Abstract
Histone methyl transferase EZH2 (Enhancer of Zeste Homolog 2) is generally associated with H3K27 methylation and gene silencing, as a member of the polycomb repressor 2 (PRC2) complex. Immunoprecipitation and mass spectrometry of the EZH2–protein interactome in estrogen receptor positive, breast cancer-derived MCF7 cells revealed EZH2 interactions with subunits of chromatin remodeler SWI/SNF complex and TRIM28, which formed a complex with EZH2 distinct from PRC2. Unexpectedly, transcriptome profiling showed that EZH2 primarily activates, rather than represses, transcription in MCF7 cells and with TRIM28 co-regulates a set of genes associated with stem cell maintenance and poor survival of breast cancer patients. TRIM28 depletion repressed EZH2 recruitment to chromatin and expression of this gene set, in parallel with decreased CD44hi/CD24lo mammosphere formation. Mammosphere formation, inhibited by EZH2 depletion, was rescued by ectopic expression of EZH2 but not by TRIM28 expression or by EZH2 mutated at the region (pre-SET domain) of TRIM28 interaction. These results support PRC2-independent functions of EZH2 and TRIM28 in activation of gene expression that promotes mammary stem cell enrichment and maintenance.
INTRODUCTION
EZH2 (Enhancer of Zeste Homolog 2), the major, cellular H3K27 tri-methyltransferase, catalyzes addition of methyl groups at lysine 27 of histone H3 (H3K27me3).1 EZH2 is primarily characterized in repression of transcription, as a subunit of the canonical polycomb repressor 2 (PRC2) complex, along with EED, SUZ12 and RBBP4.1 The mammalian PRC2 complex does not bind genomic DNA directly in a sequence-specific manner; however, multiple DNA-binding proteins, including JARID2,2,3 AEBP24 and ATRX,5 are implicated in recruitment of the PRC2 complex to a variety of target gene loci to silence gene expression. Previous studies, using MCF7 and other breast cancer-derived cells, showed that the PRC2 complex has oncogenic roles that center on aberrant silencing of genes that normally activate apoptosis and, when associated with histone H2A ubiquitin ligase TRIM37, tumor suppressor genes.6–8
EZH2 is overexpressed in multiple cancers, such as prostate, melanoma, lymphoma, endometrial and breast, and ectopic expression of EZH2 promotes neoplastic transformation of immortalized human breast epithelial cells.9,10 Small molecule EZH2 inhibitors that target its catalytic SET domain are now in clinical trials. However, recent evidence suggests that the catalytic activity of EZH2 may not be sufficient to account for all functions of EZH2 in cancer, which may prove to be non-responsive to SET-domain inhibitors.11 In contrast to its well-studied role in repression of transcription, EZH2 also acts as a transcriptional co-activator of estrogen- and WNT-regulated proliferation pathways.12 EZH2 overexpression promotes expansion of a sub-population of malignant cells, known as breast cancer initiating cells or breast cancer stem cells, by activating RAF1-β-catenin signaling13 or, as shown elsewhere, by activating NOTCH signaling in an H3K27me3-independent manner.14 In addition, mammary gland-specific overexpression of EZH2 in transgenic mice induces hyperplasia of mammary epithelium but is not sufficient for tumor initiation and progression.15 Taken together, these studies suggest that EZH2 relies on multiple mechanisms to intersect with aberrant signaling pathways that have a role in tumorigenesis.
TRIM28/KAP1 is well studied as a co-repressor of transcription, acting in multiple pathways.16–18 Like EZH2, TRIM28 does not bind to DNA in a sequence-specific manner; it is generally recruited to DNA via sequence-specific binding proteins that have KAP1 repressor -associated binding domains.19 TRIM28 expression was previously shown to promote breast cancer proliferation and metastatic progression, although the mechanisms were not delineated.20 Interestingly, RNA interference screens revealed that TRIM28 is required for maintenance of mouse embryonic stem cell pluripotency and self-renewal,21 which may involve TRIM28 interactions with OCT-4 and subunits of the SWI/SNF2 chromatin remodeling complex, such as Smarcad1, Brg-1 and BAF155, to induce embryonic stem cell-specific genes.22 In the studies described here, we show that TRIM28 interacts with SWI/SNF and EZH2, in a PRC2-independent manner, to coordinately regulate genes with roles in breast stem cell maintenance. A broader understanding of EZH2-mediated regulation and intersection with epigenetic regulators in cancers and stem cells is needed to effectively target therapy-resistant cancers and address the challenges of tumor heterogeneity.
RESULTS
TRIM28 interacts with EZH2 in a PRC2-independent complex
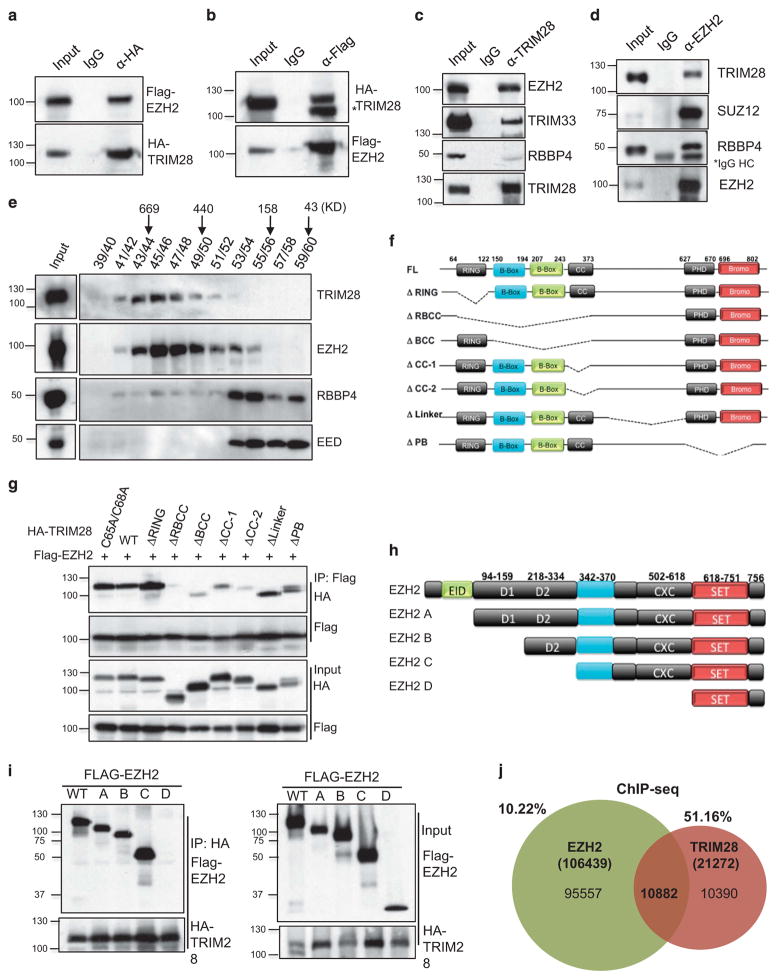
EZH2 is associated with neoplastic transformation of breast epithelial cells;10 however, the regulatory mechanisms involved in EZH2-mediated transformation are not well understood. We used MCF7 cells, as a model of human estrogen receptor (ER) positive, luminal-type breast cancers that comprise a majority of all breast cancers,23,24 and determined interacting protein partners of EZH2. Immunoprecipitation of endogenous EZH2, followed by liquid chromatography-tandem mass spectrometry/mass spectrometry analysis, identified known members of EZH2 complexes, such as SUZ12, EED, PHF1, AEBP2 and DNMT1,2 as well as multiple proteins not previously associated with PRC2 (Supplementary Figure S1A). EZH2 co-purified with proteins, which have four or more peptides identified by liquid chromatography-tandem mass spectrometry/mass spectrometry, of known functions in binding, remodeling and/or modification of chromatin structure, for example, BRG1, SMARCC2, SMARCA5, ARID1A, p300, CHD4 and TRIM28. As EZH2 is primarily associated with repression of transcription, we assessed its potential functions as a partner of TRIM28 co-repressor protein.16 We validated interaction between EZH2 and TRIM28 by co-immunoprecipitation of exogenously expressed Flag-EZH2 and HA-TRIM28 (Figures 1a and b), as well as endogenous EZH2 in HEK293T cells (Supplementary Figure 1B). Endogenous TRIM28 in MCF7 cells likewise interacts with endogenous EZH2, as well as TRIM33, previously reported as interacting with TRIM2825 (Figure 1c). Interestingly, although EZH2 associated with TRIM28, there was minimal interaction with PRC2 complex member RBBP4 (Figure 1c). Antibody enrichment of endogenous EZH2 also co-precipitated TRIM28, as well as previously reported PRC2 complex members SUZ12 and RBBP4 (Figure 1d). To assess protein complex composition, we performed gel filtration chromatography and found two distinct complexes of TRIM28 and RBBP4/EED, which co-fractionated with EZH2 (Figure 1e). Although EED and RBBP4 are canonical PRC2 complex members, previously shown as essential for EZH2-mediated H3K27 methylation,26–28 the majority of EZH2 in MCF7 cells co-fractionated with TRIM28 in a higher molecular weight complex (Figure 1e).
