Abstract
Immunity against microbes depends on recognition of pathogen-associated molecular patterns by innate receptors. Signaling pathways triggered by Brucella abortus DNA involves TLR9, AIM2 and STING. In this study, we observed by microarray analysis that several type I IFN associated genes, such as IFN-β and guanylate-binding proteins (GBPs), are down-regulated in STING knockout (KO) macrophages infected with Brucella or transfected with DNA. Additionally, we determined that STING and cGAS are important to engage the type I IFN pathway but only STING is required to induce IL-1β secretion, caspase-1 activation and GBP2 and GBP3 expression. Further, we determined that STING but not cGAS is critical for host protection against Brucella infection in macrophages and in vivo. This study provides evidence of a cGAS-independent mechanism of STING-mediated protection against an intracellular bacterial infection. Additionally, infected IRF-1 and IFNAR KO macrophages had reduced GBP2 and GBP3 expression and these cells were more permissive to Brucella replication compared to wild-type control macrophages. Since GBPs are critical to target vacuolar bacteria, we determined whether GBP2 and GBPchr3 affect Brucella control in vivo. GBPchr3 but not GBP2 KO mice were more susceptible to bacterial infection, and siRNA treated macrophages showed reduction in IL-1β secretion and caspase-1 activation. Finally, we also demonstrated that Brucella DNA co-localizes with AIM2, and AIM2 KO mice are less resistant to B. abortus infection. In conclusion, these findings suggest that the STING-dependent type I IFN pathway is critical for the GBP-mediated release of Brucella DNA into the cytosol and subsequent activation of AIM2.
Introduction
The innate immune system is important as the first line of defense to sense invading pathogens (1). Nucleic acids represent critical pathogen signatures that trigger a host proinflammatory immune response (2). Much progress has been made in understanding how DNA and RNA trigger host defense countermeasures; however, several aspects of how cytosolic nucleic acids are sensed remain unclear. Microbial nucleic acids commonly find their way into subcellular compartments of the infected cells to be sensed by innate receptors. Several DNA sensors have been identified that recognize cytosolic DNA. These include RNA polymerase III (3), DNA-dependent activator of IFN-regulatory factors (4), Lrrfip1 (5), Ifi204 (6), Mre11 (7), Ddx41 (8), LSm14A (9), AIM2 (10), among others. The large numbers of DNA sensors identified in the host suggest that they may play redundant roles during infection.
One important DNA sensor is STING, a signaling molecule associated with the endoplasmic reticulum that is critical to control the transcription of numerous host defense genes, including type I interferons in response to invading DNA viruses, bacteria or transfected DNA (11). STING leads to activation of TBK1, which phosphorylates IRF3, a transcription factor required for the induction of IFN-β expression (12). Actually, STING functions as a direct sensor of bacterial-derived cyclic dinucleotides (CDNs) as well as an adaptor molecule in DNA recognition (13). Recent studies have demonstrated that cytosolic DNA species trigger STING activation after binding to the enzyme cyclic GMP-AMP synthase (cGAS) generating the second-messenger cyclic GMP-AMP (cGAMP) (14, 15). CDNs produced by cGAS bind to STING in the ER inducing a conformational change in the molecule that leads to relocation of STING and TBK1 to the perinuclear region of the cell resulting in activation (16).
During intracellular bacterial infections, activation of STING can be accomplished via two different mechanisms. First, STING can directly recognize bacterial CDNs and thus function as a primary pattern recognition receptor (PRR) (13). Alternatively, DNA sensing via cGAS triggers the synthesis of cGAMP which then engages STING as a secondary receptor (15). Listeria monocytogenes actively secretes the bacterial second messenger c-di-AMP that binds directly to STING and activates the production of IFN-β in mice (17). However, in contrast to L. monocytogenes, Mycobacterium tuberculosis, Legionella pneumophila and Chlamydia trachomatis appear to activate this same STING-dependent pathway but via the DNA sensor cGAS (18, 19).
Brucella abortus is a Gram-negative facultative intracellular bacterium, that causes brucellosis, with pathological manifestations of arthritis, endocarditis, and meningitis in humans, and in cattle it leads to abortion and infertility resulting in serious economic losses to the livestock industry (20). This pathogen resists killing by neutrophils, and replicates inside macrophages and dendritic cells (DCs) maintaining a long lasting interaction with host cells. Recently, we have identified Brucella DNA as a major bacterial component that induces type I IFN and IL-1β. Our study revealed that Brucella DNA operates through a mechanism dependent on STING and AIM2 (21, 22). However, currently it is unclear whether cGAS participates as a Brucella DNA sensor and what the contribution of cGAS and STING are in protective immunity and possible cooperation with AIM2.
In this study, we demonstrate that the DNA sensor STING detects B. abortus infection and trigger a type I IFN response and IRF-1-dependent signaling cascade leading to the upregulation of several genes including the GBPs. Furthermore, GBPs were critical to induce inflammasome activation and IL-1β secretion, and GBPchr3 KO mice were more susceptible to Brucella infection compared to wild-type animals. Importantly, cGAS and STING were required to induce type I IFN responses; however, STING but not cGAS played the major role in controlling bacterial infection in macrophages and in vivo.
Materials and Methods
Mice
Wild-type C57BL/6 and 129 Sv/Ev mice were purchased from the Federal University of Minas Gerais (UFMG). STING−/−, cGAS−/−, AIM2−/−, MAVS−/−, GBP2−/−, GBPchr3−/−, IRF-1−/− and IFNAR−/− mice were described previously (10, 11, 23–26). The animals were maintained at UFMG and used at 6–8 weeks of age. All animal experiments were preapproved by the Institutional Animal Care and Use Committee of UFMG (CETEA #128/2014).
Bacterial strains
Bacteria used in this study included B. abortus virulent strain S2308 obtained from our laboratory collection and a variant that constitutively expresses GFP (Brucella-GFP). The Brucella c-di-GMP guanylate cyclase mutant strain (Δ1520) was generated in our laboratory using the constructs previously described (27). Before being used for cell infection or DNA extraction, bacteria were grown in Brucella broth medium (BD Pharmingen, San Diego, CA) for 3 days at 37˚C under constant agitation.
Measurement of c-di-GMP in Brucella wild-type and mutant strain
To measure relative levels of intracellular c-di-GMP, a luciferase (lux) reporter vector containing the Vc2 riboswitch was used as previously demonstrated (28). Briefly, the c-di-GMP responsive riboswitch was cloned from 348bp upstream of the Vibrio cholerae VC1722 gene through the VC1722 start codon and inserted into pBBR1MCS-4 (GenBank U25060), upstream of a promoterless luxCDABE operon. To confirm the levels of c-di-GMP produced, the parental wild-type strain and Δ1520 mutant were transformed with this plasmid containing the c-di-GMP-responsive riboswitch driving lux expression. Bioluminescence was assayed on a Veritas Microplate Luminometer 100 (Promega) 24 hrs post-transfection.
Additionally, we used a chemical c-di-GMP inhibitor, termed Ebselen. Mouse (C57 BL/6) bone marrow-derived macrophages (BMDMs) were plated on 6-well tissue culture plates at 5 × 105/well in 2ml/well culture media (RPMI + 10% FBS) and cultured overnight at 37°C in a 5% CO2 incubator. The next day, cells were pretreated with Ebselen (50μM) for 30 min, then they were uninfected (medium) or infected with 100 MOI of B. abortus (strain 2308) for 24 hrs in the presence or absence of Ebselen. Supernatant was then harvested and assayed for mouse IFN-β using the LEGEND MAX™ Mouse IFN-β ELISA kit (BioLegend) following the manufacturer’s instructions.
Cell culture
BMDMs were generated and cultured in DMEM medium as described (22). Briefly, bone marrow cells were differentiated for 10 days in DMEM (Gibco) containing 10% FBS (HyClone, USA), 1% HEPES (Gibco), 1% penicillin G sodium (100 U/ml), and streptomycin sulfate (100 mg/ml), 10% L929 cell-conditioned medium (LCCM), as the source of M-CSF for macrophages at 37°C in a 5% CO2. A day prior to stimulation of infection, macrophages were harvested and seeded onto 24 well plates at the density of 5 × 105 cells/well (for cytokine and western blot analysis) or 1 × 105 cells/well over a sterile coverslip (for microscopy analysis). BMDMs were infected with B. abortus virulent strain 2308 or B. abortus Δ1520 with indicated MOI (see figure legends). WT and STING−/− murine embryonic fibroblasts (MEFs) were provided by Dr Glen N. Barber (University of Miami, Florida, USA). Cells were maintained in high-glucose DMEM (Gibco) supplemented with 10% fetal bovine serum (Gibco), 10 mmol/L glutamine, 100 units/mL penicillin, and 100 μg/mL streptomycin (Life Technologies Invitrogen) at 37°C in 5% CO2/95% air in a humidified incubator. MEFs were seeded on 24-well plates containing sterile cover-slips at 1 × 104 cells/well a day before the experiment, and kept on normal growth medium.
Purification of B. abortus DNA and transfection experiments
B. abortus were grown for 3 days at 37°C under constant agitation and the DNA was purified using the Illustra bacteria genomic Prep Mini Spin Kit (GE Healthcare) according to the manufacturer’s instructions. Transient transfections of BMDMs and MEFs were carried out using Lipofectamine 2000 (Invitrogen) ratio (in ml) of 1:0.25, following the manufacturer’s directions. Cells were cultured in DMEM and transfected with Brucella DNA (1 μg/well), dsDNA90 (3 μg/mL) and 2′,3′-cGAMP (3 μg/well) (Invivogen).
In vivo and in vitro infection with B. abortus
Mice were infected via intraperitoneal (i.p.) injection of 1 × 106 CFU of virulent B. abortus strain S2308. Animals were kept for different periods of time and then killed for CFU counting and pathology. Cultured cells were infected in vitro with virulent B. abortus strain 2308, B. abortus-GFP, or B. abortus Δ1520 in DMEM supplemented with 1% FBS (macrophages) or 10% FBS (MEFs). Different MOI of bacteria were used as challenge for different analysis, as described below.
