SUMMARY
The optic neuroepithelial continuum of vertebrate eye develops into three differentially growing compartments – the retina, ciliary margin (CM), and retinal pigment epithelium (RPE). The neurofibromin 2 (Nf2) is strongly expressed in slowly expanding RPE and CM compartments, and the loss of mouse Nf2 causes hyperplasia in these compartments, replicating the ocular abnormalities seen in human NF2 patients. The hyperplastic ocular phenotypes were largely suppressed by heterozygous deletion of Yap and Taz, key targets of the Nf2-Hippo signaling pathway. We further found that, in addition to feedback transcriptional regulation of Nf2 by Yap/Taz in the CM, activation of Nf2 expression by Mitf in the RPE and suppression by Sox2 in retinal progenitor cells are necessary for the differential growth of the corresponding cell populations. Together, our findings reveal that Nf2 is a key player that orchestrates the differential growth of optic neuroepithelial compartments during vertebrate eye development.
eTOC
Moon et al. identify a mechanism underlying the differential growth of optic neuroepithelial compartments in the mouse eye. Differential transcriptional regulation of the tumor suppressor Neurofibromin 2 (Nf2) in each neuroepithelial compartment is necessary for the differential growth of the tissue.
INTRODUCTION
Development of distinctive tissues and organs with defined sizes and shapes in multicellular organisms requires a series of compartmentalization steps (Zhu and Skoultchi, 2001). Each compartment shows unique growth rate as well as identity compared with its neighbors to construct a 3-dimensional structure with multiple tissue domains (Stanger, 2008). Therefore, perturbation of the growth of individual compartments causes tissue hypertrophy and hypotrophy, and often results in organ malformation.
The orchestrated growth of each tissue compartment is particularly evident in vertebrate eye development. Optic neuroepithelium in the ventral lateral diencephalic region evaginates to form the optic vesicle (OV), which then invaginates to form a double-layered optic cup structure (Chow and Lang, 2001; Heavner and Pevny, 2012). The retinal pigment epithelium (RPE) develops in the outer layer of the optic cup, whereas the neural retina arises in the inner layer. The ciliary margin (CM) is specified at the rim of the optic cup and becomes the border between the RPE and the retina. The non-pigmented inner CM (ICM) layer is continuous with the retina, while the pigmented outer CM (OCM) layer is continuous with the RPE (Beebe, 1986; Napier and Kidson, 2005). The CM neuroepithelium matures into non-pigmented ciliary epithelium (NPCE) in the inner layer and pigmented ciliary epithelium (PCE) in the outer layer to constitute the ciliary body (CB) and iris in the mature eye. It also gives rise to neurons in the peripheral retina via retinal progenitor cells (RPCs) in an intermediate structure termed the ciliary marginal zone (CMZ) (Belanger et al., 2017; Marcucci et al., 2016).
The neuroepithelium in each optic compartment has distinct characteristics in terms of cell shape, stiffness, and proliferative potential (Chow and Lang, 2001). The cell population in the future retina has a higher proliferative potential than cells in the other two compartments, and is capable of differentiating into neurons and Müller glia. In contrast, the RPE is composed of post-mitotic epithelial cells that form a monolayer sheet. CM cells are also proliferative, but their growth rate is lower than the retinal growth rate. However, CM cells retain their proliferative potential until post-natal stages, when the majority of retinal cells become post-mitotic. The regional diversification of proliferative potential therefore might be related with the differential growth of these optic compartments during eye development; however, the underlying molecular mechanisms are largely unknown.
Neurofibromin 2 (NF2; also known as Merlin and Schwannomin) is the product of the NF2 gene, which is broadly expressed in vertebrate nervous tissues. Various mutations in the NF2 gene are associated with a human disease called neurofibromatosis type-2, which is characterized by the development of benign tumors in neural tissues (Asthagiri et al., 2009). NF2 is homologous to ezrin/radixin/moesin (ERM) family actin-binding proteins, and serves as a linker between plasma membrane proteins and the actin cytoskeleton (Bretscher et al., 2002). NF2 has been known to primarily function as a tumor suppressor by mediating the contact-dependent inhibition of cell proliferation (Curto et al., 2007). Recent studies have further shown that NF2 is a multifunctional protein that merges extracellular and intracellular signals to regulate cell proliferation, fate determination, and survival by interacting with various cellular proteins (Curto and McClatchey, 2007; Stamenkovic and Yu, 2010). Notably, NF2 has been also shown to act as an upstream regulator of the Hippo pathway, which regulates tissue growth and homeostasis (Yu and Guan, 2013).
In the developing mouse eye, Nf2 is strongly enriched in the cornea, lens and RPE, but is hardly detectable in the retina (Huynh et al., 1996)(Figure 1C). Homozygous loss of Nf2 specifically in embryonic lens epithelium in mice, thus, results in the failure of lens formation (Wiley et al., 2010). Consistent with these observations, human patients afflicted with NF2 syndrome develop ocular lesions including juvenile cataracts (McLaughlin et al., 2007b; Ragge et al., 1997). In addition to lens defects, hypertrophies in the posterior parts of the eye, manifesting as retinal detachment, hyperpigmentation of the RPE, intraocular neurilemmoma, epiretinal membrane, combined pigment epithelial and retinal hamartoma (CPERH), congenital glaucoma, optic nerve glioma and Lisch nodules, have also been reported in human NF2 patients (Baser et al., 1999; Good et al., 1991; Sisk et al., 2010). These observations therefore suggest that NF2 might be crucial for the development and/or maintenance of these ocular structures; however, the functions of NF2 in these ocular tissues are poorly understood.
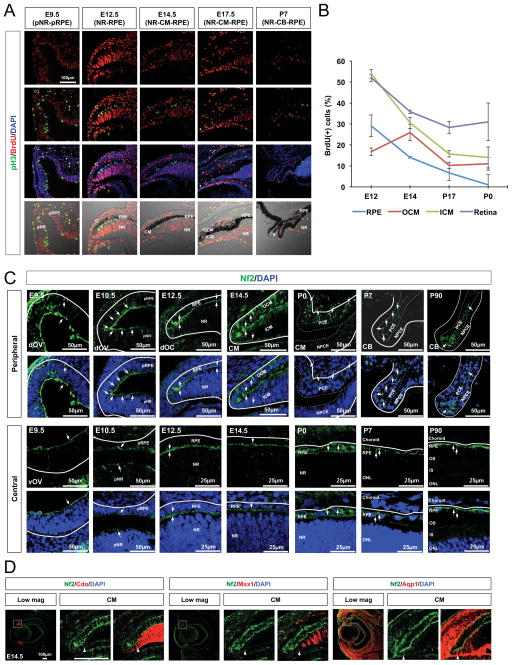
Figure 1. Expression of Nf2 in differentially growing optic neuroepithelial compartments.
(A) Sections of mouse embryonic heads and post-natal eyes (C57BL/6J) were immunostained to identify the cells possess mitotic cell cycle marker pH3 and DNA-labeled with BrdU. Nuclei are visualized by DAPI staining. NR, neural retina; pNR, presumptive neural retina; RPE, retinal pigmented epithelium; pRPE, presumptive retinal pigmented epithelium; CM, ciliary margin; ICM, inner ciliary margin; OCM, outer ciliary margin; CB, ciliary body. (B) Quantification of BrdU-positive cell population in the each optic compartment at indicated ages. Error bars in the graphs represent mean ± SEM (n = 6 from 4 independent litters). (C) Expression of Nf2 in the mouse eyes at the indicated ages was examined by immunostaining. Arrows point the Nf2 immunostaining signals. dOV, dorsal optic vesicle; vOV, ventral optic vesicle; LV, lens vesicle; IS, inner segment; OS, outer segment; dOS, dorsal optic stalk; vOS, ventral optic stalk; PCE, pigmented ciliary epithelium; NPCE, non-pigmented ciliary epithelium. (D) Nf2 distribution in the entire ICM (marked by Cdo, left), proximal ICM (marked by Msx1, center), and distal ICM (marked by Aqp1, right) was determined by co-immunodetection of Nf2 and representative ICM markers. Arrowheads indicate the proximal margins of Nf2 staining signals.
RESULTS
Each optic neuroepithelial compartment exhibits unique proliferative potential during development
The proliferative potential of the optic neuroepithelium becomes divergent along the body axes to form a double-layered optic cup in mouse embryo. Proliferative potential, determined by examining the numbers of proliferating cells that incorporated with bromodeoxyuridine (BrdU) into their DNA and mitotic cells that express phosphorylated histone H3 (pH3), was largely uniform in the mouse neuroepithelium of the OV at embryonic day 9.5 (E9.5) (Figure 1A, leftmost column). During invagination of the OV between E10.5 and E12.5, the neuroepithelium in the inner optic cup layer, which develops into the retina, underwent more intensive proliferation than that in the outer layer, which is specified to the RPE (Figure 1A [second column from the left], 1B). In E14.5 mouse eyes, the proliferation rate in the retina was decreased moderately compared with that at E12.5, but was significantly compromised in the RPE (Figure 1A [center column], 1B). The proliferation of the CM neuroepithelium was intermediate between those in the retina and RPE at this stage. This tripartite proliferation pattern was maintained during embryogenesis. As eyes matured in post-natal days, a majority of cells in the retina withdrew permanently from the cell cycle and differentiated, leaving a minor proliferating population in the peripheral region (Figure 1A [second column from the right], 1B). CM cells, however, continued to proliferate and became integrated into epithelial sheets composing the CB and iris until post-natal day 7 (P7), while the RPE population remained quiescent (Figure 1A [rightmost column], 1B).
Nf2 is expressed predominantly in the RPE and CM of the mouse eye
The divergent changes in the proliferative potentials of the optic neuroepithelium suggest the presence of regulator(s) that are differentially expressed or activated in each optic neuroepithelial compartment. The promoter activities of human NF2 are specifically enriched in the outer optic cup layer (Akhmametyeva et al., 2006), which is hypoproliferative relative to the inner layer. These intriguing observations led us to hypothesize that Nf2 could be one of the factors accounting for the lower cell proliferation in the outer optic cup layer. We, thus, further examined the expression pattern of Nf2 in mouse eyes at various developmental stages by immunostaining. Nf2 was detectable in the apical side of the optic neuroepithelium throughout the entire OV at E9.5, and started to decay in ventral and proximal OV regions from E10.5 (Figure 1C, two leftmost columns). Nf2 was maintained at high levels in the RPE and CM of the mouse embryos at mid-gestation (i.e., E12.5 and E14.5), while it decayed in the retina (Figure 1C, third and fourth columns from the left).
Nf2 expression in the ICM was more prominent in the aquaporin-1 (Aqp1)-positive distal part than the Msh homeobox 1 (Msx1)-positive proximal part (Figure 1D), indicating a gradual decrease in Nf2 expression along the distal-proximal (i.e., peripheral-central) axis of the inner optic cup. In the post-natal mouse eye, Nf2 was found predominantly in pigmented cells, including the RPE and PCE, in the outer optic cup layer (Figure 1C, three rightmost columns). These results therefore suggest that Nf2 might control the growth of the mouse optic cup by limiting the proliferation of the RPE and CM neuroepithelium.
Nf2 loss leads to hyperplasia of the pigmented population and expansion of the RPC population in the mouse eye
To investigate the roles of Nf2 in mouse eye development, we generated Nf2f/f;Nes-Cre mice lacking Nf2 in the entire optic neuroepithelium by crossing Nf2-flox (Nf2f/f) mice with a neuroepithelium-specific Nestin-Cre mice (McClatchey and Giovannini, 2005; Tronche et al., 1999). The Cre recombinase activity, which can be determined based on Cre-sensitive β-galactosidase (β-gal) expression from a lacZ reporter gene knocked in the ROSA26 locus (R26R) (Soriano, 1999), was detectable throughout the entire optic neuroepithelium (Figure S1A). Thus, Nf2 was no longer detectable in the R26R-positive E14.5 Nf2f/f;Nes-Cre mouse optic cup, but was expressed in the R26R-negative ocular tissues including cornea (Figure S1B).
Adult Nf2f/f;Nes-Cre mice exhibited various oculo-facial dysplasias, characterized by microphthalmia, cataracts, and iris malformation together with craniofacial defects (Figure 2A – 2D). Symptoms noted in the retinas and RPE of human NF2 patients, including CPERH, epiretinal membrane and optic disc glioma, were also displayed by the Nf2f/f;Nes-Cre mice (Figure 2C and 2E). A histological examination of Nf2f/f;Nes-Cre mouse eyes revealed that microphthalmia was evident as early as E14.5, at which point the pigmented cells started to expand across the borders between the RPE and retina and the RPE and optic stalk (Figure S1C – S1E).
