Abstract
TGFBIp is a constituent of the extracellular matrix in many human tissues including the cornea, where it is one of the most abundant proteins expressed. TGFBIp interacts with Type I, II, IV, VI and XII collagens as well as several members of the integrin family, suggesting it plays an important role in maintaining structural integrity and possibly corneal transparency as well. Significantly, more than 60 point mutations within the TGFBI gene have been reported to result in aberrant TGFBIp folding and aggregation in the cornea, resulting in severe visual impairment and blindness. Several studies have focused on targeting TGFBIp in the cornea as a therapeutic approach to treat TGFBI-linked corneal dystrophies, but the effect of this approach on corneal homeostasis and matrix integrity remained unknown. In the current study, we evaluated the histological and proteomic profiles of corneas from TGFBI-deficient mice as well as potential redundant functions of the paralogous protein POSTN. The absence of TGFBIp in mouse corneas did not grossly affect the collagen scaffold, and POSTN is unable to compensate for loss of TGFBIp. Proteomic comparison of wildtype and TGFBI−/− mice revealed 11 proteins were differentially regulated, including Type VI and XII collagens. However, as these alterations did not manifest at the macroscopic and behavioral levels, these data support partial or complete TGFBI knockdown as a potential therapy against TGFBI-linked corneal dystrophies. Lastly, in situ hybridization verified TGFBI mRNA in the epithelial cells but not in other cell types, supportive of a therapy directed specifically at this lineage.
Keywords: Cornea, ECM, iTRAQ, LC-MS/MS, POSTN, TGFBIp
Graphical abstract
TGFBIp is a major constituent of the human cornea and is involved in the development of TGFBI-linked corneal dystrophy. The corneal proteome of TGFBIp knock-out mice showed only minor changes and no apparent alterations of the ultrastructure. The data support TGFBI gene suppression as a potential therapy to TGFBI-linked corneal dystrophies.
Introduction
Visual impairment has a negative impact on quality of life and is estimated to affect approximately 285 million people worldwide [1]. While some visual disorders such as myopia can be treated noninvasively by corrective lenses, other diseases require surgical intervention. The latter category includes 5q31-linked transforming growth factor beta-induced (TGFBI) corneal dystrophies, a heterogeneous group of genetic disorders in which mutations within the TGFBI gene result in a progressive accumulation and aggregation of the secreted extracellular TGFBI protein (TGFBIp, UniProt entry Q15582) in the cornea [2]. Treatment of TGFBI-linked corneal dystrophies relies on surgical removal of the affected area using laser photoablation or ultimately replacing the affected cornea with a donor cornea. However, donor shortage and the recurrence of protein accumulation indicate the need to develop new treatment strategies.
Several studies have suggested that downregulating corneal TGFBIp expression may be a way to treat dystrophic patients, using either siRNA-based methods [3–5] or drugs targeting the molecular pathways involved in TGFBIp expression [6]. However, the practical aspects of drug delivery and the potential side effects of the partial or complete elimination of one of the most abundant proteins in the human cornea remain unclear [7]. In addition, TGFBIp is known to interact with a wide range of extracellular macromolecules, including the proteoglycans decorin and biglycan [8] and Type I, II, IV, VI and XII collagens [9–12], and is thought to link cells to the extracellular matrix (ECM) through a number of integrin binding sites [13–19]. Additionally, TGFBIp is considered a matricellular protein, meaning that TGFBIp is rapidly turned over and could play a regulatory role via sequestration and modulation of specific growth factors [20]. Thus, changing the expression level of TGFBIp in the cornea may have unforeseen consequences on the structural integrity of the cornea and aggravate the disease condition.
In this study, we investigated the role of TGFBIp in maintaining the structural integrity of corneal cells and the collagenous matrix using a TGFBI-deficient mouse model (TGFBI−/−). A histological and ultrastructural examination demonstrated that TGFBIp deficiency has little to no effect on the gross structure of the cornea, which remained transparent, suggesting the existence of functional redundancy. Comparing the corneal proteomes of wildtype (TGFBI+/+) and TGFBI−/− mice resulted in the identification of 11 differentially expressed proteins; including Type VI and XII collagens, annexin A2, fibromodulin and fibulin-5 which are all known for their roles in maintaining corneal structural integrity. Thus, even though expression changes were observed at the molecular level, the macroscopic, microscopic and ultrastructural appearance of the TGFBI−/− mouse cornea was unaffected, indicating that partial or complete TGFBI knockdown would be a feasible treatment option for TGFBI-linked corneal dystrophies.
Results
Corneal origin of TGFBIp expression
Systemic TGFBI-null mice were generated as previously described [21], and these mice are fully viable, fertile and indistinguishable from wildtype littermates. The corneal transparency and mouse behavior remained unaffected throughout adulthood, suggesting that TGFBI−/− mice have normal vision. To verify that TGFBIp protein deposition is completely absent in adult TGFBI−/− mouse corneas, we examined TGFBI mRNA and TGFBIp protein expression using in situ hybridization and immunoblotting (Figs. 1, 2). In situ hybridization demonstrated that TGFBI mRNA is specifically expressed within the corneal epithelium (Fig. 1A) and not seen in the stromal embedded keratocytes nor in the endothelium. Moreover, in situ hybridization confirmed that TGFBI mRNA is absent in TGFBI−/− mouse eyes (negative data not shown). Immunoblotting verified that TGFBIp was undetectable in the corneal tissue of TGFBI−/− mice and that a single 68-kDa TGFBIp band was present in isolated wildtype adult corneas (Fig. 2C). In addition, no obvious changes in the total protein expression profile were observed between TGFBI+/+ and TGFBI−/− corneal tissues subjected to SDS-PAGE followed by Coomassie brilliant blue staining (Fig. 2B), indicating that there were no major alterations in protein expression in TGFBI−/− mice.
Figure 1. TGFBIp and POSTN expression in the cornea.
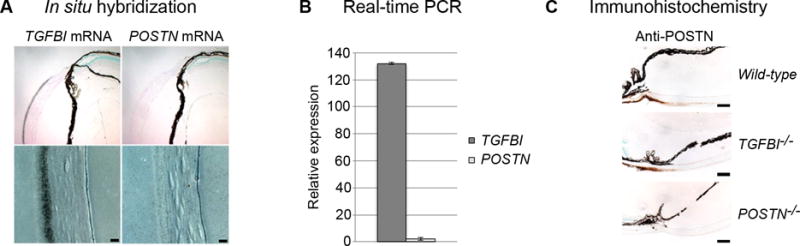
(A) In situ hybridization analysis of TGFBI and POSTN mRNA in wildtype mouse corneas. A strong signal (indicated via silver grains) for TGFBI mRNA is restricted to the corneal epithelium, whereas no apparent signal is observed for POSTN mRNA (scale bar = 20μm). (B) Real-Time PCR supports the presence of TGFBI and the lack of POSTN mRNA in isolated corneal tissues (n = 3). Error bars represents the SD. (C) Immunohistochemically staining using a POSTN antibody verifies the absence of POSTN in the central cornea and reveals that POSTN protein is localized at the corneal periphery. Moreover, the extent of POSTN deposition in wildtype and TGFBI−/− mice corneas is unchanged, whereas no POSTN was detected in POSTN−/− corneas (scale bars = 100μm).
Figure 2. TGFBI−/− corneas show no aberrant changes.
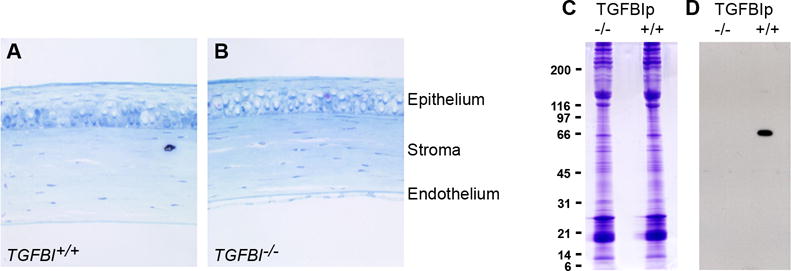
(A, B) No morphological changes are observed throughout the corneal in sections of (A) TGFBI+/+ and (B) TGFBI−/− corneas examined by light microscopy. Tissue sections are stained with toluidine blue; a 4x objective was used. (C) SDS-PAGE of TGFBI−/− corneas shows no major differences in protein expression when compared to control TGFBI+/+ corneas. (D) Western blotting using TGFBIp anti-serum confirms complete knockout in the TGFBI-deficient mouse eyes.