B. abortus counts in spleens
Five mice from each group of C57BL/6, STING−/−, cGAS−/−, AIM2−/−, GBP2−/− and GBPchr3−/− were infected with B. abortus as described above and killed at mentioned time intervals. To count residual Brucella CFU, the spleen collected from each animal was macerated in 10 ml saline, serially diluted, and plated in duplicate on Brucella Broth agar. After 3 days of incubation at 37 ° C, the number of CFU was determined as described previously (29).
Cytokine analysis
BMDMs were stimulated by Brucella infection (MOI 100:1) or DNA transfection as described above. Where indicated, cells were treated with 100 U/ml of IFN-β 18 hrs prior the course of infection or were untreated. After 17 hrs, supernatants from cell culture were harvested and assayed for the production of murine IL-1β, CXCL10/IP-10, IL-6 and TNF-α by ELISA (R&D Systems), in accordance with the manufacturer’s instructions. Human CXCL10 was measured in the supernatants of hTERT transfected cells by ELISA (R&D Systems), in accordance with the manufacturer’s instructions.
Western blot analysis
BMDMs were lysed with M-PER Protein Extraction Reagent (Thermo Scientific) supplemented with 1:100 protease inhibitor mixture (Sigma-Aldrich). Equal amounts of protein were loaded onto 12% SDS polyacrylamide gel, transferred to nitrocellulose membranes (Amersham Biosciences) and were blocked 1 hr at room temperature with TBS containing 0.1% Tween-20 and 5% nonfat dry milk. The following primary antibodies were incubated overnight at 4°C: rabbit mAb IRF-1 #8478 (Cell Signaling); rabbit mAb β-Actin #4970 (Cell Signaling) anti-Caspase-1 (p20, mouse mAb #AG-20B-0042, Adipogen) and IL-1β (mouse mAb #3A6, Cell Signaling). For testing siRNA knock down efficiency on hTERT cells, similar procedure was performed using rabbit anti-STING polyclonal antibody 1:5000 and rabbit anti-cGAS (#D1D3G, Cell Signaling) 1:1000. Subsequently, membranes were incubated for 1 hr at room temperature with anti-rabbit IgG HRP-conjugated (#7074, Cell Signaling) or anti-mouse IgG HRP-conjugated (Cell Signaling) antibodies. Proteins were visualized using Luminol chemiluminescent HRP substrate (EMD Millipore) in an Amersham Imager 600 (GE Healthcare).
Gene Array Analysis
Transcripts were profiled for Brucella-infected and bacterial DNA-transfected BMDMs from STING KO and C57BL/6 mice. Total RNA was isolated from BMDMs with the RNeasy RNA extraction kit (QIAGEN) and analyzed by Bioanalyzer RNA 6000 Nano (Agilent Technologies). Gene array analysis was examined by Illumina Sentrix BeadChip Array (Mouse WG6 version 2 for RNA extracted from BMDM) (Affymetrix) at the Oncogenomics Core Facility, University of Miami. Microarray data based on Affymetrix Mouse 2.0 ST platform were normalized using the Robust MultiChip Averaging (RMA) algorithm as implemented in the Bioconductor package Affy. The probes were annotated using Bioconductor annotation package mogene20sttranscriptcluster.db. Fold change was used to compare each pair of microarray samples. The heatmap was generated by R package ggplot2. Microarray analysis was performed at the Center of Computational Science, University of Miami, Florida, USA. GEO accession number is GSE96071(https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=uvcjkweipvyzhyb&acc=GSE96071).
Real-time RT-PCR
RNA was extracted from BMDMs with TRIzol reagent (Invitrogen) to isolate total RNA in accordance with the manufacturer’s instructions. Reverse transcription of 2μg of total RNA was performed using Illustra Ready-To-Go RT-PCR Beads (GE Healthcare) according to the manufacturer’s directions. Real-time RT-PCR was performed using 2x SYBR Green PCR Master Mix (Applied Biosystems) on ABI 7900 real-time PCR instrument (Applied Biosystems). The appropriate primers were used to amplify a specific fragment corresponding to specific gene targets as follows: β-actin F: 5′-GGC TGT ATT CCC CTC CAT CG-3′, β-actin R: 5′-CCA GTT GGT AAC AAT GCC ATG T-3′, IFN-β F: 5′-GCC TTT GCC ATC CAA GAG ATG C-3′, IFN-β R: 5′-ACA CTG TCT GCT GGT GGA GTT C-3′, CXCL10 F: 5′-CCTGCCCACGTGTTGAGAT-3′, CXCL10 R: 5′-TGATGGTCTTAGATTCCGGATTC-3′, GBP2 F: 5′-CTG CAC TAT GTG ACG GAG CTA-3′, GBP2 R: 5′-CGG AAT CGT CTA CCC CAC TC-3′, GBP3 F: 5′-CTG ACA GTA AAT CTG GAA GCC AT-3′, GBP3 R: 5′-CCG TCC TGC AAG ACG ATT CA-3′, GBP4 F: 5′-GGA GAA GCT AAC GAA GGA ACA A-3′, GBP4 R: 5′-TTC CAC AAG GGA ATC ACC ATT TT-3′, GBP5 F: 5′-CTG AAC TCA GAT TTT GTG CAG GA-3′, GBP5 R: 5′-CAT CGA CAT AAG TCA GCA CCA G-3′, IRGB10 F: 5′-TAA TGC CCT TCG GGG AAT AGG-3′, IRGB10 R: 5′-CTG GTT TGA AGT TAG TTG TCC CA-3, MX1 F: 5′-GGGGAGGAAATAGAGAAAATGAT-3′, Mx1 R: 5′-GTTTACAAAGGGCTTGCTTGCT-3′, PYDC3 F: 5′GCCTGATGGAAGCTTGGGAA-3′, PYDC3 R: 5′CTGGGGAGTCAGTGGTTCAC-3′, PYHIN1 F: 5′TCTGGACCCTCCAGTGTCTT-3′, PYHIN1 R: 5′ACCTTGCTGGTGACCATTTT-3′, CXCL11 F: 5′-AGGAAGGTCACAGCCATAGC-3′, CXCL11 R: 5′-CGATCTCTGCCATTTTGACG-3′, TNFSF10 F: 5′-CAACGAGCTGAAGCAGAT-3′, TNFSF10 R: 5′-GGGTCCCAATAACTGTCATC-3′, NOS2 F: 5′-AGC ACT TTG GGT GAC CAC CAG GA-3′, NOS2 R: 5′-AGC TAA GTA TTA GAG CGG CGG CA-3′, ARG1 F: 5′-TGA CAT CAA CAC TCC CCT GAC AAC-3′, ARG1 R: 5′-GCC TTT TCT TCC TTC CCA GCA G-3′. All data are presented as relative expression units after normalization to the β-actin gene, and measurements were conducted in triplicate.
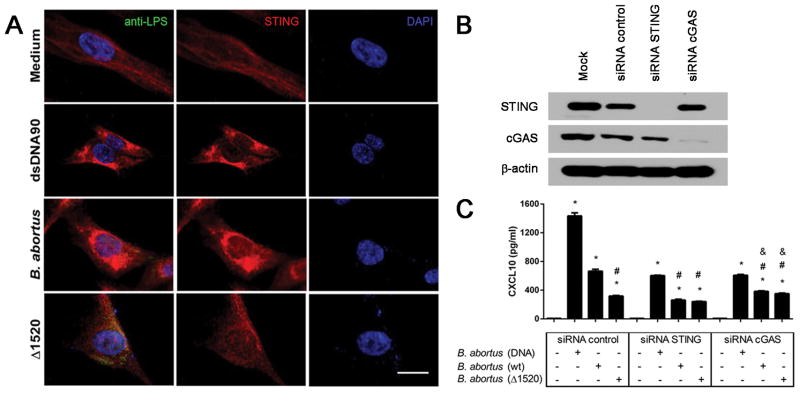
Knockdown via small interfering RNA
BMDMs were transfected with small interfering RNA (siRNA) from siGENOME smart pools (Dharmacon) with the GenMute siRNA Transfection Reagent according to the manufacturer’s instructions (SignaGen). siGENOME SMARTpool siRNA specific for mouse GBP2 (M-040199-00-0005), GBP3 (M-063076-01-0005), GBP5 (M-054703-01-0005), were used in this study. A control siRNA pool was used (D-001206-14-05). Forty-eight hrs after transfection, cells were infected with B. abortus (MOI 100:1) for 24 hrs as described above. hTERT cells were transfected with small interfering RNA (siRNA) from siGENOME smart pools siRNA STING (J-024333-20-0002), siRNA cGAS (L-015607-02-0005) or siRNA Control (D-001810-10-05) at 80μM siRNA and 1μl Lipofectamine RNAiMAX (ThermoFisher) in 100 μl of Opti-MEM media. Culture medium was replaced 48 hrs after transfection with DMEM supplemented with 10% heat-inactivated fetal bovine serum, pen-strep, 20% 199 media, sodium pyruvate, L-glutamine solution and the cells were incubated for additional 24 hrs. After 72 hours of the transfection process, hTERT cells were infected with B. abortus or mutant strain Δ1520 (MOI 100) or transfected with bacterial DNA (1μg) and the supernatant was collected after 17 hours to measure human CXCL10 by ELISA.
Labeling of Brucella DNA in macrophages
Click-iT EdU Imaging kit (Molecular Probes) was used to provide specific labeling of bacterial DNA in infected macrophages. Brucella was grown as described above and Edu solution was added (20μM) to the growth medium for 6 hrs before macrophage infection. Bacteria were then washed 3 times in PBS before being added to macrophages to prevent carryover of unbound Edu. Detection of incorporated Edu with Alexa Fluor 488 was performed in 24 hrs infected macrophages following manufacturer’s instructions (see below). Additionally, Prolong Gold with DAPI mounting medium (Invitrogen) was used to label eukaryotic as well bacterial DNA in all slides prepared for microscopy.