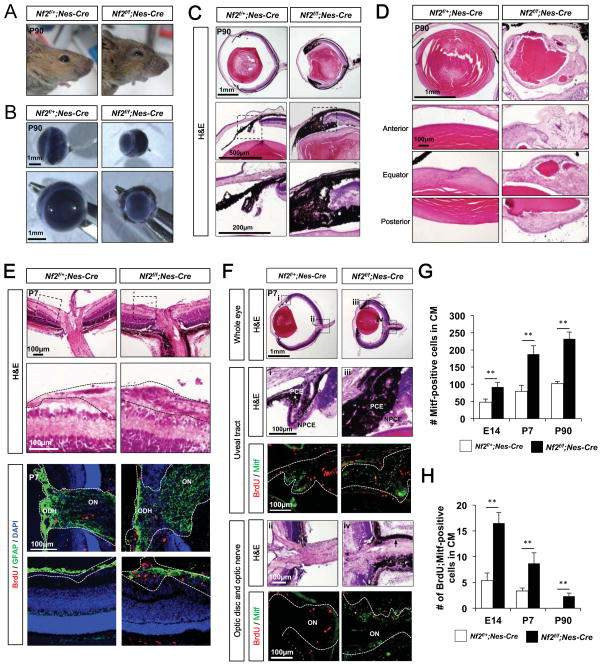
Figure 2. Loss of Nf2 in neuroepithelium results in the expansion of pigmented cells in the microphthalmic eyes.
Images of heads (A) and isolated eyes (B) of P90 Nf2f/+;Nes-Cre and Nf2f/f;Nes-Cre littermate mice. (C) Hamatoxylin and eosin (H&E) staining images of eye sections of P90 Nf2f/+;Nes-Cre and Nf2f/f;Nes-Cre littermate mice. The images in the bottom row are the magnified versions of the areas marked by dot-lines in the middle row. (D) H&E staining images of the lenses isolated from P90 Nf2f/+;Nes-Cre and Nf2f/f;Nes-Cre littermate mouse eyes (E) H&E staining images of the optic disc head in P7 littermate mouse eyes (top). Images in the second row shows magnified version of the areas surrounded by dot-line boxes in top row. Dot-lines outline the GCL and optic nerve. Astrocytes and proliferating cells in the optic disc head area are shown by immunostaining of GFAP and BrdU, respectively. ODH, optic disk head; ON, optic nerve. (F) Pigmented cells in the uveal tract (i and iii) and the optic nerve (ii and iv) of P7 Nf2f/+;Nes-Cre and Nf2f/f;Nes-Cre littermate mouse eyes sections in top row are magnified in the second and fourth rows with corresponding numbers. Immunostaining images in the third and fifth rows show proliferating cell incorporated with BrdU and pigmented cells expressing Mitf. (G) Quantification of Mitf-positive pigmented cells in the CM of Nf2f/+;Nes-Cre and Nf2f/f;Nes-Cre eyes at E14, P7 and P90. (H) Quantification of BrdU-positive cell population in Mitf-positive pigmented cells in Nf2f/+;Nes-Cre and Nf2f/f;Nes-Cre mouse eyes at E14, P7 and P90 (n = 6 from 3 independent litters). P-values (p) were obtained by Student’s two-tailed unpaired t-test (**, p < 0.01).
In support of this, the number of cells expressing a pigment cell marker microphthalmia-associated transcription factor (Mitf) was significantly increased in the retina-RPE border of Nf2f/f;Nes-Cre mice by E14.5, and came to more than double the number of such cells in Nf2f/+;Nes-Cre littermates by P7 (Figure 2F and 2G; Figure S1F, S1I, and S1M [left columns]; quantified in Figure S1H, S1L, and S1P). The number of pigmented cells in the central part of Nf2f/f;Nes-Cre embryonic and post-natal mouse optic cup was also significantly increased, and these cells do not form monolayer sheet (Figure S1F, S1I, and S1M[right columns]; quantified in Figure S1H, S1L, and S1P). The expanded pigmented cell population expressed Otx2, which is specifically expressed in RPE and OCM cells but not in choroidal melanocytes (Figure S1G, S1J, and S1N), implying genuine expansion of an optic-neuroepithelium-derived pigmented cell population in the Nf2f/f;Nes-Cre mouse eyes. The BrdU-positive proliferative population among those pigmented cells was also increased in the Nf2f/f;Nes-Cre mouse eyes compared with that in Nf2f/+;Nes-Cre littermates (Figure 2F and 2H; Figure S1F, S1K, and S1O; quantified in Figure S1H, S1L, and S1P).
In contrast to the disorganized layer structures of P90 Nf2f/f;Nes-Cre mouse retinas (Figure 2C), three retinal nuclear layers were exhibited in P7 Nf2f/f;Nes-Cre mouse retinas (Figure 2E). However, Nf2f/f;Nes-Cre mouse retinas failed to express markers for late-born retinal cells, such as protein kinase C-α (PKCα; a marker for rod bipolar cells), and glutamine synthase (GS; a marker for Müller glia), but did express markers for early-born retinal cells, including retinal ganglion cells (RGCs; positive for Brn3b), amacrine cells (positive for syntaxin), and horizontal cells (positive for calbindin) (data not shown). Interestingly, a significant number of cells in the inner nuclear layer (INL) co-expressed the RPC markers, Pax6, Vsx2 and Sox2, which are normally segregated into amacrine, bipolar and Müller glial cells, respectively, in the mouse retina (Figure S1Q and S1R). Furthermore, those ectopic RPCs in the P7 Nf2f/f;Nes-Cre mouse retina were still capable of proliferating, as evidenced by incorporation of BrdU, whereas cells in Nf2f/+;Nes-Cre littermate mouse retinas did not proliferate (Figure S1S and S1T). Collectively, these results suggest that Nf2 supports mouse eye development by inhibiting the proliferation of the pigmented cell population in the outer optic cup layer. They also propose that Nf2 prevents abnormal expansion of the RPC population, which is normally destined to exit the cell cycle and differentiate into bipolar cells and Müller glia in the post-natal mouse retina.
Nf2 loss in the RPE and CM results in hyperplastic malformation of these compartments
Next, we further investigated the roles of Nf2 in optic cup growth by deleting Nf2 in the RPE and CM compartments, where Nf2 expression is maintained from embryo to adult (Figure 1C). Nf2f/f mice were bred with tyrosinase-related protein 1 (TRP1)-Cre mice, which express Cre recombinase in the primitive RPE of the invaginating OV at E10.5 and in the RPE and CM cells from E12.5 (Mori et al., 2002). Nf2 immunoreactivity was eliminated in the RPE and CM of E14.5 Nf2f/f;TRP1-Cre mouse eyes, indicating successful deletion of the Nf2 gene in these Cre recombinase-active cell populations (Figure 3A). Interestingly, the Cre recombinase reporter R26R covered a broader region of these microphthalmic Nf2f/f;TRP1-Cre mouse retinas compared with the R26R coverage in Nf2f/+;TRP1-Cre littermate mouse retinas (Figure 3A, images at low magnification; Figure S3A), implying the expansion of the Nf2-deficient CM cell population into the Nf2f/f;TRP1-Cre mouse retina.
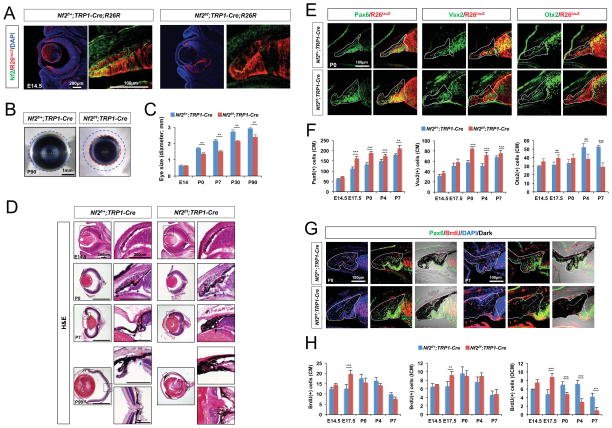
Figure 3. Nf2 loss in the CM and RPE resulted in the hyperplasia of the compartments.
(A) Nf2 immunoreactivity and R26lacZ (R26R) Cre recombinase activity reporter expression in the eyes of E14.5 Nf2f/+;TRP1-Cre;R26R and Nf2f/f;TRP1-Cre;R26R littermates. (B) Microscopic images of eyes isolated from P90 Nf2f/+;TRP1-Cre and Nf2f/f;TRP1-Cre littermates. (C) Quantification of eye sizes of Nf2f/+;TRP1-Cre and Nf2f/f;TRP1-Cre littermates at indicated ages (n = 6 from 3 independent litters). (D) H&E staining images of eye sections of Nf2f/+;TRP1-Cre and Nf2f/f;TRP1-Cre littermates at indicated ages. Images in right columns are magnified versions of dot-line box areas in left columns. (E) Sections of P0 Nf2f/+;TRP1-Cre;R26R and Nf2f/f;TRP1-Cre;R26R littermate mouse eyes were stained with antibodies against a pan-CM marker Pax6, an ICM and RPC marker Vsx2, and an OCM and RPE marker Otx2. CM areas are outlined by solid lines, and the borders between ICM and OCM are marked by dot-lines. (F) Quantification of Pax6-positive (left), Vsx2-positive (center), and Otx2-positive (right) CM cells in the staining images of Nf2f/+;TRP1-Cre and Nf2f/f;TRP1-Cre mouse eyes at indicated ages (n = 6 from 3 independent litters). (G) Nf2f/+;TRP1-Cre and Nf2f/f;TRP1-Cre eyes from P0 and P7 mice were analyzed to detect proliferating cells, which are positive to BrdU and Pax6 in the CM areas (outlined by solid lines). (H) Quantification of BrdU-positive cells in the whole CM (left), ICM (middle), and OCM (right) of Nf2f/+;TRP1-Cre and Nf2f/f;TRP1-Cre eyes (n = 6 from 3 independent litters). *, p < 0.05; **, p < 0.01; ***, p < 0.005.
Nf2f/f;TRP1-Cre mice also developed microphthalmia (Figure 3B and 3C), and exhibited most of the histological defects observed in Nf2f/f;Nes-Cre mouse eyes, except for cataracts (Figure 3D). Most prominently, the pigmented cells were accumulated in the anterior part of the eyes, where the CB and iris failed to develop properly (Figure 3D, rightmost column). These pigmented cells were identified as Otx2-positive RPE and OCM populations (Figure S2A). The number of Otx2-positive cells was increased in the OCM of embryonic Nf2f/f;TRP1-Cre mouse eyes compared with that in littermate Nf2f/+;TRP1-Cre mouse eyes, but was decreased in post-natal Nf2f/f;TRP1-Cre eyes (Figure 3E and 3F [graph at right]; Figure S2A). Conversely, the number of Vsx2-positive cells in the ICM was increased only in post-natal Nf2f/f;TRP1-Cre mouse eyes (Figure 3E and 3F [center graph]; Figure S2A). The number of Vsx2-positive cells in mouse retinas was also increased, and were found to be RPCs, which co-express Pax6 and Sox2 (Figure S3C), as evident in P7 Nf2f/f;Nes-Cre mouse retinas (Figure S1Q). Different from the Nf2f/f;Nes-Cre mouse retinas, rod bipolar cell marker PKCα and Müller glial cell marker GS could be seen in Nf2f/f;TRP1-Cre mouse retinas; however, these cells were R26R-negative wild-type cells (Figure S3A and S3B), implicating that Nf2 deficiency impairs neuronal differentiation of RPCs autonomously.
The Pax6;BrdU-positive proliferating CM cell population was transiently increased in E17.5 Nf2f/f;TRP1-Cre mouse eyes (Figure 3G and 3H [graph at left]), despite consistent increase in total CM cell numbers in embryonic and post-natal Nf2f/f;TRP1-Cre mouse eyes (Figure 3E and 3F [graph at left]; Figure S2A). Hyperproliferation was observed in both the Vsx2-positive ICM and Otx2-positive OCM of E17.5 Nf2f/f;TRP1-Cre mouse retinas (Figure 3G and 3H [graphs in the center and right]; Figure S2A). However, the number of Otx2;BrdU-positive cells in the OCM was lower in the post-natal Nf2f/f;TRP1-Cre mouse eye compared with that in Nf2f/+;TRP1-Cre littermates (Figure 3G and 3H [graph at right]), suggesting that hyperproliferation of Nf2-deficient cells in the OCM occurred only during the embryonic period. The number of Vsx2;BrdU-positive cells in the ICM of post-natal Nf2f/f;TRP1-Cre mouse eyes was not significantly different from that of Nf2f/+;TRP1-Cre littermates (Figure 3G and 3H [center graph]). Instead, BrdU-positive cells were elevated in P7 Nf2f/f;TRP1-Cre mouse retinas (Figure S3E and S3F), and those were Sox2-positive RPCs, which likely co-express Vsx2 and Pax6 (Figure S3C and S3D).