Figure 1.
The RBCC domain of TRIM28 interacts with the pre-SET region of EZH2. (a) Flag-EZH2 and HA-TRIM28 were co-transfected into HEK293T cells and either HA (a) or Flag was immunoprecipitated (b) followed by immunoblotting with Flag or HA antibody (*non-specific band). (c) Endogenous TRIM28 in MCF7 cells was immunoprecipitated and immunoblotted to detect EZH2. (d) Endogenous EZH2 in MCF7 cells was immunoprecipitated, and immunoblotted to detect TRIM28 and other PRC2 members. (e) Gel filtration assay. MCF7 lysate was fractionated by Superose 6 chromatography, followed by immunoblotting for EZH2, EED, RBBP4 and TRIM28. (f) Schematic representation of various deletion mutants of HA-TRIM28 used for co-interaction analyses. (g) Flag-EZH2 and HA-TRIM28 full-length and deletion mutants were ectopically expressed; followed by Flag-immunoprecipitations and immunoblotting with Flag and HA antibodies. Inputs are shown in bottom panels. (h) Schematic representation of various deletion mutants of Flag-EZH2 used for co-interaction analyses (i) HA-TRIM28 was co-transfected with various Flag-EZH2 mutated plasmids followed by HA-immunoprecipitations and immunoblotting with Flag and HA antibodies. Inputs are shown in panels on the right. (j) Venn diagram of intersection of genomewide EZH2-binding sites in HMEC cells with TRIM28-binding sites in MCF7 cells obtained from ChIP-Seq analysis.
We determined that the interaction between EZH2 and TRIM28 was direct by pull-down assays with bacterially expressed, recombinant proteins MBP-EZH2 and GST-TRIM28 (Supplementary Figure 1C). Further, we delineated the domains of interaction between EZH2 and TRIM28 by mutation and expression of Flag-EZH2 and HA-TRIM28 constructs. A series of TRIM28 site-specific RING mutation (C65A, C68A) and deletion mutants, which removed the RING, RBCC, CC, B-BOX, PHD/Bromo domains or the linker region (Figure 1f), were co-expressed with Flag-EZH2 in HEK293T cells for affinity precipitation and analysis (Figure 1g and Supplementary Figure 1C). Removal of the multi-domain RBCC fragment led to loss of EZH2-TRIM28 interaction, whereas separation of the B-boxes and Coiled-Coil domains decreased binding of EZH2 and TRIM28, showing that multiple interactions with EZH2 likely occur across the RBCC domains (Figure 1g). Similarly, EZH2 was serially fragmented as EZH2 A, B, C, D clones from amino- to carboxy-termini (Figure 1h). Expression and co-immunoprecipitation of these mutated forms of EZH2 with full-length TRIM28 indicate that the SET domain of EZH2 is insufficient for TRIM28 interactions (Figure 1i). These results, combined with assays of GST-EZH2 and interaction with endogenous TRIM28 (Supplementary Figure 1D), narrowed binding to the carboxyl-terminal amino acids of 385–618 that encompass the pre-SET domain. To investigate EZH2 and TRIM28 co-association across the whole genome, we performed TRIM28 chromatin immunoprecipitation (ChIP)-Seq in MCF7 cells. Intersection of this genome-wide binding profile of TRIM28 in MCF7 cells with a publically available EZH2 ChIP-seq data set of cultured human mammary epithelial cells (see Materials and Methods)29 showed that ~ 51% of all TRIM28-binding sites and 10% of EZH2 sites overlap (Figure 1j) (Supplementary Figure 1E and Supplementary Table S1). BED-TOOLS (v2.25.0) analysis showed that a majority of TRIM28/EZH2-binding sites overlap across a 1 kb region (Supplementary Figure 1F). Taken together, these results suggest that distinct complexes of EZH2 associate with chromatin, some of which encompass a substantial proportion of TRIM28 chromatin enrichment across the genome.
EZH2 and TRIM28 co-regulate genes associated with differentiation and morphogenesis
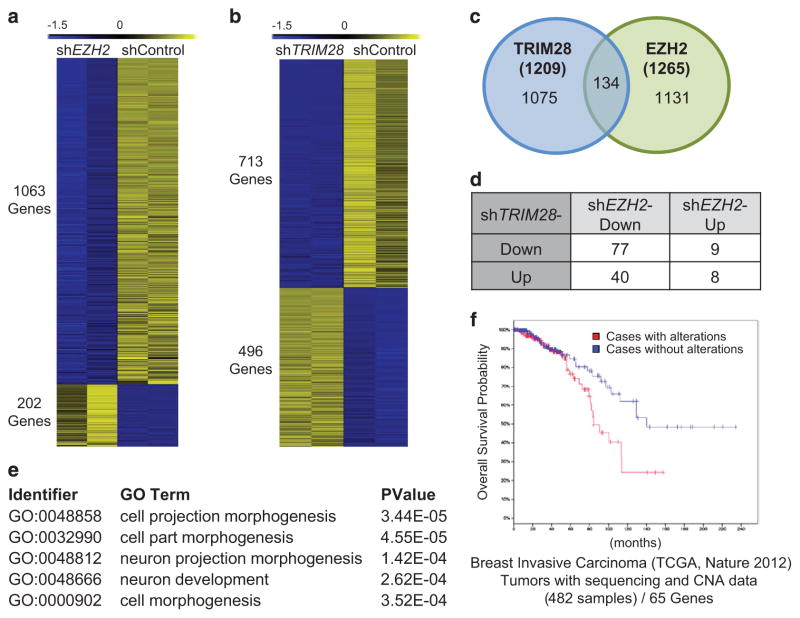
To understand the impact of EZH2 and TRIM28 on gene expression in ER-positive, breast cancer cells, we profiled EZH2-and TRIM28-dependent transcriptomes in MCF7 cells. We generated stable EZH2-depleted MCF7 cells using short-hairpin RNAs and confirmed knockdown by assessing mRNA and protein levels of EZH2 (clonal line #4 of Supplementary Figure 2A). Genome-wide expression profiling (RNA-sequencing; twofold cutoff, false discovery rate<0.01) showed that EZH2 activates considerably more genes (1063 genes, repressed in shEZH2) than it represses (202 genes, activated in shEZH2) in MCF7 cells (Figure 2a). Functional annotation analyses (gene ontology or GO functions) revealed that EZH2 activates differentiation and morphogenesis-related genes (Supplementary Figure 2C and Supplementary Tables S2 and S3), which are associated by KEGG pathway analysis with ligand–receptor interaction molecules and pathways in cancer (Supplementary Figure 2G). The top biological processes (GO functions) of genes repressed by EZH2 are collectively associated with the unfolded protein response (Supplementary Figure 2D and Supplementary Table S3).
Figure 2.
EZH2 and TRIM28 co-regulate genes associated with differentiation and morphogenesis. (a, b) Heatmap showing expression data for EZH2-dependent (a), and TRIM28-dependent (b) genes in MCF7 cells. The 0 to − 1.5 range represents the standard scores of RNA-seq expressions for each gene. (c) Intersection of TRIM28- and EZH2-regulated transcriptomes reveal 134 co-regulated genes. The P-value for the Venn diagram is 4.3e-11 (Fisher’s Exact Test). (d) Distribution of genes differentially co-regulated by TRIM28 and EZH2. (e) Functional annotation analysis of genes coordinately activated by EZH2 and TRIM28 (top five categories shown). (f) Impact of altered gene expression for 65 protein-coding genes, co-activated by EZH2 and TRIM28, illustrated by Kaplan–Meier survival plots using the TCGA 2012 breast invasive carcinoma data set (from http://www.cbioportal.org/).