POSTN is located at the corneal periphery
Additionally, we sought to determine if periostin (POSTN), a closely related gene family member of TGFBI [22], was similarly expressed. Although POSTN is widely expressed in adult organs under normal and pathological conditions [23], wildtype mouse corneas do not express POSTN mRNA. Using both in situ hybridization and real-time PCR data, we found that POSTN mRNA is absent in adult corneas (Figs. 1A, B), and thus is not co-expressed with TGFBI. Likewise, immunohistochemistry revealed that POSTN protein is deposited at the corneal periphery, where ciliary body/scleral venous sinus are (Fig. 1C). As the same degree of POSTN deposition was observed for wildtype and TGFBI-deficient mouse corneas, this also suggests a non-compensatory role for POSTN. The apparent discrepancy between site of POSTN mRNA expression and protein deposition indicates that corneal surrounding tissues are responsible for active POSTN expression, which then translocates to the corneal periphery. This is supported by a previous study showing that human POSTN is expressed by limbal stem cells which differentiate into the corneal epithelium [24].
Gross structure of the TGFBI−/− cornea appears normal
To assess major effects of TGFBI knockout in mouse corneas, tissue sections of TGFBI+/+ and TGFBI−/− corneas were histologically compared (Fig. 2). No significant structural changes were observed in the stroma, or the Descemet’s membrane. Likewise, the epithelial and endothelial cell layers appeared unaffected in the TGFBI−/− corneas. Therefore, we conclude from these data that the absence of TGFBIp does not affect the gross architecture of the corneal structure.
Proteomic analysis of TGFBI+/+ and TGFBI−/− corneas
The histological observations described above led us to hypothesize that either TGFBIp is not essential for maintaining the structure of the developed cornea or functional redundancy exists due to the expression of other proteins to compensate for the lack of TGFBIp. To test these alternatives, a comparative analysis of the TGFBI+/+ and TGFBI−/− mouse cornea proteomes was performed. The iTRAQ MS analysis resulted in the quantification of 877 proteins across all datasets, of which 516 proteins were quantified in all three biological samples of both the TGFBI+/+ and TGFBI−/− groups (Figs. 3 and Table SI). Of the 516 proteins quantified in all samples, 11 were significantly mis-regulated using a significance threshold of 0.01 and a fold change of at least 0.5 on the log2 scale (Table 1, Fig. 3).
Figure 3. The ECM of TGFBI-deficient corneas is affected at the protein level.
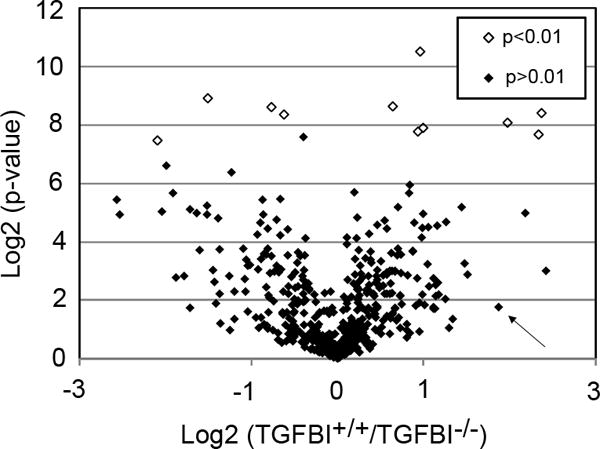
The relative quantification of TGFBI+/+ and TGFBI−/− corneas using the iTRAQ methodology followed by LC-MS/MS resulted in 516 proteins being quantified in all three biological replicates of both the TGFBI+/+ and TGFBI−/− groups. The volcano plot shows the TGFBI+/+/TGFBI−/− ratio distribution of quantified proteins using the log2 scale compared to the log2 of the p-values. Using a p-value cut-off of 0.01, we identified 11 proteins (Table 1) that were differentially expressed in the TGFBI−/− cornea (empty dots). Six proteins (Type VI collagen, Type XII collagen, annexin, fibromodulin, prolargin and fibulin-5) known for their role in ECM arrangement were among the significantly mis-regulated proteins identified, suggesting that the ECM is altered in the TGFBI−/− cornea. The arrow indicates POSTN, a TGFBIp paralog, that shows a 1.9-fold upregulation on the log2 scale in the TGFBI−/− cornea; however, it was not significantly mis-regulated at p<0.01.
Table 1.
Differential expressed proteins in TGFBI−/− corneas
| AccessionNr | Name | Mass kDa | P – value | Log2 (−/−/+/+) |
|---|---|---|---|---|
| P40124 | Adenylyl cyclase-associated protein 1 | 51.565 | 0.003 | −0.76 |
| P07356 | Annexin A2 | 38.676 | 0.004 | 1.00 |
| Q60847 | Collagen alpha-1(XII) chain | 340.214 | 0.004 | 1.98 |
| Q02788 | Collagen alpha-2(VI) chain | 110.334 | 0.005 | 0.94 |
| P50608 | Fibromodulin | 43.055 | 0.003 | 2.38 |
| Q9WVH9 | Fibulin-5 | 50.193 | 0.005 | 2.34 |
| P14602 | Heat shock protein beta-1 | 23.014 | 0.002 | −1.51 |
| Q9Z331 | Keratin, type II cytoskeletal 6B | 60.322 | 0.001 | 0.96 |
| Q9JK53 | Prolargin | 43.293 | 0.003 | 0.64 |
| Q9R1P1 | Proteasome subunit beta type-3 | 22.965 | 0.003 | −0.62 |
| P07724 | Serum albumin | 68.693 | 0.006 | −2.09 |
Table shows mis-regulated proteins in TGFBI−/− compared to TGFBI+/+ mice corneas with a p value below 0.01 and a minimum 0.5 fold mis-regulation on the log2 scale.
POSTN does not compensate for TGFBIp deficiency in the cornea
Of note, even though POSTN was above the significance threshold of 0.01 (p = 0.3), the protein was upregulated in TGFBI−/− mice, with a log2 fold change of 1.9 in the iTRAQ MS analysis (Table S1). Although POSTN is a TGFBIp paralog that exhibits high sequence similarity, and the two are often co-expressed, we show that POSTN mRNA is absent within the wildtype cornea (Fig. 1A, B). Increased POSTN protein levels in TGFBI−/− mice would support the idea that one or more proteins could compensate for the functional loss of TGFBIp. To test this, we performed a targeted quantitative selected reaction monitoring (SRM) analysis of POSTN in the three biological TGFBI+/+ and TGFBI−/− replicates in each group (Fig. 4). However, the LC-SRM-MS analysis supported the iTRAQ data and immunohistochemical staining and revealed no significant upregulation of POSTN protein in the TGFBI−/− mice, thus disproving the hypothesis that POSTN is upregulated to compensate for the lack of TGFBIp in the TGFBI−/− corneas.
Figure 4. The TGFBIp paralog POSTN does not compensate for the lack of TGFBIp expression.
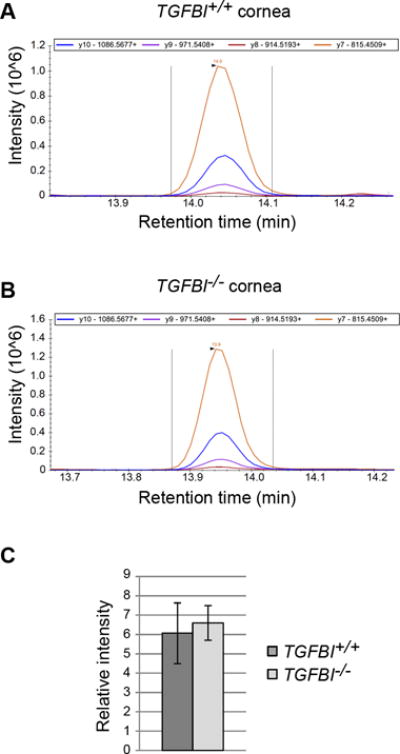
(A, B) Targeted LC-SRM-MS assay was developed to assess whether the TGFBIp paralog POSTN is upregulated in the TGFBI−/− cornea to compensate for loss of TGFBIp. POSTN was one of the most regulated proteins in the iTRAQ analysis; however, its change in expression was not significant at p<0.01. The SRM assay was designed to monitor four proteotypic peptides for POSTN, including four transitions per proteotypic peptide. Each represent one of the biological replicates (n = 3) and show the intensity of the four transitions for the tryptic POSTN peptide IIDGVPVEITEK in TGFBI+/+ (A) and TGFBI−/− (B). (C) No significant difference between the TGFBI+/+ and TGFBI−/− groups (n = 3) was observed using a Student’s t-test and a p-value cut-off of 0.01, thus rejecting the hypothesis that the functional loss of TGFBIp in the TGFBI-deficient cornea is compensated by POSTN upregulation. Error bars represents the SD.