Immunofluorescence and microscopy analysis
Intracellular localization of AIM2, STING, GBP2, IRF3 and NF-κB-p65 was analyzed by immunofluorescence in macrophages or MEFs infected with Brucella-GFP, or transfected with Brucella DNA as follows: a) localization of STING and nuclear translocation of transcription factors was analysed in MEFs infected as described above with Brucella-GFP (MOI 1000:1) or transfected with Brucella DNA for 1, 2, 3 and 4 hours.; b) intracellular localization of AIM2 was analyzed in macrophages infected with Edu-incorporated Brucella (MOI 100:1) for 24 hrs; c) intracellular localization of GBP2 was analyzed in macrophages infected with Brucella-GFP (MOI 100:1) for 24 hours. At specific times, cells were washed twice with phosphate-buffered saline (PBS) and fixed in 4% paraformaldehyde, pH 7.4, at room temperature for 30 min. After fixation, coverslips were washed 3X with PBS and kept at 4°C until immunofluorescence was performed. Permeabilization was done in PBS containing 0.3% Triton X-100 for 15 min, and cells were subsequently blocked for 1 hr with 1% BSA in PBS at room temperature prior to incubation with anti-IRF3, anti-NF-κB-p65, anti-GBP2 or anti-AIM2 primary antibodies at 4°C overnight. Anti-rabbit conjugated with AlexaFluor-546 was used for detection of primary antibodies. Coverslips were mounted in slides using Prolong Gold with DAPI mounting medium (Invitrogen). Confocal microscopy analysis was performed in a Zeiss 880 confocal system. Three coverslips were analyzed per sample and photographs were taken using a 40X objective for 6 random areas of each coverslip providing information for an average of 100 cells/coverslip. The differences in intensity of anti-GBP2 labelling in infected versus uninfected macrophages were quantified to assess alterations in GBP2 intracellular localization. Briefly, the intensity of grey levels (pseudo coloured in red in the image) was measured using ImageJ from anti-GBP2 images in three circular areas of 3μm diameter for each cell. This was done to obtain an estimation of mean intensity of GBP2 staining in at least 9μm2 of each cell. Regions of brightest antibody signal on each cell (perinuclear regions) were preferentially chosen. Antibodies were purchased from the following sources: anti-GBP2 (Proteintech), anti-IRF3 (FL-425) (Santa Cruz Biotechnology), anti-NF-κB-p65 (Cell signaling), anti-AIM2 (Santa Cruz Biotechnology). Anti-mouse and anti-rabbit secondary antibodies conjugated with AlexaFluor-488 or AlexaFluor-546 were purchased from Jackson Immuno Research. Rabbit polyclonal antibody against STING was described previously (11). For evaluation of STING activation by Brucella abortus mutant strain Δ1520, MEFs or hTERT cells were infected with Brucella abortus S2308 or Brucella abortus Δ1520 (MOI 1000:1) or transfected with dsDNA90 (3μg/ml) or cGAMP (1μg/well) for 4 hrs. Cells were processed for immunofluorescence as described above and incubated with anti-STING or anti-Brucella LPS (1:100) antibodies for bacterial staining. Coverslips were mounted in slides using Prolong Gold with DAPI mounting medium (Invitrogen) and microscopy analysis was performed like described above.
Estimation of intracellular Brucella numbers by confocal microscopy
BMDMs were infected as described above with Brucella-GFP (MOI 10:1). Such lower bacterial load at the beginning of infection ensured that macrophages from more susceptible mouse strains would not have an overgrowth of intracellular Brucella and could be analyzed. Bacteria number was assessed in cells infected for 24, 48 and 72 hrs. Cells infected for only 24 hrs were kept in the absence of antibiotics during the whole experiment. After the first 24 hrs, gentamicin (10μg/ml) was added to the medium of cells infected for longer periods to prevent secondary infections. Fixation and permeabilization of the cells were performed as for immunofluorescence. Staining of the actin cytoskeleton with Rhodamine-phalloidin (0,04μM in PBT, ThermoFisher) was performed to visualize cell shape. Preparation of slides and acquisition of microscopy data was performed as described for immunofluorescence. Counts of intracellular bacteria were performed manually by visualization of individual GFP-expressing Brucella.
Measurement of Brucella CFU in macrophages
For the measurement of viable intracellular bacteria using colony forming units (CFUs), after transfection with siRNA and infection with Brucella abortus strain 2308 or B. abortus Δ1520 at different MOIs (see figure legends), cells were washed twice with PBS then lysed for 10 min at room temperature in 800 μl of PBS containing 0.1% Triton X-100 under manual agitation. Lysates were diluted from 10 to 1,000 times in PBS and plated on Petri dishes containing Brucella Broth Agar. Petri dishes were incubated for three to four days at 37°C before CFU counting.
Transmission Electron Microscopy of infected macrophages
BMDMs (1×106) from C57BL/6 and GPBcrh3 KO mice were derived as described previously, infected with B. abortus (MOI 100:1) for 6 hrs at 37°C and washed three times with phosphate buffer (1 M). The cells were then fixed with glutaraldehyde (2.5% in 1 M phosphate buffer) for 24 hours at 4°C, washed three times with phosphate buffer (1 M) and the samples were sent to the Microscopy Center at UFMG for dehydration, treatment with osmium tetroxide and uranyl acetate and Transmission Electron Microscopy (TEM) analysis. To evaluate the percentage of ruptured BCV membranes, we evaluated 30 macrophages per group. Each macrophage was evaluated in relation to the total number of bacteria in 14500x magnification. Then, the membrane integrity of each BCV was carefully evaluated in a higher magnification (60000x). After analysis, we calculate the percentage of ruptured BCVs in relation to the total number of bacteria counted.
Flow cytometry analysis
C57BL/6, STING−/− and cGAS−/−mice were infected with B. abortus 1×106 CFU. Seven days after infection, the spleen cells were harvested and washed twice with sterile PBS. After washing, the cells were adjusted to 1×106 cells in RPMI medium supplemented with 10% fetal bovine serum, 150 U penicillin G sodium and 150 μg streptomycin sulfate per well in a 96-well plate. All cells were stimulated with ConA (5 μg/mL) and 1 μg of brefeldin A was added per well, and the cultures were incubated at 37°C for 4 hrs. After the incubation period, the cells were centrifuged at 1,500 rpm for 7 min at 4°C and washed with PBS containing 1% bovine serum albumin (PBS/BSA). Then, the cells were incubated with anti-CD16/CD32 (FcBlock) (1:30 diluted in PBS/BSA) for 20 min at 4°C, washed in PBS/BSA and incubated for 20 min at 4°C with a mixture of the following antibodies: hamster IgG anti-murine CD3 conjugated to biotin (clone 500A2; 1:200) and rat IgG2b anti-murine CD4 conjugated to APC-Cy7 (clone GK 1.5; 1:200). All antibodies were obtained from BD Bioscience. After that, splenocytes were washed again with PBS/BSA and incubated with streptavidin conjugated to PE-Cy5.5 (1:30) for 20 min at 4°C, and fixed and permeabilized using BD Cytofix/Cytoperm™ reagent (BD Bioscience, San Diego, CA, USA) according to the manufacturer’s instructions. The cells were then incubated with rat IgG1 anti-murine IFN-γ conjugated to PE (clone XMG1.2; 1:30; BD) or with rat IgG1 anti-murine IL-4 conjugated to PE (clone 11B11; 1:30; BD) 30 min at 4°C. Finally, the cells were washed three times, suspended in PBS buffer and evaluated using Attune Acoustic Focusing® equipment (Life Technologies, Carlsbad, CA, USA). The results were analyzed using FlowJo software (Tree Star, Ashland, OR, USA).
Statistical analysis
The results of this study were analyzed using ANOVA, as indicated, with GraphPad Prism 5 computer software (GraphPad Software). The level of significance in the analysis was p < 0.05 (*), p < 0.01 (**) or p < 0.001 (***).
Results
STING is critical for Brucella DNA-mediated innate immune signaling
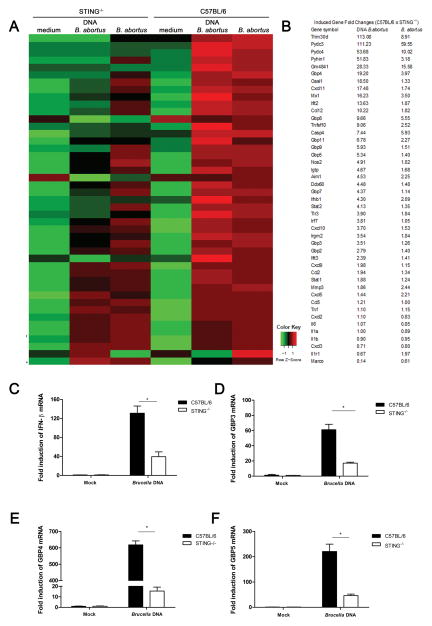
To test the role of STING during B. abortus infection in vitro, primary murine bone marrow-derived macrophages (BMDMs) were isolated from STING KO and C57BL/6 mice. These cells were infected with virulent Brucella abortus S2308 or transfected with B. abortus S2308 DNA and microarray analysis performed. Microarray analysis revealed that several type I IFN related genes are down-regulated in STING KO cells either infected with Brucella or transfected with bacterial DNA as observed in Figure 1A and 1B. To confirm that the induction of these type I IFN related mRNAs were STING-dependent genes, we performed qPCR analysis. Macrophages from C57BL/6 transfected with Brucella DNA robustly activate an array of genes, including IFN-β, guanylate-binding protein 3 (GBP3), GBP4 and GBP5 (Figure 1 C–F). However, the expression of these genes were dramatically reduced in STING KO cells. GBPs are known to co-localize with vacuolar bacterial pathogens such as S. typhimurium and Mycobacterium bovis and to recruit anti-microbial effector mechanisms (30, 31). Additionally, other STING-dependent genes were also validated by qPCR such as CXCL11, Mx1, TNFSF10, PYDC3 and PYHIN1 (Supplementary Figure 1). Taken together, our data demonstrated that both Brucella infection and bacterial DNA induces the expression of several innate immune genes in a STING-dependent fashion.
Figure 1. STING-mediated Brucella-induced innate immune activation gene profile.
BMDM C57BL/6 and STING−/− were transfected with bacterial genomic DNA (1 μg/well) or infected with B. abortus strain 2308 (MOI 100:1) for 17 hrs. (A) Total RNA was purified and examined for gene expression by Illumina Sentrix BeadChip Array (Mouse WG6 version 2). Highest variable genes were selected. Rows represent individual genes; columns represent individual samples. Pseudo-colors indicate transcript levels below, equal to, or above the mean (green, black, and red, respectively). The scale represents the intensity of gene expression (log10 scale ranges between −1 and 1). (B) Fold-change values of the highest-variable genes analyzed by microarray are shown. qPCR analysis of BMDMs from STING−/− transfected with Brucella DNA compared to wild-type macrophages for the following genes: (C) IFN-β, (D) GBP3, (E) GBP4 and (F) GBP5. Data are representative of at least three independent experiments. Significant differences comparing WT versus STING are denoted by an asterisk (two-way ANOVA, p< 0.05).
