Abstract
Background
Panel-based genetic testing has identified increasing numbers of patients with pancreatic ductal adenocarcinoma (PDAC) who carry germline mutations. However, small sample sizes or number of genes evaluated limit prevalence estimates of these mutations. We estimated prevalence of mutations in PDAC patients with positive family history.
Methods
We sequenced 25 cancer susceptibility genes in lymphocyte DNA from 302 PDAC patients in the Mayo Clinic Biospecimen Resource for Pancreatic Research Registry. Kindreds containing at least two first-degree relatives with PDAC met criteria for Familial Pancreatic Cancer (FPC), while the remaining were familial, but not FPC.
Results
Thirty-six patients (12%) carried at least one deleterious mutation in one of 11 genes. Of FPC patients, 25/185 (14%) were carriers, while 11/117 (9%) non-FPC patients with family history were carriers. Deleterious mutations (n) identified in PDAC patients were BRCA2 (11), ATM (8), CDKN2A (4), CHEK2 (4), MUTYH/MYH (3 heterozygotes, not biallelic), BRCA1 (2), and 1 each in BARD1, MSH2, NBN, PALB2, and PMS2. Novel mutations were found in ATM, BARD1, and PMS2.
Conclusions
Multiple susceptibility gene testing in PDAC patients with family history of pancreatic cancer is warranted regardless of FPC status, and will inform genetic risk counseling for families.
Keywords: Familial pancreatic cancer (FPC), pancreatic ductal adenocarcinoma (PDAC), pathogenic variant (PV), variants of uncertain significance (VUS)
INTRODUCTION
Pancreatic cancer is a devastating disease and the fourth leading cause of cancer-related death in the United States. The absolute number of new cases and deaths due to pancreatic cancer has increased steadily since 20041 and Rahib et al. predict that pancreatic cancer will be the second most common cause of cancer mortality by 2030.2 Approximately 95% of pancreatic neoplasms are ductal adenocarcinomas (PDAC), which are the most challenging to treat. The rapid progression of pancreatic cancer and its high rate of mortality underlie the importance of improved risk stratification and early detection.
Numerous studies have demonstrated that family history of pancreatic cancer in a first degree relative is associated a 2- to 3-fold increased risk of pancreatic cancer.3 Previous epidemiologic studies and case reports have demonstrated pancreatic cancer clustering in families. “Familial pancreatic cancer” (FPC) has been defined as kindreds with at least a pair of first degree relatives affected with pancreatic adenocarcinoma4 and accounts for approximately 5 to 10 percent of patients with pancreatic adenocarcinoma.5 This standardized definition is now widely used and has facilitated research addressing a variety of questions, including genetic susceptibility.
Increased risk of pancreatic cancer is associated with several inherited syndromes for which the predisposing genes have been identified. The most prominent syndromes are hereditary breast-ovarian cancer syndrome, particularly due to germline mutations in BRCA2, and familial atypical mole and melanoma syndrome, due to mutations in CDKN2A. In the most comprehensive analysis to characterize the genetic variation in FPC patients to date, the Pancreatic Cancer Genetic Epidemiology (PACGENE) Consortium found that mutations in the HBOC genes (BRCA1, BRCA2) and CDKN2A were identified in 7.4% of FPC probands.6 Pancreatic cancer risk is also increased in Lynch syndrome (MLH1, MSH2, PMS2, and MSH6)7–9 and hereditary pancreatitis.10 In addition, germline mutations in PALB26,11,12 and ATM7,12,13 have been identified among FPC probands using next-generation sequencing, extending the catalog of predisposing genes. Functional roles for identified germline mutations in these two genes were further supported by loss of heterozygosity of the wild-type allele in the pancreatic tumor of the mutation carriers.11,12
Pancreatic cancer thus appears to be a genetically heterogeneous disease which is associated with a number of syndromes. Although the American College of Gastroenterology (ACG) provides some guidance about what genes may be appropriate for testing, these guidelines are limited to patients who meet criteria for FPC.14 Importantly, germline mutations in genes with known pancreatic cancer risk are associated with increased risk of other cancers. Hence, panel testing of multiple cancer-risk genes is likely to increase etiologic information and potentially enhance genetic risk assessment of pancreatic cancer patients who report a positive family history not limited to FPC.
Previously, kindreds treated at the Mayo Clinic that contained at least two affected biological relatives with pancreatic cancer were assessed for mutations in four PDAC-risk genes.6 In light of the genetic heterogeneity of pancreatic cancer, here we more comprehensively assessed the prevalence of genetic variants in this cohort. This was done by analyzing 25 cancer susceptibility genes in one affected member per kindred. This included individuals with a personal and family history of pancreatic cancer not limited to FPC. Genes with known pancreatic cancer risk were included, including the HBOC genes, Lynch syndrome genes, and familial melanoma genes. Additional high and moderate penetrance genes associated with increased risk of other cancers, including breast, ovarian, and colorectal, were also included.
PATIENTS AND METHODS
Patients
Institutional review board approval was granted at Mayo Clinic, and written consent was obtained from all patients in order to be included in the study. Affected patients were recruited into the Biospecimen Resource for Pancreatic Research registry between the years 2000 and 2013 at the time they sought clinical care at Mayo Clinic. Demographic, clinical, and pathological data are maintained in the registry. A diagnosis of PDAC in affected patients was established via biopsy or clinical documentation. Individuals with hereditary pancreatitis were excluded. Family history information was self-reported in questionnaires, including reports of cancers in family members. Kindreds with at least one pair of first degree relatives who were affected with PDAC were considered FPC; kindreds with at least two affected blood relatives that did not meet the FPC definition were considered “familial non-FPC”.