In parallel, we generated stable TRIM28-depleted MCF7 cells using multiple short-hairpin RNAs (Supplementary Figure 2B). Global expression profiling of stable shTRIM28 MCF7 cells (TRIM28 shRNA#1) showed that TRIM28 depletion led to increased expression of 496 genes and decreased expression of 713 genes (Figure 2b). The GO functions of genes activated by TRIM28 (repressed in shTRIM28) include cell projection organization, morphogenesis and development (Supplementary Figures 2E and 2H; Supplementary Tables S4 and S5), whereas those repressed by TRIM28 (upregulated in shTRIM28) are associated with extracellular matrix organization, structure, secretion and adhesion (Supplementary Figure 2F and Supplementary Tables S4 and S5).
To determine whether EZH2 and TRIM28 regulate a shared set of genes in MCF7 cells, we intersected EZH2 and TRIM28 transcriptome data. This intersection revealed 134 genes that are differentially expressed in both shEZH2 and shTRIM28 MCF7 cells (Figure 2c and Supplementary Table S6). Forty EZH2-activated genes are repressed by TRIM28, and only eight genes are co-repressed by both EZH2 and TRIM28 (Supplementary Table S6). Of the 134 differentially EZH2/TRIM28 co-regulated genes, a subset of 77 is downregulated in both shTRIM28 and shEZH2 MCF7 cells (Figure 2d and Supplementary Table S6). Biological process analysis revealed that these 77 genes, activated by both TRIM28 and EZH2, share GO functions of cell projection, morphogenesis and development that are associated with stem cells and metastasis (Figure 2e and Supplementary Table S7). Examination of breast cancer patient data, available for 65 of the 77 TRIM28/EZH2-activated genes, show that altered expression of the 65 co-activated genes, 62 of which exhibit gene amplification,30–32 is correlated with a poor probability of overall survival for breast cancer patients with invasive carcinomas (Figure 2f).
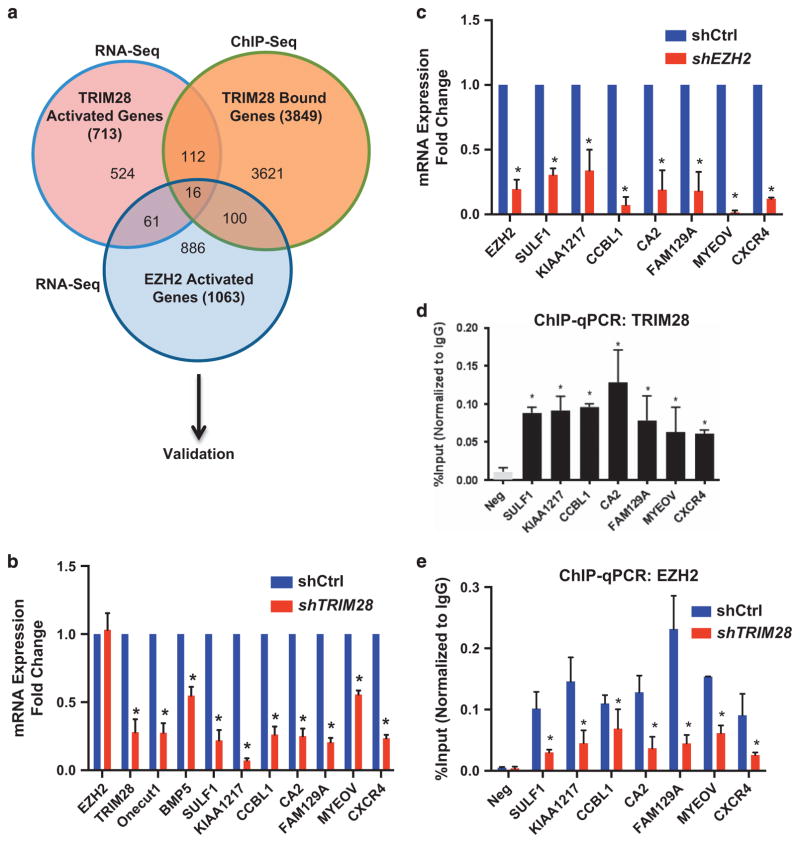
TRIM28 binding occurred at ~ 21 272 sites, with 4.2% in promoter regions (transcription start site ± 3 kb), 2.1% in coding gene exons, 37% of binding sites in intron and 54.10% of binding sites in intergenic space (including enhancers, unannotated IncRNA transcription units) (Supplementary Figure 3A and Supplementary Table S8). To identify the directly regulated, target genes of TRIM28 in breast cancer cells, we integrated TRIM28 ChIP-Seq data (Supplementary Table S9) with our RNA-seq data and found 47 genes, within the subset of 134 TRIM28/EZH2 differentially expressed genes, had overlapping TRIM28/EZH2-binding sites within 10 kb of the TSS. Sixteen of the TRIM28 target genes depend on both TRIM28 and EZH2 for transcription activation (Figure 3a and Supplementary Table S10) (Figure 3a). To validate that these genes are directly regulated by interaction of both TRIM28 and EZH2 with chromatin, we performed ChIP-PCR to determine TRIM28 and EZH2 chromatin enrichment and real-time RT-PCR of RNA isolated from shTRIM28, shEZH2 and shControl MCF7 cells. These analyses revealed that 7 of the 16 genes are both significantly activated and directly bound by both EZH2 and TRIM28 within 10 kb of each gene’s transcription start site (Figures 3b–e; Supplementary Figures 3B and C). Importantly, TRIM28 binding was essential for EZH2 association with chromatin at the regulatory regions of these EZH2/TRIM28 co-activated genes, which were previously reported to be associated with various aspects of cancer promotion, metastasis and stem cells (Figure 3e, Supplementary Figure 2I).
Figure 3.
EZH2 and TRIM28 directly co-activates a subset of genes. (a) Venn diagram showing the comparison of TRIM28- and EZH2-dependent activated genes obtained from RNA-Seq with TRIM28 ChIP-Seq. (b, c) Expression levels of selected TRIM28/EZH2 co-activated genes were analyzed in shCtrl and shTRIM28 (b), and in shCtrl and shEZH2 (c) MCF7 cells by RT-qPCR. Error bars represent s.e.m. from three independent experiments (*P<0.05). (d) ChIP-qPCR analysis of TRIM28 occupancy at SULF1, KIAA1217, CCBL1, CA2, FAM129A, MYEOV and CXCR4 promoters in MCF7 cells. Error bars represent s.e.m. from three independent experiments. (*P<0.05). (e) ChIP-qPCR analysis of EZH2 occupancy at selected gene promoters in shCtrl and shTRIM28 MCF7 cells. Error bars represent s.e.m. from three independent experiments (*P<0.05).