Brucella and its DNA induce STING translocation and activation of IRF-3 and NF-κB
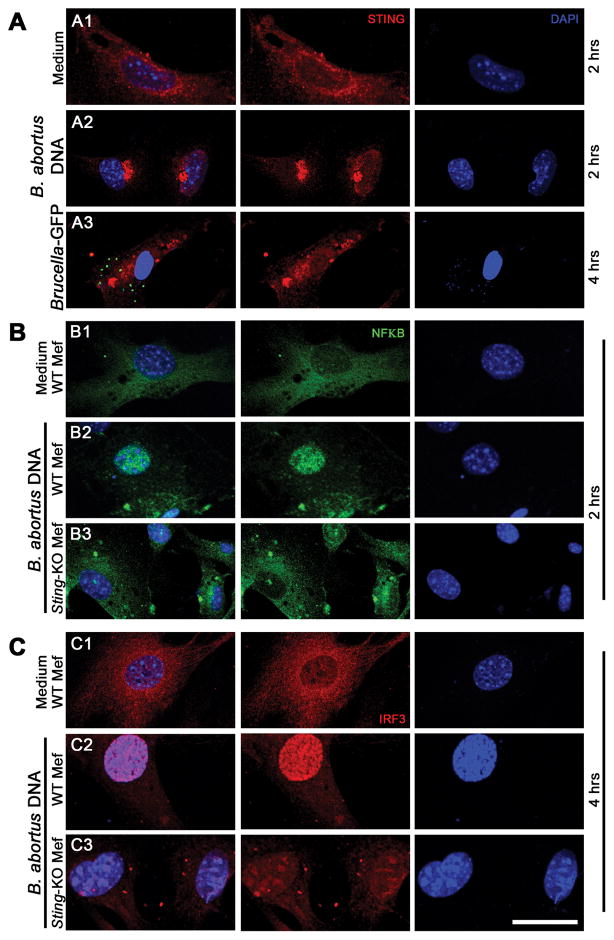
Given the importance of STING in regulating cytoplasmic DNA signaling events, we infected murine embryonic fibroblasts (MEFs) with Brucella-green fluorescent protein (GFP) or transfected with Brucella DNA and observed STING translocation through confocal microscopy. After 4 hrs of infection with Brucella and 2 hrs of transfection with bacterial DNA, STING rapidly underwent trafficking from the ER to the perinuclear-associated endosomal regions of the cell (Figure 2A). This event usually accompanies STING phosphorylation and degradation, likely to avoid sustained STING-activated cytokine production (32). Additionally, after transfection of wild-type MEFs with Brucella DNA, IRF3 and the p65 subunit of NF-κB became phosphorylated and translocated into the nucleus; however, these events did not occur in STING KO cells (Figure 2B and 2C). These data indicate that the pathway of IRF-3 and NF-κB activation induced by bacterial DNA is STING-dependent.
Figure 2. Brucella abortus DNA induces STING activation and translocation of NF-κB and IRF3.
MEFs from wild-type (WT) or STING KO were transfected with B. abortus DNA (1 μg/well, for 2 or 4 hrs as indicated) and cells from WT mice were infected with B. abortus S2308-GFP+ for 4 hrs (MOI 1000:1), fixed and subjected to immunofluorescence microscopy analysis of STING (A), NF-κB (B) or IRF3 (C). Pronounced translocation of STING was observed as aggregated speck formation in the perinuclear region 2 hrs after cells were transfected with bacterial DNA or 4 hrs after B. abortus S2308-GFP infection. STING-dependent-NFκB and -IRF3 activation was induced in wild-type MEFs by Brucella DNA transfection but not in KO cells. Antibody staining is shown in middle panels for STING (A), NF-κB (B) and IRF3 (C) and nuclei staining (DAPI) is shown in blue on the right panels. Left panels are merged images from those shown on middle and right panels. Data are representative of three independent experiments and three replicates in each experimental group. Size bar shown corresponds to 30 μm in all panels.
STING is required for production of proinflammatory mediators in vitro
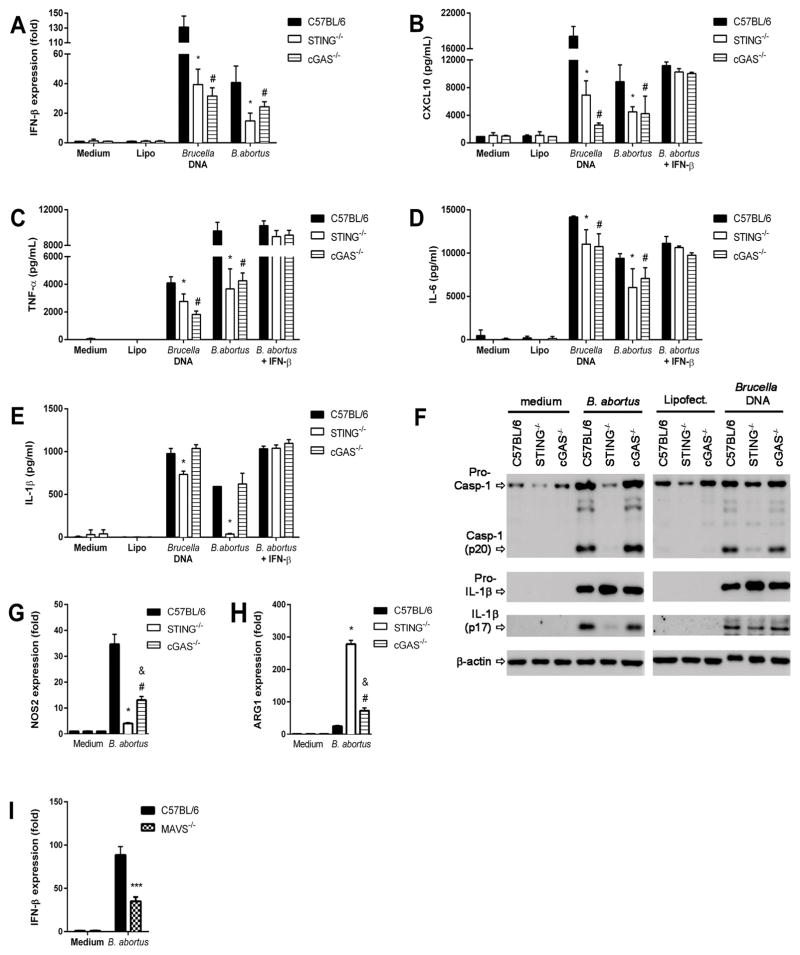
To investigate the roles of STING and cGAS during cytosolic sensing of Brucella DNA or bacterial infection in inducing proinflammatory cytokine production, BMDMs were isolated from STING and cGAS KO mice. Brucella DNA or bacterial infection triggered IFN-β expression was partially dependent on STING and cGAS (Figure 3A). However, the level of IFN-β expression induced by transfected bacterial DNA was much higher than Brucella infection. Delivery of the STING ligand cGAMP via transfection bypassed the cGAS requirement for IFN-β expression and CXCL10 production in agreement with previous reports establishing that cGAS functions upstream of STING (15) (Supplementary Figure 2). We also observed that CXCL10 production, a surrogate cytokine for type I IFN expression, was reduced in STING and cGAS KO BMDMs infected or transfected with Brucella DNA (Figure 3B). Since STING activation is required for NF-κB translocation induced by bacterial DNA (Figure 2), we also measured TNF-α and IL-6 production by STING and cGAS KO BMDMs transfected with bacterial DNA or infected and compared to wild-type (WT) cells. Either Brucella infected or DNA transfected macrophages from cGAS and STING KO mice produced a reduction in TNF-α and a modest decrease in IL-6 compared to WT cells (Figure 3C and D). These data suggest that the STING pathway is required for full production of TNF-α and IL-6. However, other Brucella ligands or bacterial DNA may activate NF-κB in a cGAS/STING-independent pathway, for example through the TLR pathway. We have previously demonstrated that IFN-β can be partially produced by innate cells activated with Brucella components via TLR7 (33). Furthermore, we have shown here using MAVS KO macrophages that type I IFN expression depends in part of the RIG-I pathway independently of STING/cGAS axis (Figure 3I).
Figure 3. Proinflammatory cytokine production induced by Brucella is partially dependent on STING pathway.
BMDM derived from C57BL/6, STING−/− and cGAS−/− mice were transfected with DNA purified from B. abortus (1 μg/well) encapsulated with lipofectamine or infected with B. abortus (MOI 100:1). Total RNA was extracted and qPCR was performed to measure IFN-β (A) expression. Culture supernatants were harvested 17 hrs after treatment to measure CXCL10 (B), TNF-α (C), IL-6 (D) and IL-1β (E) by ELISA assay. Where indicated, cells were treated with 100 U/ml IFN-β 18 hrs prior the course of infection or were untreated. (F) The same culture supernatants or cell lysates were harvested 17 hrs after infection and pro-IL-1β (cell lysates), IL-1β (supernatant) and caspase-1 processing were determined by Western blot. Equal loading was controlled by measuring β-actin in the corresponding cell lysates. (G) NOS2 and (H) Arg1 gene expression was determined in macrophages infected with Brucella abortus (MOI 100:1) for 24 hours by qPCR. (I) Macrophages from C57BL/6 and MAVS−/− mice were stimulated with B. abortus S2308 (MOI 100:1) for 24 hrs and qPCR analysis was performed for IFN-β expression. Data are representative of at least three independent experiments and three replicates in each experimental group. Significant differences comparing WT versus STING are denoted by an asterisk, WT versus cGAS are denoted by # and STING versus cGAS are denoted by & (two-way ANOVA, p< 0.05). *** P< 0.0001 for statistically significant differences from MAVS−/− compared to C57BL/6.