Unrelated affected patients in FPC or familial non-FPC kindreds who provided a blood sample were eligible for the study. Although the vast majority of patients were included in our prior report of 4 genes (300/302, 99.3%)6, 21 additional genes were tested in this study. This included 16 genes known to be associated with PDAC (APC, ATM, BMPR1A, BRCA1, BRCA2, CDK4, CDKN2A, EPCAM, MLH1, MSH2, MSH6, PALB2, PMS2, SMAD4, STK11, and TP53) and 9 genes with no known association with PDAC (BARD1, BRIP1, CDH1, CHEK2, MUTYH/MYH, NBN, PTEN, RAD51C, and RAD51D). All genes included in the ACG guidelines for pancreatic cancer genetic testing are included in this panel.14 Each patient’s frozen buffy coat sample was de-identified and labeled with a unique identifier, and all samples were sent to Myriad Genetic Laboratories, Inc. for DNA extraction and mutation testing.
Next Generation Sequencing (NGS) Assay
Sample preparation for next generation sequencing (NGS) was performed from DNA extracted from buffy coats using the RainDance microdroplet polymerase chain reaction (PCR) system (RainDance Technologies, Billerica, Massachusetts).15 Briefly, PCR products representing exons and proximal splicing elements of patient DNA were amplified in merged droplets consisting of fragmented patient DNA and select target enrichment primers. These PCR products were subsequently tagged with barcodes and sequencing adaptors for NGS on the Illumina HiSeq platform. To circumvent highly homologous pseudogenes, modified sample preparation with long-range and nested PCR followed by NGS on the Illumina MiSeq platform was used for portions of the CHEK2 and PMS2 genes. All clinically actionable variants identified by NGS, and regions that did not meet our pre-set NGS quality metrics were independently confirmed with orthogonal site-specific Sanger sequencing.
To detect exonic deletions and duplications, NGS dosage, microarray comparative genomic hybridization (CGH), multiplex ligation-dependent probe amplification (MLPA) or a combination of these analyses was performed, with all positive results confirmed by an orthogonal method.15
Variant Classification
Variants were classified using American College of Medical Genetics and Genomics recommendations, with supporting linkage, biochemical, clinical, functional and statistical data used for specific missense and intronic alterations.16,17 Pathogenic variants (PVs) are those that received a laboratory classification of deleterious or suspected deleterious. Variants for which the pathogenicity could not be determined were categorized as variants of uncertain significance (VUS). Novel, previously unreported mutations discovered in this study were defined as those not previously identified by the testing laboratory (Myriad Genetic Laboratories, Inc.).
Data Analysis
Analytic methods used in this study have been previously described.6 Briefly, prevalence of PV and VUS of the 25 genes studied were compared between individuals of FPC and familial non-FPC kindreds using odds ratios (OR) and 95% confidence intervals (CI). Descriptive statistics and mutation rates were calculated. Comparisons of the mutation prevalence between groups were measured using either chi-square or Fisher’s exact tests, depending on sample sizes. Differences in family history of other cancers by mutation carrier status were quantified using chi-square or Fisher’s exact tests. Statistical analyses were completed using SAS 9.4 (SAS Institute, Cary, NC).
RESULTS
Patient characteristics and variant results of the 302 study participants are presented in Table 1; 185 (61.3%) were FPC, and 117 (38.7%) were familial non-FPC. Median age of diagnosis was 65 years (range 37–93 years), and 53.6% were males. The tested patients were almost entirely White/Caucasian (97.7%), and 8 (2.7%) were of Ashkenazi Jewish descent. The distribution of pancreatic cancer stage was similar to what would be expected in an unselected pancreatic cancer patient population, with roughly equal proportions of resectable, locally advanced, and metastatic cancer. There were no substantial differences between the two familial groups. Most kindreds (79.2%) contained 2 affected members with pancreatic cancer, 14.6% reported 3 affected individuals, and 6.2% contained 4 or more affected individuals. Patients in the FPC group reported a higher number of PDAC per kindred relative to the familial non-FPC group.
Table 1.
Clinical and demographic characteristics, total and by FPC status
| Total (n=302) | FPC (n=185) | Familial non-FPC (n=117) | ||||
|---|---|---|---|---|---|---|
| Median age at PDAC (range) | 65 (37–93) | 66 (37–93) | 64 (38–87) | |||
|
| ||||||
| N | % | N | % | N | % | |
|
| ||||||
| Sex, male | 162 | 53.5 | 98 | 53.0 | 64 | 54.7 |
| Race | ||||||
| White | 293 | 97.7 | 180 | 98.4 | 113 | 96.6 |
| African American | 4 | 1.3 | 3 | 1.6 | 1 | 0.9 |
| American Indian/Alaska Native | 1 | 0.3 | 0 | 0 | 1 | 0.9 |
| Native Hawaiian | 1 | 0.3 | 0 | 0 | 1 | 0.9 |
| Multiracial | 1 | 0.3 | 0 | 0 | 1 | 0.9 |
| Missing | 2 | - | 2 | - | 0 | - |
| Ethnicity | ||||||
| Non-Hispanic/non-Latino | 291 | 99.7 | 176 | 100 | 115 | 99.1 |
| Hispanic/Latino | 1 | 0.3 | 0 | 0 | 1 | 0.9 |
| Missing | 10 | - | 9 | - | 1 | - |
| Ashkenazi Jewish heritage | ||||||
| No | 288 | 97.3 | 173 | 95.6 | 115 | 100 |
| Yes | 8 | 2.7 | 8 | 4.4 | 0 | 0 |
| Missing | 6 | - | 4 | - | 2 | - |
| Number PDAC per kindred | ||||||
| 2 | 239 | 79.2 | 128 | 69.2 | 111 | 94.9 |
| 3 | 44 | 14.6 | 38 | 20.5 | 6 | 5.1 |
| 4+ | 19 | 6.2 | 19 | 10.2 | 0 | 0 |
| PDAC stage | ||||||
| Resectable | 88 | 31.7 | 48 | 29.1 | 40 | 35.4 |
| Locally advanced | 77 | 27.7 | 46 | 27.9 | 31 | 27.4 |
| Metastatic | 113 | 40.6 | 71 | 43.0 | 42 | 37.2 |
| Missing | 24 | 20 | - | 4 | - | |
Table 2 summarizes germline PV prevalence among the 302 tested patients. The aggregate prevalence was 36/302 (11.9%) for all cases with any positive PDAC family history. When results were stratified by familial group, the probability that any mutation was identified in an FPC patient was 13.5%, compared to 9.4% among familial non-FPC patients (OR=1.5; 95% CI=0.7–3.2; p=0.28).