TGFBIp absence does not affect corneal ultra-structure
The macroscopic appearance and animal behavior of the TGFBI−/− mice seem normal, although the proteomic analysis indicates several changes in expression of ECM proteins known to be expressed and involved in corneal stroma collagenous matrix organization. Consequently, we analyzed potential ultrastructural differences between TGFBI+/+ and TGFBI−/− mouse corneas using transmission electron microscopy (TEM), to quantify subepithelial collagen fibril diameters and interfibrillar spacing (Fig. 5). Collagen fibril diameters were similar for both TGFBI+/+ and TGFBI−/− corneas (mean: 30±3 vs. 31±3nm). However, the mean values of the center-to-center interfibrillar spacing were 58±10nm and 62±11nm in the TGFBI+/+ and TGFBI−/− corneas, respectively. This supports the proteomic data and indicates that the collagenous matrix organization is marginally affected in TGFBI−/− corneas, resulting in a less densely packed collagen fibril corneal arrangement.
Figure 5. Stromal interfibrillar spacing is slightly affected in TGFBI−/− corneas.
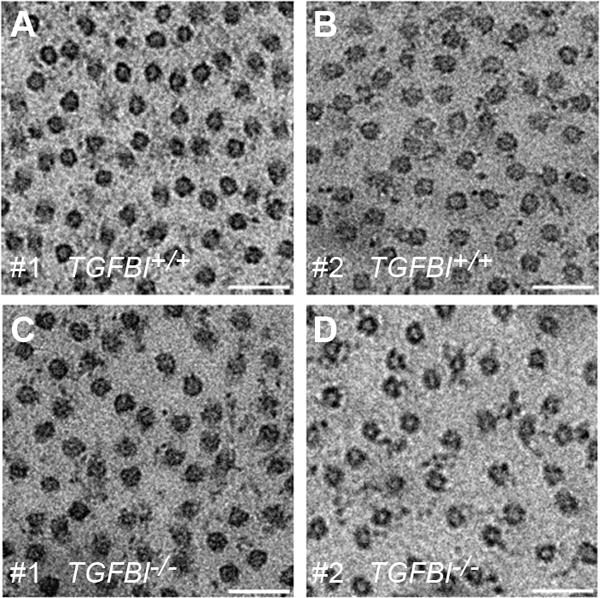
(A-D) TGFBIp is known to interact with Type VI and XII collagens, which were both mis-regulated in the iTRAQ analysis (Table 1). To investigate the effect of TGFBI deficiency on the delicate collagen architecture of the cornea, TEM micrographs were taken of TGFBI+/+ (A, B) and TGFBI−/− corneas (C, D). All micrographs taken represent the central anterior stroma, and the subsequent morphometric analysis relied on five randomly chosen TEM micrographs per cornea. Collagen fibril diameter (n = 250) and center-to-center interfibrillar spacing (n = 250) were measured and compared between the TGFBI+/+ and TGFBI−/− groups. The morphometric analysis showed no change in collagen fibril diameter (mean: 30±3 vs. 31±3nm), whereas interfibrillar spacing increased slightly in the TGFBI-deficient cornea (58±10nm vs. 62±11). Scale bar = 500nm.
Humans express elevated corneal TGFBIp amounts compared to mouse
Unpublished observations suggest differential expression of TGFBIp between species. Mice appear to express substantial lower corneal levels of TGFBIp compare to human. In order to determine the degree of expression levels we performed a relative label-free MS1 quantification between corneal TGFBIp in humans and mice. This analysis showed that human corneal TGFBIp levels exceed mice by more than 10-fold (Table S2). Thus, completely removing TGFBIp as a therapy against corneal dystrophies in humans may result in a larger effect on the protein expression profile than observed in mice mutants.
Discussion
TGFBIp is a major constituent of adult cornea
We have previously demonstrated that TGFBIp is one of the most abundant proteins in the human cornea [7]. We have also shown that TGFBIp and POSTN can exhibit both overlapping and mutually exclusive gene expression patterns [22], and that TGFBI null mice are viable and live a normal lifespan [21]. Our expression analysis (Fig. 1) revealed that only TGFBIp is robustly expressed in the adult corneal epithelium, suggesting continual secretion and deposition of TGFBIp from the epithelium into the corneal stroma. Additionally, we show that it is unlikely that the TGFBIp paralogue, POSTN, is able to compensate for the systemic loss of TGFBI (Fig. 1). However, future analysis of viable double TGFBI:POSTN knockout mice [25], will be helpful in determining whether low level corneal POSTN, as observed in both wildtype and TGFBI−/− corneas via iTRAQ MS analysis (Fig. 3), is sufficient to maintain normal corneal morphology and definitively conclude whether TGFBIp and POSTN indeed share any redundant and/or parallel functions in the adult cornea.
Preservation of corneal integrity in TGFBI−/− mice
Studies using mouse models lacking proteins important for proper corneal collagen architecture have demonstrated changes to the cornea including stromal thickness, disorganization of collagen fibrils and loss of transparency [26–29]. TEM of corneas from keratocan-null mice showed less organized collagen fibers than those of wildtype mice [27]. Corneas of lumican-null mice exhibited major changes in expression of ECM proteins when analyzed by a similar quantitative MS approach as performed in this study [28]. Severe disruption in collagen fibril structure and organization was observed in the corneas of a decorin and biglycan double-deficient mouse model [29], whereas fibromodulin was shown to be involved in and affect collagen fibrillogenesis in the peripheral cornea in fibromodulin-deficient mice [26]. As TGFBIp binds several ECM proteins involved in collagen organization, it was anticipated that knocking out TGFBI would introduce changes in the collagenous matrix similar to that observed for keratocan-, lumican-, decorin/biglycan-, or fibromodulin-deficient mice. However, our examination of TGFBI+/+ and TGFBI−/− mice did not reveal structural changes in the corneas of TGFBI-null mice. This is in agreement with observations of the corneas from another TGFBI-deficient mouse model, where no histological changes or differences in animal development or behavior were observed [30].
In addition to histological examination, our analysis of the ultrastructure using TEM only showed minor changes in collagen fibril diameter and interfibrillar spacing. This is in sharp contrast to the profound changes in the arrangement of collagen fibrils observed in the keratocan-, lumican-, decorin/biglycan- and fibromodulin-null mouse corneas [26–29]. However, the small changes in interfibrillar spacing seen in our TEM data support the findings in our quantitative proteomic analysis, which identified 11 proteins as being differentially expressed in the TGFBI-deficient mice (Table 1). Six of these proteins are known to be involved in the assembly of the collagen scaffold and cell adhesion to the ECM. The exact functional consequences of this expression profile alteration remain unclear, but it is likely that TGFBIp functions as a regulatory mediator between the ECM and the embedded corneal cell lineages. Whether TGFBIp directly interacts with these mis-expressed ECM components and if they could themselves contribute to altered TGFBIp protein aggregation will be key in the development of therapeutic agents capable of dissolving and/or inhibiting the formation of mutant TGFBIp aggregates in the cornea. The recently published high resolution crystal structure of TGFBIp [31] (PDB entry: 5NV6) will provide new insight into the functional role of TGFBIp and its interaction to the extracellular matrix and may be a help to interpret and link to the differential expressed proteins identified in this study.
The TGFBIp paralog POSTN is well characterized as a matricellular protein [32], a term that may also be applied to TGFBIp. Matricellular proteins are a diverse group of non-structural molecules present in the ECM that are vital for maintaining the integrity of the ECM in health and disease. In addition to being important components of the ECM itself, matricellular proteins also play crucial regulatory roles and can modulate cellular responses to extracellular signals induced during tissue remodeling and repair as well as in numerous disease states [33]. Interestingly, matricellular protein-deficient mice models often show mild changes in phenotypic appearance [33], which correlates well with the observations of the mouse model presented in the current study.