EZH2, SWI/SNF and TRIM28 regulate a CXCR4 signaling pathway
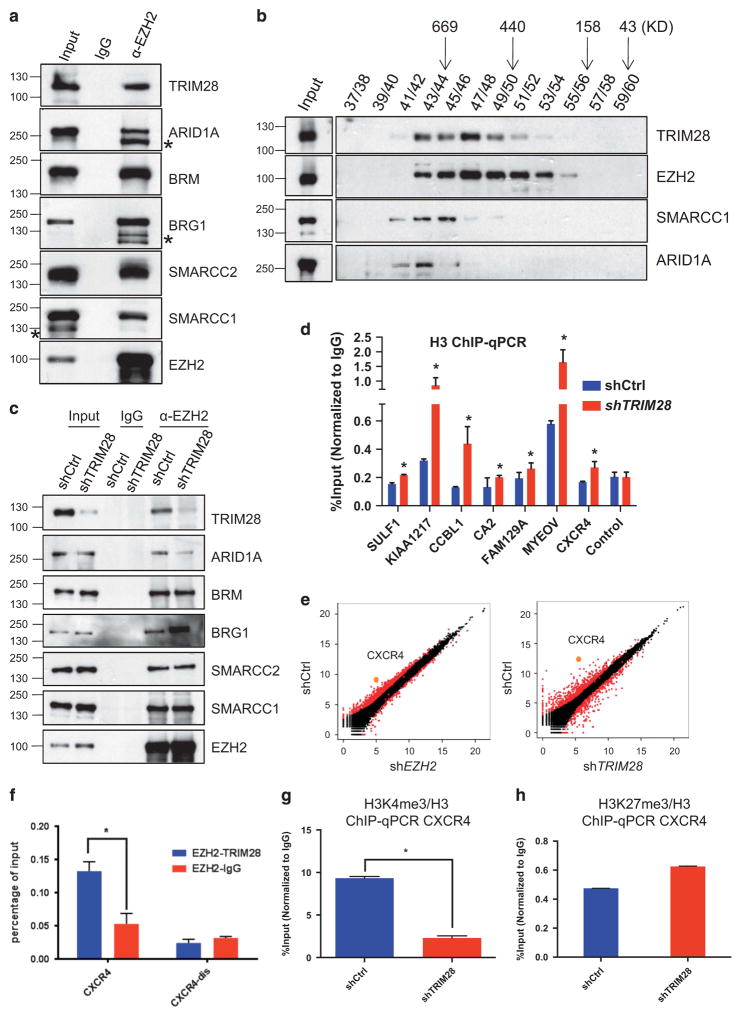
To further illuminate a mechanism for gene activation versus repression by an EZH2/TRIM28 regulatory axis, we confirmed that multiple subunits of the SWI/SNF complex: BRG1, SMARCC2, SMARCA5, BAF180, ARID1A, interacted with EZH2 and TRIM28 (Figure 4a), as observed by mass spectrometry analysis of EZH2-bound proteins (Supplementary Figure 1A). EZH2 likewise associated with TRIM28 and SWI/SNF complex subunits in breast cancer-derived, ER-positive T47D cells, which like MCF7 are considered Luminal A-like23,24 (Supplementary Figure 4). Gel filtration and size fractionation of MCF7 extracts showed that SWI/SNF subunits co-elute with TRIM28 and EZH2 as a likely sub-complex of proteins associated with EZH2 (Figure 4b) and distinct from PRC2 subunits RBBP4 and EED fractionation (Figure 1e). Interestingly, we saw that EZH2 associated more with the ATPase BRG1, compared with the BRM ATPase, when TRIM28 was depleted from MCF7 cells (shTRIM28; Figure 4c). This enrichment of BRG1 was even more highly EZH2-specific in T47D cells, where EZH2/BRM interaction was undetectable (Supplementary Figure 4). BRM- versus BRG1-containing SWI/SNF complexes have been implicated in tissue- and gene-specific functions, and multiple subunits of SWI/SNF are mutated in human cancers.33 TRIM28-dependent enrichment of specific SWI/SNF subunits with EZH2 was likewise supported by parallel depletion of TRIM28 and ARID1A in MCF7 cells (Figure 4c). In T47D cells, ARID1A is undetectable in whole cell lysates but is highly enriched by association and co-immunoprecipitation with EZH2 (Supplementary Figure 4). The interaction between ARID1A and EZH2 in breast cancer-derived cells is noteworthy: ARID1A (SMARCF1, BAF250a), the most frequently mutated SWI/SNF subunit in human cancers, is a major effector of SWI/SNF complex stability and associated with cellular proliferation and DNA repair.34,35
Figure 4.
TRIM28 and EZH2 regulate CXCR4 gene expression. (a) Endogenous EZH2 in MCF7 cells was immunoprecipitated, and immunoblotted to detect TRIM28 and members of SWI/SNF complex. (* non-specific band). (b) Gel filtration assay. MCF7 lysate was fractionated by Superose 6 chromatography, followed by immunoblotting for EZH2, SMARCC1, ARID1A and TRIM28. (c) Endogenous EZH2 was immunoprecipitated from lysates of shControl, shTRIM28 MCF7 cells, followed by western blotting using SWI/SNF complex members antibodies as indicated. (d) ChIP-qPCR analysis of histone H3 occupancy at selected gene promoters in shCtrl and shTRIM28 MCF7 cells. Error bars represent s.e.m. from three independent experiments (*P<0.05). (e) Scatter plots of gene expression levels measured as FPKM (fragments per kilobase of transcript per million mapped reads) values in shCtrl and shEZH2 (left), and in shCtrl and shTRIM28 (right) MCF7 cells. CXCR4 is highlighted as an example of TRIM28 and EZH2 co-activated gene (f) Sequential ChIP analysis in MCF7 cells: Chromatin fragments enriched by EZH2 were used in a sequential ChIP with antibody recognizing TRIM28 or IgG (negative control) at CXCR4 promoter and distal regions (CXCR4-dis), Error bars represent s.e.m. from three independent experiments (*P<0.05). (g, h) ChIP-qPCR analysis of H3K4me3 (g) and H3K27me3 (h) occupancy normalized to H3 at the promoter of CXCR4 gene.
SWI/SNF chromatin remodeling complexes function in positioning, sliding, exchanging or evicting nucleosomes, which impact chromatin structure-dependent processes, including transcription, replication and DNA repair essential for cellular homeostasis.33 We analyzed histone H3 enrichment by ChIP-PCR as a gross assessment of chromatin state across the sites of TRIM28/EZH2 interactions at their differentially regulated target genes in MCF7 cells (Figure 4d). TRIM28-dependent decreases in histone H3 occupancy ranged from minor to substantial with higher H3 levels associated with chromatin at specific genes in response to TRIM28 depletion, although all were statistically significant (Figure 4d). These findings suggest that TRIM28/EZH2-mediated interactions with SWI/SNF complexes alter chromatin structure of their shared, activated target genes.
KEGG analysis of the TRIM28/EZH2 co-activated 77 genes showed that the cytokine–cytokine receptor interaction pathway (CXCR4, CXCL12, EGFR, KIT) is the top pathway represented among differentially TRIM28/EZH2-activated genes (Supplementary Table S6). Each TRIM28/EZH2-regulated gene in this pathway, CXCR4,36,37 CXCL12,36,37 EGFR38 and KIT,39 has been identified as a regulator of cancer stem cells. CXCR4 is a highly and differentially regulated gene when either EZH2 or TRIM28 is depleted in MCF7 cells, as shown in scatter plots of EZH2 and TRIM28 transcriptomes (Figure 4e) and by graphical display of RNA expression (RNA-Seq) alongside TRIM28 and EZH2 ChIP-Seq (Supplementary Figure 3B).40 Real-time quantitative RT-PCR analysis confirmed that EZH2 and TRIM28 depletion significantly decreased the mRNA expression levels of CXCR4 in MCF7 cells (Figures 3b and c). Consistent with the activation of CXCR4 as a direct result of both TRIM28 and EZH2 interaction with chromatin, sequential ChIP or Re-ChIP of EZH2 and TRIM28 showed enrichment of both EZH2 and TRIM28 at the CXCR4 promoter but not distal of the transcription start site (Figure 4f). In support of chromatin modification independently of PRC2, we found that H3K4me3 (Figure 4g) decreases but H3K27me3 enrichment is unchanged at the CXCR4 promoter when TRIM28 is depleted (Figure 4h). We conclude that TRIM28 modulates association of a stable SWI/SNF complex between EZH2 and chromatin to regulate EZH2-activated genes, such as CXCR4, independently of PRC2-dependent, EZH2-mediated H3K27 methylation.