Nf2 is necessary for the coordinated growth of two CM layers
Expansion of the RPC population in Nf2f/f;TRP1-Cre mouse retinas might not only be the result of autonomous hyperproliferation of RPCs, it could also be the reflection of a non-cell–autonomous event influenced by the Nf2-deficient RPE and OCM. Conversely, the alterations of Otx2-positive OCM population in the Nf2f/f;TRP1-Cre mouse eyes could be resulted from an autonomous increase/decrease in proliferative potential and/or a non-autonomous regulation by Nf2-deficient ICM cells.
To investigate these possibilities, we crossed Nf2f/f mice with two different Cre mice, in which Nf2 is deleted selectively in the inner or outer optic cup layer. Cre recombinase in Chx10-Cre mice is active in RPCs in the embryonic retina from E11.5, and then is restricted to the bipolar cells of mature retina (Rowan and Cepko, 2004). R26R-positive cells were detectable in the ICM as well as the retina of E14.5 Chx10-Cre;R26R mouse eyes (Figure 4A), allowing us to selectively eliminate Nf2 in the inner optic cup layer. As expected, Nf2 immunostaining was lost in the ICM of E14.5 Nf2f/f;Chx10-Cre mice, but was maintained in the OCM and RPE (Figure 4A). Adult Nf2f/f;Chx10-Cre mice exhibited microphthalmic eyes, which lacked a detectable CB and iris (data not shown). However, both CM layers were detectable in P0 Nf2f/f;Chx10-Cre mouse eyes (Figure 4B and 4C), and its ICM was thicker and contained more Vsx2-positive cells than the corresponding layer in Nf2f/+;Chx10-Cre littermates (Figure 4D [center graph]; Figure S2B). The number of Vsx2-positive cells remained higher in P7 Nf2f/fChx10-Cre mouse retinas, and these cells were turned out as RPCs that co-express Sox2 and Pax6 (data not shown), consistent with the results obtained in P7 Nf2f/f;Nes-Cre and Nf2f/f;TRP1-Cre mouse retinas (Figure S1Q and S3C).
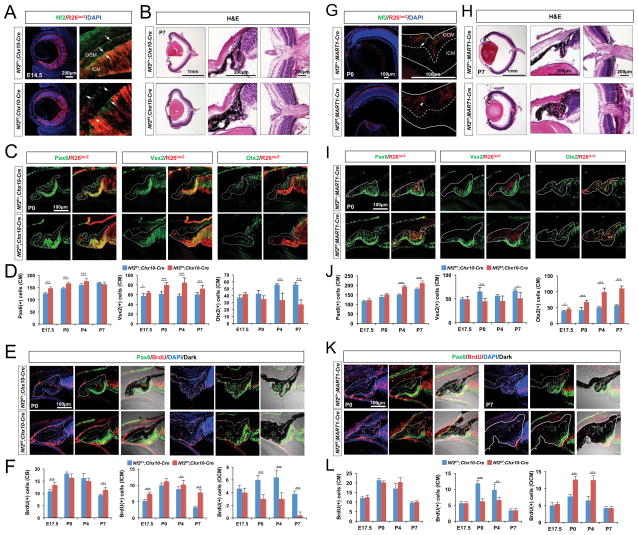
Figure 4. Autonomous expansion of the CM population in the absence of Nf2.
(A and G) Nf2 immunoreactivity and R26lacZ Cre recombinase activity reporter expression in the eyes of E14.5 Nf2f/+;Chx10-Cre;R26R and Nf2f/f;Chx10-Cre;R26R littermates (A) and P0 Nf2f/+;MART1-Cre;R26R and Nf2f/f;MART1-Cre;R26R littermates (G) were examined. Nf2 immunoreactivity is eliminated in R26R-positive ICM cells (arrowheads in A) of E14.5 Nf2f/f;Chx10-Cre eyes, but it remains in R26R-negative OCM cells (arrow in A). Nf2 immunoreactivity is eliminated in R26R-positive OCM cells (arrowheads in G) of P0 Nf2f/f;MART1-Cre eyes. (B and H) H&E staining images of eye sections of P7 Nf2f/+;Chx10-Cre and Nf2f/f;Chx10-Cre littermates (B) and P7 Nf2f/+;MART1-Cre and Nf2f/f;MART1-Cre littermates (H). (C and I) Sections of P0 Nf2f/+;Chx10-Cre;R26R and Nf2f/f;Chx10-Cre;R26R mouse eyes (C) and P0 Nf2f/+;MART1-Cre;R26R and Nf2f/f;MART1-Cre;R26R mouse eyes (I) are stained with antibodies against Pax6, Vsx2, and Otx2. CM areas are outlined by solid lines, and the borders between ICM and OCM are marked by dot-lines. (D and J) Quantification of Pax6-positive (left), Vsx2-positive (center), and Otx2-positive (right) cells in the Nf2f/+;Chx10-Cre;R26R and Nf2f/f;Chx10-Cre;R26R mouse eyes (D) and Nf2f/+;MART1-Cre;R26R and Nf2f/f;MART1-Cre;R26R mouse eyes (J) at indicated ages (n = 6 from 3 independent litters). (E and K) Nf2f/+;Chx10-Cre and Nf2f/f;Chx10-Cre (E) and Nf2f/+;MART1-Cre and Nf2f/f;MART1-Cre (K) mouse eyes from P0 and P7 mice were analyzed to detect proliferating CM cells, which are positive to BrdU and Pax6. (F and L) Quantification of BrdU-positive cells in the whole CM (left), ICM (middle), and OCM (right) of Nf2f/+;Chx10-Cre and Nf2f/f;Chx10-Cre (F) and Nf2f/+;MART1-Cre and Nf2f/f;MART1-Cre (L) mouse eyes (n = 6 from 3 independent litters).
In contrast to the transient increase of BrdU-incorporation in late embryonic ICM of Nf2f/f;TRP1-Cre mice (Figure 3G and 3H), the number of BrdU;Vsx2 double-positive cells in the Nf2f/f;Chx10-Cre mouse ICM remained higher from embryo to post-natal stages (Figure 4E and 4F [center graph]). Conversely, the number of Otx2-positive cells in the OCM of post-natal Nf2f/f;Chx10-Cre mouse eyes was significantly decreased compared with that in Nf2f/+;Chx10-Cre littermate mouse eyes (Figure 4C and 4D [graph at right]; Figure S2B). This phenotype was associated with a significant decrease in BrdU-incorporation into Otx2-positive OCM cells of Nf2f/f;Chx10-Cre mice (Figure 4E and 4F [graph at right]). These results suggest that the loss of Nf2 in the ICM and retina promotes the proliferation of RPCs autonomously, but interferes with the proliferation of pigmented cells in the adjacent OCM non-autonomously.
As an alternative strategy, we deleted Nf2 only in the outer optic cup layer by crossing Nf2-flox mice with MART1 (melanoma antigen recognized by T cells 1)-Cre mice, in which Cre recombinase activity is confined to pigmented cells in the RPE and OCM in addition to melanocytes in the skin and choroid (Aydin and Beermann, 2011). Nf2 signals were absent in the RPE and a majority of OCM cells in P0 Nf2f/f;MART1-Cre mouse eyes (Figure 4G; Figure S2D). Nf2f/f;MART1-Cre adult mice also exhibited microphthalmia, albeit less severe than that in Nf2f/f;TRP1-Cre mouse eyes (data not shown). They also showed expansion of pigmented cells without forming CB folds in the anterior ocular segments, but did have an RPE monolayer in the posterior region (Figure 4H). These excessive pigmented cells in the eyes were positive for the R26R reporter and expressed Otx2, identifying them as Nf2-deficient RPE and/or OCM cells (Figure 4I).
Unlike Nf2f/f;TRP1-Cre mouse eyes, which showed an increase in Otx2-positive pigmented epithelium only in the embryonic OCM (Figure 3E and 3F [graph at right]; Figure S2A), Nf2f/f;MART1-Cre mouse eyes exhibited a continuously elevated number of Otx2-positive pigmented cells in post-natal stages (Figure 4I and 4J [graph at right]; Figure S2C). The increased OCM cell population in Nf2f/f;MART1-Cre mice kept proliferating in the post-natal period (Figure 4K and 4L [graph at right]; Figure S2C), when the proliferation of Vsx2-positive cells in the ICM became lower than that in Nf2f/+;MART1-Cre littermates (Figure 4K and 4L [center graph]; Figure S2C). These results suggest that the loss of Nf2 in the OCM promotes the proliferation of pigmented cells autonomously, but inhibits the proliferation of neighboring ICM cells non-autonomously.
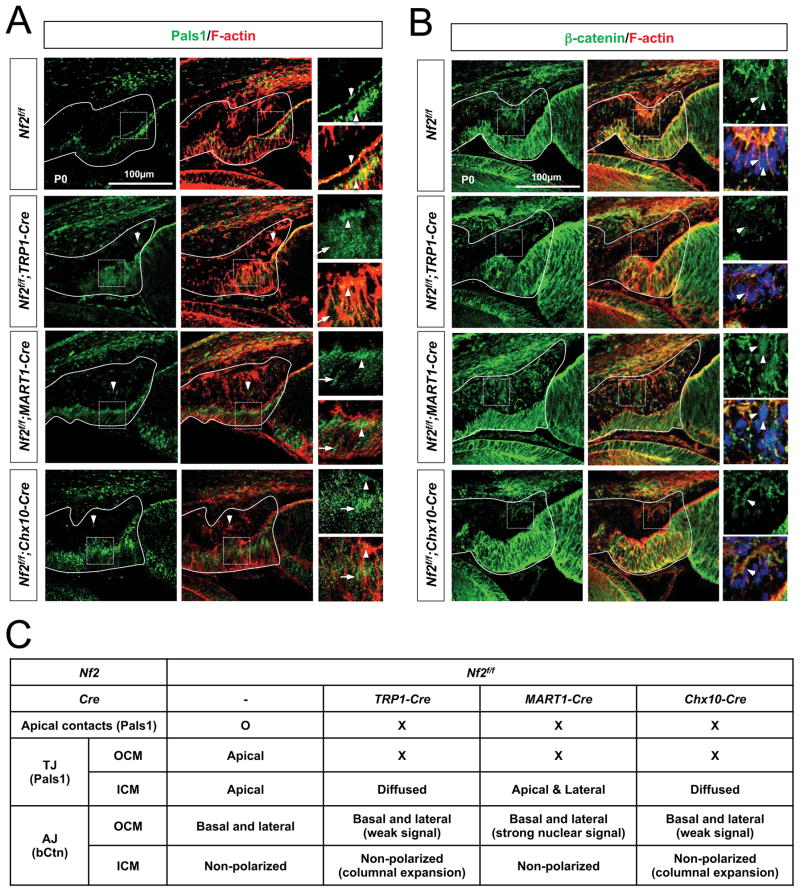
Loss of polarity in CM cells lacking Nf2 autonomously and non-autonomously
Nf2 maintains cellular architecture by linking the cortical actin cytoskeleton to proteins in the apical membrane of the epithelium (Bretscher et al., 2002; Gladden et al., 2010). We thus investigated whether the loss of Nf2 changed CM cell proliferation in Nf2f/f;TRP1-Cre, Nf2f/f;Chx10-Cre, and Nf2f/f;MART1-Cre mice by affecting the polarity of the CM neuroepithelia. In contrast to the discrete localization of Pals1 (protein associated with Lin-7) at apical tight junctions (TJs) in the CM neuroepithelia of Nf2f/f mouse eyes, Pals1 was diffused in the ICM cells of P0 Nf2f/f;TRP1-Cre mouse eyes while it was significantly decreased in the OCM cells (Figure 5A [second row] and 5C). Nf2-deficient cells in the OCM of P0 Nf2f/f;MART1-Cre mouse eyes and those in the ICM of P0 Nf2f/f;Chx10-Cre mouse eyes showed a common decrease in Pals1 localization at apical junctions (Figure 5A [third and fourth rows] and 5C). Pals1 also failed to become concentrated in the apical compartments of the counterpart wild-type ICM and OCM cells in those Nf2f/f;MART1-Cre and Nf2f/f;Chx10-Cre mouse eyes, respectively. These results suggest that Pals1 is distributed in apical junctions in the CM neuroepithelium only when Nf2 is expressed properly in both CM layers.