Differential expression of collagen scaffold proteins
The cornea accounts for approximately 70% of the refractive power of the eye and together with the aqueous humor and the lens protects the retina from UV light [34, 35]. Corneal transparency is dependent on the highly ordered collagen network found in the stroma comprising approximately 90% of the corneal thickness. The corneal collagen network consists of approximately 200 interwoven lamellae, and each lamella consists of collagen Types I and V heterotypic fibrils arranged in a hexagonal pattern [36]. Collagen Types VI, XII and XIV together with the proteoglycans decorin, lumican, keratocan, mimecan, biglycan and fibromodulin provide structural support for proper collagen fibril assembly and spacing [37]. Notably, Types VI and XII collagens were upregulated in our LC-MS/MS analysis, and both collagen types have previously been shown to covalently bind TGFBIp [9, 12]. This suggests that one of the important physiological roles of TGFBIp is to bind Type VI and XII collagen and thereby participate in matrix organization and regulation of collagen constituent composition. TGFBIp was found to be upregulated in the cartilage of a collagen Type IX-deficient mouse model [38]. Both Type IX and XII collagens belong to the FACIT (Fibril Associated Collagens with Interrupted Triple helices) collagen family and may therefore share similar functionalities. In addition to increased levels of TGFBIp in the cartilage of the Type IX collagen-deficient mouse, a change in TGFBIp degradation was observed. This is noteworthy because corneal TGFBIp is similarly proteolytically processed [39–41], implying that TGFBIp degradation is sensitive to changes in its matrix environment.
In addition to increased expression of Type VI collagen, we observed annexin A2 upregulation. Annexin A2 has been suggested to mediate secretion of Type VI collagen in epithelial cells [42], and we found that these two proteins were upregulated to a comparable extent, supporting a similar connection in our mouse mutant model.
Furthermore, our iTRAQ analysis indicated highest upregulation of fibromodulin, one of the proteoglycans involved in accurate matrix assembly. Fibromodulin is expressed in the developing cornea but is located only peripherally in the mature cornea [26]. Even though no interaction between fibromodulin and TGFBIp has been reported in the literature, the fibromodulin homologs decorin and biglycan have both been shown to bind TGFBIp in a non-covalent manner [8]. Hence, fibromodulin may possess unappreciated binding properties similar to those of decorin and biglycan. In our previous quantitative analysis of corneal proteins, decorin and biglycan are among the most abundant proteins in the corneal stroma and endothelium [7]. Moreover, TGFBIp has been shown in vitro to enhance the effect of biglycan and decorin on Type VI collagen aggregation, and decorin/biglycan double-deficient mice display major defects in collagen fibril morphology [29]. However, decorin and biglycan interactions with TGFBIp may be less pronounced in the mouse cornea, as neither protein was represented on our list of differentially regulated proteins (Table 1), and no major alterations in collagen fibril diameter or spacing was observed in the TGFBI-deficient mouse corneas, as indicated by our TEM analysis.
Prolargin, like lumican, decorin and biglycan, is a small leucine-rich repeat proteoglycan. Prolargin has been shown to be a major proteoglycan of both the sclera and the cornea [7, 43] and can bind Type I collagen and function as an anchor between the ECM and basement membranes. Thus, the fact that TGFBI knockout results in an upregulation of prolargin in the mouse cornea suggests that TGFBIp may play a role in anchoring the stromal collagen scaffold to the epithelium basement membrane.
Fibulin-5 has been shown to be important for elastic fiber organization but does not affect collagen assembly [44], and we have previously identified it as one of the more abundant proteins in the Descemet’s membrane [45] and endothelial layer in the cornea [7]. It contains the RGD integrin-binding motif that is also found in TGFBIp [46]. This motif mediates binding to cells; hence, the significant fibulin-5 upregulation we observed may act to compensate for loss of TGFBIp cell adhesion. Whether fibulin-5 is able to form a complex with TGFBIp via the RGD motif and why fibulin-5 is upregulated in the absence of TGFBIp may be important for our understanding of normal cell adhesion in the cornea.
TGFBI knockdown as a therapeutic treatment in corneal dystrophies
The genetic origins of TGFBI-associated corneal dystrophies are heterogeneous, with new disease-causing mutations being reported on a regular basis. Therefore, a therapeutic strategy with a broad effect is desirable. This strategy could rely on suppression of the disease-causing protein. Several groups have addressed this using ex vivo and in vitro systems. Courtney et al. used siRNA targeting only the mutant allele of TGFBI in heterozygous primary corneal epithelial cells from a patient with lattice corneal dystrophy [3]. As this allele-specific strategy only knocked down the mutant protein, the patient still expressed wildtype TGFBIp in the cornea and therefore presumably exhibited a non-affected cornea. The same group later demonstrated the use of mutant allele-specific suppression of keratin 12 using CRISPR/Cas9 in a model of Meesmann’s epithelial corneal dystrophy. They concluded that a similar approach may be applicable to a substantial number of the TGFBI mutations reported to date [47]. However, the allele-specific approach can only be applied to heterozygous patients, whereas for homozygous patients, complete TGFBI knockdown will be required. Combined, our data indicate that it is possible to maintain structural integrity and to sustain protein expression following a complete removal of TGFBIp in the mouse cornea. This opens up possibilities for therapeutic strategies relying on complete elimination of TGFBIp. Yuan et al. and Yellore et al. both address this possibility using siRNA for total TGFBI knockdown in human corneal cell lines [4, 5], whereas Choi et al. used lithium treatment on corneal fibroblasts to inhibit TGFBIp expression [6]. In the latter case, lithium treatment affected the upstream signaling pathway involved in TGFBIp expression, which likely affects the expression of many other proteins including TGFBIp, giving rise to undesirable side-effects. Another consideration in the design of therapies, is which corneal cell to target for TGFBIp suppression or knockdown to achieve the desired effect. Our in situ hybridization data (Fig. 1) show that TGFBI mRNA is restricted to the corneal epithelium and does not appear to be transcribed by the corneal keratocytes embedded in the stroma or in the innermost endothelial layer of the cornea. This suggests that generation of a therapy against TGFBI-related corneal dystrophies can be achieved by targeting only the corneal epithelium, which in terms of therapeutic delivery would most likely be less complicated than targeting all corneal cell types. Of note, Allaman-Pillet et al. showed that corneal TGFBIp was mainly present in the corneal stroma [30], suggesting that TGFBIp is expressed and secreted by the epithelium and diffuses throughout the underlying cornea. This may also partly explain why the epicenter of TGFBIp deposition in the corneal stroma appears to be different depending on the site and type of mutation in the TGFBI gene [48]. Recently, Liu et al. using quantitative PCR showed TGFBI mRNA expression in human keratocytes [49], however the authors did not compare mRNA expression levels between corneal cell types and the contribution by keratocytes may be significant lower than seen for the epithelium as supported from our in situ hybridization data. Our unpublished observation of human corneas maintained in culture medium where the epithelial layer has detached, shows unusual low levels of TGFBIp in the stroma when analyzed by mass spectrometry than normally seen for corneas without pre-detachment of the epithelium. Hence, all together we interpret these data as that the majority of TGFBIp originate from the corneal epithelium. This assumption is also supported by in situ hybridization experiments on rabbit corneas only detecting TGFBI mRNA in the epithelium [50]. Lastly, it should be noted with care that corneal TGFBIp expression levels in humans and mice differ substantially. The relative comparison between TGFBIp expression levels shows that it is 10-fold more abundant in humans than in mice (Table S2), which may amplify the minor changes observed in ECM arrangement in our TGFBI-null mouse to a more severe situation in humans.
Several potential non-invasive therapeutic strategies to treat TGFBI-linked corneal dystrophy have been published, all relying on partial or complete knockdown of TGFBIp without considering the consequences of this on corneal homeostasis and integrity. In this study, we examined TGFBIp expression and characterized the gross structural and proteomic changes in mice deficient in TGFBIp compared to wildtype mice. Our data revealed only minor changes in the structure of the corneas in the TGFBI-null mice. In addition, the expression levels of only a few proteins were affected upon removal of TGFBIp. Hence, these data suggest that total TGFBIp knockdown in corneas is a viable therapeutic strategy for the treatment of TGFBI-linked dystrophic patients.