TRIM28 regulates MCF7 mammosphere formation and stem cell maintenance
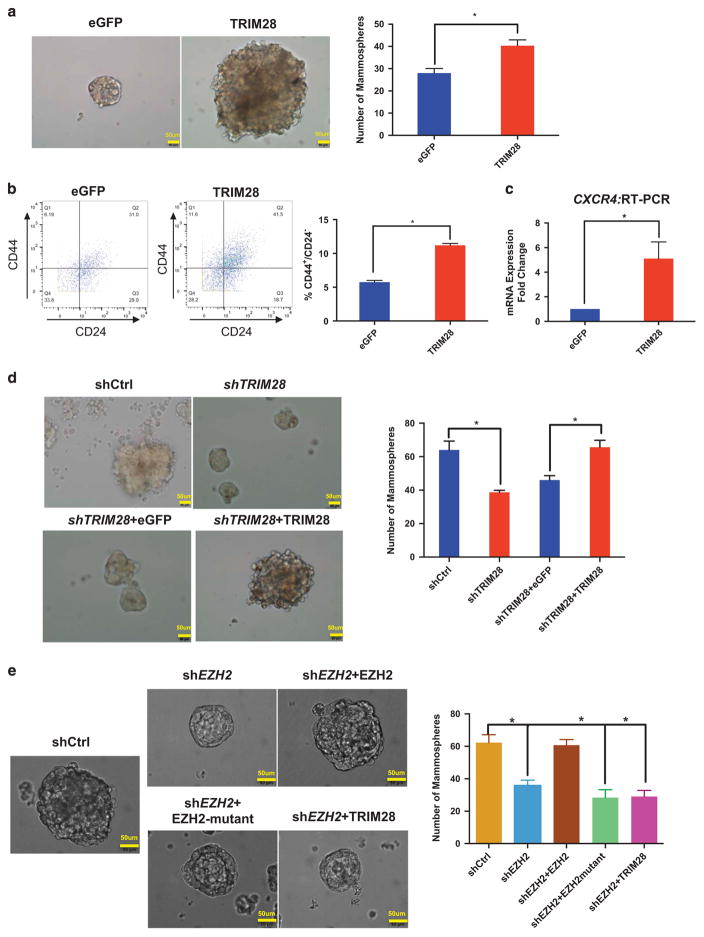
Activation of CXCR4 expression is a reported indicator of a cancer stem cell population.36 Therefore, we used MCF7 sphere-forming or mammosphere assays,41 as a surrogate for cancer stem cell assessment and to determine whether a TRIM28/EZH2 regulatory axis has a role in breast stem cell maintenance. Ectopically expressed TRIM28 in MCF7 cells increased size and efficiency of mammosphere formation, compared to eGFP expression (Figure 5a). Breast stem cell populations are characterized by cell surface marker expression patterns of CD44+ and CD24− or CD24-low.42,43 We determined levels of these indicators by mass cytometry (cytometry by time of flight analysis, Fluidigm, South San Francisco, CA, USA)44 of MCF7 cells with ectopic expression of TRIM28 or eGFP (Figure 5b). The population of CD44+/CD24− cells in MCF7 cells that over express TRIM28 is twofold greater than control MCF7 plus eGFP (Figure 5b). CXCR4 gene expression is also increased in mammospheres with ectopic TRIM28 expression (Figure 5c). In contrast, the number of mammospheres formed was significantly decreased by depletion of TRIM28 and rescued by exogenous expression of short-hairpin RNA-resistant TRIM28 (Figure 5d). Similarly, T47D cells readily formed mammospheres but, when depleted of TRIM28 (shTRIM28) (Supplementary Figure 5C), showed decreased mammosphere formation (Supplementary Figure 5A), as well as a decrease in the population of CD44+/CD24− cells (Supplementary Figure 5B) and CXCR4 mRNA expression compared with shControl T47D cells (Supplementary Figure 5C). Together, these various assays indicate that TRIM28 increases the enrichment and maintenance of mammosphere-forming cells derived from MCF7 and T47D cultures, as previously reported for EZH2 function in breast cancer stem cells.13,14
Figure 5.
TRIM28 promotes mammosphere formation. (a) Brightfield images showing that mammosphere formation ability of MCF7 cells is increased by ectopic expression of TRIM28. Bar graphs are average sphere numbers ±s.e.m. per 2 × 103 plated cells from three independent experiments (*P<0.05). Mammopheres of size >60 μm were counted. (b) CD44+/CD24− populations analyzed by mass cytometry of control eGFP- and ectopic TRIM28-expressing MCF7 cells with quantified results shown in the graph. Error bars represent s.e.m. from three independent experiments: Number of events counted were 3838, 3082 and 2668 for TRIM28 overexpression; and 2453, 1697 and 2078 for eGFP control, respectively (*P<0.05). (c) CXCR4 expression analyzed by RT-qPCR in mammospheres of MCF7 cells overexpressing TRIM28. Error bars represent s.e.m. from three independent experiments (*P<0.05). (d) Brightfield images of mammospheres in TRIM28-depleted MCF7 cells ectopically expressing eGFP or shRNA-resistant TRIM28. Bar graphs are average sphere numbers ±s.e.m. per 2 × 103 plated cells from three independent experiments (*P<0.05). Mammopheres of size >60 μm were counted. (e) Brightfield images of mammospheres in EZH2-depleted MCF7 cells ectopically expressing TRIM28 or shRNA-resistant wild type or mutant EZH2. Bar graphs are average sphere numbers ±s.e.m. per 2 × 103 plated cells from three independent experiments (*P<0.05). Mammopheres of size >60 μm were counted.
We showed that EZH2 and TRIM28 interact via the pre-SET domain of EZH2 (Figures 1h and i; Supplementary Figure 2). Ectopic expression of mutant EZH2, deleted at the pre-SET domain and invulnerable to shEZH2, failed to rescue EZH2-dependant mammosphere formation (Figure 5e). Full-length TRIM28 also failed to rescue EZH2-dependant mammosphere formation (Figure 5e). These results suggested that EZH2 plays a critical role in mammosphere formation that relies on interaction between EZH2 and TRIM28. Taken together with our global expression analyses, which indicated that TRIM28 and EZH2 coordinately regulated expression of genes with biological functions in differentiation and morphogenesis (Figure 2e), our findings support a role for TRIM28 in chromatin enrichment of EZH2 and alterations of chromatin structure by interactions with SWI/SNF, both of which facilitate EZH2-mediated functions in stem cell maintenance.
DISCUSSION
Numerous studies illustrate a conserved function for EZH2, as the catalytic subunit of the PRC2 complex, in repression of transcription. Gene silencing by di- and tri-methylation of H3K27 is mediated by the four core subunits of PRC2: EZH2, EED, SUZ12 and RBBP4.1,26–28,45 PRC2 is normally associated with multiple biological processes, including differentiation, cell identity, proliferation and stem cell plasticity.46 Oncogenic mutations or overexpression of EZH2 with subsequent increases in H3K27 methylation are correlated with tumor development and progression, which led to the development of small molecule inhibitors of the SET domain of EZH2 to ablate its catalytic functions.47,48 However, recent studies suggest that EZH2 in the absence of its PRC2 partners aberrantly functions in activation of transcription, which has been correlated with treatment resistance in prostate cancers.49 Previous studies of EZH2 in breast cancers showed that β-catenin signaling promoted EZH2-mediated activation of transcription, independently of H3K27 methylation, by mechanisms that depend on the ER-status of the cancers. In ER-positive breast cancers, EZH2 interacts with and promotes TCF/Wnt/β-catenin interactions with activated ER at gene regulatory sites.15 In ER-negative breast cancer cells, EZH2 is implicated in pro-inflammatory signaling by promoting NFkB RELA/RELB activities and expression of IL6 and TNF.50 Wnt-signaling is further implicated in EZH2-PRC2-independent activities in colon cancers by proliferating cell nuclear antigen-associated factor, which by its dissociation from proliferating cell nuclear antigen and interactions with EZH2 and β-catenin activates Wnt-target genes and promotes proliferation and transformation.51 These findings underscore the need to understand the mechanisms of EZH2-PRC2-independent functions, which may or may not be responsive to SET-domain inhibition and are likely context-dependent.
Here, we present a newly identified mechanism for EZH2 in ER-positive, breast cancer-derived cells, where EZH2 activates transcription of specific genes in the absence of PRC2. Using mass spectrometry to identify protein partners of EZH2, we uncovered interactions between EZH2 and TRIM28 in a complex that includes SWI/SNF subunits but lacks canonical PRC2 subunits. TRIM28 acts at the level of chromatin recruitment by interactions between the RBCC domain of TRIM28 and the pre-SET domain of EZH2, and promotes stable SWI/SNF association with EZH2. Decreased histone H3 enrichment and post-translational modifications associated with activation of target genes, such as increased H3K4me3 versus repressive EZH2-mediated H3K27me3, occur at targeted binding sites in a TRIM28- and EZH2-dependent manner.
Both EZH2 and TRIM28 are generally considered as chromatin-associated, repressors of transcription.16,46 However, depletion of either TRIM28 or EZH2 showed that these epigenetic factors were mainly activators of gene expression in MCF7 cells, independently of conserved PRC2 subunits. A significant number of the TRIM28 and EZH2 coordinately activated genes cluster in differentiation and morphogenesis and include genes in the cytokine–cytokine receptor (CXCR4/CXCL12) pathway that is highly correlated with cancer stem cells.36 EZH2 itself has been previously implicated in maintenance of breast cancer stem cells by mechanisms initiated by either Wnt- or Notch signaling.13,14 Previous unbiased RNA interference-based screens showed that TRIM28 functions in maintenance of mouse embryonic stem cell pluripotency.21 EZH2 and TRIM28 coordinately activate CXCR4, promote expansion of a CD44+/CD24− population of cells and have critical roles in mammosphere formation.