Fig. 5. Junctional polarity of CM neuroepithelium is lost in the absence of Nf2 in either CM layer.
Immunostaining of a TJ marker Pals1 (A) and an AJ marker β-catenin (B) in the CM neuroepithelium of P0 Nf2f/f, Nf2f/f;TRP1-Cre, Nf2f/f;MART1-Cre, Nf2f/f;Chx10-Cre mouse eyes. F-actin was visualized by staining with Alexa647-conjugated Phalloidin. Arrowheads point Pals1 localization in the TJs and β-catenin in the AJs, while arrows indicate diffused Pals1. (C) Summary of junctional protein distribution in wild-type (Nf2f/f) and Nf2-deficient mouse CM neuroepithelia.
The β-catenin, a marker for adherens junction (AJ) of epithelium, is not polarized in ICM cells and was unchanged by deletions of Nf2 in any CM layers (Figure 5B and 5C). On the contrary, β-catenin signals were accumulated in the basolateral membranes of OCM cells, and became detectable at higher level in the nuclei of P0 Nf2f/f;MART1-Cre mouse OCM cells (Figure 5B [third row] and 5C), which proliferate persistently (Figure 4K and 4L). However, β-catenin signals in OCM cells of Nf2f/f;TRP1-Cre and Nf2f/f;Chx10-Cre mouse eyes were not elevated, but instead were decreased (Figure 5B [second and fourth rows] and 5C), suggesting a potential relationship between reduced β-catenin level and hypoplasia of post-natal OCM cells in these mouse eyes (Figure 3H and 4L).
These data therefore suggest that Nf2 is necessary for the establishment of polarity in a CM neuroepithelium, which in turn supports polarization of the CM neuroepithelium in the other layer. Thus, the loss of Nf2 in one CM neuroepithelium simultaneously impairs the polarity of the neuroepithelium in the other layer, thereby disrupting coordinated growth of the two CM layers. The Nf2-deficient CM cells hyperproliferate autonomously, whereas wild-type cells in the other CM layer hypoproliferate (Figure 3H and 4L), although cell polarity is disrupted in both CM layers (Figure 5C). These results therefore suggest that inhibition of CM cell hyperproliferation is not simply mediated by cell polarity maintenance, but it might be more responsible by intracellular events regulated by Nf2.
Inactivation of the Hippo signaling pathway induces hyperplasia of pigment epithelium
Next, we investigated the molecular mechanism underlying the hyperplasia of Nf2-deficient CM neuroepithelium. Although the functions of Nf2 are highly varied and cell-type specific, overwhelming evidence has indicated that Nf2 acts as an upstream activator of the Hippo signaling pathway to mediate contact-dependent inhibition of cell proliferation in vitro and regulate tissue homeostasis in vivo. Nf2-activated Hippo signaling maintains the transcription co-activators, Yap and Taz (Yap/Taz), in the cytoplasm and inhibits cell proliferation induced by Yap/Taz target genes (Lavado et al., 2013; Serinagaoglu et al., 2015; Zhang et al., 2010; Zhu et al., 2015). Moreover, Yap/Taz-dependent transcription is elevated in uveal melanoma induced by a gain-of-function mutation in Gq/11, which inactivates the Hippo pathway through the activation of Rho GTPase (Yu et al., 2014), suggesting the importance of anti-proliferative Nf2-Hippo pathway in the pigmented cells.
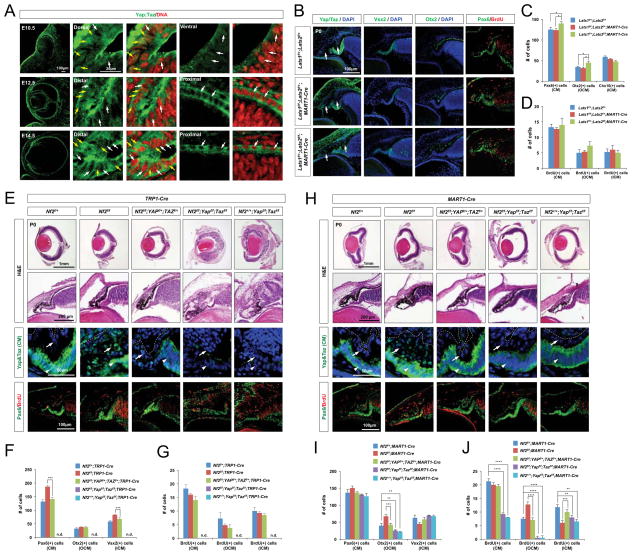
Interestingly, Yap/Taz expression in the mouse optic neuroepithelia largely coincided with the pattern of Nf2 expression. Yap/Taz expression was detected in the entire OV, and became more concentrated to the RPE and CM of the optic cup (Figure 6A, top row). Yap/Taz were strongly accumulated in the nuclei of cells in the invaginating OV neuroepithelia at E10.5, suggesting a role for Yap/Taz in RPE specification in the dorsal OV (Cabochette et al., 2015; Kim et al., 2016; Miesfeld and Link, 2014). Yap/Taz proteins were localized to both the cytoplasm and nucleus in embryonic ICM and OCM cells, whereas they were primarily detected in the cytoplasm in embryonic RPE cells (Figure 6A, second and third rows). Post-natal OCM cells showed reduced Yap/Taz expression, whereas ICM cells continued to exhibit strong Yap/Taz expression in the cytoplasm and nucleus (Figure 6E and 6H, leftmost columns).
Figure 6. Yap/Taz haplodeficiency rescues CM hyperplasia caused by Nf2 deficiency.
(A) Sections of mouse eyes at indicated ages were stained with an antibody against Yap/Taz. Single focal plane images are provided to identify subcellular distribution of Yap/Taz. Note that RPE cells show very limited amount of Yap/Taz expression in the nucleus (white arrows), whereas CM cells exhibits expression in the nucleus as well as cytoplasm (yellow arrows). (B) P0 Lats1f/+;Lats2f/+, Lats1f/f;Lats2f/+;MART1-Cre and Lats1f/+;Lats2f/f;MART1-Cre littermate mouse eye sections were stained with antibodies recognizing corresponding markers. Arrows point Yap/Taz in the OCM. (C and D) The numbers of CM cells expressing corresponding markers in the mouse eyes were quantified (n = 4 from 2 independent litters). (E and H) Sections of P0 mouse eyes with indicated genotypes were stained with H&E and antibodies against corresponding markers. The levels of Yap/Taz are higher in the ICM (E, arrowheads in third row) than the OCM of P0 wild-type mouse eyes. Yap/Taz are elevated and strongly accumulated in the nuclei of the OCM and RPE cells in P0 Nf2f/f;TRP1-Cre mice (E, arrows in third row). Yap/Taz were strongly accumulated in the nuclei of the OCM of P0 Nf2f/f;MART1-Cre mice (H, arrows in third row). (F and I) Pax6-positive total CM cells, Otx2-positive OCM cells, and Vsx2-positive ICM cells in P0 mouse eyes with indicated genotypes are quantified and shown in a graph (n = 5 from 2 independent litters). (G and J) Quantification of BrdU;Pax6-positive cells in total CM cells, OCM, and ICM area of P0 mouse eyes with indicated genotypes (n = 5 from 2 independent litters). n.d., not detectable.
Immunostaining intensities of Yap/Taz proteins were remarkably increased in the nuclei of OCM and RPE cells of P0 Nf2f/f;TRP1-Cre and Nf2f/f;MART1-Cre mouse eyes (Figure 6E and 6H, second columns from left), suggesting that hyperactivation of Yap/Taz in these pigmented cells in the absence of Nf2. Yap/Taz were also expressed in Vsx2;Pax6;Sox2-positive ectopic RPCs, which likely arose from Nf2-deficient ICM, in the post-natal Nf2f/f;TRP1-Cre mouse retinas, whereas these proteins were detectable only in Müller glia of littermate Nf2f/+;TRP1-Cre mouse retinas (Figure S3G). These results suggest that Yap/Taz activation, which is likely induced by inactivation of the Hippo pathway, might be responsible for the expansion of CM populations in Nf2-mutant mouse eyes.
To delineate the regulatory roles of the Hippo pathway in the proliferation of pigmented epithelia, we deleted Lats1 and -2 in mouse RPE and OCM. Lats proteins are core kinases of the Hippo pathway that phosphorylates Yap/Taz, resulting in cytoplasmic retention of the corresponding phosphorylated proteins (Halder and Johnson, 2011). Unfortunately, Lats1f/f;Lats2f/f;MART1-Cre mice died in utero (data not shown); thus, we were unable to investigate the effects of complete inactivation of the Hippo pathway on post-natal mouse eye development. Upon immunohistochemical examination of P0 Lats1f/+;Lats2f/f;MART1-Cre mouse eyes, we found that Otx2-positive pigmented population in the OCM of was already enlarged significantly and their BrdU incorporation rate was higher than those in Lats1f/f;Lats2f/+;MART1-cre or Lats1f/+;Lats2f/+ littermates (Figure 6B – 6D), sharing the phenotypes of P0 Nf2f/f;MART1-Cre mice (Figure 4G – 4L). These results therefore suggest that Nf2 might act through the Hippo pathway to restrain pigmented cell proliferation.
Yap/Taz haplodeficiency rescues hyperplasia of Nf2-deficient pigmented epithelium
To test whether the increase in nuclear Yap/Taz in CM cells is responsible for the ocular phenotypes of Nf2-mutant mice, we next co-deleted Yap and Taz together with Nf2 in a combinatorial manner. Heterozygous deletion of Yap and Taz in the CM and RPE of Nf2f/f;TRP1-Cre (Nf2f/f;Yapf/+;Tazf/+;TRP1-Cre) mice caused a remarkable rescue of the ocular phenotypes (Figure 6E – 6G; Figure S4A). Not only the number of Pax6-positive total CM cells, but also the numbers of Otx2-positive pigmented and Vsx2-positive non-pigmented cells in the CM were normalized in P0 Nf2f/f;Yapf/+;Tazf/+;TRP1-Cre mouse eyes (Figure 6E [center column] and 6F; Figure S4A). The number of Pax6;Vsx2;Sox2-positive RPCs in P7 Nf2f/f;Yapf/+;Tazf/+;TRP1-Cre mouse retinas was also remarkably decreased, and each transcription factor was expressed separately in amacrine cells (Pax6), bipolar cells (Vsx2) and Müller glia (Sox2) (Figure S3B – S3D).
However, the complete loss of Yap and Taz in the CM and RPE resulted in ocular malformation that was more severe than that in Nf2f/f;TRP1-Cre mice. The RPE was undetectable in P0 Yapf/f;Tazf/f;TRP1-Cre and Nf2f/f;Yapf/f;Tazf/f;TRP1-Cre mouse eyes, and was transformed into ectopic retina, which failed to form distinct retinal layers (Figure 6E, two right columns; Figure S4A). Strangely, R26R-Cre reporter signals were almost undetectable in those ectopic retinas (Figure S4A, top row), suggesting that Yap/Taz-deficient cells failed to expand and/or to survive during eye development, allowing wild-type cells to expand into the entire optic cup area.
Heterozygous deletion of Yap/Taz in the RPE of Nf2f/f;MART1-Cre mice (Nf2f/f;Yapf/+;Tazf/+;MART1-Cre) also significantly neutralized the hyperplasia of pigmented cells in the OCM and the hypoplasia of the adjacent ICM (Figure 6H – 6J; Figure S4B). However, homozygous deletions of Yap and Taz (Yapf/f;Tazf/f;MART1-Cre and Nf2f/f;Yapf/f;Tazf/f;MART1-Cre) caused hypoplasia of the OCM, with a significant decrease in BrdU-incorporation regardless of their Nf2 genotypes (Figure 6H [bottom row] and 6J). Homozygous deletions of Yap and Taz in the retinas of Nf2f/f;Chx10-Cre mice also caused hypoplasia of the ICM (Figure S5), implicating cell-autonomous growth supportive function of Yap/Taz in the CM. Collectively, our results suggest that Yap/Taz in the neuroepithelia in the OV and early optic cup play a role in specifying and maintaining RPE fate, as suggested by previous reports (Cabochette et al., 2015; Kim et al., 2016; Miesfeld and Link, 2014), and subsequently support the proliferation of CM cells in the optic cup. The inference is that Yap/Taz activity in the optic cup compartments is regulated by the Nf2-Hippo pathway; otherwise, CM cells would persistently proliferate without differentiating to post-mitotic retinal neurons in the inner optic cup and to the post-mitotic RPE in the outer optic cup (Figure 3 and 4; Figure S3E).