Experimental Procedures
Transgenic mice
Mice with null genetic deletions of POSTN or TGFBI were generated and genotyped as previously described [21, 51]. After euthanization, eyes from TGFBI+/+ and TGFBI−/− adult mice (8-10 weeks) and POSTN−/− adult (8–10 weeks) were harvested in cold phosphate-buffered saline (PBS) and either placed immediately in liquid nitrogen or fixed in 4% paraformaldehyde or 4% paraformaldehyde/2% glutaraldehyde. Equal ratios of male and female mice were used in all experiments, and the mice are maintained on a C57BL/6J background. The mice were housed in standard barrier rack cages supplied with Purina Rodent Chow 5001 with automatic watering dispensers. All procedures were performed with the approval of the Institutional Animal Care and Use Committee at Indiana University School of Medicine (Indianapolis, IN; protocol 10807).
RNA Isolation and Quantitative PCR
mRNA from micro-dissected 10 week old adult wildtype corneas (n = 6 corneas from 3 mice) was isolated using RNAEasy (Qiagen, Venlo, The Netherlands) kit. To compare TGFBI and POSTN mRNA expression levels within isolated cornea, we used quantitative PCR (qPCR) on cDNA synthesized using a Superscript-II kit (Invitrogen, Waltham, MA). cDNA was amplified within the linear range using primers (TGFBI forward 5′-ACCATGGACCGGATGTTGAC-3′ and reverse 5′- GGCCACCAGCATGCTAAAAC-3′; POSTN forward 5′-AAGGAAAAGGGTCATACACGTACTTC-3′ and reverse 5′-CCTCTGCGAATGTCAGAATCC-3′ from Qiagen) and normalized to GAPDH (PPM02946E-200), as described [52]. qPCR reactions were carried out using SyberGreenER (Invitrogen) and run on the same plate. All qPCR results are representative of three separate experiments (n = 3) and ΔΔCt based fold-changes were calculated using GAPDH as a housekeeping standard.
In situ hybridization and Immunohistochemistry
Murine 8-10 week old eyes were harvested, fixed in 4% paraformaldehyde overnight at 4°C, dehydrated through a graded ethanol series, cleared in xylenes, and processed for paraffin embedding and sectioning at 10μm thickness. Sense and anti-sense [35S]UTP-radiolabeled TGFBI and POSTN cDNA probes were transcribed for in situ hybridization, as previously described [22]. Following de-waxing and in situ analysis, a specific signal pattern was observed only with the anti-sense probe and was detected in at least three consecutive wildtype eye sections (n = 3). Subsequently, following de-waxing, whole wildtype, TGFBI−/− and POSTN−/− eye sections were used to examine POSTN protein deposition. POSTN was detected using a rabbit polyclonal antibody (1:10,000 dilution) and ABC kit (Vectorstain, Burlingame, CA) with DAB and hydrogen peroxide as a chromogen, as described [53]. Serial sections probed with the POSTN antibody were examined for signal detection in consecutive sections in at least three eyes of each genotype (n = 3 per genotype).
SDS PAGE
Equal amounts of TGFBI+/+ and TGFBI−/− corneal tissue were boiled for 10-min in SDS sample buffer under reducing condition (5mM dithiotreitol), and separated on 5-15% (w/v) gradient polyacrylamide sodium dodecyl sulfate gels, cast in-house [54].
Western blotting
The SDS-PAGE gels were equilibrated with blotting buffer consisting of 10mM CAPS, pH 11, 10% methanol (VWR chemicals) and transferred to polyvinylidene difluoride (PVDF) membranes (Immobilon-P, Millipore, Billerica, MA) by electroblotting at 0.5A for 15min. The membranes were blocked using 5% milk (Milex 240) in TBS-T for an hour at 23°C followed by incubation overnight at 4°C with rabbit polyclonal anti-TGFBIp serum [12] diluted 1:10,000 in 2% skim milk TBS-T. The following day, the membrane was washed three times in TBS-T before incubation with secondary antibodies against rabbit (peroxidase conjugated anti-rabbit IgG, Sigma Aldrich, Missouri, US) diluted 1:25,000 in 2% skim milk TBS-T for two hours at 23°C. Finally, the membrane was developed using Amersham Hyperfilm and the enhanced chemiluminescence (ECL) kit (GE healthcare Life Sciences, Chalfont, UK).
Light microscopy
Formaldehyde-fixed and paraffin-embedded TGFBI+/+ and TGFBI−/− corneal tissue sections 7μm thick were cut perpendicular to the epithelium. The sections were deparaffinized and stained with toluidine blue using standard histological procedures. Histological examinations (n = 6 eyes from each genotype) were performed using light microscopy.
Preparation of corneas for mass spectrometry
Eyes from TGFBI+/+ control mice and TGFBI−/− mice (n = 6 eyes from each genotype) were removed and immediately frozen in liquid nitrogen. The corneas were subsequently excised using a 2mm biopsy needle (Miltex, Tuttlingen, Germany) and washed three times in 200μL of ice cold PBS and once in 1mL of PBS. The corneas from two mice were pooled, resulting in three biological replicates within each group, and lyophilized in a Speed-Vac concentrator (Thermo Scientific, Waltham, MA) for 18h. Proteins were then chemically cleaved by adding 100μL of 0.66M CNBr in 70% trifluoroacetic acid and incubating at 23°C for 20h. The digest was then lyophilized and washed twice in 500μL of Milli-Q H2O with intermittent drying in the Speed-Vac. The washed samples were then dissolved in 100μL of 8M urea, 0.2M Tris-HCl, pH 8.3, and the peptide concentrations were determined using a 2-D Quant Kit (GE Healthcare Life Sciences, Little Chalfont, United Kingdom). Disulfide bridges were reduced by incubating with 10mM dithiothreitol for 1h at 23°C. The reduced cysteines were then alkylated by adding 30mM iodoacetamide and incubating for 1h in the dark at 23°C. The samples were then diluted five times in 0.1M Tris-HCl, pH 8.3, and 10μg was incubated overnight at 37°C with 200ng of trypsin (Promega, Madison, WI). The samples were centrifuged for 5min at 17,000 × g to remove precipitates, and the supernatants were desalted using POROS R2 C8/C18 resin (Life Technologies, Paisley, UK) packed in 200μL gel-loading pipet tips.
iTRAQ labeling and strong cation exchange (SCX) fractionation
For each biological sample, 2.5μg of desalted peptide was pooled in a reference “master mix” to be labeled with iTRAQ 114, and 7.5μg of each sample was dissolved in 25μL of iTRAQ dissolution buffer (Life Technologies, Carlsbad, CA, US). The samples were labeled with 4plex iTRAQ (115, 116, or 117) reagents according to the manufacturer’s protocol, resulting in two iTRAQ sets to be analyzed by LC-MS/MS. Briefly, each iTRAQ label was diluted with 50μL of ethanol, mixed with the sample and incubated for 2h at 23°C. The samples were then mixed, lyophilized and desalted using POROS R2 C8/C18 resin as described above. The two iTRAQ sets were then applied to an SCX column (PolyLC Inc., Columbia, MD) equilibrated in 30% acetonitrile, 5mM KH2PO4, pH 2.7 and eluted with a linear gradient from 0mM to 500mM KCl in 30% acetonitrile, 5mM KH2PO4, pH 2.7. Eluted peptides were collected in 12 fractions including the flow-through for each iTRAQ set. Each fraction was desalted using POROS R2 micro purification and lyophilized.
LC-MS/MS of iTRAQ samples
Samples were dissolved in 0.1% formic acid and analyzed by nanoflow LC-MS/MS on an EASY-nLC II system (Thermo Scientific, Waltham, MA, US) coupled to a TripleTOF 5600 mass spectrometer (Sciex, Framingham, MA, US). The samples were loaded on a 0.1x21mm C18 trap column and a 0.075x100mm C18 analytical column (NanoSeparations, Nieuwkoop, Netherlands). Peptides were eluted and electrosprayed directly into the mass spectrometer using a 50min gradient from 5-40% acetonitrile in 0.1% formic acid at a flow rate of 250nL/min. The data were acquired using an ion spray voltage of 2.3kV, a curtain gas setting of 30 and an interface heater temperature of 150°C.