Table 2.
Germline pathogenic variants (PV) prevalence by type of familial patient (patients meeting Familial Pancreatic Cancer [FPC] criteria vs familial Non-FPC), categorized by genes associated and not associated with PDAC. Genes with no PV included APC, BMPR1A, BRIP1, CDH1, CDK4, EPCAM, MLH1, MSH6, PTEN, RAD51C, RAD51D, SMAD4, STK11, and TP53.
| Total (n=302) | FPC (n=185) | Non-FPC (n=117) | p-value** | ||||
|---|---|---|---|---|---|---|---|
| N | % | N | % | N | % | ||
| All Genes | 36 | 11.9 | 25* | 13.5 | 11 | 9.4 | 0.29 |
|
| |||||||
| Genes Associated with PDAC | |||||||
|
| |||||||
| ATM | 8 | 2.6 | 6 | 3.2 | 2 | 1.7 | |
| BRCA1 | 2 | 0.7 | 2 | 1.1 | 0 | 0 | |
| BRCA2 | 11 | 3.6 | 8 | 4.3 | 3 | 2.6 | |
| CDKN2A (p16) | 4 | 1.3 | 4 | 2.2 | 0 | 0 | |
| MSH2 | 1 | 0.3 | 1 | 0.5 | 0 | 0 | |
| PALB2 | 1 | 0.3 | 1 | 0.5 | 0 | 0 | |
| PMS2 | 1 | 0.3 | 1 | 0.5 | 0 | 0 | |
| Subtotal* | 27 | 8.9 | 22* | 11.9 | 5 | 4.3 | 0.02 |
|
| |||||||
| Genes Not Associated with PDAC | |||||||
|
| |||||||
| BARD1 | 1 | 0.3 | 1 | 0.5 | 0 | 0 | |
| CHEK2 | 4 | 1.3 | 1 | 0.5 | 3 | 2.6 | |
| MUTYH/MYH | 3 | 1.0 | 0 | 0 | 3 | 2.6 | |
| NBN | 1 | 0.3 | 1 | 0.5 | 0 | 0 | |
| Subtotal | 9 | 3.0 | 3 | 1.6 | 6 | 5.1 | 0.09 |
One FPC case carried two PV: one in BRCA1 and one in BRCA2.
P-value compares prevalence of mutations in FPC versus familial non-FPC probands.
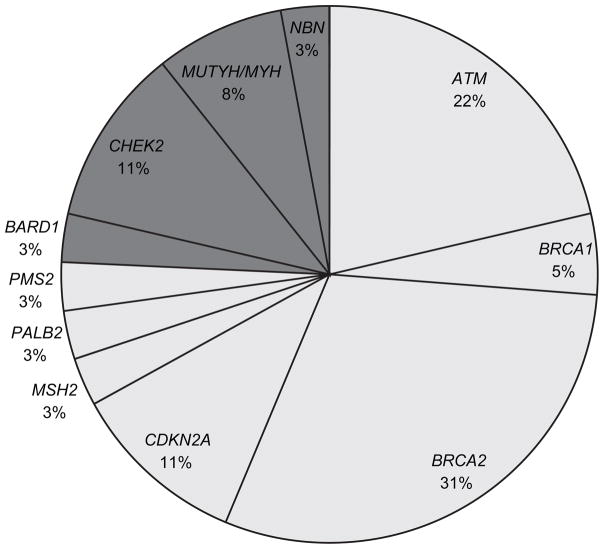
The distribution of PVs is presented in Figure 1. PVs were identified in 11 genes, including 7 PDAC-associated genes (ATM, BRCA1, BRCA2, CDKN2A, MSH2, PALB2, and PMS2) and 4 genes with no known PDAC association (BARD1, CHEK2, MUTYH/MUY, NBN). Overall, 75% of PVs detected were in genes known to be associated with PDAC. There was a statistically significant higher proportion of PVs in PDAC-associated genes in the FPC group (11.9%) compared to the non-FPC group (4.3%) (OR=3.0; 95% CI=1.1–8.2; p=0.02). There were no statistically significant differences in the PV positive-rate for genes with no known PDAC association between the FPC and non-FPC groups (1.6% vs 5.1%; p=0.09), although the number of PVs was small.
Figure 1.
Distribution of pathogenic variants in 302 patients with pancreatic ductal adenocarcinoma (PDAC) with positive family history. Genes associated with PDAC are colored in dark gray; genes not known to be associated with PDAC are colored in light gray.
Twenty-seven individuals were found to carry a PV in a gene associated with increased risk of breast cancer (ATM, BARD1, BRCA1, BRCA2, CHEK2, PALB2, NBN). A larger proportion breast cancer PV carriers had a first degree relative with breast cancer (13/27, 48.1%) versus non-carrier patients (75/275 (27.3%); p=0.02). When limited to only female patients, the proportions were 9/12 (75%) for breast cancer-PV carriers and 38/128 (29.7%) for non-carriers (p=0.003).
Table 3 lists patient characteristics of the 36 patients who tested positive for at least one germline PV. All but one patient who tested positive for a PV carried a mutation in only one tested gene. The single exception was an FPC patient of Ashkenazi Jewish heritage who carried mutations in both BRCA1 (187delAG) and BRCA2 (6174delT); this double mutation carrier has been previously reported.6
Table 3.