Mitf regulates the expression of Nf2 in the RPE
It has been shown that Nf2 is a target of TEAD (TEA domain family member) transcription factors, which tightly interact with Yap/Taz transcriptional co-activators (Dai et al., 2015; Moroishi et al., 2015). Thus, the coincident expression of Yap/Taz and Nf2 in mouse eyes suggests a feedback regulation of Nf2 expression in the CM and RPE by a Yap/Taz-TEAD transcription complex. In support of this idea, expression of Nf2 was lost in the OCM of P0 Yapf/f;Tazf/f;MART1-Cre mice (Figure S4C, left column). However, Nf2 was still maintained in the Yapf/f;Tazf/f;MART1-Cre mouse RPE (Figure S4C, right column). Together with the cytoplasmic localization of Yap/Taz in the RPE (Figure 6A), these results suggest that Nf2 expression in the RPE is regulated in a Yap/Taz-independent manner.
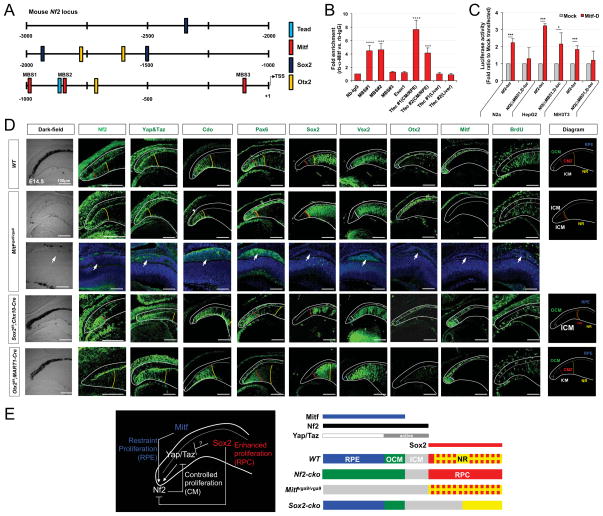
Previous studies have shown that a −2.4-kb sequence of the human NF2 gene can function as a promoter to direct the expression of a β-galactosidase reporter in the outer optic cup layer of mouse embryo (Akhmametyeva et al., 2006), thereby recapitulating endogenous Nf2 expression (Figure 1C). To identify transcription factors responsible for Nf2 gene expression in the mouse CM and RPE, we searched potential transcription factor target sequences within a −2.5-kb upstream region of the mouse Nf2 gene (Figure 7A). Notably, we found that this region contains multiple E-box and M-box sequences, which can be recognized by MITF, a member of the helix-loop-helix transcription factor family (Hemesath et al., 1994). MITF is a key transcription factor supporting RPE fate, and various loss-of-function mutations of human MITF have been shown to result in defects in the development of RPE that ultimately lead to anophthalmia and microphthalmia (Hodgkinson et al., 1993). Mouse Mitf is also expressed primarily in pigmented cell populations of the eye, including the RPE and OCM in the optic cup as well as melanocytes in the choroid (Martinez-Morales et al., 2004), suggesting that Mitf is a potential transcription factor that induces Nf2 expression in the RPE and OCM.
Figure 7. Antagonistic regulation of Nf2 expression by Mitf and Sox2 is necessary for differential growth of optic neuroepithelial compartments.
(A) Schematic distribution of putative transcription factor target sequences within −2.5kb upstream region of mouse Nf2 gene. TSS, transcription start site. (B) ChIP qPCR analyses of adult mouse optic cup devoid of the retina revealed the enrichment of Mitf transcription factor in its target M-box sequences (MBS) in the upstream region of Nf2 gene. The Tfec promoter sequences, which are know as targets of Mitf, were used for positive controls in each ChIP experiment (see details in Methods). (C) N2a, HepG2, and NIH3T3 cell lines were transfected with Nf2_luc reporters alone or together the RPE-enriched Mitf-D isoform and the effects of Mitf on luciferase expression were examined by detecting chemoluminoscence emitted by cell lysates (n=6). (D) Expression of Nf2 and Yap/Taz in P0 WT, Mitfmi-vga9/mi-vga9, Sox2f/f;Chx10-Cre, Otx2f/f;MART1-Cre mouse eyes was examined by immunostaining (second and third columns from the left). Distribution of CM (marked by Pax6), OCM/RPE (marked by Otx2 and Mitf), ICM (marked by Cdo and Vsx2), and RPC (marked by Vsx2 and Sox2) was also examined by immunostaining. Schematic diagrams showing the distribution of optic compartments in each mouse line are given in the rightmost column. Yellow dot-line indicates a border between the retina and ICM; red dot-line indicates a border between the proximal CM and distal CM; white dot-line indicate a border between the ICM and OCM. (E) Schematic diagram depicts regulation of Nf2 expression by transcription factors expressed in each optic neuroepithelial compartment.
To test whether Mitf can bind to the promoter sequences of the Nf2 gene in vivo, we performed chromatin immunoprecipitation (ChIP) assay that isolates Mitf bound to genomic DNA in adult mouse optic cup devoid of cornea, lens, and retina. Quantitative polymerase chain reaction (qPCR) analyses revealed that Mitf bound to M-box sequences (MBSs) starting at −997 bp (MBS1) and −828 bp (MBS2) positions relative to the transcription start site of mouse Nf2 gene (Figure 7B). By contrast, we failed to amplify those M-box sequences in Mitf-ChIP DNA isolated from Mitfmi-vga9/mi-vga9 mouse eyes (data not shown), which do not express any Mitf isotypes (Hodgkinson et al., 1993). We also found that Mitf induces luciferase expression, which is dependent of mouse Nf2 promoter activity, in cultured cell-lines (Figure 7C). However, it induces luciferase expression less effectively from the reporter constructs lacking the MBS1 and MBS2, suggesting that Mitf binds to these M-box sequences and induces Nf2 expression in the pigmented cells in the outer optic cup layer.
However, Nf2 was still detectable in the outer optic cup layer of Mitfmi-vga9/mi-vga9 mouse eyes (Figure 7D, second column from the left), implicating a redundant Nf2 transcription potentially by other transcription factors, including Yap/Taz-TEAD complex. In support of this idea, the cells in the outer optic cup layer of the Mitfmi-vga9/mi-vga9 mice showed elevated nuclear Yap/Taz expression. Interestingly, the cells having the elevated Yap/Taz in the central outer optic cup layer of Mitfmi-vga9/mi-vga9 mice were not maintained in a monolayer (Figure 7D, third row from the top; pointed by arrowheads; Figure S6A). Furthermore, these cells expressed an ICM-specific marker Cdo, but not an OCM/RPE marker Otx2 and a RPC marker Sox2, suggesting RPE-to-ICM fate transformation in Mitfmi-vga9/mi-vga9 mouse eyes. Cells in the OCM of these mouse eyes were also positive for the ICM marker Cdo, implying fate transformation of the entire outer optic cup layer to ICM that expresses Nf2 in a Mitf-independent manner (Figure 7D, second row; Figure S6A and S6B).
Compartment-specific transcriptional regulation of Nf2 in developing mouse eyes
In addition to those M-box sequences and the TEAD target site, the upstream region of the Nf2 gene also contains conserved sequences for various transcription factors expressed in the RPE, CM, and retina. These include target sequences for transcription factors specific for the RPE/OCM (Otx2) and retina (Sox2) (Figure 7A). However, Nf2 was still detectable in the RPE and OCM of Otx2f/f;MART1-Cre mice, suggesting that Otx2 is dispensable for the expression of Nf2 (Figure 7D, bottom row).
Sox2 is expressed in RPC population in the retina and Msx1-positive CMZ, but not in Aqp1-positive cells in the distal ICM (Figure S7). It has also been reported to bind to a target sequence in the mouse Nf2 promoter to suppress the transcription of Nf2 in a murine osteocarcinoma cell-line (Basu-Roy et al., 2015). In support of this idea, deletion of Sox2 in the retinas of Sox2f/f;Chx10-Cre mice resulted in expansion of the Nf2-positive cell population (which co-expresses the ICM marker Cdo) into the retinal territory (Figure 7D, fourth row from the top). The expanded population of ICM cells in the retina also expresses Yap/Taz (Figure 7D), which are mutually exclusive to Sox2 in the retina (Figure S7). These results also suggest that Yap/Taz in the expanded ICM of the Sox2f/f;Chx10-Cre mouse retina might support Nf2 expression in parallel to the absence of Sox2-dependent suppression of Nf2 expression.
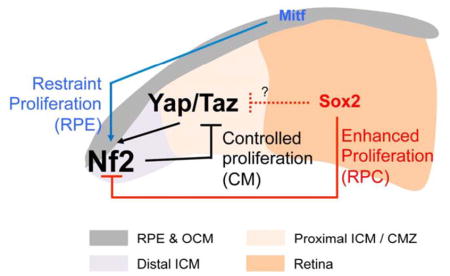
Collectively, our results suggest the following model of region-specific transcriptional regulation of Nf2 in the mouse optic cup: (1) Mitf, but not Yap/Taz and Otx2, might be responsible for the expression of Nf2 in the RPE (Figure 7A – D [second and third rows]), given the low nuclear content of Yap/Taz (Figure 6A) and the presence of Nf2 in Yap/Taz- or Otx2-deficient RPE (Figure 7D; Figure S4C). (2) Mitf might also support the expression of Nf2 in pigmented cells in the OCM. In contrast to the post-mitotic RPE, cells in the OCM maintain their proliferative potential by maintaining Yap/Taz in their nuclei (Figure 6A). Therefore, Yap/Taz and Mitf might act redundantly to induce Nf2 expression in the OCM. (3) Yap/Taz might play a major role in the expression of Nf2 in the ICM, thus Nf2 was accordingly increased in the ectopic Yap/Taz-positive ICM population in the Mitfmi-vga9/mi-vga9 and Sox2f/f;Chx10-Cre mouse eyes (Figure 7D). (4) Sox2 might repress Nf2 expression directly or indirectly by inhibiting Yap/Taz expression in the RPC. In conclusion, the specific regulation of Nf2 expression in three domains – the post-mitotic RPE (positively by Mitf), slowly dividing OCM (positively by Mitf and Yap/Taz) and ICM (positively by Yap/Taz), and rapidly dividing RPC (negatively by Sox2) – may differentiate the growth of each optic neuroepithelium during eye morphogenesis (schematic diagram in Figure 7E).
DISCUSSION
An anti-proliferative function of Nf2 has been described in various progenitor populations, and has been thought to be mediated by the Hippo pathway (Lavado et al., 2013; Serinagaoglu et al., 2015; Zhang et al., 2010). In addition to Nf2, multiple components of the Hippo pathway, including Drosophila expanded (Ex) and the mammalian warts homolog Lats2, have been identified as transcription targets of Yap/Taz (Dai et al., 2015; Moroishi et al., 2015; Park et al., 2016). These observations might establish a mechanism for feedback inhibition of Yap/Taz activity by these Yap/Taz targets that would prevent constitutive proliferation of progenitors. However, the physiological events subjected to such feedback regulation have not been clearly identified.
In this study, we provide several lines of evidence to suggest that an Nf2-Hippo-Yap/Taz feedback circuit governs the limited, but sustained, growth of the CM population, which is a long-lasting multipotent progenitor population in the eye (Belanger et al., 2017; Marcucci et al., 2016). Upon genetic inactivation of the feedback inhibition component Nf2, the PCE and RPE progenitors in the OCM of mouse eyes became hyperproliferative with strong nuclear accumulation of Yap/Taz (Figure 6E and 6H). However, the Nf2-deficient RPE in the central optic cup remained in a post-mitotic state, despite elevated Yap/Taz expression (data not shown), suggesting that Yap/Taz-dependent cell proliferation might be antagonized by other factors enriched in the RPE. Mitf, which also binds to the Nf2 promoter and induces Nf2 expression (Figure 7B and 7C), could be one of such factors, because Mitf-deficient RPE and OCM were transformed into the ICM with elevated Yap/Taz expression (Figure 7D; Figure S6).