Processing of iTRAQ data
All 12 wiff files generated for either iTRAQ set 1 or 2 were searched simultaneous against the SwissProt Mus musculus database (2016_8) using ProteinPilot v.4.5 (Sciex, Framingham, MA) and the Paragon algorithm v. 4.5.0.0. Trypsin was specified as the digestion enzyme, allowing up to 5 missed cleavages. Iodoacetamide was entered as a fixed modification of cysteine, and the default standard workup modification set was used as the variable modifications. The instrument setting was specified as TripleTOF 5600, defining the default charge state and mass accuracy of the instrument, and the iTRAQ 4plex (peptide labeled) protocol was selected as the sample type. Quantitative data were normalized using the bias and background corrections to correct for any experimental and technical bias. Thorough ID and false discovery rate (FDR) analysis were both selected. The searches were imported to and parsed using MS Data Miner v. 1.3.0 [55], using a 1% global FDR at the protein level. To find significantly mis-regulated proteins, Student’s t-test with a significance threshold of 0.01 was performed for proteins quantified in all 3 biological replicates of both the TGFBI+/+ and TGFBI−/− groups. Proteins with a p-value less than 0.01 and a minimum fold change of 0.5 on the log2 scale were considered up- or downregulated in TGFBI−/− mice. TGFBIp was excluded from the data processing, as it is not possible to obtain a ratio between TGFBI+/+ and TGFBI−/− samples.
Targeted MS analysis of POSTN – LC-SRM-MS
The SRM assay was developed using Skyline 1.4.0.4421 [56] and based on the optimization of 4 stable isotope-labeled POSTN analog peptides (SpikeTides, JPT Peptide Technologies GmbH, Germany) (DQGPNVCALQQILGTK (+2), NGVIHLIDEVLIPDSAK (+2), LLYPADIPVGNDQLLELLNK (+2), and IIDGVPVEITEK (+2)). The final optimized assay included 4 transitions per peptide monitored, including both light and heavy peptide variants. New digests of TGFBI+/+ and TGFBI−/− corneas were prepared as described above. The LC-SRM-MS analysis was performed on an EASY-nLC II system (Thermo Scientific, Waltham, MA, US) connected in-line to a Qtrap 6500 mass spectrometer (Sciex) equipped with a NanoSpray III source (AB Sciex) and operated under Analyst 1.6.1 control. Peptides were eluted at a flow rate of 250nL/min using a 12min gradient from 5% to 35% phase B (0.1% formic acid and 90% acetonitrile), followed by re-equilibration for 10min to the starting conditions. The Qtrap 6500 was used in positive ion mode with an ion spray voltage of 25kV, a curtain gas setting of 30, an ion source gas setting of 5 and an interface heater temperature of 150°C. The eluted peptides were measured with a scheduled (3min window) SRM method using a 20ms dwell time for all transitions (32 in total) and a target scan time of 0.8s.
Relative comparison of human and mouse corneal TGFBIp amounts
Human tissue was used with full ethical approval and in accordance with the Declaration of Helsinki. Equal amounts of post-mortem healthy human cornea (n = 2, female, age 86; and male, 79) and wildtype mouse cornea (n = 2, 8–10 weeks) were processed as described above using a combination of CNBr chemical cleavage and trypsin digestion. The digested samples were normalized according to the OD280 value, and equal amounts were subjected to LC-MS/MS analysis using a method allowing for post-acquisition label-free quantification at the MS1 level. Wiff files were imported to Skyline 1.4.0.4421 [56], and four TGFBIp peptides (NNVVSVNK, QHGPNVCAVQK, STVISYECCPGYEK, SPYQLVLQHSR) identical in sequence between species were used for a relative MS1 quantitative comparison (Table S2).
Transmission electron microscopy (TEM)
Corneas from TGFBI−/− and age-matched TGFBI+/+ controls (n = 2 corneas from each genotype), which had all been stored at −20 °C before use, were fixed in 4% paraformaldehyde, 2% glutaraldehyde, and 0.1M sodium cacodylate, pH 7.2 and postfixed with 1% osmium tetroxide followed by 0.5% uranyl acetate. The corneas were then dehydrated using an ethanol series and propylene oxide. The corneal specimens were infiltrated and embedded using an EMbed 812 Kit according to the manufacturer’s protocol (Electron Microscopy Sciences, Hatfield, PA.). Thin sections of 40–60nm on average were cut using a Leica Ultracut UCT (Leica Mikrosysteme, Vienna, Austria) with a 45° diamond knife (Diatome, Biel, Switzerland) and post-stained with saturated aqueous uranyl and lead citrate [57]. The samples were imaged on a Tecnai G2 Spirit transmission electron microscope (FEI, Hillsboro, Oregon) equipped with a TemCam F416 CCD camera (Tvips, Gauting, Germany), operating at 120kV. All micrographs used in the morphometric analysis represent the central anterior corneal stroma and were acquired at 11,000× magnification.
Morphometric analysis of collagen fibrils
TEM micrographs were analyzed using ImageJ v.1.42q software (imagej.nih.gov). Collagen fibril diameter (n = 250) and center-to-center interfibrillar spacing (n = 250) were measured for TGFBI−/− and TGFBI+/+ corneas (n = 2 corneas from each genotype). Each measurement relied on 5 randomly chosen TEM micrographs per cornea, all representing the central anterior stroma.
Supplementary Material
Table S1: Proteins quantified in all biological replicates of both TGFBIp wildtype and deficient mice corneas.
Table S2: Relative comparison of human and mouse corneal TGFBIp.
Acknowledgments
We would like to express our sincere gratitude to the late Dr. Gordon K. Klintworth, who sadly passed away before publication of this paper. Dr. Klintworth was involved in initial histological examination of corneal tissue described in this paper.
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE [58] partner repository with the dataset identifier PXD006494.
This study was supported by the Danish Council for Independent Research – Medical Sciences (DFF-4004-00471), the Lundbeck Foundation (R164-2013-15912), the VELUX FOUNDATION, and Fight for Sight, Denmark and supported, in part, by NIH grant R01 HL115619 (SJC).
Abbreviations
- ECM
extracellular matrix
- iTRAQ
isobaric tags for relative and absolute quantitation
- LC-MS/MS
liquid chromatography tandem mass spectrometry
- POSTN
periostin gene
- SRM
selected reaction monitoring
- TEM
transmission electron microscopy
- TGFBI
transforming growth factor beta-induced protein
Footnotes
Author contributions:
ETP, KR, SJC and JJE designed the study, analyzed the results and wrote the paper. ETP, KR, NSN and MVL performed LC-MS/MS analyses, MS data processing, SDS-PAGE, and WB experiments. PS, OS and SJC were responsible for animal work, qPCR, IHS and the ISH analyses. KT and ETP performed the TEM analysis. HV was responsible for human tissue samples and involved in the histological examination. All authors approved the final version of the manuscript.
Conflict of interest: The authors declare that they have no conflicts of interest with the contents of this article.