Our studies of stem cell activities underscore the cell-type specificity of EZH2 functions and offer a mechanism by which TRIM28 promotes interactions between EZH2 and chromatin remodeler SWI/SNF to activate chromatin structure. Previous studies showed that specific cancer-derived cell lines with mutations of SWI/SNF subunits were dependent on PRC2 subunits for proliferation and growth; however, EZH2 acted only to stabilize PRC2 not to methylate H3K27.52 Thus, even when non-catalytic functions of EZH2 are uncovered, which render specific cancers inert to SET-domain targeting therapeutics, the mechanisms of action and selectivity of aberrant EZH2 activities may be highly cancer-specific or even restricted to a sub-population of cancer cells. The newly defined SWI/SNF-TRIM28-EZH2 regulatory axis in breast cancer cells and the identified domains of interaction between TRIM28 and EZH2 may open new avenues for therapeutic development that target a cancer stem cell population.
MATERIALS AND METHODS
Cell culture, vectors, mutants and protein purification
The HEK293T, and breast cancer cell lines MCF7 and T47D were obtained from the American Type Culture Collection and grown in suggested media conditions. HA-TRIM28 vector was a gift from Dr Patrick Ryan Potts.53 Cloning of MBP-EZH2, GST-TRIM28, Flag-TRIM28 and HA-TRIM28 mutated forms was performed as described.54,55 EGFP, TRIM28 WT lentiviral plasmids were generated via the Gateway system in a pLX304 lentiviral backbone. ShTRIM28-resistant TRIM28 was generated by inducing silent mutations at three nucleotides (Supplementary Table S11), targeted by shTRIM28 RNA. MBP and GST proteins were purified from Escherichia Coli with MBP beads and GST beads (GE Healthcare, BioSciences, Pittsburgh, PA, USA), respectively.56
Lentiviral infections, establishment of stable knockdown cell lines
EZH2 and TRIM28 knockdowns were achieved using stable pGIPZ short-hairpin interfering RNA in lentivirus (clone: V2LHS_17507 for EZH2, clones of shTRIM28, Clone #1: V3LHS_358497, Clone #2: V3LHS_358502, GE Dharmacon, Lafayette, CO, USA). A lentiviral scrambled pGIPZ plasmid (GE Dharmacon) was used as control for transfection and virus packaging. The protocol for virus packaging and generation of stable cell lines was followed as described previously.54 In brief, the pGIPZ-shEZH2, pGIPZ-shTRIM28, eGFP, shTRIM28-resistant TRIM28 lentiviral plasmid was co-transfected with packaging plasmids psPAX2, and pMD.2G (Addgene, Cambridge, MA, USA) into HEK293T cells to generate virus. After 48 h, the supernatant was collected, filtered and transferred into targeted cell lines for infection for 2–3 days, followed by selection with 2 μg/ml Puromycin (Sigma, St Louis, MO, USA) and growth of stable resistant clones.
Western blots
Western blots were performed following established protocols.57 List of antibodies used: EZH2 mAb #39875(Active Motif, Carlsbad, CA, USA) was used for IP and ChIP, pAb #39933 (Active Motif), and Rabbit mAb#5246 (Cell Signaling, Danvers, MA, USA) was used for western blotting; TRIM28, Ab10483 (Abcam, Cambridge, UK), Santa Cruz (Dallas, TX, USA), SC-33186 (H300) were used for western blotting; other antibodies used are SUZ12, Abcam ab12073; EED, GTX628007; RBBP4 GTX62136; TRIM33, Abnova (Walnut, CA, USA), #51992-M01; HA, Roche Life Science-US (Indianapolis, IN, USA) 12CA5; Flag, Sigma; Myc Santa Cruz, SC-40 (9E10), SWI/SNF antibody kit, Cell Signaling.
Immunoprecipitation-mass spectrometry, gel flitration chromatography
MCF7, T47D cells were lysed in buffer (25 mM Tris-HCl pH 7.5, 150 mM NaCl, 5 mM ethylenediaminetetraacetic acid, 0.5% NP-40) for 30 min at 4 °C, 500 μg cell lysate was used to incubate with either EZH2, TRIM28 antibodies overnight. Then proteinase A Sepharose beads (GE Health) were added for additional 2 h incubation. After extensive washing, 35 μl sodium dodecyl sulfate loading dye was added into the beads, the beads were boiled for 5 min to elute the protein complex. Elutes were then subject to western blot in various percentage of sodium dodecyl sulfate polyacrylamide gel electrophoresis gels. Gel was cut and sent out for mass spec at UTMB proteomics core.57 For gel filtration assay, chromatography was performed at 0.4 ml/min, collecting 0.5 ml fractions in cold room. The fractions were analyzed by sodium dodecyl sulfate polyacrylamide gel electrophoresis gels followed by western blot with various antibodies as indicated in the figures.
Mammosphere assay
Single-cell association for mammosphere formation was performed, following established protocols,43 with MCF7 cells and derived mutant cell lines at a density of 1 × 104 cells/ml. Mammospheres were cultured in MammoCult Human Basal Medium with added proliferation Supplement (Stem Cell Technologies, BC, Canada) on Costar Ultra Low Attachment tissue culture plates. After 7 days, mammosphere sizes and numbers were determined using an inverted microscope. Size was measured as the widest diameter with the scale bar. Only mammospheres exhibiting greater than 60 μm diameter were counted. All experiments were done in triplicates.
RNA extraction, RNA-sequencing analysis, DAVID pathway analysis
Total RNA was extracted using Trizol reagent from shControl, shEZH2 and shTRIM28 MCF7 cell lines. Replicates of RNA were used for RNA-sequencing library preparation using Illumina Tru-Seq RNA library prep kit V2; sequencing was performed using Illumina HiSeq 2500 to obtain paired-end 150 mer reads. Raw reads were aligned to human reference genome (hg19) by Tophat v2.0.10,58 reads count for each transcript was obtained by htSeq.59 Differentially expressed gene lists were generated using edgeR,60 with adjusted P-value set to be <0.01. The differentiated gene lists were uploaded into the online DAVID functional annotation tool to analyze GO functions, biological pathways and annotations.61
Chromatin immunoprecipitation
These methods were performed as previously reported.54 ChIP and Re-ChIP were performed as described62 with antibodies for EZH2 and TRIM28. In brief, MCF7 cells were cross-linked with 1% formaldehyde for 15 min. The cross-linking reaction was stopped with 0.125 M glycine. The cross-linked cells were washed with phosphate-buffered saline three times and stored at − 80 °C before use. The fragmented, precleared chromatin lysate was incubated overnight with EZH2 antibody, and Protein A beads were used to collect the EZH2 ChIP’d chromatin. Three duplicates of EZH2 ChIP’d chromatin were combined and followed by a second ChIP with an antibody recognizing TRIM28 or negative control imunoglobulin G only. Pelleted chromatin was treated with RNase, Proteinase K and reverse cross-linked overnight. ChIP’d DNA was extracted by Phenol/chloroform. To analyze specific, antibody- and protein-bound DNA, qPCR was conducted in a 7500 FAST ABI instrument. Primers are listed in Supplementary Table S12.
ChIP-sequencing analysis
ChIP-seq raw reads were aligned to human reference genome (hg19) using Bowtie v1.0.0,63 allowing up to one mismatch per read. Only uniquely mapped reads were kept for downstream analysis. Peaks were called by MACS v1.3.7,64 using P-value cutoff 1e-5. The ChIP-seq intensities were normalized to 10 millions reads per sample. Overlaps between TRIM28 peaks and EZH2 peaks were defined using BEDTOOLS (v2.25.0) with default parameters. The genomic location of ChIP-seq peaks were analyzed using in-house python scripts. Publically available data set used for EZH2 ChIP-Seq in human mammary epithelial cells were: (https://www.encodeproject.org/experiments/ENCSR000ARE) (GEO:GSM1003501). The ChIP-seq snapshots were visualized by the UCSC genome browser.40
Mass cytometry sample processing and analysis cytometry by time of flight, and flow cytometry
MCF7 cells were washed with phosphate-buffered saline then disassociated with 0.05% Trypsin (Sigma-Aldrich, St Louis, MO, USA). Live/dead cisplatin staining and subsequent fixation and sample processing was done according to the methods described previously.65 Antibody labeling of cells was performed in a buffer of 0.5% bovine serum albumin (Sigma-Aldrich) and 0.02% sodium azide (Sigma-Aldrich) in phosphate-buffered saline at room temperature using a concentration of 4 × 105 cells/μl with CD24 and CD44 antibodies (Fluidigm). Cells were analyzed using a cytometry by time of flight mass cytometer run at a concentration of 5 × 106 cells/ml (Fluidigm). EQ Four Element Calibration Beads (Fluidigm) were added to the samples immediately prior to sample loading. Data were normalized based on bead signal using the included cytometry by time of flight software. Initial data processing and gating were performed using FlowJo vX10.0. Beads were gated off of the samples and data were gated on singlets, based upon Ir193 and Event Length parameters. Live/dead cell discrimination was performed based upon the Pt198 channel. Flow cytometry was performed as previously reported.66 In brief, single-cell suspensions of shCtrl, and shTRIM28 T47D cells were stained with antibodies to CD44 (eBioscience), and CD24 (eBioscience, San Diego, CA, USA). Cells were acquired on a flow cytometry analyzer (LSR II; BD Biosciences) and data were analyzed with DIVA software (BD Biosciences, San Jose, CA, USA).