Additionally, we have demonstrated that inflammasome activation is important to induce protective immunity against Brucella infection (22). However, the molecular mechanisms that govern assembly of the DNA sensor AIM2 are less clear than those described for the NLRP3 inflammasome. Since type I IFN contributes to activation of the AIM2 inflammasome in response to F. novocida (34), we determined the role of STING in IL-1β secretion and caspase-1 activation during B. abortus infection. Macrophages from STING KO mice transfected with bacterial genomic DNA or infected with Brucella showed a reduction in IL-1β secretion and caspase-1 activation when compared to cGAS KO or WT cells (Figure 3E and 3F). Therefore, only STING deficiency leads to a reduction in IL-1β and caspase-1 activation when compared to cGAS KO macrophages. These findings are consistent with reduction of IL-1β but not pro-IL-1β observed in STING KO cells in the western blot (Figure 3F) what suggest that lack of STING affects caspase-1 processing. Furthermore, addition of exogenous rIFN-β increased the levels of CXCL10, TNF-α, IL-6 and IL-1β in Brucella-infected macrophages what demonstrates the role of this molecule in modulating pro-inflammatory cytokine production.
Finally, NOS2 and Arg1 have been identified as markers for M1 and M2 macrophages, respectively (35). In this study, we observed that STING KO macrophages had an increased in Arg1 expression and decreased in NOS2 expression compared to wild-type and cGAS KO cells (Figure 3G and 3H). This profile is suggestive of a M2-type macrophages (alternatively activated macrophages) that are typically associated with bacterial persistence.
Taken together, these results demonstrate that even though cGAS and STING are important sensors involved in the production of inflammatory cytokines, STING plays the predominant role during Brucella infection.
Brucella produces c-di GMP that induces STING-dependent type I IFN responses
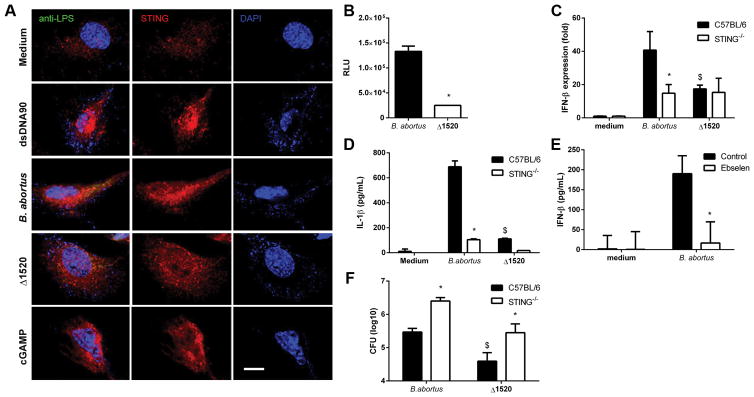
Bacterial c-dinucleotide levels are regulated by the opposing activities of cyclases and phosphodiesterases. To determine the levels of c-di-GMP produced by Brucella wild-type strain versus c-di-GMP guanylate cyclase mutant (Δ1520), we transformed all strains with a plasmid containing a c-di-GMP-responsive riboswitch that drives lux expression as previously demonstrated (28). Brucella Δ1520 mutant displayed a phenotype showing lower levels of c-di-GMP when compared to wild-type strain S2308 (Figure 4B). Furthermore, we performed confocal microscopy analysis in wild-type MEFs infected with Brucella Δ1520 mutant and wild-type S2308 strains. Cells infected with wild-type bacteria or transfected with DNA or cGAMP showed a specific STING-activation profile characterized by an aggregate speck formation in the perinuclear region of the cell; however, we did not observe this activation phenotype in Brucella Δ1520 mutant infected MEFs (Figure 4A). Additionally, we infected BMDMs from STING KO and C57BL/6 mice with Brucella Δ1520 mutant or wild-type S2308 strains and measured IFN-β expression and IL-1β production. Our results demonstrated that the Brucella Δ1520 mutant which produces lower levels of c-di-GMP, induced lower expression levels of IFN-β and IL-1β secretion compared to the virulent strain S2308 (Figure 4C and 4D) and equivalent to production in the STING KO macrophages. To confirm the Brucella Δ1520 mutant findings, we tested a chemical inhibitor of c-di-GMP, termed Ebselen (36). Macrophages treated with Ebselen and infected with wild-type Brucella produced much less IFN-β compared to cells untreated and infected (Figure 4E). The greater difference observed in type I IFN responses between the use of Ebselen compared to the Δ1520 mutant strain might be related to the fact that Ebselen inhibits the binding of c-di-GMP to receptors containing an RxxD domain including PelD and diguanylate cyclases with a broader action and potential off-target effects (36). In contrast, the Δ1520 mutant strain has a deletion on a single Brucella diguanylate cyclase. Taken together, these results suggest that c-di-GMP produced by Brucella is a key metabolite to induce type I IFN responses. Furthermore, we measured CFU counts of Brucella Δ1520 mutant and wild-type S2308 strains after 48 hrs of infection in macrophages. Brucella Δ1520 mutant showed reduced bacterial numbers compared to wild-type bacteria in either C57BL/6 or STING KO cells (Figure 4F).
Figure 4. Deletion of the Brucella guanylate cyclase reduces cellular cyclic di-GMP levels and IFN-β expression.
(A) MEFs from wild-type (WT) mice were transfected with dsDNA90 (3μg/ml) or cGAMP (1μg/well) or infected with Brucella abortus S2308 or Brucella abortus Δ1520 (MOI 1000:1) for 4 hrs, fixed and subjected to immunofluorescence microscopy analysis of STING. Pronounced translocation of STING was observed as aggregated speck formation in the perinuclear region 4 hrs after cells were transfected with dsDNA90, cGAMP or infected with Brucella abortus S2308, but not after infection with Brucella abortus Δ1520. Antibody staining is shown in middle panels for STING and nuclei staining (DAPI) is shown in blue on the right panels. Left panels are merged images from those shown on middle and right panels including staining with anti-Brucella LPS in green. Data are representative of three independent experiments. Size bar shown corresponds to 25 μm in all panels. (B) Wild-type Brucella or Δ1520 mutant strains were transfected with a c-di-GMP regulated lux reporter system. Bacteria were grown to stationary phase and relative luminescence (RLU) units measured (n=4 replicates). Significant differences between wild-type virulent Brucella compared to Δ1520 mutant is denoted by an asterisk (p<0.05). Macrophages derived from C57BL/6 and STING−/− mice were infected with B. abortus wild-type or Δ1520 mutant strain (MOI 100:1). After 17 hrs, total RNA was purified and the gene expression of IFN-β (C) was analyzed by qPCR and IL-1β (D) was measured in the supernatant by ELISA. (E) BMDMs from wild-type mice were treated or not-treated with Ebselen. Cells were either not-infected (medium) or infected (MOI 100:1) with B. abortus for 24 hrs with or without Ebselen (50μM). Supernatants were then harvested and IFN-β production measured by ELISA. (F) BMDMs from C57BL/6 and STING−/− mice were infected with B. abortus wild-type or Δ1520 mutant strain (MOI 10:1) for 48 hrs and bacterial CFU counts were analyzed. Data are representative of three independent experiments and three replicates in each experimental group. Significant differences comparing C57BL/6 mice versus STING KO is denoted by an asterisk (*) and between wild-type Brucella strain 2308 versus Δ1520 mutant is denoted by an $ (two-way ANOVA, p< 0.05).
To confirm our findings in the human system, we used human fibroblast cells (hTERT). After infection of hTERT with Brucella Δ1520 mutant or wild-type S2308 strain, we observed STING activation by confocal microscopy in the cells infected with wild-type bacteria but not with the mutant strain (Figure 5A). Additionally, we performed siRNA silencing of STING and cGAS in hTERT cells. The siRNA knock down efficiency is demonstrated in Figure 5B. The results observed in Figure 5A were correlated to CXCL10 production that was reduced in siRNA control cells infected with Brucella Δ1520 mutant compared to wild-type bacteria (Figure 5C). Additionally, cells treated with cGAS or STING siRNA produced diminished levels of CXCL10 compared to siRNA control when they were infected with either Brucella strain or with bacterial DNA.
Figure 5. STING activation in human fibroblasts following B. abortus infection.
hTERT cells were transfected with dsDNA90 (3μg/mL) or infected with B. abortus strain 2308 or B. abortus Δ1520 mutant (MOI 1000:1) for 4 hrs, fixed and subjected to immunofluorescence microscopy analysis of STING. (A) Pronounced translocation of STING was observed as aggregated speck formation in the perinuclear region 4 hrs after cells were transfected with dsDNA90 or infected with B. abortus strain 2308 but not after infection with B. abortus Δ1520 mutant. Antibody staining is shown in the middle panels for STING (red) and nuclei staining (DAPI) is shown in blue on the right panels. Left panels are merged images from those shown on middle and right panels including staining with anti-Brucella LPS in green. Size bar shown corresponds to 25 μm in all panels. (B) hTERT cells were transfected with mock, control siRNA (non specific), STING siRNA or cGAS siRNA for 3 days. The efficiency of cGAS and STING silencing was demonstrated by immunoblotting, with β-actin serving as a loading control. (C) hTERT cells were transfected with B. abortus DNA, or infected with B. abortus strain 2308 or B. abortus Δ1520 mutant for 24 hrs and supernatants were collected for CXCL10 measurement by ELISA. Data are representative of three independent experiments and three replicates in each experimental group. Significant differences compared to untreated cells are denoted by an asterisk, compared to siRNA control plus Brucella are denoted by # and compared to siRNA STING plus Brucella are denoted by & (two-way ANOVA, p< 0.05).
Collectively, these results suggest that Brucella is able to produce its own second messenger to activate the STING pathway directly. Herein, we provide strong evidence that the bacterial c-di-GMP is important in activating STING in mouse macrophages and human cells.