Characteristics of pancreatic ductal adenocarcinoma (PDAC) patients with germline pathogenic variants (PV), categorized by genes associated and not associated with PDAC and by family status. Note: Pt 6373 has two PV.
| Pt | Sex | Age PC | Race/ethnicity | Personal history of other cancer (age) | Family status | # PDAC in family* | Family history of other cancers in first degree relative (n if more than 1) | Pathogenic variant | |
|---|---|---|---|---|---|---|---|---|---|
| Genes Associated with PDAC | |||||||||
|
| |||||||||
| 75 | M | 79 | Non-Hispanic white | Prostate (69) | FPC | 2 | Breast | ATM c.1978del (p.Met660Trpfs*4) | |
| 3124 | F | 50 | Non-Hispanic white | None | FPC | 2 | None | ATM c.741dup (p.Arg248Serfs*6) | |
| 3489 | F | 59 | Non-Hispanic white | None | FPC | 3 | Breast, esophageal | ATM c.7630-2A>C | |
| 6828 | M | 52 | Non-Hispanic white | None | FPC | 2 | Colorectal, head/neck | ATM c.1564_1565del (p.Glu522Ilefs*43) | |
| 7832 | M | 61 | Non-Hispanic white | None | FPC | 2 | None | ATM c.8264dup (p.Tyr2755*) | |
| 8089 | M | 58 | Non-Hispanic white | None | FPC | 2 | Brain, lung, myeloma | ATM c.3245_3247delinsTGAT (p.His1082Leufs*14) | |
| 485 | M | 60 | Non-Hispanic white | Bladder (51) | Non-FPC | 2 | Breast, prostate, sarcoma | ATM c.3802del (p.Val1268*) | |
| 4155 | M | 59 | Non-Hispanic white | None | Non-FPC | 2 | None | ATM c.5932G>T (p.Glu1978*) | |
| 6133 | M | 77 | Non-Hispanic white | Prostate (63) | FPC | 2 | Gynecologic (non-ovarian) | BRCA1 c.4382_4388dup (p.Tyr1463*) 4507ins7 | |
| 6373 | M | 60 | Non-Hispanic white, Ashkenazi Jewish | None | FPC | 2 | Breast |
BRCA1 c.68_69del (p.Glu23Valfs*17) 187delAG; BRCA2 c.5946del (p.Ser1982Argfs*22) 6174delT |
|
| 746 | F | 56 | Non-Hispanic white | None | FPC | 2 | Breast, sarcoma | BRCA2 c.961C>T (p.Gln321*) Q321X (1189C>T) | |
| 3268 | M | 93 | Non-Hispanic white, Ashkenazi Jewish | None | FPC | 3 | Breast (3), lung, prostate | BRCA2 c.5946del (p.Ser1982Argfs*22) 6174delT | |
| 4983 | F | 49 | Non-Hispanic white | None | FPC | 3 | Breast (2), leukemia | BRCA2 c.3744_3747del (p.Ser1248Argfs*10) 3972del4 | |
| 6245 | F | 73 | Non-Hispanic black | None | FPC | 2 | Head/neck | BRCA2 c.3407_3408insAlu | |
| 6533 | M | 49 | Non-Hispanic white | None | FPC | 2 | Lung, ovarian | BRCA2 c.9867del (p.Phe3289Leufs*24) 10095delT | |
| 6588 | M | 81 | Non-Hispanic white, Ashkenazi Jewish | Prostate (71) | FPC | 2 | None | BRCA2 c.5946del (p.Ser1982Argfs*22) 6174delT | |
| 8607 | F | 51 | Non-Hispanic white | None | FPC | 3 | Gynecologic (non-ovarian), prostate | BRCA2 c.6373dupA (p.Thr2125Asnfs*4) 6601insA | |
| 1 | F | 42 | Non-Hispanic white | None | Non-FPC | 2 | Prostate | BRCA2 c.5722_5723del (p.Leu1908Argfs*2) 5950delCT | |
| 1116 | M | 59 | Non-Hispanic white | None | Non-FPC | 2 | Gynecologic (non-ovarian) | BRCA2 c.4965C>G (p.Tyr1655*) Y1655X (5193C>G) | |
| 1279 | F | 64 | Non-Hispanic white | Breast (47) | Non-FPC | 2 | Brain, breast (2), sarcoma | BRCA2 c.1813dupA (p.Ile605Asnfs*11) 2041insA | |
| 883 | M | 64 | Non-Hispanic white | None | FPC | 2 | Bladder | CDKN2A/p16 c.286del (p.Val96Cysfs*50) 286delG | |
| 1359 | M | 56 | Non-Hispanic white | None | FPC | 5 | Gynecologic (non-ovarian) (3) | CDKN2A/p16 c.457G>T (p.Asp153Tyr) D153Y (457G>T) | |
| 3986 | F | 61 | Non-Hispanic white | None | FPC | 2 | None | CDKN2A/p16 c.-34G>T 5′UTR-34G>T | |
| 6888 | M | 76 | Non-Hispanic white | Melanoma (39) | FPC | 2 | Colorectal, lung | CDKN2A/p16 c.301G>T (p.Gly101Trp) G101W (301G>T) | |
| 703 | F | 54 | Non-Hispanic white | None | FPC | 3 | Colorectal, leukemia, prostate | MSH2 c.1046C>T (p.Pro349Leu) P349L (1046C>T) | |
| 6547 | F | 45 | Non-Hispanic white | None | FPC | 2 | Breast, prostate | PALB2 del exon 13 | |
| 571 | F | 85 | Non-Hispanic white | None | FPC | 2 | Colorectal | PMS2 c.2175-1G>A IVS12-1G>A | |
|
| |||||||||
| Genes Not Associated with PDAC | |||||||||
|
| |||||||||
| 4819 | M | 67 | Non-Hispanic white | None | FPC | 5 | None | BARD1 c.632T>A (p.Leu211*) | |
| 5994 | M | 55 | Non-Hispanic white | None | FPC | 2 | Breast, ovarian | CHEK2 c.1100del (p.Thr367Metfs*15) | |
| 590 | M | 72 | Non-Hispanic white | None | Non-FPC | 2 | None | CHEK2 c.1100del (p.Thr367Metfs*15) | |
| 6213 | F | 68 | Non-Hispanic white | None | Non-FPC | 3 | Ovarian, prostate | CHEK2 c.1100del (p.Thr367Metfs*15) | |
| 8142 | F | 59 | Non-Hispanic white | None | Non-FPC | 3 | Bladder, breast, prostate | CHEK2 c.1100del (p.Thr367Metfs*15) | |
| 4910 | M | 86 | Non-Hispanic white | Prostate (68) | Non-FPC | 2 | Breast, prostate | MUTYH/MYH c.1145G>A (p.Gly382Asp) G382D (1145G>A) | |
| 5297 | M | 51 | Non-Hispanic white | None | Non-FPC | 2 | None | MUTYH/MYH c.494A>G (p.Tyr165Cys) Y165C (494A>G) | |
| 5700 | F | 63 | Non-Hispanic white | Breast (58) | Non-FPC | 2 | Head/neck, stomach | MUTYH/MYH c.1145G>A (p.Gly382Asp) G382D (1145G>A) | |
| 3312 | F | 63 | Non-Hispanic white | None | FPC | 2 | None | NBN c.657_661del (p.Lys219Asnfs*16) | |
# PDAC in family includes tested individual and is not limited to first degree relative
Supplemental Table 1 summarizes the prevalence of VUSs by patient’s family history status. The aggregate prevalence is 90/302 (29.8%) for all cases with any positive family history. The probability that a patient carried a VUS in any gene was 31.4% and 27.4% for FPC versus familial non-FPC, respectively; these rates were not statistically significantly different. There were no statistically significant differences in the PDAC-associated or non-PDAC-associated VUS rates by family status.