References
- 1.Pascolini D, Mariotti SP. Global estimates of visual impairment: 2010. The British journal of ophthalmology. 2012;96:614–8. doi: 10.1136/bjophthalmol-2011-300539. [DOI] [PubMed] [Google Scholar]
- 2.Klintworth GK. Corneal dystrophies. Orphanet journal of rare diseases. 2009;4:7. doi: 10.1186/1750-1172-4-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Courtney DG, Atkinson SD, Moore JE, Maurizi E, Serafini C, Pellegrini G, Black GC, Manson FD, Yam GH, Macewen CJ, Allen EH, McLean WH, Moore CB. Development of allele-specific gene-silencing siRNAs for TGFBI Arg124Cys in lattice corneal dystrophy type I. Investigative ophthalmology & visual science. 2014;55:977–85. doi: 10.1167/iovs.13-13279. [DOI] [PubMed] [Google Scholar]
- 4.Yellore VS, Rayner SA, Aldave AJ. TGFB1-induced extracellular expression of TGFBIp and inhibition of TGFBIp expression by RNA interference in a human corneal epithelial cell line. Investigative ophthalmology & visual science. 2011;52:757–63. doi: 10.1167/iovs.10-5362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yuan C, Zins EJ, Clark AF, Huang AJ. Suppression of keratoepithelin and myocilin by small interfering RNAs (siRNA) in vitro. Molecular vision. 2007;13:2083–95. [PubMed] [Google Scholar]
- 6.Choi SI, Kim BY, Dadakhujaev S, Jester JV, Ryu H, Kim TI, Kim EK. Inhibition of TGFBIp expression by lithium: implications for TGFBI-linked corneal dystrophy therapy. Investigative ophthalmology & visual science. 2011;52:3293–300. doi: 10.1167/iovs.10-6405. [DOI] [PubMed] [Google Scholar]
- 7.Dyrlund TF, Poulsen ET, Scavenius C, Nikolajsen CL, Thogersen IB, Vorum H, Enghild JJ. Human cornea proteome: identification and quantitation of the proteins of the three main layers including epithelium, stroma, and endothelium. Journal of proteome research. 2012;11:4231–9. doi: 10.1021/pr300358k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reinboth B, Thomas J, Hanssen E, Gibson MA. Beta ig-h3 interacts directly with biglycan and decorin, promotes collagen VI aggregation, and participates in ternary complexing with these macromolecules. The Journal of biological chemistry. 2006;281:7816–24. doi: 10.1074/jbc.M511316200. [DOI] [PubMed] [Google Scholar]
- 9.Hanssen E, Reinboth B, Gibson MA. Covalent and non-covalent interactions of betaig-h3 with collagen VI. Beta ig-h3 is covalently attached to the amino-terminal region of collagen VI in tissue microfibrils. The Journal of biological chemistry. 2003;278:24334–41. doi: 10.1074/jbc.M303455200. [DOI] [PubMed] [Google Scholar]
- 10.Hashimoto K, Noshiro M, Ohno S, Kawamoto T, Satakeda H, Akagawa Y, Nakashima K, Okimura A, Ishida H, Okamoto T, Pan H, Shen M, Yan W, Kato Y. Characterization of a cartilage-derived 66-kDa protein (RGD-CAP/beta ig-h3) that binds to collagen. Biochimica et biophysica acta. 1997;1355:303–14. doi: 10.1016/s0167-4889(96)00147-4. [DOI] [PubMed] [Google Scholar]
- 11.Kim JE, Park RW, Choi JY, Bae YC, Kim KS, Joo CK, Kim IS. Molecular properties of wild-type and mutant betaIG-H3 proteins. Investigative ophthalmology & visual science. 2002;43:656–61. [PubMed] [Google Scholar]
- 12.Runager K, Klintworth GK, Karring H, Enghild JJ. The insoluble TGFBIp fraction of the cornea is covalently linked via a disulfide bond to type XII collagen. Biochemistry. 2013;52:2821–7. doi: 10.1021/bi400212m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bae JS, Lee SH, Kim JE, Choi JY, Park RW, Yong Park J, Park HS, Sohn YS, Lee DS, Bae Lee E, Kim IS. Betaig-h3 supports keratinocyte adhesion, migration, and proliferation through alpha3beta1 integrin. Biochemical and biophysical research communications. 2002;294:940–8. doi: 10.1016/S0006-291X(02)00576-4. [DOI] [PubMed] [Google Scholar]
- 14.Kim HJ, Kim IS. Transforming growth factor-beta-induced gene product, as a novel ligand of integrin alphaMbeta2, promotes monocytes adhesion, migration and chemotaxis. The international journal of biochemistry & cell biology. 2008;40:991–1004. doi: 10.1016/j.biocel.2007.11.001. [DOI] [PubMed] [Google Scholar]
- 15.Kim JE, Jeong HW, Nam JO, Lee BH, Choi JY, Park RW, Park JY, Kim IS. Identification of motifs in the fasciclin domains of the transforming growth factor-beta-induced matrix protein betaig-h3 that interact with the alphavbeta5 integrin. The Journal of biological chemistry. 2002;277:46159–65. doi: 10.1074/jbc.M207055200. [DOI] [PubMed] [Google Scholar]
- 16.Kim JE, Kim SJ, Lee BH, Park RW, Kim KS, Kim IS. Identification of motifs for cell adhesion within the repeated domains of transforming growth factor-beta-induced gene, betaig-h3. The Journal of biological chemistry. 2000;275:30907–15. doi: 10.1074/jbc.M002752200. [DOI] [PubMed] [Google Scholar]
- 17.Kim MO, Yun SJ, Kim IS, Sohn S, Lee EH. Transforming growth factor-beta-inducible gene-h3 (beta(ig)-h3) promotes cell adhesion of human astrocytoma cells in vitro: implication of alpha6beta4 integrin. Neuroscience letters. 2003;336:93–6. doi: 10.1016/s0304-3940(02)01260-0. [DOI] [PubMed] [Google Scholar]
- 18.Nam JO, Kim JE, Jeong HW, Lee SJ, Lee BH, Choi JY, Park RW, Park JY, Kim IS. Identification of the alphavbeta3 integrin-interacting motif of betaig-h3 and its anti-angiogenic effect. The Journal of biological chemistry. 2003;278:25902–9. doi: 10.1074/jbc.M300358200. [DOI] [PubMed] [Google Scholar]
- 19.Ohno S, Noshiro M, Makihira S, Kawamoto T, Shen M, Yan W, Kawashima-Ohya Y, Fujimoto K, Tanne K, Kato Y. RGD-CAP ((beta)ig-h3) enhances the spreading of chondrocytes and fibroblasts via integrin alpha(1)beta(1) Biochimica et biophysica acta. 1999;1451:196–205. doi: 10.1016/s0167-4889(99)00093-2. [DOI] [PubMed] [Google Scholar]
- 20.Viloria K, Hill NJ. Embracing the complexity of matricellular proteins: the functional and clinical significance of splice variation. Biomolecular concepts. 2016;7:117–32. doi: 10.1515/bmc-2016-0004. [DOI] [PubMed] [Google Scholar]
- 21.Ahlfeld SK, Wang J, Gao Y, Snider P, Conway SJ. Initial Suppression of Transforming Growth Factor-beta Signaling and Loss of TGFBI Causes Early Alveolar Structural Defects Resulting in Bronchopulmonary Dysplasia. Am J Pathol. 2016;186:777–93. doi: 10.1016/j.ajpath.2015.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lindsley A, Li W, Wang J, Maeda N, Rogers R, Conway SJ. Comparison of the four mouse fasciclin-containing genes expression patterns during valvuloseptal morphogenesis. Gene expression patterns: GEP. 2005;5:593–600. doi: 10.1016/j.modgep.2005.03.005. [DOI] [PubMed] [Google Scholar]
- 23.Conway SJ, Izuhara K, Kudo Y, Litvin J, Markwald R, Ouyang G, Arron JR, Holweg CT, Kudo A. The role of periostin in tissue remodeling across health and disease. Cellular and molecular life sciences: CMLS. 2014;71:1279–88. doi: 10.1007/s00018-013-1494-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Qu Y, Chi W, Hua X, Deng R, Li J, Liu Z, Pflugfelder SC, Li DQ. Unique expression pattern and functional role of periostin in human limbal stem cells. PloS one. 2015;10:e0117139. doi: 10.1371/journal.pone.0117139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schwanekamp JA, Lorts A, Sargent MA, York AJ, Grimes KM, Fischesser DM, Gokey JJ, Whitsett JA, Conway SJ, Molkentin JD. TGFBI functions similar to periostin but is uniquely dispensable during cardiac injury. PloS one. 2017;12:e0181945. doi: 10.1371/journal.pone.0181945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen S, Oldberg A, Chakravarti S, Birk DE. Fibromodulin regulates collagen fibrillogenesis during peripheral corneal development. Developmental dynamics: an official publication of the American Association of Anatomists. 2010;239:844–54. doi: 10.1002/dvdy.22216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu CY, Birk DE, Hassell JR, Kane B, Kao WW. Keratocan-deficient mice display alterations in corneal structure. The Journal of biological chemistry. 2003;278:21672–7. doi: 10.1074/jbc.M301169200. [DOI] [PubMed] [Google Scholar]
- 28.Shao H, Chaerkady R, Chen S, Pinto SM, Sharma R, Delanghe B, Birk DE, Pandey A, Chakravarti S. Proteome profiling of wild type and lumican-deficient mouse corneas. J Proteomics. 2011;74:1895–905. doi: 10.1016/j.jprot.2011.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang G, Chen S, Goldoni S, Calder BW, Simpson HC, Owens RT, McQuillan DJ, Young MF, Iozzo RV, Birk DE. Genetic evidence for the coordinated regulation of collagen fibrillogenesis in the cornea by decorin and biglycan. The Journal of biological chemistry. 2009;284:8888–97. doi: 10.1074/jbc.M806590200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Allaman-Pillet N, Oberson A, Bustamante M, Tasinato A, Hummler E, Schorderet DF. Tgfbi/Bigh3 silencing activates ERK in mouse retina. Experimental eye research. 2015;140:159–70. doi: 10.1016/j.exer.2015.09.004. [DOI] [PubMed] [Google Scholar]