The loss of Nf2 in the RPE did not cause as significant a local overgrowth as that observed in the OCM. Instead, RPCs derived from Nf2-deficient ICM expanded themselves rigorously into the retina by virtue of Yap/Taz accumulation, but they did not differentiate into retinal neurons and Müller glia (Figure S3). This made the retina hypoplastic, suggesting that the transition of ICM progenitors to RPCs must be accompanied by the downregulation of Yap/Taz activity to allow proper generation of the retinal repertoire subsequently. Given the elevation of Yap/Taz as well as Nf2 in Sox2-deficient cells in the Sox2f/f;Chx10-Cre mouse retina, Sox2 could be a candidate for inhibiting Yap/Taz during the transition to the RPC fate. However, Sox2 has rather been shown to bind to a promoter sequence in the mouse Yap gene to activate Yap expression in immature osteoblasts (Seo et al., 2013). Thus, Sox2 could suppress Yap expression as part of transcription repressor complexes in RPCs, but might associate with transcription activators to induce Yap in osteoblasts.
Hyperproliferation of Nf2-deficient pigmented cells was observed only in the embryonic Nf2f/f;TRP1-Cre mouse eyes; in contrast, pigmented cells persistently proliferated in the post-natal Nf2f/f;MART1-Cre mouse eyes (Figure 3 and 4). This difference might be related with the developmental competence of pigmented cells. TRP1-Cre affects the primitive RPE as early as E10 before the optic cup forms, whereas MART1-Cre starts to act in post-mitotic cells in the outer optic cup after E12 (Aydin and Beermann, 2011; Mori et al., 2002). TRP1-Cre-active PCE/RPE progenitors in the OCM expressed Yap/Taz in the nucleus as well as the cytoplasm, thereby maintaining their proliferation potential, whereas MART1-Cre-active post-mitotic RPE cells have Yap/Taz mainly in the cytoplasm (Figure 6A). Thus, the TRP1-Cre-active OCM progenitors might have adapted to divide slowly by a Yap/Taz-dependent feedback activation of Hippo pathway. In contrast, MART1-Cre-active post-mitotic OCM/RPE cells might have lost the growth-regulatory circuit centered on Yap/Taz, and thus continue to proliferate during post-natal development upon the loss of Nf2 by virtue of the presence of Yap/Taz in their nuclei. This might be comparable to the difference between RPCs, which have adapted to Yap/Taz-independent rapid cell proliferation, and ICM cells, which divide slowly because of the Nf2-Hippo-Yap/Taz feedback growth-regulating circuit (model diagram in Figure 7E). Collectively, our data indicate that Yap/Taz might not only prevent progenitors from exiting the cell cycle, they also prevent them from transitioning to rapidly dividing cells. This mechanism could be also applied other tissues, which express Nf2 in Yap/Taz in progenitor population as well as post-mitotic cells.
Yap/Taz activity is not solely regulated by Hippo signaling; it is also subjected to many signals that act independent of the Hippo pathway. For instance, non-canonical Wnt signaling induces Yap-dependent expression of canonical Wnt antagonists, dickkopf-related protein 1 (Dkk1) and secreted frizzled-related protein 3 (Sfrp-3), via a Rho GTPase that is activated by Gq/11 (Park et al., 2015). Multiple Wnt ligands are reported to be expressed in embryonic mouse eyes (Fuhrmann, 2008; Liu et al., 2003); these include canonical Wnt2 in the cornea, Wnt2b in the OCM, and Wnt3 in the retina; and non-canonical Wnt5a in the ICM, and Wnt5b and Wnt7a in the lens. Thus, strong Yap/Taz activity in the CM could be induced by these non-canonical Wnt ligands produced by the CM and adjacent lens.
Canonical Wnt signaling activity visualized by a Tcf/Lef transcription factor reporter is also strongest in the CM region (Fuhrmann et al., 2009). Moreover, canonical Wnt signaling has been suggested to play an essential role in the specification of CM and RPE (Cho and Cepko, 2006; Fujimura et al., 2009; Westenskow et al., 2009), as does Yap/Taz (Cabochette et al., 2015; Kim et al., 2016; Miesfeld and Link, 2014). Therefore, the canonical Wnt pathway and Yap/Taz might not simply antagonize each other in vivo, as proposed based on the results in cultured cells, but instead might act cooperatively. Indeed, a feed-forward circuit consisting of Wnt/Wg and Yap/Yki is proposed to play roles in Drosophila wing growth (Zecca and Struhl, 2010). Therefore, the relationship between the canonical Wnt pathway and Yap/Taz in vertebrate development should also be investigated carefully in future studies.
Another important cue that affects CM growth is bone morphogenetic proteins (Bmps). Bmp4 and -7 are strongly expressed in the CM and are necessary for development of the CB (Zhao et al., 2002). The Yap-TEAD complex was shown to induce Bmp4 expression in zebrafish endothelial cells (Uemura et al., 2016), suggesting that Yap/Taz could be a potential regulator of Bmp4 (and -7) expression in the CM. However, inverse effects of Bmp signaling on the nuclear accumulation of Yap/Taz have not yet been demonstrated. In addition to the role of these known morphogens in CB development, various external factors, which act on GPCRs mediating Gq/11 to inactivate Lats2 via Rho or GPCRs mediating Gs to activate Lats1 via PKA (Kim et al., 2013; Yu et al., 2014), might also play roles in Yap/Taz-dependent gene expression in the CM. Combining effects of those external factors might accumulate more Yap/Taz in the nucleus of the CM cells than the RPE and maintain the growth at the CM.
Interestingly, the hyperproliferation of Nf2-deficient ICM and OCM cells was accompanied by hypoproliferation of CM cells in the other layer (Figures 3 and 4). Nf2 maintains structural integrity in several tissue types by establishing junctional complexes at the apical epithelium (Gladden et al., 2010; McClatchey and Giovannini, 2005; McLaughlin et al., 2007a; Wiley et al., 2010). Disruption of the junctional polarities of the CM neuroepithelium was a common feature in P0 Nf2f/f;TRP1-Cre, Nf2f/f;MART1-Cre, and Nf2f/f;Chx10-Cre mice (Figure 5). Adhesion between two apical membranes of the inner and outer CM neuroepithelium is necessary for contact-dependent signaling, which supports the coordinated growth of the inner and outer CM (Napier and Kidson, 2005; Zhou et al., 2013). Thus, the altered polarity and defective intercellular adhesion between those two CM neuroepithelial layers in Nf2-mutant mouse eyes may also affect the intercellular signaling events. For instance, Notch2 activation in OCM/PCE by Jagged-1 (Jag1), expressed in ICM/NPCE, was shown to be necessary for the coordinated growth of the CB in post-natal mouse eyes (Zhou et al., 2013). Given the RPE-to-RPC Notch signaling in the central optic cup (Ha et al., 2017), a potential PCE-to-NPCE Notch signaling might also occur during CB growth. In support of these ideas, levels of the Notch target, Hes1, in wild-type PCE and NPCE, which are adjacent to the Nf2-deficient NPCE of P7 Nf2f/f;Chx10-Cre mouse eyes and PCE of P7 Nf2f/f;Chx10-Cre mouse eyes, were significantly decreased, respectively (data not shown). Therefore, these results suggest that Nf2 might support reciprocal Notch signaling between the two CM layers during post-natal CB growth by maintaining the apical junctional complexes.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Animals
Information of mouse strains used in the experiments is provided in KEY RESOURCES TABLE (above, Experimental Models: Organisms/Strains). To obtain the conditional knockout (cko) mice, the floxed mice were crossed with the mice expressing corresponding Cre recombinases. Cell population underwent Cre-mediated recombination in the cko mouse eyes was tracked by analyzing Cre-dependent β-galactosidase expression after breeding the mice with the R26R mice. All experiments were performed under the authorization of the Institutional Animal Care and Use Committee (IACUC) at KAIST (KA2011-37).
Cell culture and luciferase assay
Mouse fibroblast NIH/3T3 cells, mouse neuroblastoma N2a cells, and human liver cancer HepG2 cells were grown in Dulbecco’s Modified Eagle Medium (DMEM; Invitrogen) supplemented with 10% fetal bovine serum and 1% Penicillin-Streptomycin. The cells were transfected with plasmid DNA encodes human MITF-D together with pGL3-Nf2(-2.5kb) (pNF2_luc) or its deletion variant reporter DNA constructs using GeneJet transfection reagent (Signagene Laboratory). A CMV-β-gal plasmid (Promega) was co-transfected to normalize transfection efficiencies of individual samples. Cells were harvested 48 hours after transfection and luminescence was evaluated as previously described (Kim et al., 2017).
METHOD DETAILS
Immunohistochemistry and microscopy
After anaesthetizing pregnant mice or post-natal mice by intraperitoneal injection of tribromoethanol (Avertin, Sigma), the mice were perfused with 4% paraformaldehyde (PFA) in phosphate buffered saline (PBS, pH7.5), prior to the isolation of the eyes. The embryos were collected and then fixed in 4% PFA/PBS solution at 4°C for 2 hours (h) prior to the cryo-protection in 20% sucrose/PBS solution at 4°C for 1 6 h and subsequent sample freezing in the Tissue-Tek OCT medium (Sakura Finetek). Histologic examination was performed with hematoxylin- and eosin (H&E) staining of tissue sections (10 – 14 μm). For immunohistochemistry, the sections were blocked with 5% normal donkey serum in PBS/TritonX-100 0.2% prior to incubating with appropriate primary antibodies KEY RESOURCES TABLE (above, Antibodies) at 4°C for 16 h. The samples were further stained with Alexa488-, Cy3-, or Alexa647-conjugated secondary antibodies (Jackson ImmunoResearch Laboratories) against the primary antibodies and subsequently were analyzed with confocal microscopy (Olympus FV1000).
For BrdU labeling, pregnant or postnatal mice were injected with 5-bromo-2′-deoxyuridine (BrdU; 30 mg/kg, Sigma) into their peritoneal cavity at 2 h prior to the sample collection. The sections were post-fixed in 4% PFA/PBS for 5 min, and washed three times with PBS with TritonX-100 0.2% before treatment with 2N HCl for 30 min. The samples were then neutralized by immersing in 0.1 M borate buffer (pH 8.0) for 5 min (three times), and then were subjected to the immunohistochemistry procedures using primary anti-BrdU antibody.
Chromatin immunopreciptation (ChIP)
The ChIP assays were performed as described previously with minor modification (Kim et al., 2017). Briefly, 40 mouse eyes were dissected to remove the cornea, lens, and retina. Remained eye cups were incubated in 1.5% formaldehyde in DMEM for 15 mins at room temperature to allowing the cross-linking of DNA and proteins in the optic cells. Sonicated nuclear extracts of the cells were subjected to immunoprecipitation with 2 μg rabbit anti-Mitf IgG for 16 h at 4°C to isolate Mitf and DNA fragmen ts cross-linked to the proteins. Control precipitation was also carried out with 2 μg of pre-immune rabbit IgG for each biological replicate. The immune complexes were then captured by Protein A Sepharose beads (Sigma) and eluted from the beads repeatedly. DNA fragments were released from the immunoprecipitated proteins by de-cross-linking at 65°C for 16 h, and then purified for quantitative PCR (qPCR) analysis using SYBR Green PCR Master Mix (Applied Biosystems). The fold enrichment of each transcription factor was calculated as 2 to difference of cycle threshold (2ΔCt) from pre-immune rabbit IgG. The relative enrichment of each genomic region was measured as the difference from the 2ΔCt value of IgG. The sequence information of the qPCR primers is provided in KEY RESOURCES TABLE (above, Oligonecleotides).
