors in intestinal tumorigenesis. Gut. 60:166–176. doi: 10.1136/gut.2009.204719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Knoop KA, McDonald KG, Kulkarni DH, Newberry RD. Antibiotics promote inflammation through the translocation of native commensal colonic bacteria. Gut. 2016;65:1100–U1160. doi: 10.1136/gutjnl-2014-309059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Eckburg PB, et al. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Koenig JE, et al. Succession of microbial consortia in the developing infant gut microbiome. Proc Natl Acad Sci U S A. 2011;108(Suppl 1):4578–4585. doi: 10.1073/pnas.1000081107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yatsunenko T, et al. Human gut microbiome viewed across age and geography. Nature. 2012;486:222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Qin J, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kobata R, et al. High levels of growth factors in human breast milk. Early human development. 2008;84:67–69. doi: 10.1016/j.earlhumdev.2007.07.005. [DOI] [PubMed] [Google Scholar]
- 47.Nojiri T, et al. Clinical significance of amphiregulin and epidermal growth factor in colostrum. Archives of gynecology and obstetrics. 2012;286:643–647. doi: 10.1007/s00404-012-2365-8. [DOI] [PubMed] [Google Scholar]
- 48.Matsuoka Y, Idota T. The concentration of epidermal growth factor in Japanese mother’s milk. Journal of nutritional science and vitaminology. 1995;41:241–251. doi: 10.3177/jnsv.41.241. [DOI] [PubMed] [Google Scholar]
- 49.Britton JR, George-Nascimento C, Koldovsky O. Luminal hydrolysis of recombinant human epidermal growth factor in the rat gastrointestinal tract: segmental and developmental differences. Life Sci. 1988;43:1339–1347. doi: 10.1016/0024-3205(88)90299-8. [DOI] [PubMed] [Google Scholar]
- 50.Ballard O, Morrow AL. Human milk composition: nutrients and bioactive factors. Pediatr Clin North Am. 2013;60:49–74. doi: 10.1016/j.pcl.2012.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McCuskey RS, Nishida J, McDonnell D, Williams C, Koldovsky O. Effect of milk-borne epidermal growth factor on the hepatic microcirculation and Kupffer cell function in suckling rats. Biology of the neonate. 1997;71:202–206. doi: 10.1159/000244418. [DOI] [PubMed] [Google Scholar]
- 52.Gale SM, Read LC, George-Nascimento C, Wallace JC, Ballard FJ. Is dietary epidermal growth factor absorbed by premature human infants? Biology of the neonate. 1989;55:104–110. doi: 10.1159/000242903. [DOI] [PubMed] [Google Scholar]
- 53.Kim KS, et al. Dietary antigens limit mucosal immunity by inducing regulatory T cells in the small intestine. Science. 2016 doi: 10.1126/science.aac5560. [DOI] [PubMed] [Google Scholar]
- 54.Ohnmacht C, et al. The microbiota regulates type 2 immunity through RORgammat+ T cells. Science. 2015 doi: 10.1126/science.aac4263. [DOI] [PubMed] [Google Scholar]
- 55.Sefik E, et al. Individual intestinal symbionts induce a distinct population of RORgamma+ regulatory T cells. Science. 2015;349:993–997. doi: 10.1126/science.aaa9420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shaw SY, Blanchard JF, Bernstein CN. Association between the use of antibiotics in the first year of life and pediatric inflammatory bowel disease. The American journal of gastroenterology. 2010;105:2687–2692. doi: 10.1038/ajg.2010.398. [DOI] [PubMed] [Google Scholar]
- 57.Marra F, et al. Antibiotic use in children is associated with increased risk of asthma. Pediatrics. 2009;123:1003–1010. doi: 10.1542/peds.2008-1146. [DOI] [PubMed] [Google Scholar]
- 58.Metsala J, et al. Mother’s and offspring’s use of antibiotics and infant allergy to cow’s milk. Epidemiology. 2013;24:303–309. doi: 10.1097/EDE.0b013e31827f520f. [DOI] [PubMed] [Google Scholar]