Statistical analyses
GraphPad Prism6 software (GraphPad Software, Inc., La Jolla, CA, USA) was used for statistical analysis. The two-tailed paired student’s t-test was employed to verify whether the difference between two groups is significant. P-values <0.05 were considered to be statistically significant.
Data access: TRIM28, EZH2 RNA-seq and TRIM28 ChIP-seq data can be accessed at NCBI Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) under accession no. GSE76271.
Supplementary Material
Acknowledgments
This work was supported in part by CPRIT RP110471 to MCB, WL and SYD. JL received support by a Gigli Family Endowed Scholarship of the UT-Graduate School of Biomedical Sciences at Houston, TX. AKJ was supported by The Laura and John Arnold Foundation and Odyssey Program fellowship. We thank Dr M-C Hung for the Myc-EZH2 plasmid, Dr J-I Park for Flag-EZH2 plasmids and Dr PR Potts for HA-TRIM28, GST-TRIM28 full-length and RBCC only plasmids. This study used the Science Park NGS Core, supported by CPRIT Core Facility Support Grant RP120348, and the Flow Cytometry and Cellular Imaging Facility, which is supported in part by the National Institutes of Health through MD Anderson’s Cancer Center Support Grant CA016672.
Footnotes
CONFLICT OF INTEREST
The authors declare no conflict of interest.
Supplementary Information accompanies this paper on the Oncogene website (http://www.nature.com/onc)
References
- 1.Shen X, Liu Y, Hsu YJ, Fujiwara Y, Kim J, Mao X, et al. EZH1 mediates methylation on histone H3 lysine 27 and complements EZH2 in maintaining stem cell identity and executing pluripotency. Mol Cell. 2008;32:491–502. doi: 10.1016/j.molcel.2008.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li G, Margueron R, Ku M, Chambon P, Bernstein BE, Reinberg D. Jarid2 and PRC2, partners in regulating gene expression. Genes Dev. 2010;24:368–380. doi: 10.1101/gad.1886410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pasini D, Cloos PA, Walfridsson J, Olsson L, Bukowski JP, Johansen JV, et al. JARID2 regulates binding of the Polycomb repressive complex 2 to target genes in ES cells. Nature. 2010;464:306–310. doi: 10.1038/nature08788. [DOI] [PubMed] [Google Scholar]
- 4.Kim H, Kang K, Kim J. AEBP2 as a potential targeting protein for Polycomb Repression Complex PRC2. Nucleic Acids Res. 2009;37:2940–2950. doi: 10.1093/nar/gkp149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sarma K, Cifuentes-Rojas C, Ergun A, Del Rosario A, Jeon Y, White F, et al. ATRX directs binding of PRC2 to Xist RNA and Polycomb targets. Cell. 2014;159:869–883. doi: 10.1016/j.cell.2014.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bhatnagar S, Gazin C, Chamberlain L, Ou J, Zhu X, Tushir JS, et al. TRIM37 is a new histone H2A ubiquitin ligase and breast cancer oncoprotein. Nature. 2014;516:116–120. doi: 10.1038/nature13955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Squazzo SL, O’Geen H, Komashko VM, Krig SR, Jin VX, Jang SW, et al. Suz12 binds to silenced regions of the genome in a cell-type-specific manner. Genome Res. 2006;16:890–900. doi: 10.1101/gr.5306606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tan J, Yang X, Zhuang L, Jiang X, Chen W, Lee PL, et al. Pharmacologic disruption of Polycomb-repressive complex 2-mediated gene repression selectively induces apoptosis in cancer cells. Genes Dev. 2007;21:1050–1063. doi: 10.1101/gad.1524107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Changchien YC, Tatrai P, Papp G, Sapi J, Fonyad L, Szendroi M, et al. Poorly differentiated synovial sarcoma is associated with high expression of enhancer of zeste homologue 2 (EZH2) J Transl Med. 2012;10:216. doi: 10.1186/1479-5876-10-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kleer CG, Cao Q, Varambally S, Shen R, Ota I, Tomlins SA, et al. EZH2 is a marker of aggressive breast cancer and promotes neoplastic transformation of breast epithelial cells. Proc Natl Acad Sci USA. 2003;100:11606–11611. doi: 10.1073/pnas.1933744100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim W, Bird GH, Neff T, Guo G, Kerenyi MA, Walensky LD, et al. Targeted disruption of the EZH2-EED complex inhibits EZH2-dependent cancer. Nat Chem Biol. 2013;9:643–650. doi: 10.1038/nchembio.1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shi B, Liang J, Yang X, Wang Y, Zhao Y, Wu H, et al. Integration of estrogen and Wnt signaling circuits by the polycomb group protein EZH2 in breast cancer cells. Mol Cell Biol. 2007;27:5105–5119. doi: 10.1128/MCB.00162-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chang CJ, Yang JY, Xia W, Chen CT, Xie X, Chao CH, et al. EZH2 promotes expansion of breast tumor initiating cells through activation of RAF1-beta-catenin signaling. Cancer Cell. 2011;19:86–100. doi: 10.1016/j.ccr.2010.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gonzalez ME, Moore HM, Li X, Toy KA, Huang W, Sabel MS, et al. EZH2 expands breast stem cells through activation of NOTCH1 signaling. Proc Natl Acad Sci USA. 2014;111:3098–3103. doi: 10.1073/pnas.1308953111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li X, Gonzalez ME, Toy K, Filzen T, Merajver SD, Kleer CG. Targeted overexpression of EZH2 in the mammary gland disrupts ductal morphogenesis and causes epithelial hyperplasia. Am J Pathol. 2009;175:1246–1254. doi: 10.2353/ajpath.2009.090042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Iyengar S, Farnham PJ. KAP1 protein: an enigmatic master regulator of the genome. J Biol Chem. 2011;286:26267–26276. doi: 10.1074/jbc.R111.252569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rowe HM, Kapopoulou A, Corsinotti A, Fasching L, Macfarlan TS, Tarabay Y, et al. TRIM28 repression of retrotransposon-based enhancers is necessary to preserve transcriptional dynamics in embryonic stem cells. Genome Res. 2013;23:452–461. doi: 10.1101/gr.147678.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wolf D, Goff SP. TRIM28 mediates primer binding site-targeted silencing of murine leukemia virus in embryonic cells. Cell. 2007;131:46–57. doi: 10.1016/j.cell.2007.07.026. [DOI] [PubMed] [Google Scholar]
- 19.Quenneville S, Verde G, Corsinotti A, Kapopoulou A, Jakobsson J, Offner S, et al. In embryonic stem cells, ZFP57/KAP1 recognize a methylated hexanucleotide to affect chromatin and DNA methylation of imprinting control regions. Mol Cell. 2011;44:361–372. doi: 10.1016/j.molcel.2011.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Addison J, Koontz C, Fugett JH, Creighton CJ, Chen D, Farrugia MK, et al. KAP1 promotes proliferation and metastatic progression of breast cancer cells. Cancer Res. 2014;75:344–355. doi: 10.1158/0008-5472.CAN-14-1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hu G, Kim J, Xu Q, Leng Y, Orkin SH, Elledge SJ. A genome-wide RNAi screen identifies a new transcriptional module required for self-renewal. Genes Dev. 2009;23:837–848. doi: 10.1101/gad.1769609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Seki Y, Kurisaki A, Watanabe-Susaki K, Nakajima Y, Nakanishi M, Arai Y, et al. TIF1beta regulates the pluripotency of embryonic stem cells in a phosphorylation-dependent manner. Proc Natl Acad Sci USA. 2010;107:10926–10931. doi: 10.1073/pnas.0907601107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Neve RM, Chin K, Fridlyand J, Yeh J, Baehner FL, Fevr T, et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 2006;10:515–527. doi: 10.1016/j.ccr.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 25.Herquel B, Ouararhni K, Khetchoumian K, Ignat M, Teletin M, Mark M, et al. Transcription cofactors TRIM24, TRIM28, and TRIM33 associate to form regulatory complexes that suppress murine hepatocellular carcinoma. Proc Natl Acad Sci USA. 2011;108:8212–8217. doi: 10.1073/pnas.1101544108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hansen KH, Bracken AP, Pasini D, Dietrich N, Gehani SS, Monrad A, et al. A model for transmission of the H3K27me3 epigenetic mark. Nat Cell Biol. 2008;10:1291–1300. doi: 10.1038/ncb1787. [DOI] [PubMed] [Google Scholar]