Supplemental Table 2 summarizes the PVs and VUSs in the 25 genes for this sample; novel variants are highlighted. The 5 novel PVs detected were ATM c.741dup (p.Arg248Serfs*6), ATM c.1978del (p.Met660Trpfs*4), ATM c.8264dup (p.Tyr2755*), BARD1 c.632T>A (p.Leu211*), and PMS2 c.2175-1G>A, IVS12-1G>A. The 7 novel VUSs detected were APC c.4462T>G (p.Leu1488Val), L1488V (4462T>G), CDH1 c.258A>C (p.Lys86Asn), CDKN2A c.329del (p.Gly110Valfs*62), NBN c.1490C>T (p.Thr497Ile), PMS2 c.2453T>C (p.Ile818Thr), I818T (2453T>C), RAD51C c.1103G>A (p.Arg368Gln), and RAD51D c.287G>T (p.Gly96Val).
DISCUSSION
In this study, we tested newly extracted lymphocyte DNA from 302 pancreatic cancer patients who reported a positive family history using a genetic testing panel of 25 cancer predisposition genes. We found 11.9% of patients carried a PV in at least one of 11 of genes included on the panel. The majority of PVs were in genes previously known to be associated with pancreatic cancer (ATM, BRCA1, BRCA2, CDKN2A, MSH2, PALB2, and PMS2). We observed mutations in genes not currently included in the catalog of genes associated with pancreatic cancer susceptibility (BARD1, CHEK2, MUTYH/MYH, and NBN). When analyzed by FPC status, we found only a slightly higher overall prevalence of PVs among FPC (13.5%) patients versus non-FPC patients (9.4%). However, when analyzing PDAC-associated variants, the prevalence of PV among FPC patients was higher than among non-FPC patients (11.9% vs 4.3%), which is consistent with our previous finding when only 4 PDAC-associated genes were analyzed.6
When results of germline genetic mutations in pancreatic cancer across multiple studies are considered, there is the expected observation that more mutations are detected when the number of tested genes increases. We found that the overall mutation prevalence of 11.9% was higher than the previous report assessing mutations in 4 genes in this same cohort (10.4%).6 Increasing the gene panel from 4 to 25 genes thus increased the PV identification rate by 1.5%. The combined mutation prevalence of BRCA1 and BRCA2 genes in our current study (12/302, 4.0%), is also similar to that reported by Holter et al.18 in an unselected series (14/306, 4.6%). However, it could be argued that the incremental difference in the overall positive rate suggests that the detection rate may be saturated even when more genes are interrogated. For example, the overall positive mutation rate here is slightly lower than that reported by Hu et al. In study of 21 genes in 96 consecutive PDAC cases unselected for family history, Hu et al. found that 13.5% of patients were found to carry a pathogenic mutation.19 It should be noted that two patients included in the Hu et al. study were also included here, including one BRCA2 positive patient. In another study, Grant et al. examined 13 genes in 290 PDAC cases reported a mutation prevalence of only 3.8% (11/290).7 This lower mutation prevalence is likely related to the fact that the cohort assessed by Grant et al. was lower risk, with nearly two-thirds of cases reporting no family history of pancreatic, breast, or ovarian cancer.
On the other hand, we also observed PVs in four genes that are not considered to be PDAC-associated. This observation, combined with discovery of novel PV in ATM, BARD1, and PMS2, and that 80/266 (30.1%) of patients carried VUS but not PV (and will require further investigation to determine whether they are deleterious or not) opens new areas for investigation. Interestingly, 10/36 (28%) PV carriers also carried a VUS as well, though not necessarily in the same gene in which the PV occurred (only one ATM PV carrier also had a VUS in that gene; and only two CDKN2A PV carriers also had a VUS in that gene).