STING but not cGAS is required for in vitro and in vivo control of Brucella infection
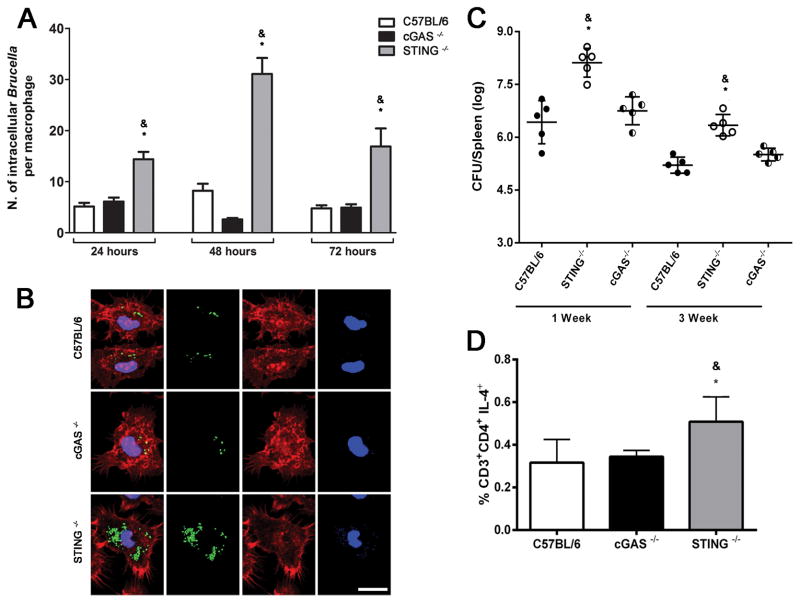
To determine whether STING or cGAS restricts Brucella growth in macrophages and in vivo, we infected WT, STING and cGAS KO BMDMs and mice, and quantified Brucella-GFP+ in vitro at 24, 48, and 72 hrs by confocal microscopy and in vivo at 1- and 3-weeks postinfection. Wild-type macrophages and cGAS KO infected with MOI 100:1 efficiently controlled intracellular replication whereas STING KO BMDMs contained larger numbers of B. abortus at all time intervals studied (Figure 6A and 6B). Since the cGAS/STING axis is important for stimulating type I IFN, we investigated whether these DNA sensors also play a role in host defense in vivo. We infected age- and sex-matched WT, cGAS and STING KO mice i.p. with 1 ×106 CFU of B. abortus virulent strain 2308. STING KO displayed a significantly higher bacterial burden at 1- and 3-weeks postinfection compared to WT animals. However, cGAS KO had no defect in overall resistance to B. abortus, as we observed similar bacterial numbers in spleens of these mice compared to WT (Figure 6C). Since Th1-responses are crucial for efficient control of B. abortus, we measured the frequency of CD4+ T cells producing IFN-γ or IL-4 in WT, cGAS and STING KO mice after infection. As demonstrated in Figure 6D, the frequency of CD4+ T cells producing IL-4 is higher in STING KO animals compared to WT and cGAS KO mice. Regarding the percentage of CD4+ T cells producing IFN-γ, similar levels were detected in all mouse strains tested (data not shown). Furthermore, the presence of markers associated with M2 macrophages shown previously also coincided with enhanced frequency of CD4+ T cells producing IL-4 and bacterial loads in STING KO mice compared to cGAS KO and wild-type control.
Figure 6. STING but not cGAS is required for Brucella control in macrophages and in mice.
BMDMs derived from C57BL/6, cGAS and STING KO mice were infected with Brucella-GFP+ (MOI 10:1) for 24, 48 or 72 hrs and processed for fluorescent microscopy analysis. The number of GFP-expressing bacteria/cell was counted on 200 cells for each mouse strain, for all three times points assessed. Results are shown in (A) as the average number of Brucella/macrophage. Images shown in (B) were taken from macrophages infected with Brucella-GFP+ for 72 hrs and are representative of all experiments analyzed. Mouse strains are indicated on the left. GFP-expressing bacteria are shown in green, phalloidin staining of the actin cytoskeleton for cell shape determination is shown in red, and DAPI (DNA) is shown in blue. Size bar shown on the lower left panel corresponds to 10 μm in all panels. (C) Residual B. abortus CFU in the spleen of wild-type, STING and cGAS KO mice (n=5) were determined at 1 and 3 wks post-infection. (D) C57BL/6, cGAS and STING KO mice were infected with B. abortus and 1 week after infection, splenocytes were submitted to flow cytometry analysis. Cells were assessed for CD3+CD4+ producing IL4. Data are the mean ± SD of five mice/group. Significant differences comparing STING KO versus wild-type mice are denoted by an asterisk and STING versus cGAS KO are indicated by & (p<0.05, two-way ANOVA). The graphs are representative of three independent experiments and three replicates in each experimental group.
Taken together, these findings suggest that cGAS appears to be dispensable for bacterial resistance in vivo and in vitro. These results confirmed that STING is critical to host defense against B. abortus in vitro and in vivo.
STING regulates liver granuloma during B. abortus infection
Infection with B. abortus results in the formation of liver and spleen granulomas, where inflammatory cells aggregate to restrain bacterial growth. Since only STING KO mice were more susceptible to infection in vivo, we analyzed the role of STING in regulating liver pathology. At one and six-weeks after infection, STING KO mice displayed a significant reduction in granuloma number and size when compared to WT counterparts (Supplementary Figure 3). These data suggest that STING modulates host liver pathology at early and late stages of Brucella infection.
IRF-1 and type I IFN expression are required to control Brucella replication in macrophages
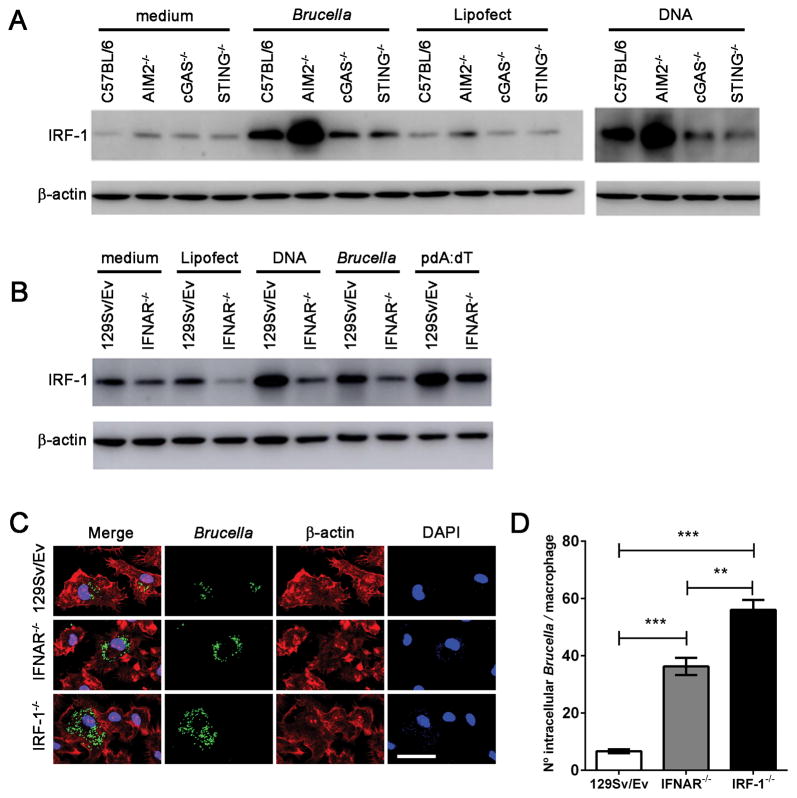
The transcription factor IRF-1 impacts adaptive immune responses by regulating MHC class I expression and influencing development of NK and T cells (37). Additionally, IRF-1 was identified by its binding to DNA sequences that are common to the promoters of IFNα/β genes (38). Thus, IRFs were proposed to be the regulators of type I IFN responses. Herein, we evaluated the level of IRF-1 protein expression in macrophages of WT, STING, cGAS and AIM2 KO animals following infection with Brucella or transfection with bacterial DNA. We observed a major increase in IRF-1 expression in WT cells after bacterial infection or DNA transfection compared to the negative control (Figure 7A). However, this upregulation was partially dependent on the STING/cGAS pathway as we observed reduced IRF-1 expression in cGAS and STING KO macrophages. Conversely, lack of AIM2 robustly augmented IRF-1 expression in macrophages infected with Brucella or DNA transfected, demonstrating that AIM2 regulates type I IFN responses. Furthermore, we tested whether IRF-1 expression was dependent on IFNAR signaling pathway. As shown in Figure 7B, IFNAR KO cells transfected with bacterial DNA or infected with Brucella had diminished levels of IRF-1 when compared to WT macrophages.
Figure 7. IRF-1 and type I IFN signaling are required to restrict Brucella replication in macrophages.
Macrophages derived from (A) C57BL/6, STING, cGAS and AIM2 KO or (B) 129Sv/Ev and IFNAR KO mice were infected with B. abortus (at MOI 100:1) or transfected with bacterial DNA (1μg/well) or poly dA:dT (1μg/well) encapsulated with lipofectamine or lipofectamine alone as control. Cell lysates were harvested 17 hrs after treatment and processing by western blot to determine the levels of IRF-1. (C) BMDMs derived from WT (129 Sv/Ev), IFNAR−/−, or IRF-1−/− mice were infected with Brucella-GFP (MOI 10:1) for 24 hrs and processed for fluorescent microscopy analysis. GFP-expressing bacteria are shown in green, phalloidin staining of the actin cytoskeleton for cell shape determination is shown in red, and DAPI (DNA) is shown in blue. Images show infected 129Sv/Ev, IFNAR−/−, or IRF-1−/− macrophages on top, middle and lower panels, respectively, as indicated on the left. Size bar shown on the lower-right panel in B corresponds to 30 μm in all panels. (D) The number of GFP-expressing bacteria was assessed for each cell, and 200 cells were analyzed for each mouse strain. Significant differences in average number of Brucella/cell comparing WT versus IFNAR and IRF-1 KO is denoted by *** (p<0.001) and IRF-1 versus IFNAR KO is denoted by ** (p<0.01 two-way ANOVA). Data are representative of three independent experiments and three replicates in each experimental group.
Since others demonstrated that IRF-1 KO animals are more susceptible to Brucella infection in vivo (39), we investigated the hypothesis that type I IFN signaling and IRF-1 affect the ability of macrophages to control Brucella replication. Therefore, we infected wild-type, IFNAR KO and IRF-1 KO BMDMs with Brucella-GFP+ and quantified bacteria overtime, Figure 7C. Single-cell analysis revealed that wild-type macrophages restricted bacterial replication after 24 hrs of infection, whereas IFNAR and IRF-1 KO cells failed to control bacterial replication and harbored a significantly larger number of bacteria compared to wild-type BMDMs, Figure 7D. The greater susceptibility to Brucella replication observed in IRF-1 KO cells suggests that IRF-1 is important in multiple antimicrobial defense mechanisms.
