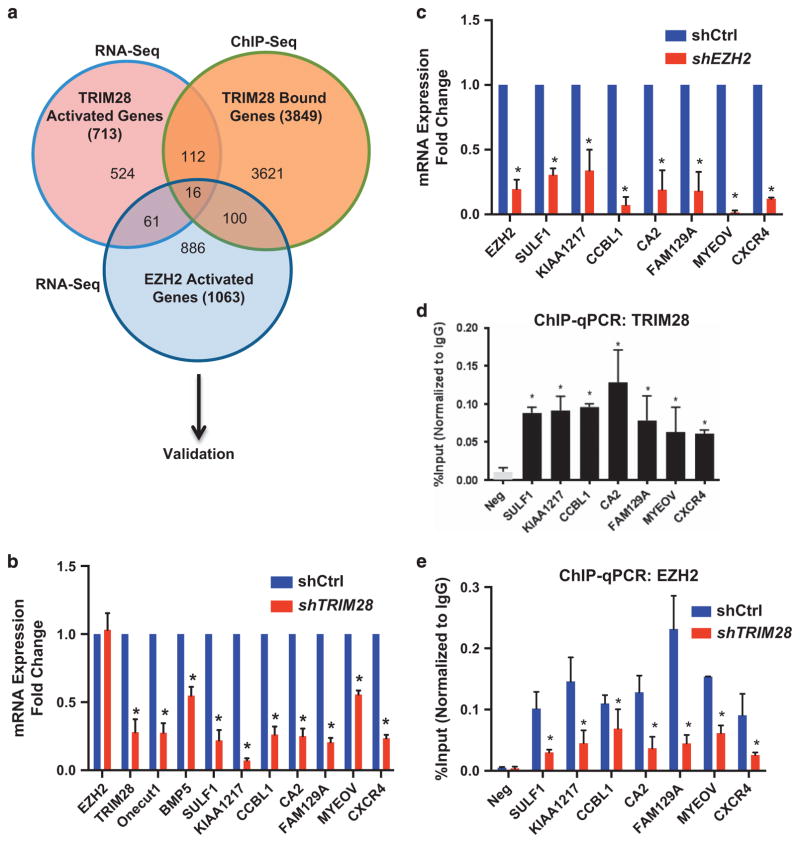
Figure 3.
EZH2 and TRIM28 directly co-activates a subset of genes. (a) Venn diagram showing the comparison of TRIM28- and EZH2-dependent activated genes obtained from RNA-Seq with TRIM28 ChIP-Seq. (b, c) Expression levels of selected TRIM28/EZH2 co-activated genes were analyzed in shCtrl and shTRIM28 (b), and in shCtrl and shEZH2 (c) MCF7 cells by RT-qPCR. Error bars represent s.e.m. from three independent experiments (*P<0.05). (d) ChIP-qPCR analysis of TRIM28 occupancy at SULF1, KIAA1217, CCBL1, CA2, FAM129A, MYEOV and CXCR4 promoters in MCF7 cells. Error bars represent s.e.m. from three independent experiments. (*P<0.05). (e) ChIP-qPCR analysis of EZH2 occupancy at selected gene promoters in shCtrl and shTRIM28 MCF7 cells. Error bars represent s.e.m. from three independent experiments (*P<0.05).