Overall, 27 individuals were found to carry mutations in genes associated with an increased risk of breast cancer, the most common of which was BRCA2. Although there was an increased family incidence of breast cancer among these individuals, only 48% of these individuals reported a history of breast cancer in a first degree relative. However, a previous study by Rosenthal et al. showed that only 25% of women tested with a multi-gene panel who had a mutation in a breast cancer-risk gene had a family history consistent with an increased risk of breast cancer.20 These findings were observed for high penetrance genes, such as BRCA1 and BRCA2, as well as moderate penetrance genes. This suggests that family history may not be apparent in all individuals at increased risk. In addition, the PVs identified in these genes represent clinically actionable findings, with gene-specific management guidelines to minimize risk of breast and/or ovarian cancer for patients and their family members.21
After BRCA2, ATM was the most common breast cancer risk gene in which PVs were identified. Eight individuals were identified as carrying a PV in ATM, six of whom were FPC. While there is growing consensus in the pancreatic cancer genetics literature that germline mutations in ATM can predispose to pancreatic cancer, there are also caveats. ATM is a large gene, and little is known about the genetic epidemiology of ATM with respect to PDAC. ATM localizes to chromosome 11q and was named for its association with ataxia telangiectasia.22 It belongs to the protein family of PI3K-related protein kinases and plays an important role in DNA repair. In 2012, ATM was reported as a predisposition gene for familial pancreatic adenocarcinoma.13 Heterozygous, constitutional ATM mutations were identified in whole-genome and whole-exome sequencing performed on 2 kindreds with familial pancreatic cancer. When the analysis was expanded to consider an additional 166 familial pancreatic cancer patients, an additional 4 patients were found to have deleterious mutations of ATM, compared to none in 190 spouse controls (p=0.046).13 Along with others,7,12 our study has found numerous VUSs in ATM, and this may indicate that these do not represent putative disease-causing variants.
We also identified four individuals with PVs in CHEK2. The CHEK2 gene plays a role in checkpoint response in the setting of DNA damage and mutation of this gene has been associated with elevated breast cancer23 and prostate cancer risk.24 The protein is, however, found in a wide range of tissues and there is some suggestion that mutation may predispose to thyroid and kidney cancer as well.25 In the study by Hu et al. of unselected PDAC patients, one patient with no family history was found to carry a PV in CHEK2.19 Here, all four patients were found to carry CHEK2 c.1100del, the common European founder mutation.26 Given that the majority of our population reported European ancestry, this may be consistent with the expected population frequency of this mutation. However, in combination with previous reports, further study is warranted to investigate the possible association with PDAC.
Five individuals were found to carry PVs in genes associated with hereditary colorectal cancer. This includes 3/302 (0.99%) patients who were found to carry a monoallelic mutation in MUTYH/MYH. The human MutY homologue (MUTYH/MYH) is a DNA glycosylase that is implicated in base excision repair. Mutations of this gene have been shown to be affiliated with familial colorectal adenoma and carcinoma in a syndrome termed MUTYH-associated polyposis (MAP).27 Extra-colonic manifestations of this gene mutation are still being investigated, but early reports suggest it may predispose to endometrial28 and duodenal cancers.29 Although this is the first report of a MUTYH/MYH germline mutation in PDAC, this positive rate may be similar to the population frequency of monoallelic MUTYH/MYH mutations. A previous study by Tung et al. in women with breast cancer tested with a multi-gene panel reported a 1.8% prevalence of monoallelic MUTYH/MYH mutations.30 In comparison, an incidence of 2.1% was reported in a study of patients with colorectal cancer, which is associated with monoallelic MUTYH/MYH mutations.31 Collectively, this indicates that additional studies would be necessary to determine whether MUTYH/MYH is associated with PDAC or if these represent incidental findings. However, these do represent clinically actionable findings, as professional society guidelines recommend increased screening for individuals with no personal or family history of colorectal cancer who carry a monoallelic MUTYH mutation.32
We also identified two individuals with PVs in genes associated with Lynch syndrome. One individual was found to carry a PV in PMS2 here, which is characterized primarily by predisposition to colorectal and endometrial cancer. Another individual was found to carry a PV in MSH2. While mutations of several genes can result in Lynch syndrome, mutation of MSH2 appears to carry higher risk than other genes for development of cancer at any site.33 In addition to colorectal cancer, MSH2 mutation has been linked to endometrial, ovarian, gastric, biliary tract, and urinary tract malignancies.8 Previous studies have also identified mutations in the mismatch repair genes among patients with PDAC. Kastrinos et al. showed that over 20% of families with known mutations in the mismatch repair genes reported at least one case of PDAC.9 In addition, Yang et al. genotyped Lynch Syndrome genes in 66 PDAC patients and identified PV in PMS2 in 2 patients.34 Previous studies have also shown that families with MSH2 mutations do seem to be at increased risk for PDAC.35 Collectively, this supports the association of PDAC with Lynch syndrome and demonstrates the value in including these genes in routine PDAC genetic screening.
This study also identified germline mutations in with limited previous observations, including BARD1 and NBN. BRCA1-associated ring domain (BARD1) is a protein that complexes with BRCA1 and localizes to regions of DNA damage, and is thought to play a role in the DNA checkpoint response. It has been speculated that BARD1 mutations may contribute to breast and/or ovarian cancer susceptibility. 27,28 There has also been some limited observation of PVs in BARD1 among patients with PDAC: Smith et al. observed a PDAC patient with a BARD1 PV36 and Hu et al. found a PV in a PDAC patient with a positive family history.19 Heterozygous PVs in NBN are also associated with increased breast cancer risk, as well as other malignancies.37,38 The study by Hu et al. also reported one patient with no family history of PDAC who was found to carry a PV in NBN.19 In combination with the observations reported here, this suggests that these genes may be associated with an increased pancreatic cancer risk in the setting of FPC.
There were some limitations to this study. The cohort included here was almost entirely European. As such, it is unclear whether the findings presented here are applicable to patients of non-European ancestry. In addition, this study was limited to patients with a personal and family history of PDAC. In the absence of a control group, additional studies would be necessary to investigate causality for PVs identified in genes with no current known association with PDAC.
The foregoing discussion underscores the importance of the need for further research to characterize the genetic risk for PDAC and other cancers in families that may be segregating PV. Importantly, this genetic predisposition expends beyond families who meet the definition of FPC. While our study found that family history of cancer is an important factor in risk assessment and selection for genetic testing, PVs were identified across many genes associated with different cancer genetic syndromes. This highlights the potential advantage of multi-gene panel testing over syndrome-specific sequential testing approaches in PDAC patients. We anticipate that as more PDAC patients are screened by next generation sequencing, the challenges for health care providers will increase, particularly when interpreting results in the absence of family history. Our results underscore the importance of comprehensive family history assessment combined with multi-gene panel testing to enhance risk assessment among pancreatic cancer patients and their families.