- 31.Garcia-Castellanos R, Nielsen NS, Runager K, Thogersen IB, Lukassen MV, Poulsen ET, Goulas T, Enghild JJ, Gomis-Ruth FX. Structural and Functional Implications of Human Transforming Growth Factor beta-Induced Protein, TGFBIp, in Corneal Dystrophies. Structure. 2017 doi: 10.1016/j.str.2017.09.001. http://dx.doi.org/10.1016/j.str.2017.09.001. [DOI] [PubMed]
- 32.Walker JT, McLeod K, Kim S, Conway SJ, Hamilton DW. Periostin as a multifunctional modulator of the wound healing response. Cell and tissue research. 2016;365:453–65. doi: 10.1007/s00441-016-2426-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bornstein P, Sage EH. Matricellular proteins: extracellular modulators of cell function. Current opinion in cell biology. 2002;14:608–16. doi: 10.1016/s0955-0674(02)00361-7. [DOI] [PubMed] [Google Scholar]
- 34.Sliney DH. How light reaches the eye and its components. International journal of toxicology. 2002;21:501–9. doi: 10.1080/10915810290169927. [DOI] [PubMed] [Google Scholar]
- 35.Reddy VN, Giblin FJ, Lin LR, Chakrapani B. The effect of aqueous humor ascorbate on ultraviolet-B-induced DNA damage in lens epithelium. Investigative ophthalmology & visual science. 1998;39:344–50. [PubMed] [Google Scholar]
- 36.Meek KM, Knupp C. Corneal structure and transparency. Progress in Retinal and Eye Research. 2015;49:1–16. doi: 10.1016/j.preteyeres.2015.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Massoudi D, Malecaze F, Galiacy SD. Collagens and proteoglycans of the cornea: importance in transparency and visual disorders. Cell and tissue research. 2016;363:337–49. doi: 10.1007/s00441-015-2233-5. [DOI] [PubMed] [Google Scholar]
- 38.Brachvogel B, Zaucke F, Dave K, Norris EL, Stermann J, Dayakli M, Koch M, Gorman JJ, Bateman JF, Wilson R. Comparative proteomic analysis of normal and collagen IX null mouse cartilage reveals altered extracellular matrix composition and novel components of the collagen IX interactome. The Journal of biological chemistry. 2013;288:13481–92. doi: 10.1074/jbc.M112.444810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Karring H, Runager K, Valnickova Z, Thogersen IB, Moller-Pedersen T, Klintworth GK, Enghild JJ. Differential expression and processing of transforming growth factor beta induced protein (TGFBIp) in the normal human cornea during postnatal development and aging. Experimental eye research. 2010;90:57–62. doi: 10.1016/j.exer.2009.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Karring H, Poulsen ET, Runager K, Thogersen IB, Klintworth GK, Hojrup P, Enghild JJ. Serine protease HtrA1 accumulates in corneal transforming growth factor beta induced protein (TGFBIp) amyloid deposits. Molecular vision. 2013;19:861–76. [PMC free article] [PubMed] [Google Scholar]
- 41.Poulsen ET, Runager K, Risor MW, Dyrlund TF, Scavenius C, Karring H, Praetorius J, Vorum H, Otzen DE, Klintworth GK, Enghild JJ. Comparison of two phenotypically distinct lattice corneal dystrophies caused by mutations in the transforming growth factor beta induced (TGFBI) gene. Proteomics Clinical applications. 2014;8:168–77. doi: 10.1002/prca.201300058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dassah M, Almeida D, Hahn R, Bonaldo P, Worgall S, Hajjar KA. Annexin A2 mediates secretion of collagen VI, pulmonary elasticity and apoptosis of bronchial epithelial cells. Journal of cell science. 2014;127:828–44. doi: 10.1242/jcs.137802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Johnson JM, Young TL, Rada JA. Small leucine rich repeat proteoglycans (SLRPs) in the human sclera: identification of abundant levels of PRELP. Molecular vision. 2006;12:1057–66. [PubMed] [Google Scholar]
- 44.Yanagisawa H, Schluterman MK, Brekken RA. Fibulin-5, an integrin-binding matricellular protein: its function in development and disease. Journal of Cell Communication and Signaling. 2009;3:337–347. doi: 10.1007/s12079-009-0065-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Poulsen ET, Dyrlund TF, Runager K, Scavenius C, Krogager TP, Hojrup P, Thogersen IB, Sanggaard KW, Vorum H, Hjortdal J, Enghild JJ. Proteomics of Fuchs’ endothelial corneal dystrophy support that the extracellular matrix of Descemet’s membrane is disordered. Journal of proteome research. 2014;13:4659–67. doi: 10.1021/pr500252r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Runager K, Enghild JJ, Klintworth GK. Focus on molecules: Transforming growth factor beta induced protein (TGFBIp) Experimental eye research. 2008;87:298–9. doi: 10.1016/j.exer.2007.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Courtney DG, Moore JE, Atkinson SD, Maurizi E, Allen EH, Pedrioli DM, McLean WH, Nesbit MA, Moore CB. CRISPR/Cas9 DNA cleavage at SNP-derived PAM enables both in vitro and in vivo KRT12 mutation-specific targeting. Gene therapy. 2016;23:108–12. doi: 10.1038/gt.2015.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lakshminarayanan R, Chaurasia SS, Anandalakshmi V, Chai SM, Murugan E, Vithana EN, Beuerman RW, Mehta JS. Clinical and genetic aspects of the TGFBI-associated corneal dystrophies. The ocular surface. 2014;12:234–51. doi: 10.1016/j.jtos.2013.12.002. [DOI] [PubMed] [Google Scholar]
- 49.Liu Y, Huang H, Sun G, Alwadani S, Semba RD, Lutty GA, Yiu S, Edward DP. Gene Expression Profile of Extracellular Matrix and Adhesion Molecules in the Human Normal Corneal Stroma. Current eye research. 2016:1–8. doi: 10.1080/02713683.2016.1200099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rawe IM, Zhan Q, Burrows R, Bennett K, Cintron C. Beta-ig. Molecular cloning and in situ hybridization in corneal tissues. Investigative ophthalmology & visual science. 1997;38:893–900. [PubMed] [Google Scholar]
- 51.Rios H, Koushik SV, Wang H, Wang J, Zhou HM, Lindsley A, Rogers R, Chen Z, Maeda M, Kruzynska-Frejtag A, Feng JQ, Conway SJ. periostin null mice exhibit dwarfism, incisor enamel defects, and an early-onset periodontal disease-like phenotype. Molecular and cellular biology. 2005;25:11131–44. doi: 10.1128/MCB.25.24.11131-11144.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Snider P, Simmons O, Wang J, Hoang CQ, Conway SJ. Ectopic Noggin in a Population of Nfatc1 Lineage Endocardial Progenitors Induces Embryonic Lethality. Journal of cardiovascular development and disease. 2014;1:214–236. doi: 10.3390/jcdd1030214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kruzynska-Frejtag A, Wang J, Maeda M, Rogers R, Krug E, Hoffman S, Markwald RR, Conway SJ. Periostin is expressed within the developing teeth at the sites of epithelial-mesenchymal interaction. Developmental dynamics: an official publication of the American Association of Anatomists. 2004;229:857–68. doi: 10.1002/dvdy.10453. [DOI] [PubMed] [Google Scholar]
- 54.Bury AF. Analysis of protein and peptide mixtures. Journal of Chromatography A. 1981;213:491–500. [Google Scholar]
- 55.Dyrlund TF, Poulsen ET, Scavenius C, Sanggaard KW, Enghild JJ. MS Data Miner: a web-based software tool to analyze, compare, and share mass spectrometry protein identifications. Proteomics. 2012;12:2792–6. doi: 10.1002/pmic.201200109. [DOI] [PubMed] [Google Scholar]
- 56.MacLean B, Tomazela DM, Shulman N, Chambers M, Finney GL, Frewen B, Kern R, Tabb DL, Liebler DC, MacCoss MJ. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics (Oxford England) 2010;26:966–8. doi: 10.1093/bioinformatics/btq054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Reynolds ES. The use of lead citrate at high pH as an electron-opaque stain in electron microscopy. The Journal of cell biology. 1963;17:208–12. doi: 10.1083/jcb.17.1.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vizcaino JA, Csordas A, del-Toro N, Dianes JA, Griss J, Lavidas I, Mayer G, Perez-Riverol Y, Reisinger F, Ternent T, Xu QW, Wang R, Hermjakob H. 2016 update of the PRIDE database and its related tools. Nucleic acids research. 2016;44:D447–56. doi: 10.1093/nar/gkv1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1: Proteins quantified in all biological replicates of both TGFBIp wildtype and deficient mice corneas.
Table S2: Relative comparison of human and mouse corneal TGFBIp.