STING and type I IFN signaling are required for GBP expression in Brucella-infected macrophages
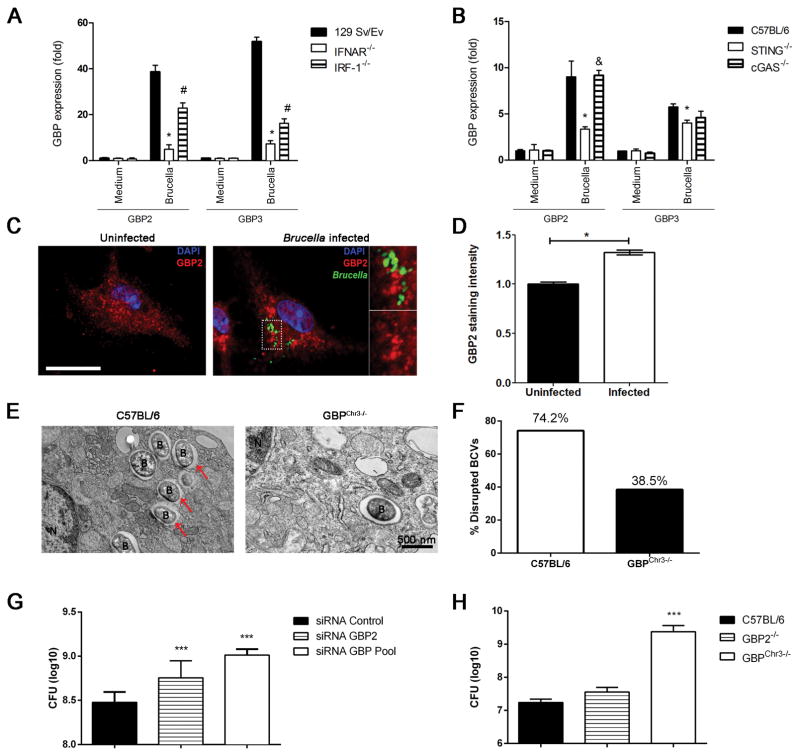
Guanylate-binding proteins are interferon-inducible GTPases that exert anti-microbial effects (24). In addition, GBPs encoded by genes on mouse chromosome 3 (GBP1, GBP2, GBP3, GBP5 and GBP7) promote recognition of the vacuolar bacterium Salmonella typhimurium leading to the escape of the bacteria into the cytosol (31). Herein, we detected by qPCR analysis the downregulation of GBP2 and GBP3 genes in the absence of IRF-1 and IFNAR in macrophages infected with Brucella (Figure 8A). However, the residual expression of GBP2 and GBP3 in IRF-1 KO macrophages indicate that an alternative pathway independent of IRF-1 exists to induce GBPs expression. To address that, we infected IFNAR KO macrophages with B. abortus and found that lack of type I IFN signaling robustly reduced GBP2 and GBP3 expression (Figure 8A). Additionally, we observed that GBP2 and GBP3 expression is partially dependent on the STING pathway, probably via IFN-β production (Figure 8B). Interestingly, reduction of GBP2 and GBP3 expression following Brucella infection in macrophages was only dependent on STING and not cGAS. Furthermore, we observed by confocal microscopy that Brucella infection induces the formation of GBP2 aggregates (37% increase compared to uninfected cells) located in close proximity of bacterial containing compartments in macrophages (Figures 8C and D). Additionally, we performed electron microscopy analysis and we detected that 74.2% of Brucella-containing vacuole (BCV) was disrupted in C57BL/6 macrophages when compared to 38.5% in GBPchr3 KO cells (Figure 8E and 8F). This result suggests that the GBP machinery is important to target the BCV to release bacterial components to host cell cytosol.
Figure 8. Type I IFN induced by STING activation are required for GBP expression that controls bacterial replication.
BMDMs from C57BL/6, 129/SvEv and STING, cGAS, IRF-1 and IFNAR KO mice were infected with B. abortus (MOI 100:1) and total RNA was extracted 17 hrs after infection. Analysis of GBP2 and GBP3 expression by qPCR of macrophages from (A) 129Sv/Ev, IRF-1 and IFNAR KO or (B) C57BL/6, STING and cGAS KO. The results are shown as mean ± SD of fold induction and normalized to β-actin gene. Data are representative of at least three independent experiments. Significant differences comparing WT versus STING or WT versus IFNAR are denoted by an asterisk, WT versus IRF-1 are denoted by #, and STING versus cGAS or IFNAR versus IRF-1 are denoted by & (two-way ANOVA, p< 0.05). (C) B. abortus infection induces aggregation of GBP2 protein. Macrophages derived from C57BL/6 mice were infected with B. abortus-GFP (green) for 24 hrs, fixed and subjected to immunofluorescence of GBP2 (in red). GBP2 localization in an uninfected cell is shown on the left panel and evident clustering of GBP2 can be observed in cells infected with Brucella-GFP (right panel). Quantification of grey levels (pseudo coloured in red in this image) from anti-GBP2 signal was performed on an area of approximately 9μm2 of each cell and mean values were plotted in (D). Significant differences comparing uninfected versus infected cells are denoted by an asterisk (p<0.05). Data are representative of three independent experiments. Size bar shown on the left panel in A corresponds to 10μm in all panels. (E) Bone marrow derived macrophages from C57BL/6 and GBPcrh3 mice were infected with B. abortus for 6 hrs and the integrity of BCV membranes were evaluated by TEM. The red arrows indicate regions of BCV membrane rupture. The percentage of disrupted BCV membranes was evaluated and is represented in (F). N = nucleus, B = B. abortus and size bar shown corresponds to 500 nm. (G) BMDMs from C57BL/6 mice were transfected with small interfering RNA (siRNA) from siGENOME smart pools (Dharmacon) for GBP2 and GBPpool (GBP2, 3 and 5) for 48 hrs and infected with B. abortus (MOI 100:1) for 24 hrs and bacterial CFU counts were analyzed. Significant differences in relation to siRNA control are denoted by *** (p<0.001). (H) C57BL/6 and GBP2 and GBPchr3 KO were intraperitoneally inoculated with 106 CFU of B. abortus strain S2308 and 1 week postinfection, the CFU were determined in spleens. Data are expressed as mean ± SD of five animals. Significant differences comparing GBPchr3 KO in relation to C57BL/6 and GBP2 KO are denoted by *** (p<0.001). Data are representative of three independent experiments and three replicates in each experimental group.
Together these findings also provide evidence that GBP2 and GBP3 are produced in response to B. abortus infection, and that GBP2 and GBP3 are under the control of type I IFN signaling at least partially via the STING pathway.
GBPs control Brucella abortus replication in vitro and in vivo
GBPs can target vacuolar bacteria such as Salmonella and Francisella and induce the recruitment of antimicrobial peptides to kill the bacteria (25, 31). To investigate whether GBPs contained on mouse chromosome 3 (GBPchr3) or GBP2 alone directly affected the viability of bacteria in macrophages, we treated wild-type BMDMs with GBP2 and GBP pool (containing GBP2, GBP3 and GBP5) siRNA and infected them with B. abortus and bacteria were quantified after 24 hrs postinfection. Analysis of CFU counts revealed that WT macrophages restricted bacterial replication whereas GBP pool and GBP2 siRNA treated cells failed to control Brucella growth intracellularly and harbored a significantly larger number of bacteria (Figure 8G). We further analyzed the role of GBP2 and GBPchr3 in controlling bacterial infection in vivo by infecting GBP2 and GBPchr3 KO and WT mice and measuring CFU in spleen after 1 week postinfection. As shown in Figure 8H, only GBPchr3 KO mice were more susceptible to Brucella infection when compared to GBP2 KO or WT animals. These findings demonstrated that GBPs mediate Brucella control in vitro and in vivo.
Activation of inflammasome during Brucella infection partially requires GBPs
Since GBPs play a role in releasing bacteria from vacuoles and thus enabling greater access to cytosolic sensors, we sought to address the role of GBPs in inflammasome activation during Brucella infection. Brucella-infected macrophages from C57BL/6 mice treated with siRNA for GBP2 or a GBP pool were assessed for pro-IL-1β in cell lysates, IL-1β secretion and caspase-1 activation. GBP2 and GBP pool treated macrophages displayed a significant reduction in cytokine release and caspase-1 activation compared to siRNA control treated cells (Figure 9A and B). Even though, GBP2 and GBP pool siRNA treated cells had a reduced IL-1β secretion, the amount of pro-IL-1β was intact indicating that pathogen recognition receptor (PRR) and NF-κB responses were normal but an inflammasome activatory signal was affected. To confirm these findings, we used macrophages from GBP2 and GBPchr3 KO mice and transfected them with bacterial DNA or infected with Brucella and measured IL-1β secretion. The results demonstrated that GBP2 and GBPchr3 KO cells also showed a reduced secretion of IL-1β when macrophages were either transfected with DNA or infected with the bacteria, corroborating with our siRNA data (Supplementary Figure 4). Taken together, these findings are consistent with a role for GBPs in releasing Brucella components into the cytosol for inflammasome detection.
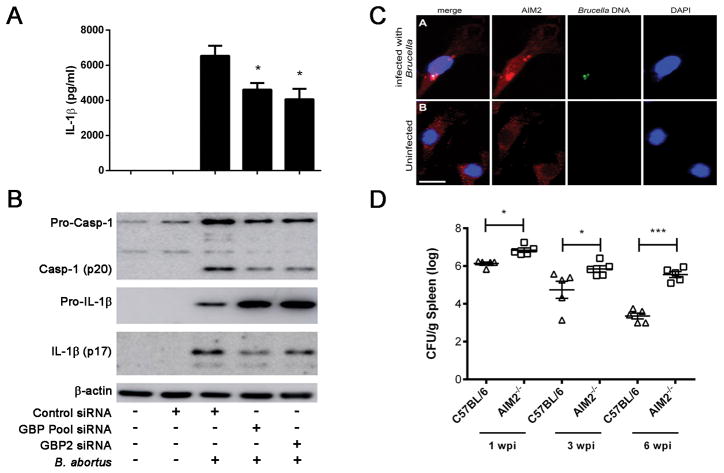
Figure 9. Inflammasome activation by Brucella partially requires functionally active guanylate binding proteins.