Conclusions
With 12% prevalence of deleterious mutations, multiple susceptibility gene testing in PDAC patients with a positive family history of pancreatic cancer is warranted regardless of meeting FPC criteria. These findings will inform genetic risk counseling for family members.
Supplementary Material
Acknowledgments
Research Support: Funding support for this study includes: National Institutes of Health NCI SPORE grant P50 CA102701 and the NCI grant R01CA97075 for the Pancreatic Cancer Genetic Epidemiology (PACGENE) Consortium.
The authors thank the patients and their families, Krystal Brown PhD, Lynne Pauley, and study assistants Ryan Wuertz, Bridget Eversman, Sarah Amundson, and Megan Reichmann for their contributions to the study. Funding support for this study includes National Cancer Institute (NCI) grant R01 CA97075 and National Institutes of Health NCI SPORE grant P50 CA102701. This work was presented at the American Society of Clinical Oncology (ASCO) annual meeting, Chicago, IL, June 4, 2016.
Footnotes
Conflict of Interest:
Kari G. Chaffee: None
Ann L. Oberg: None
Robert R. McWilliams: None
Neil Majithia: None
Brian A. Allen: Employed by Myriad Genetics Laboratories, Inc. and received salaries and stock options as compensation at the time the research project was performed.
John Kidd: Employed by Myriad Genetics Laboratories, Inc. and received salaries and stock options as compensation at the time the research project was performed.
Nanda Singh: Employed by Myriad Genetics Laboratories, Inc. and received salaries and stock options as compensation at the time the research project was performed.
Anne-Renee Hartman: Employed by Myriad Genetics Laboratories, Inc. and received salaries and stock options as compensation at the time the research project was performed.
Richard J. Wenstrup: Employed by Myriad Genetics Laboratories, Inc. and received salaries and stock options as compensation at the time the research project was performed.
Gloria M. Petersen: None
References
- 1.Society AC. Cancer Facts and Figures 2013. Atlanta: 2013. [Google Scholar]
- 2.Rahib L, Smith BD, Aizenberg R, Rosenzweig AB, Fleshman JM, Matrisian LM. Projecting cancer incidence and deaths to 2030: the unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer research. 2014 Jun 1;74(11):2913–2921. doi: 10.1158/0008-5472.CAN-14-0155. [DOI] [PubMed] [Google Scholar]
- 3.Petersen GM. Familial Pancreatic Adenocarcinoma. Hematol Oncol Clin North Am. 2015;29(4):641–653. doi: 10.1016/j.hoc.2015.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hruban RH, Petersen GM, Ha PK, Kern SE. Genetics of pancreatic cancer. From genes to families. Surgical oncology clinics of North America. 1998 Jan;7(1):1–23. [PubMed] [Google Scholar]
- 5.Petersen GM, de Andrade M, Goggins M, et al. Pancreatic cancer genetic epidemiology consortium. Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2006 Apr;15(4):704–710. doi: 10.1158/1055-9965.EPI-05-0734. [DOI] [PubMed] [Google Scholar]
- 6.Zhen DB, Rabe KG, Gallinger S, et al. BRCA1, BRCA2, PALB2, and CDKN2A mutations in familial pancreatic cancer: a PACGENE study. Genet Med. 2015 Jul;17(7):569–577. doi: 10.1038/gim.2014.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grant RC, Selander I, Connor AA, et al. Prevalence of germline mutations in cancer predisposition genes in patients with pancreatic cancer. Gastroenterology. 2015 Mar;148(3):556–564. doi: 10.1053/j.gastro.2014.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lin KM, Shashidharan M, Thorson AG, et al. Cumulative incidence of colorectal and extracolonic cancers in MLH1 and MSH2 mutation carriers of hereditary nonpolyposis colorectal cancer. J Gastrointest Surg. 1998;2(1):67–71. doi: 10.1016/s1091-255x(98)80105-4. [DOI] [PubMed] [Google Scholar]
- 9.Kastrinos F, Mukherjee B, Tayob N, et al. Risk of pancreatic cancer in families with Lynch syndrome. JAMA. 2009 Oct 28;302(16):1790–1795. doi: 10.1001/jama.2009.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Klein AP. Genetic susceptibility to pancreatic cancer. Molecular carcinogenesis. 2012 Jan;51(1):14–24. doi: 10.1002/mc.20855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jones S, Hruban RH, Kamiyama M, et al. Exomic sequencing identifies PALB2 as a pancreatic cancer susceptibility gene. Science. 2009 Apr 10;324(5924):217. doi: 10.1126/science.1171202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roberts NJ, Norris AL, Petersen GM, et al. Whole Genome Sequencing Defines the Genetic Heterogeneity of Familial Pancreatic Cancer. Cancer discovery. 2016;6(2):166–175. doi: 10.1158/2159-8290.CD-15-0402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roberts NJ, Jiao Y, Yu J, et al. ATM mutations in patients with hereditary pancreatic cancer. Cancer discovery. 2012;2(1):41–46. doi: 10.1158/2159-8290.CD-11-0194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Syngal S, Brand RE, Church JM, Giardiello FM, Hampel HL, Burt RW. ACG clinical guideline: Genetic testing and management of hereditary gastrointestinal cancer syndromes. Am J Gastroenterol. 2015 Feb;110(2):223–262. doi: 10.1038/ajg.2014.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Judkins T, Leclair B, Bowles K, et al. Development and analytical validation of a 25-gene next generation sequencing panel that includes the BRCA1 and BRCA2 genes to assess hereditary cancer risk. BMC cancer. 2015;15(1):215. doi: 10.1186/s12885-015-1224-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eggington JM, Bowles KR, Moyes K, et al. A comprehensive laboratory-based program for classification of variants of uncertain significance in hereditary cancer genes. Clinical genetics. 2014 Nov 8;86(3):229–237. doi: 10.1111/cge.12315. [DOI] [PubMed] [Google Scholar]
- 17.Richards S, Aziz N, Bale S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genetics in medicine : official journal of the American College of Medical Genetics. 2015 May;17(5):405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Holter S, Borgida A, Dodd A, et al. Germline BRCA Mutations in a Large Clinic-Based Cohort of Patients With Pancreatic Adenocarcinoma. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2015;33(28):3124–3129. doi: 10.1200/JCO.2014.59.7401. [DOI] [PubMed] [Google Scholar]
- 19.Hu C, Hart SN, Bamlet WR, et al. Prevalence of Pathogenic Mutations in Cancer Predisposition Genes among Pancreatic Cancer Patients. Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2016;25(1):207–211. doi: 10.1158/1055-9965.EPI-15-0455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rosenthal ET, Evans B, Kidd J, et al. Increased Identification of Candidates for High-Risk Breast Cancer Screening Through Expanded Genetic Testing. J Am Coll Radiol. 2016 Dec 20; doi: 10.1016/j.jacr.2016.10.003. [DOI] [PubMed] [Google Scholar]
- 21.Daly M, Pilarski R, Berry M, et al. Genetic/Familial High-Risk Assessment: Breast and Ovarian. NCCN Clinical Practice Guidelines in Oncology. 2016 (Version 2.2017). http://www.nccn.org/professionals/physician_gls/pdf/genetics_screening.pdf.