- 59.Birchenough GM, et al. Postnatal development of the small intestinal mucosa drives age-dependent, regio-selective susceptibility to Escherichia coli K1 infection. Sci Rep. 2017;7:83. doi: 10.1038/s41598-017-00123-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Birchenough GM, et al. Altered innate defenses in the neonatal gastrointestinal tract in response to colonization by neuropathogenic Escherichia coli. Infect Immun. 2013;81:3264–3275. doi: 10.1128/IAI.00268-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bergstrom A, et al. Nature of bacterial colonization influences transcription of mucin genes in mice during the first week of life. BMC Res Notes. 2012;5:402. doi: 10.1186/1756-0500-5-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ermund A, Schutte A, Johansson ME, Gustafsson JK, Hansson GC. Studies of mucus in mouse stomach, small intestine, and colon. I. Gastrointestinal mucus layers have different properties depending on location as well as over the Peyer’s patches. Am J Physiol Gastrointest Liver Physiol. 2013;305:G341–347. doi: 10.1152/ajpgi.00046.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nelson RW, Rajpal MN, Jenkins MK. The Neonatal CD4+ T Cell Response to a Single Epitope Varies in Genetically Identical Mice. J Immunol. 2015;195:2115–2121. doi: 10.4049/jimmunol.1500405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Carey AJ, et al. Rapid Evolution of the CD8+ TCR Repertoire in Neonatal Mice. J Immunol. 2016;196:2602–2613. doi: 10.4049/jimmunol.1502126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Prescott SL, et al. Transplacental priming of the human immune system to environmental allergens: universal skewing of initial T cell responses toward the Th2 cytokine profile. J Immunol. 1998;160:4730–4737. [PubMed] [Google Scholar]
- 66.Adkins B. Peripheral CD4+ lymphocytes derived from fetal versus adult thymic precursors differ phenotypically and functionally. J Immunol. 2003;171:5157–5164. doi: 10.4049/jimmunol.171.10.5157. [DOI] [PubMed] [Google Scholar]
- 67.Barnden MJ, Allison J, Heath WR, Carbone FR. Defective TCR expression in transgenic mice constructed using cDNA-based alpha- and beta-chain genes under the control of heterologous regulatory elements. Immunol Cell Biol. 1998;76:34–40. doi: 10.1046/j.1440-1711.1998.00709.x. [DOI] [PubMed] [Google Scholar]
- 68.Lindquist RL, et al. Visualizing dendritic cell networks in vivo. Nat Immunol. 2004;5:1243–1250. doi: 10.1038/ni1139. [DOI] [PubMed] [Google Scholar]
- 69.Hou B, Reizis B, DeFranco AL. Toll-like receptors activate innate and adaptive immunity by using dendritic cell-intrinsic and -extrinsic mechanisms. Immunity. 2008;29:272–282. doi: 10.1016/j.immuni.2008.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rose MF, Ahmad KA, Thaller C, Zoghbi HY. Excitatory neurons of the proprioceptive, interoceptive, and arousal hindbrain networks share a developmental requirement for Math1. Proc Natl Acad Sci U S A. 2009;106:22462–22467. doi: 10.1073/pnas.0911579106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lee TC, Threadgill DW. Generation and validation of mice carrying a conditional allele of the epidermal growth factor receptor. Genesis. 2009;47:85–92. doi: 10.1002/dvg.20464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Adachi O, et al. Targeted disruption of the MyD88 gene results in loss of IL-1- and IL-18-mediated function. Immunity. 1998;9:143–150. doi: 10.1016/s1074-7613(00)80596-8. [DOI] [PubMed] [Google Scholar]
- 73.Jeon J, et al. A subpopulation of neuronal M4 muscarinic acetylcholine receptors plays a critical role in modulating dopamine-dependent behaviors. J Neurosci. 2010;30:2396–2405. doi: 10.1523/JNEUROSCI.3843-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.el Marjou F, et al. Tissue-specific and inducible Cre-mediated recombination in the gut epithelium. Genesis. 2004;39:186–193. doi: 10.1002/gene.20042. [DOI] [PubMed] [Google Scholar]
- 75.McDonald KG, et al. Epithelial expression of the cytosolic retinoid chaperone cellular retinol binding protein II is essential for in vivo imprinting of local gut dendritic cells by lumenal retinoids. Am J Pathol. 2012;180:984–997. doi: 10.1016/j.ajpath.2011.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.McDonald KG, McDonough JS, Dieckgraefe BK, Newberry RD. Dendritic cells produce CXCL13 and participate in the development of murine small intestine lymphoid tissues. Am J Pathol. 2010;176:2367–2377. doi: 10.2353/ajpath.2010.090723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Huijsdens XW, et al. Quantification of bacteria adherent to gastrointestinal mucosa by real-time PCR. Journal of clinical microbiology. 2002;40:4423–4427. doi: 10.1128/JCM.40.12.4423-4427.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.