QUANTIFICATION AND STATISTICAL ANALYSIS
Cells stained with each marker in the staining images were counted and quantified by using P rism software (GraphPad, v5.0). Data from statistical analysis were presented as the mean ± standard deviation (SD) or standard error of the mean (SEM). The Student’s t-test was used to determine the significant difference between two genotypes and the analyse s of variance (ANOVA) test was used to determine the significant difference among multiple g enotypes. P-values were calculated using a two-tailed unpaired t-test. P < 0.05 was considered statistically significant. *, P < 0.05; **, P < 0.01; ***, P <0.005; ****, P < 0.001.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse monoclonal Aquaporin 1 (Aqp1) | Novus biological | (NB600-749) |
| Rat monoclonal Bromodeoxyuridine (BrdU) | Abcam | (ab6326) |
| Goat polyclonal Brn3b | Santa Cruz | (sc31987) |
| Rabbit polyclonal Calbindin | Swant | (CB38a) |
| Mouse polyclonal Cdo | R&D Systems | (AF2429) |
| Rabbit polyclonal Glutamine synthase (GS) | Sigma | (G2781) |
| Guinea-pig polyclonal Hes1 | Gift from Dr. Ryuichiro Kageyama (Kyoto University) | |
| Rabbit polyclonal Mitf | Abcam | (ab122982) |
| Goat polyclonal Msx1 | R&D Systems | (AF5045) |
| Rabbit polyclonal NF2 | Sigma | (HPA003097) |
| Rabbit monoclonal Notch2 | Cell Signaling | (#5732) |
| Rabbit polyclonal Otx2 | Abgent | (AP20865c) |
| Rabbit polyclonal Pals1 | Upstate | (07-708) |
| Rabbit polyclonal Pax6 | Biolegend | (PRB-278P) |
| Rabbit polyclonal Protein kinase C-α (PKCα) | Santa Cruz | (sc208) |
| Rabbit polyclonal Recoverin | Chemicon | (AB5585) |
| Goat polyclonal Sox2 | Santa Cruz | (sc17320) |
| Rabbit polyclonal Sox9 | Millipore | (AB5535) |
| Mouse monoclonal Syntaxin | Sigma | (S0664) |
| Mouse monoclonal Tuj1 | Biolegend | (MMS-435P) |
| Sheep polyclonal Vsx2 | Abcam | (ab16142) |
| Rabbit monoclonal Yap/Taz | Cell Signaling | (#8418) |
| Rabbit polyclonal phospho-histone H3 (pH3) | Millipore | (06-570) |
| Rabbit polyclonal β-catenin | Cell Signaling | (#9562) |
| Chicken polyclonal β-galactosidase | Abcam | (ab9361) |
| Bacterial and Virus Strains | ||
| Biological Samples | ||
| Chemicals, Peptides, and Recombinant Proteins | ||
| Avertin(2,2,2-Tribromoethanol) | Sigma | T48402 |
| Normal donkey serum | Jackson Immunoresearch | 017-000-121 |
| Hematoxylin | Sigma | H9627 |
| Eosin Y solution | Sigma | 318906 |
| 5-Bromo-2′-deoxyuridine | Sigma | B5002 |
| Penicillin-Streptomycin | Gibco | 15140122 |
| Normal Rabbit IgG | Santa Cruz | sc-2027 |
| GenJet™ In Vitro DNA Transfection Reagent (Ver. II) | Signagen | SL100489 |
| Critical Commercial Assays | ||
| Deposited Data | ||
| Experimental Models: Cell Lines | ||
| Hep G2 | ATCC | HB-8065 |
| Neuro-2a(N2a) | ATCC | CCL-131 |
| NIH/3T3 | ATCC | CRL-1658 |
| Experimental Models: Organisms/Strains | ||
| Mouse: Nf2f/f | Riken BRC | Stock no.RBRC02344 |
| Mouse: Yapf/f | Provided by Dr. Eric Olson (University of Texas Southwestern Medical Center, Dallas, USA) | (Zhang et al., 2010) |
| Mouse: Tazf/f | Provided by Dr. Eric Olson (University of Texas Southwestern Medical Center, Dallas, USA) | (Acehan et al., 2009) |
| Mouse: Lats1f/f | Provided by Dr. Randy L. Johinson (University of Texas Southwestern Medical Center, Dallas, USA) | (Heallen et al., 2011) |
| Mouse: Lats2f/f | Provided by Dr. Randy L. Johinson (University of Texas Southwestern Medical Center, Dallas, USA) | (Heallen et al., 2011) |
| Mouse: Otx2f/f | Provided by Dr. Thomas Lamonerie (University of Nice Sophia Antipolis, Nice, France) | (Tian et al., 2002) |
| Mouse: Sox2f/f | Jackson laboratory | Stock no. 013093 |
| Mouse: Mitf Vga9/Vga9 | MMRRC | Stock no.009963 |
| Mouse: Nes-Cre | Jackson laboratory | Stock no. 003771 |
| Mouse: TRP1-Cre | Provided by Dr. Pierre Chambon (Institute of Genetics and Molecular and Cellular Biology (IGBMC), Strasbourg, France) | (Mori et al., 2002) |
| Mouse: MART1-Cre | Provided by Dr. Friedrich Beermann (Ecole Polytechnique Fédérale de Lausanne (EPFL), Lausanne, Switzerland) | (Aydin and Beermann, 2011) |
| Mouse: Chx10-Cre | Provided by Dr. Connie Cepko (Harvard Medical School, Boston, USA.) | (Rowan and Cepko, 2004) |
| Oligonucleotides | ||
| Mitf binding site #1 (MBS1) Forward: GCTCCAGTACCATGCATGCCA Reverse: CCAGGCACTGCTGCTGTGAAGA |
This paper | |
| Mitf binding site #2 (MBS2) Forward: CTACAAATCCCCAGAAAGTT Reverse: AAGAATTTTCATTTTACCCTC |
This paper | |
| Mitf binding site #3 (MBS3) Forward: AGTAGGAGTATACTTTAGGAACA Reverse: GAAGCGGGACTTCCGCTTCG |
This paper | |
| Over-NF2 promoter sequence Forward: CCCCTACCAAACAAAACAAC Reverse: CTGGTGTGTCTGAAAACAGC |
This paper | |
| Tfec #1 Forward: GCAATAACGGCATCAATGAGAGG Reverse: TTAAGGAAGAAGAAATGACACAGC |
This paper | |
| Tfec #2 Forward: ACTGAAAGTGGGCAAGGACAG Reverse: TGCCCCATAATAAACATTTTTCC |
This paper | |
| Recombinant DNA | ||
| pEGFP-N1-MITF-D | Addgene | Plasmid #38133 |
| pGL3_basic | Promega | E1751 |
| pSV-β-Galactosidase Control Vector | Promega | E1081 |
| Software and Algorithms | ||
| GraphPad Prism v5.0 | GraphPad software | |
| Fluoview 4.0 | Olympus Corporation | |
| Other | ||
| Tissue-Tek® O.C.T. Compound | Sakura | 4583 |
| Dulbecco Modified Eagle Medium-High glucose | Gibco | 11965084 |
| Protein A-Sepharose® 4B, Fast Flow from Staphylococcus aureus | Sigma | P9424 |
| SYBR Green PCR Master Mix | Applied Biosystems | 4309155 |
| Fetal Bovine Serum | Gibco | 12484010 |
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Jin Woo Kim (jinwookim@kaist.ac.kr).
Supplementary Material
Highlights.
Different parts of the eye neuroepithelium vary in proliferation rates
Nf2 is enriched in slowly proliferating optic neuroepithelial compartments.
Nf2 expression is regulated differentially in each optic compartment.
Nf2 mediates Hippo pathway to inhibit hyperproliferation of optic neuroepithelium.
Acknowledgments
This work was supported by the National Research Foundation of Korea (NRF) grants (NRF-2009-00424 (JWK); NRF-2013M3C7A1056566 (JWK); 2017R1A2B3002862 (JWK); and NRF-2011-0007218 (KHM)) funded by the Korean Ministry of Science and ICT (MSIP) and the National Eye Institute of United States (EY013760, EML).
Footnotes
AUTHOR CONTRIBUTIONS
K.H.M. designed, performed, and analyzed the experiments shown in all figures and supplemental figures, wrote and revised the manuscript. H.T.K., D.L., and M.R. generated the mice and performed immunostaining with the samples collected from the mice. E.M.L. and D.S.L. supervised the phenotype analysis, and commented on the manuscript. J.W.K. (corresponding author) wrote the original draft, and revised the manuscript, conceived and supervised the study, and secured funding for this study.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Acehan D, Khuchua Z, Houtkooper RH, Malhotra A, Kaufman J, Vaz FM, Ren M, Rockman HA, Stokes DL, Schlame M. Distinct effects of tafazzin deletion in differentiated and undifferentiated mitochondria. Mitochondrion. 2009;9:86–95. doi: 10.1016/j.mito.2008.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhmametyeva EM, Mihaylova MM, Luo H, Kharzai S, Welling DB, Chang LS. Regulation of the neurofibromatosis 2 gene promoter expression during embryonic development. Dev Dyn. 2006;235:2771–2785. doi: 10.1002/dvdy.20883. [DOI] [PubMed] [Google Scholar]
- Asthagiri AR, Parry DM, Butman JA, Kim HJ, Tsilou ET, Zhuang Z, Lonser RR. Neurofibromatosis type 2. Lancet. 2009;373:1974–1986. doi: 10.1016/S0140-6736(09)60259-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aydin IT, Beermann F. A mart-1::Cre transgenic line induces recombination in melanocytes and retinal pigment epithelium. Genesis. 2011;49:403–409. doi: 10.1002/dvg.20725. [DOI] [PubMed] [Google Scholar]
- Baser ME, Kluwe L, Mautner VF. Germ-line NF2 mutations and disease severity in neurofibromatosis type 2 patients with retinal abnormalities. Am J Hum Genet. 1999;64:1230–1233. doi: 10.1086/302338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu-Roy U, Bayin NS, Rattanakorn K, Han E, Placantonakis DG, Mansukhani A, Basilico C. Sox2 antagonizes the Hippo pathway to maintain stemness in cancer cells. Nat Comm. 2015;6:6411. doi: 10.1038/ncomms7411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beebe DC. Development of the ciliary body: a brief review. Transac Ophthal Soc the UK. 1986;105:123–130. [PubMed] [Google Scholar]
- Belanger MC, Robert B, Cayouette M. Msx1-Positive Progenitors in the Retinal Ciliary Margin Give Rise to Both Neural and Non-neural Progenies in Mammals. Dev Cell. 2017;40:137–150. doi: 10.1016/j.devcel.2016.11.020. [DOI] [PubMed] [Google Scholar]
- Bretscher A, Edwards K, Fehon RG. ERM proteins and merlin: integrators at the cell cortex. Nat Rev Mol Celll Biol. 2002;3:586–599. doi: 10.1038/nrm882. [DOI] [PubMed] [Google Scholar]
- Cabochette P, Vega-Lopez G, Bitard J, Parain K, Chemouny R, Masson C, Borday C, Hedderich M, Henningfeld KA, Locker M, et al. YAP controls retinal stem cell DNA replication timing and genomic stability. eLife. 2015;4:e08488. doi: 10.7554/eLife.08488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho SH, Cepko CL. Wnt2b/beta-catenin-mediated canonical Wnt signaling determines the peripheral fates of the chick eye. Development. 2006;133:3167–3177. doi: 10.1242/dev.02474. [DOI] [PubMed] [Google Scholar]
- Chow RL, Lang RA. Early eye development in vertebrates. Annu Rev Cell Dev Biol. 2001;17:255–296. doi: 10.1146/annurev.cellbio.17.1.255. [DOI] [PubMed] [Google Scholar]
- Curto M, Cole BK, Lallemand D, Liu CH, McClatchey AI. Contact-dependent inhibition of EGFR signaling by Nf2/Merlin. J Cell Biol. 2007;177:893–903. doi: 10.1083/jcb.200703010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curto M, McClatchey AI. Nf2/Merlin: a coordinator of receptor signalling and intercellular contact. Br J Cancer. 2007;98:256–262. doi: 10.1038/sj.bjc.6604002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai X, Liu H, Shen S, Guo X, Yan H, Ji X, Li L, Huang J, Feng XH, Zhao B. YAP activates the Hippo pathway in a negative feedback loop. Cell Res. 2015;25:1175–1178. doi: 10.1038/cr.2015.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuhrmann S. Wnt signaling in eye organogenesis. Organogenesis. 2008;4:60–67. doi: 10.4161/org.4.2.5850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuhrmann S, Riesenberg AN, Mathiesen AM, Brown EC, Vetter ML, Brown NL. Characterization of a transient TCF/LEF-responsive progenitor population in the embryonic mouse retina. Invest Ophthal Vis Sci. 2009;50:432–440. doi: 10.1167/iovs.08-2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimura N, Taketo MM, Mori M, Korinek V, Kozmik Z. Spatial and temporal regulation of Wnt/β-catenin signaling is essential for development of the retinal pigment epithelium. Dev Biol. 2009;334:31–45. doi: 10.1016/j.ydbio.2009.07.002. [DOI] [PubMed] [Google Scholar]