- 22.Gatti RA, Berkel I, Boder E, et al. Localization of an ataxia-telangiectasia gene to chromosome 11q22–23. Nature. 1988;336(6199):577–580. doi: 10.1038/336577a0. [DOI] [PubMed] [Google Scholar]
- 23.The CBCC-CC. CHEK2*1100delC and susceptibility to breast cancer: a collaborative analysis involving 10,860 breast cancer cases and 9,065 controls from 10 studies. American journal of human genetics. 2004;74(6):1175–1182. doi: 10.1086/421251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dong X, Wang L, Taniguchi K, et al. Mutations in CHEK2 associated with prostate cancer risk. American journal of human genetics. 2003 Feb;72(2):270–280. doi: 10.1086/346094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cybulski C, Gorski B, Huzarski T, et al. CHEK2 is a multiorgan cancer susceptibility gene. American journal of human genetics. 2004;75(6):1131–1135. doi: 10.1086/426403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meijers-Heijboer H, Wijnen J, Vasen H, et al. The CHEK2 1100delC mutation identifies families with a hereditary breast and colorectal cancer phenotype. American journal of human genetics. 2003 May;72(5):1308–1314. doi: 10.1086/375121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Al-Tassan N, Chmiel NH, Maynard J, et al. Inherited variants of MYH associated with somatic G:C-->T:A mutations in colorectal tumors. Nature genetics. 2002 Feb;30(2):227–232. doi: 10.1038/ng828. [DOI] [PubMed] [Google Scholar]
- 28.Barnetson RA, Devlin L, Miller J, et al. Germline mutation prevalence in the base excision repair gene, MYH, in patients with endometrial cancer. Clinical genetics. 2007;72(6):551–555. doi: 10.1111/j.1399-0004.2007.00900.x. [DOI] [PubMed] [Google Scholar]
- 29.Nielsen M, Poley JW, Verhoef S, et al. Duodenal carcinoma in MUTYH-associated polyposis. J Clin Pathol. 2006;59(11):1212–1215. doi: 10.1136/jcp.2005.031757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tung N, Lin NU, Kidd J, et al. Frequency of germline mutations in 25 cancer susceptibility genes in a sequential series of breast cancer patients. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2016;34(13):1460–1468. doi: 10.1200/JCO.2015.65.0747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yurgelun MB, Allen B, Kaldate RR, et al. Identification of a Variety of Mutations in Cancer Predisposition Genes in Patients With Suspected Lynch Syndrome. Gastroenterology. 2015;149(3):604–613. doi: 10.1053/j.gastro.2015.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Provenzale D, Gupta S, Ahnen DJ, et al. Genetic/Familial High-Risk Assessment: Colorectal. [Accessed 2016];NCCN Clinical Practice Guidelines in Oncology. 2016 doi: 10.6004/jnccn.2016.0108. Version 2.2016. [DOI] [PubMed]
- 33.Vasen HF, Stormorken A, Menko FH, et al. MSH2 mutation carriers are at higher risk of cancer than MLH1 mutation carriers: a study of hereditary nonpolyposis colorectal cancer families. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2001;19(20):4074–4080. doi: 10.1200/JCO.2001.19.20.4074. [DOI] [PubMed] [Google Scholar]
- 34.Yang XR, Rotunno M, Xiao Y, et al. Multiple rare variants in high-risk pancreatic cancer-related genes may increase risk for pancreatic cancer in a subset of patients with and without germline CDKN2A mutations. Human genetics. 2016;135(11):1241–1249. doi: 10.1007/s00439-016-1715-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lynch HT, Voorhees GJ, Lanspa SJ, McGreevy PS, Lynch JF. Pancreatic carcinoma and hereditary nonpolyposis colorectal cancer: a family study. British journal of cancer. 1985;52(2):271–273. doi: 10.1038/bjc.1985.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smith AL, Alirezaie N, Connor A, et al. Candidate DNA repair susceptibility genes identified by exome sequencing in high-risk pancreatic cancer. Cancer letters. 2016;370(2):302–312. doi: 10.1016/j.canlet.2015.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Seemanova E, Jarolim P, Seeman P, et al. Cancer risk of heterozygotes with the NBN founder mutation. Journal of the National Cancer Institute. 2007 Dec 19;99(24):1875–1880. doi: 10.1093/jnci/djm251. [DOI] [PubMed] [Google Scholar]
- 38.Zhang B, Beeghly-Fadiel A, Long J, Zheng W. Genetic variants associated with breast-cancer risk: comprehensive research synopsis, meta-analysis, and epidemiological evidence. The lancet oncology. 2011 May;12(5):477–488. doi: 10.1016/S1470-2045(11)70076-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.