- 27.Margueron R, Justin N, Ohno K, Sharpe ML, Son J, Drury WJ, 3rd , et al. Role of the polycomb protein EED in the propagation of repressive histone marks. Nature. 2009;461:762–767. doi: 10.1038/nature08398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Montgomery ND, Yee D, Chen A, Kalantry S, Chamberlain SJ, Otte AP, et al. The murine polycomb group protein Eed is required for global histone H3 lysine-27 methylation. Curr Biol. 2005;15:942–947. doi: 10.1016/j.cub.2005.04.051. [DOI] [PubMed] [Google Scholar]
- 29.Consortium EP. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cama A, Verginelli F, Lotti LV, Napolitano F, Morgano A, D’Orazio A, et al. Integrative genetic, epigenetic and pathological analysis of paraganglioma reveals complex dysregulation of NOTCH signaling. Acta Neuropathol. 2013;126:575–594. doi: 10.1007/s00401-013-1165-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cancer Genome Atlas N. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Masliah-Planchon J, Bieche I, Guinebretiere JM, Bourdeaut F, Delattre O. SWI/SNF chromatin remodeling and human malignancies. Annu Rev Pathol. 2015;10:145–171. doi: 10.1146/annurev-pathol-012414-040445. [DOI] [PubMed] [Google Scholar]
- 34.Kadoch C, Hargreaves DC, Hodges C, Elias L, Ho L, Ranish J, et al. Proteomic and bioinformatic analysis of mammalian SWI/SNF complexes identifies extensive roles in human malignancy. Nat Genet. 2013;45:592–601. doi: 10.1038/ng.2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu RC, Wang TL, Shih Ie M. The emerging roles of ARID1A in tumor suppression. Cancer Biol Ther. 2014;15:655–664. doi: 10.4161/cbt.28411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Korkaya H, Liu S, Wicha MS. Breast cancer stem cells, cytokine networks, and the tumor microenvironment. J Clin Invest. 2011;121:3804–3809. doi: 10.1172/JCI57099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Orimo A, Gupta PB, Sgroi DC, Arenzana-Seisdedos F, Delaunay T, Naeem R, et al. Stromal fibroblasts present in invasive human breast carcinomas promote tumor growth and angiogenesis through elevated SDF-1/CXCL12 secretion. Cell. 2005;121:335–348. doi: 10.1016/j.cell.2005.02.034. [DOI] [PubMed] [Google Scholar]
- 38.Li X, Lewis MT, Huang J, Gutierrez C, Osborne CK, Wu MF, et al. Intrinsic resistance of tumorigenic breast cancer cells to chemotherapy. J Natl Cancer Inst. 2008;100:672–679. doi: 10.1093/jnci/djn123. [DOI] [PubMed] [Google Scholar]
- 39.Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- 40.Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, et al. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dontu G, Abdallah WM, Foley JM, Jackson KW, Clarke MF, Kawamura MJ, et al. In vitro propagation and transcriptional profiling of human mammary stem/progenitor cells. Genes Dev. 2003;17:1253–1270. doi: 10.1101/gad.1061803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci USA. 2003;100:3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ponti D, Costa A, Zaffaroni N, Pratesi G, Petrangolini G, Coradini D, et al. Isolation and in vitro propagation of tumorigenic breast cancer cells with stem/progenitor cell properties. Cancer Res. 2005;65:5506–5511. doi: 10.1158/0008-5472.CAN-05-0626. [DOI] [PubMed] [Google Scholar]
- 44.Cheung RK, Utz PJ. Screening: CyTOF-the next generation of cell detection. Nat Rev Rheumatology. 2011;7:502–503. doi: 10.1038/nrrheum.2011.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pasini D, Bracken AP, Jensen MR, Lazzerini Denchi E, Helin K. Suz12 is essential for mouse development and for EZH2 histone methyltransferase activity. Embo J. 2004;23:4061–4071. doi: 10.1038/sj.emboj.7600402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Margueron R, Reinberg D. The Polycomb complex PRC2 and its mark in life. Nature. 2011;469:343–349. doi: 10.1038/nature09784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McCabe MT, Ott HM, Ganji G, Korenchuk S, Thompson C, Van Aller GS, et al. EZH2 inhibition as a therapeutic strategy for lymphoma with EZH2-activating mutations. Nature. 2012;492:108–112. doi: 10.1038/nature11606. [DOI] [PubMed] [Google Scholar]
- 48.Qi W, Chan H, Teng L, Li L, Chuai S, Zhang R, et al. Selective inhibition of Ezh2 by a small molecule inhibitor blocks tumor cells proliferation. Proc Natl Acad Sci USA. 2012;109:21360–21365. doi: 10.1073/pnas.1210371110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xu K, Wu ZJ, Groner AC, He HH, Cai C, Lis RT, et al. EZH2 oncogenic activity in castration-resistant prostate cancer cells is Polycomb-independent. Science. 2012;338:1465–1469. doi: 10.1126/science.1227604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lee ST, Li Z, Wu Z, Aau M, Guan P, Karuturi RK, et al. Context-specific regulation of NF-kappaB target gene expression by EZH2 in breast cancers. Mol Cell. 2011;43:798–810. doi: 10.1016/j.molcel.2011.08.011. [DOI] [PubMed] [Google Scholar]
- 51.Jung HY, Jun S, Lee M, Kim HC, Wang X, Ji H, et al. PAF and EZH2 induce Wnt/beta-catenin signaling hyperactivation. Mol Cell. 2013;52:193–205. doi: 10.1016/j.molcel.2013.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kim KH, Kim W, Howard TP, Vazquez F, Tsherniak A, Wu JN, et al. SWI/SNF-mutant cancers depend on catalytic and non-catalytic activity of EZH2. Nat Med. 2015;21:1491–1496. doi: 10.1038/nm.3968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Doyle JM, Gao J, Wang J, Yang M, Potts PR. MAGE-RING protein complexes comprise a family of E3 ubiquitin ligases. Mol Cell. 2010;39:963–974. doi: 10.1016/j.molcel.2010.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Akdemir KC, Jain AK, Allton K, Aronow B, Xu X, Cooney AJ, et al. Genome-wide profiling reveals stimulus-specific functions of p53 during differentiation and DNA damage of human embryonic stem cells. Nucleic Acids Res. 2014;42:205–223. doi: 10.1093/nar/gkt866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zheng L, Baumann U, Reymond JL. An efficient one-step site-directed and site-saturation mutagenesis protocol. Nucleic Acids Res. 2004;32:e115. doi: 10.1093/nar/gnh110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Braun P, Hu Y, Shen B, Halleck A, Koundinya M, Harlow E, et al. Proteome-scale purification of human proteins from bacteria. Proc Natl Acad Sci USA. 2002;99:2654–2659. doi: 10.1073/pnas.042684199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Allton K, Jain AK, Herz HM, Tsai WW, Jung SY, Qin J, et al. Trim24 targets endogenous p53 for degradation. Proc Natl Acad Sci USA. 2009;106:11612–11616. doi: 10.1073/pnas.0813177106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Anders S, Pyl PT, Huber W. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 62.Tsai WW, Wang Z, Yiu TT, Akdemir KC, Xia W, Winter S, et al. TRIM24 links a non-canonical histone signature to breast cancer. Nature. 2010;468:927–932. doi: 10.1038/nature09542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fienberg HG, Simonds EF, Fantl WJ, Nolan GP, Bodenmiller B. A platinum-based covalent viability reagent for single-cell mass cytometry. Cytometry A. 2012;81:467–475. doi: 10.1002/cyto.a.22067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kryczek I, Lin Y, Nagarsheth N, Peng D, Zhao L, Zhao E, et al. IL-22(+)CD4(+) T cells promote colorectal cancer stemness via STAT3 transcription factor activation and induction of the methyltransferase DOT1L. Immunity. 2014;40:772–784. doi: 10.1016/j.immuni.2014.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.