- Gladden AB, Hebert AM, Schneeberger EE, McClatchey AI. The NF2 tumor suppressor, Merlin, regulates epidermal development through the establishment of a junctional polarity complex. Dev Cell. 2010;19:727–739. doi: 10.1016/j.devcel.2010.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Good WV, Erodsky MC, Edwards MS, Hoyt WF. Bilateral retinal hamartomas in neurofibromatosis type 2. Br J Ophthal. 1991;75:190. doi: 10.1136/bjo.75.3.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha T, Moon KH, Dai L, Hatakeyama J, Yoon K, Park HS, Kong YY, Shimamura K, Kim JW. The Retinal Pigment Epithelium Is a Notch Signaling Niche in the Mouse Retina. Cell Rep. 2017;19:351–363. doi: 10.1016/j.celrep.2017.03.040. [DOI] [PubMed] [Google Scholar]
- Halder G, Johnson RL. Hippo signaling: growth control and beyond. Development. 2011;138:9–22. doi: 10.1242/dev.045500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heallen T, Zhang M, Wang J, Bonilla-Claudio M, Klysik E, Johnson RL, Martin JF. Hippo pathway inhibits Wnt signaling to restrain cardiomyocyte proliferation and heart size. Science. 2011;332:458–461. doi: 10.1126/science.1199010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heavner W, Pevny L. Eye development and retinogenesis. Cold Spring Harbor Persp Biol. 2012:4. doi: 10.1101/cshperspect.a008391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemesath TJ, Steingrimsson E, McGill G, Hansen MJ, Vaught J, Hodgkinson CA, Arnheiter H, Copeland NG, Jenkins NA, Fisher DE. Microphthalmia, a critical factor in melanocyte development, defines a discrete transcription factor family. Genes Dev. 1994;8:2770–2780. doi: 10.1101/gad.8.22.2770. [DOI] [PubMed] [Google Scholar]
- Hodgkinson CA, Moore KJ, Nakayama A, Steingrimsson E, Copeland NG, Jenkins NA, Arnheiter H. Mutations at the mouse microphthalmia locus are associated with defects in a gene encoding a novel basic-helix-loop-helix-zipper protein. Cell. 1993;74:395–404. doi: 10.1016/0092-8674(93)90429-t. [DOI] [PubMed] [Google Scholar]
- Huynh DP, Tran TM, Nechiporuk T, Pulst SM. Expression of neurofibromatosis 2 transcript and gene product during mouse fetal development. Cell Growth Diff. 1996;7:1551–1561. [PubMed] [Google Scholar]
- Kim JY, Park R, Lee JH, Shin J, Nickas J, Kim S, Cho SH. Yap is essential for retinal progenitor cell cycle progression and RPE cell fate acquisition in the developing mouse eye. Dev Biol. 2016;419:336–347. doi: 10.1016/j.ydbio.2016.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M, Kim M, Lee S, Kuninaka S, Saya H, Lee H, Lee S, Lim DS. cAMP/PKA signalling reinforces the LATS-YAP pathway to fully suppress YAP in response to actin cytoskeletal changes. EMBO J. 2013;32:1543–1555. doi: 10.1038/emboj.2013.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Lim S, Ha T, Song YH, Sohn YI, Park DJ, Paik SS, Kim-Kaneyama JR, Song MR, Leung A, et al. The LIM protein complex establishes a retinal circuitry of visual adaptation by regulating Pax6 alpha-enhancer activity. eLife. 2017:6. doi: 10.7554/eLife.21303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavado A, He Y, Paré J, Neale G, Olson EN, Giovannini M, Cao X. Tumor suppressor Nf2 limits expansion of the neural progenitor pool by inhibiting Yap/Taz transcriptional coactivators. Development. 2013;140:3323–3334. doi: 10.1242/dev.096537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Mohamed O, Dufort D, Wallace VA. Characterization of Wnt signaling components and activation of the Wnt canonical pathway in the murine retina. Dev Dyn. 2003;227:323–334. doi: 10.1002/dvdy.10315. [DOI] [PubMed] [Google Scholar]
- Marcucci F, Murcia-Belmonte V, Coca Y, Ferreiro-Galve S, Wang Q, Kuwajima T, Khalid S, Ross ME, Herrera E, Mason C. The ciliary margin zone of the mammalian retina generates retinal ganglion cells. Cell Rep. 2016;17:3153–3164. doi: 10.1016/j.celrep.2016.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Morales JR, Rodrigo I, Bovolenta P. Eye development: a view from the retina pigmented epithelium. Bioessays. 2004;26:766–777. doi: 10.1002/bies.20064. [DOI] [PubMed] [Google Scholar]
- McClatchey AI, Giovannini M. Membrane organization and tumorigenesis—the NF2 tumor suppressor, Merlin. Genes Dev. 2005;19:2265–2277. doi: 10.1101/gad.1335605. [DOI] [PubMed] [Google Scholar]
- McLaughlin ME, Kruger GM, Slocum KL, Crowley D, Michaud NA, Huang J, Magendantz M, Jacks T. The Nf2 tumor suppressor regulates cell–cell adhesion during tissue fusion. Proc Natl Acad Sci U S A. 2007a;104:3261–3266. doi: 10.1073/pnas.0700044104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaughlin ME, Pepin SM, Maccollin M, Choopong P, Lessell S. Ocular pathologic findings of neurofibromatosis type 2. Arch Ophthal. 2007b;125:389–394. doi: 10.1001/archopht.125.3.389. [DOI] [PubMed] [Google Scholar]
- Miesfeld JB, Link BA. Establishment of transgenic lines to monitor and manipulate Yap/Taz-Tead activity in zebrafish reveals both evolutionarily conserved and divergent functions of the Hippo pathway. Mech Dev. 2014;133:177–188. doi: 10.1016/j.mod.2014.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori M, Metzger D, Garnier JM, Chambon P, Mark M. Site-specific somatic mutagenesis in the retinal pigment epithelium. Invest Ophthal Vis Sci. 2002;43:1384–1388. [PubMed] [Google Scholar]
- Moroishi T, Park HW, Qin B, Chen Q, Meng Z, Plouffe SW, Taniguchi K, Yu FX, Karin M, Pan D, et al. A YAP/TAZ-induced feedback mechanism regulates Hippo pathway homeostasis. Genes Dev. 2015;29:1271–1284. doi: 10.1101/gad.262816.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napier HRL, Kidson SH. Proliferation and cell shape changes during ciliary body morphogenesis in the mouse. Developmental Dynamics. 2005;233:213–223. doi: 10.1002/dvdy.20302. [DOI] [PubMed] [Google Scholar]
- Park GS, Oh H, Kim M, Kim T, Johnson RL, Irvine KD, Lim DS. An evolutionarily conserved negative feedback mechanism in the Hippo pathway reflects functional difference between LATS1 and LATS2. Oncotarget. 2016;7:24063–24075. doi: 10.18632/oncotarget.8211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park HW, Kim YC, Yu B, Moroishi T, Mo JS, Plouffe SW, Meng Z, Lin KC, Yu FX, Alexander CM, et al. Alternative Wnt Signaling Activates YAP/TAZ. Cell. 2015;162:780–794. doi: 10.1016/j.cell.2015.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragge NK, Baser ME, Riccardi VM, Falk RE. The ocular presentation of neurofibromatosis 2. Eye. 1997;11:12–18. doi: 10.1038/eye.1997.3. [DOI] [PubMed] [Google Scholar]
- Reese BE. Development of the retina and optic pathway. Vis Res. 2011;51:613–632. doi: 10.1016/j.visres.2010.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowan S, Cepko CL. Genetic analysis of the homeodomain transcription factor Chx10 in the retina using a novel multifunctional BAC transgenic mouse reporter. Dev Biol. 2004;271:388–402. doi: 10.1016/j.ydbio.2004.03.039. [DOI] [PubMed] [Google Scholar]
- Seo E, Basu-Roy U, Gunaratne PH, Coarfa C, Lim DS, Basilico C, Mansukhani A. SOX2 regulates YAP1 to maintain stemness and determine cell fate in the osteo-adipo lineage. Cell Rep. 2013;3:2075–2087. doi: 10.1016/j.celrep.2013.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serinagaoglu Y, Pare J, Giovannini M, Cao X. Nf2-Yap signaling controls the expansion of DRG progenitors and glia during DRG development. Dev Biol. 2015;398:97–109. doi: 10.1016/j.ydbio.2014.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sisk RA, Berrocal AM, Schefler AC, Dubovy SR, Bauer MS. Epiretinal membranes indicate a severe phenotype of neurofibromatosis type 2. Retina. 2010;30:S51–58. doi: 10.1097/IAE.0b013e3181dc58bf. [DOI] [PubMed] [Google Scholar]
- Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- Stamenkovic I, Yu Q. Merlin, a “magic” linker between extracellular cues and intracellular signaling pathways that regulate cell motility, proliferation, and survival. Curr Prot Pept Sci. 2010;11:471–484. doi: 10.2174/138920310791824011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanger BZ. Organ size determination and the limits of regulation. Cell Cycle. 2008;7:318–324. doi: 10.4161/cc.7.3.5348. [DOI] [PubMed] [Google Scholar]
- Tian E, Kimura C, Takeda N, Aizawa S, Matsuo I. Otx2 is required to respond to signals from anterior neural ridge for forebrain specification. Dev Biol. 2002;242:204–223. doi: 10.1006/dbio.2001.0531. [DOI] [PubMed] [Google Scholar]
- Tronche F, Kellendonk C, Kretz O, Gass P, Anlag K, Orban PC, Bock R, Klein R, Schutz G. Disruption of the glucocorticoid receptor gene in the nervous system results in reduced anxiety. Nat Genet. 1999;23:99–103. doi: 10.1038/12703. [DOI] [PubMed] [Google Scholar]
- Uemura M, Nagasawa A, Terai K. Yap/Taz transcriptional activity in endothelial cells promotes intramembranous ossification via the BMP pathway. Sci Rep. 2016;6:27473. doi: 10.1038/srep27473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westenskow P, Piccolo S, Fuhrmann S. Beta-catenin controls differentiation of the retinal pigment epithelium in the mouse optic cup by regulating Mitf and Otx2 expression. Development. 2009;136:2505–2510. doi: 10.1242/dev.032136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiley LA, Dattilo LK, Kang KB, Giovannini M, Beebe DC. The tumor suppressor merlin is required for cell cycle exit, terminal differentiation, and cell polarity in the developing murine lens. Invest Ophthal Vis Sci. 2010;51:3611–3618. doi: 10.1167/iovs.09-4371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu FX, Guan KL. The Hippo pathway: regulators and regulations. Genes Dev. 2013;27:355–371. doi: 10.1101/gad.210773.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu FX, Luo J, Mo JS, Liu G, Kim YC, Meng Z, Zhao L, Peyman G, Ouyang H, Jiang W, et al. Mutant Gq/11 promote uveal melanoma tumorigenesis by activating YAP. Cancer Cell. 2014;25:822–830. doi: 10.1016/j.ccr.2014.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zecca M, Struhl G. A Feed-Forward Circuit Linking Wingless, Fat-Dachsous Signaling, and the Warts-Hippo Pathway to Drosophila Wing Growth. PLoS Biol. 2010;8:e1000386. doi: 10.1371/journal.pbio.1000386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N, Bai H, David KK, Dong J, Zheng Y, Cai J, Giovannini M, Liu P, Anders RA, Pan D. The Merlin/NF2 tumor suppressor functions through the YAP oncoprotein to regulate tissue homeostasis in mammals. Dev Cell. 2010;19:27–38. doi: 10.1016/j.devcel.2010.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S, Chen Q, Hung FC, Overbeek PA. BMP signaling is required for development of the ciliary body. Development. 2002;129:4435–4442. doi: 10.1242/dev.129.19.4435. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Tanzie C, Yan Z, Chen S, Duncan M, Gaudenz K, Li H, Seidel C, Lewis B, Moran A, et al. Notch2 regulates BMP signaling and epithelial morphogenesis in the ciliary body of the mouse eye. Proc Natl Acad Sci U S A. 2013;110:8966–8971. doi: 10.1073/pnas.1218145110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu C, Li L, Zhao B. The regulation and function of YAP transcription co-activator. Acta Bioch Biophys Sin. 2015;47:16–28. doi: 10.1093/abbs/gmu110. [DOI] [PubMed] [Google Scholar]
- Zhu L, Skoultchi AI. Coordinating cell proliferation and differentiation. Curr Opin Genet Dev. 2001;11:91–97. doi: 10.1016/s0959-437x(00)00162-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.