BMDMs from C57BL/6 mice were transfected with small interfering RNA (siRNA) from siGENOME smart pools (Dharmacon) for GBP2 and GBP pool (GBP2, 3 and 5) for 48 hrs and infected with B. abortus (MOI 100:1) for 17 hrs and IL-1β (A) secretion was measured by ELISA and pro-IL-1β (cell lysates), mature IL-1β (supernatant) and caspase-1 activation by Western blot (B). Significant differences from GBP2 and GBP pool siRNA in relation to siRNA control are denoted by an asterisk, two-way ANOVA (p<0.05). Data are representative of at least three independent experiments and three replicates in each experimental group. (C) Anti-AIM2 staining of Brucella-infected macrophages by confocal microscopy reveal aggregated speck formation of AIM2 in association with bacterial DNA. Brucella DNA was specifically labelled with click-IT kit and can be seen in green in the upper panel. DAPI staining allows visualization of bacterial DNA. Size bar shown corresponds to 20 μm in all panels. (D) C57BL/6 and AIM2 KO mice (n=5) were intraperitoneally inoculated with 106 CFU of B. abortus strain S2308. Residual B. abortus CFU in the spleen of wild-type and AIM2 KO mice were determined at 1, 3 and 6 weeks postinfection. Data are expressed as mean ± SD of five animals per time point and representative of three independent experiments. Significant difference in relation to C57BL/6 is denoted by an asterisk (p<0.05) or *** (p<0.001).
AIM2 is required for Brucella control in mice
AIM2 was found to recognize cytoplasmic dsDNA through its HIN-200 domain and ASC via its pyrin domain (10). Franciscella tularensis and L. monocytogenes DNA activate the AIM2 inflammasome by its interaction with ASC to induce caspase-1 (34, 40). For AIM2 to detect dsDNA, Brucella or its DNA must escape the Brucella-containing vacuole (BCV) and enter the cytoplasm. To determine whether AIM2 inflammasome directly senses Brucella DNA intracellularly, we infected C57BL/6 macrophages and observed that labeled AIM2 co-localized with Brucella DNA that was previously stained using the click-iT EdU Imaging kit. Indeed, co-staining of AIM2 and bacterial DNA from Brucella-infected macrophages revealed the formation of an inflammasome aggregation of AIM2 and Brucella DNA specks as result of cell activation (Figure 9C). We also determined the role of AIM2 in regulating Brucella growth in vivo. Consistent with our previous in vitro findings using macrophages identifying a critical role for AIM2 and ASC to trigger caspase-1 activation and IL-1β secretion (22), AIM2 KO mice showed a reduced resistance to Brucella at one, three and six weeks post-infection as determined by bacterial numbers in the spleens (Figure 9D). Bacterial load recovery was 0.7, 1.6, and 2.3 logs higher at respective weeks in AIM2 KO mice when compared to C57BL/6 animals. Together these results strongly suggest that inflammasomes are important to induce resistance to B. abortus infection and implicate AIM2 receptors in this process.
Discussion
Pathogenic bacteria can use many different strategies to enter and establish the infection inside the host, and the immune system has mechanisms to detect and eliminate a broad range of these bacteria. Cytosolic detection leads to activation of potent antimicrobial effector pathways such as the inflammasome and the cytosolic surveillance pathway (CSP). The cGAS/STING axis is an important CSP pathway by which innate immune cells recognize both viral and bacterial pathogens that access the cytosol but different bacteria have evolved different mechanisms to initiate this response (11, 23). Additionally, bacterial ligands must secure entry into the cytoplasm to activate cytosolic sensors; however, the mechanisms by which concealed ligands are liberated in the cytoplasm have remained unclear. Because the detection of Brucella infection by macrophages is so critical to the innate immune response, we sought to define the receptors that sense Brucella DNA in the cytosol and determine their role in the host response to infection. Previously, we have shown that AIM2 is important for sensing Brucella DNA and triggering IL-1β secretion and caspase-1 activation (22), but the role of cGAS/STING axis during this bacterial infection remained to be determined.
In this study, we observed that Brucella infection or bacteria DNA transfection triggers the expression of innate immune genes such as IFN-β and GBPs in a STING-dependent manner. Further, bacterial infection or DNA transfection induces STING activation in MEFs as observed by aggregated speck formation in the perinuclear region of the cell. IRF3 and NF-κB translocation to the nucleus was also observed in DNA transfected MEFs and shown to be STING-dependent. Given the potential positive and negative roles of type I IFN during bacterial infections, we sought to determine the overall role of cGAS and STING in B. abortus pathogenesis. Infection of cGAS and STING KO macrophages led to a partial decrease in type I IFN expression when compared to wild-type cells. Additionally, lack of STING had a partial effect on production of NF-κB-dependent cytokines, such as TNF-α and IL-6, suggesting that Brucella partially activates NF-κB through a STING-dependent pathway. However, STING but not cGAS control bacterial replication in vitro. The same phenotype was observed in infected knockout mice in vivo, demonstrating that lack of cGAS had no defect in overall resistance to B. abortus. Higher bacterial burdens were detected in spleens of STING KO mice compared to cGAS KO and wild-type animals. Additionally, STING altered the formation of hepatic granulomas in vivo that is an important host strategy to restrain bacterial growth. Furthermore, we observed a reduced secretion of IL-1β and caspase-1 activation and GBP2 and GBP3 expression in STING KO cells infected with B. abortus when compared to cGAS deficient macrophages. All together, these findings suggest lack of cGAS is less critical to the activation of innate immune effector mechanisms related to protective immunity against Brucella.
In context of animal models of bacterial infection, M1-type macrophages (classically activated macrophages) and NO production are often, but not exclusively associated with host protection (41). Conversely, M2-type macrophages are typically associated with bacterial persistence. In this study, we observed that the presence of markers associated with M2 macrophages in STING KO mice compared to cGAS KO and wild-type controls that also coincided with enhanced frequency of CD4+ T cells producing IL-4. Previously, Xavier et al (42) demonstrated that M2 macrophages support increased levels of intracellular Brucella replication during chronic infection. Taken together, these findings suggest that STING activation seems to be involved in inhibiting the differentiation of M2 macrophages in Brucella-infected cells that parallels with enhanced bacterial replication in STING KO cells.
L. monocytogenes effectively short-circuits the cGAS cytosolic surveillance pathway by providing its own second messenger, whereas M. tuberculosis and L. pneumophila generate the type I IFN signal via DNA binding to cGAS. Interestingly, Watson et al (18) demonstrated that cGAS also plays no role in controlling M. tuberculosis infection. Additionally, Salmonella typhimurium predominantly activates type I IFN via a cGAS/STING-independent mechanism, likely via TLR4/TRIF pathway (43). Herein, we provide evidence in mouse and human cells that production of Brucella c-di-GMP activates STING type I IFN signaling events. Further, STING activation leads to GBP expression and inflammasome activation.
IRF-1 has been identified as a transactivator of IFN-β (44); therefore, we utilized BMDMs from IFNAR and IRF-1 KO mice to determine their relative roles in restricting Brucella replication in macrophages. By confocal microscopy, we observed an increased number of intracellular Brucella-GFP+ per macrophage in both IFNAR and IRF-1 KO compared to WT cells. In the present study, STING was required for full expression of IRF-1 after Brucella infection in macrophages. STING activation, type I IFN signaling, and the transcription factor IRF-1 were also required for robust expression of GBP2 and GBP3 following Brucella infection. Since we observed that GBPs siRNA treated macrophages produced a major reduction in IL-1β secretion, we therefore concluded there is a specific requirement of STING and GBPs in Brucella-induced inflammasome activation.
Members of the IFN-inducible GTPase family are executioners of cell-autonomous immunity and have the ability to target the vacuolar membrane encapsulating intracellular parasites and bacteria (45, 46). To determine whether GBPs were directly involved in the activation of inflammasomes, we knocked-down by siRNA GBP2 and the pool of GBP2, 3 and 5 in C57BL/6 macrophages infected with Brucella. In this study, cells not expressing GBP2 or the pool of GBPs produced much less IL-1β than the cells transfected with control siRNA. Additionally, GBP2 and GBP pool knocked-down cells had a reduced ability to control Brucella replication intracellularly. However, Brucella infection observed in cells treated with the GBP pool was markedly augmented compared to cells treated GBP2 only siRNA, suggesting the GBP proteins work together non-redundantly to control Brucella. Further, we observed during in vivo experiments that only GBP3chr3 KO mice were more susceptible to Brucella at 1 week postinfection suggesting that GBPs other than GBP2 are more important to host protection. Recent study suggests that besides the membrane-destabilizing activity of GBPs, their bacteriolytic role could also result in release of microbe-associated molecular patterns such as DNA (47). GBP mediated bacteriolysis may release bacterial DNA to activate AIM2-inflammasome and/or amplification of type I IFN production via cGAS/STING pathway. Herein, we provide evidence by TEM experiments that GBPs from the chromosome 3 are able to target the Brucella-containing vacuole disrupting its membrane and making bacterial products available to be sensed by cytosolic receptors. Additionally, we demonstrate that Brucella DNA is co-localized with AIM2 during infection of macrophages. Further, we extended our findings to an in vivo setting and infected wild-type and AIM2 KO mice with B. abortus virulent strain and monitored their susceptibility to infection. Analysis of bacterial burdens showed that AIM2 KO mice harbored significantly greater numbers of Brucella in the spleen than did wild-type mice at 1, 3 and 6-weeks postinfection.
Bacterial ligands must secure entry into the cytoplasm to activate inflammasomes; however, the mechanisms by which concealed ligands are liberated in the cytoplasm have remained unclear. Several studies have shown that activation of AIM2 inflammasome by intracellular bacteria requires bacteriolysis and subsequent release of bacterial chromosomal DNA into the cytosol (48, 49). However, whether the bacteriolysis is accidental or it is an active GBP-directed process has to be clarified. Recently, others (50) have shown that interferon-inducible protein IRGB10 is essential for activation of the DNA-sensing AIM2 inflammasome by F. novicida. IRGB10 directly targeted cytoplasmic bacteria through a mechanism requiring GBPs. Localization of IRGB10 to the bacterial cell membrane compromised bacterial structure integrity and mediated cytosolic release of ligands for recognition by inflammasome sensors. We also observed a decreased expression of IRGB10 in Brucella-infected macrophages of STING KO; however, whether this molecule plays a role in Brucella bacteriolysis remains unclear (data not shown).
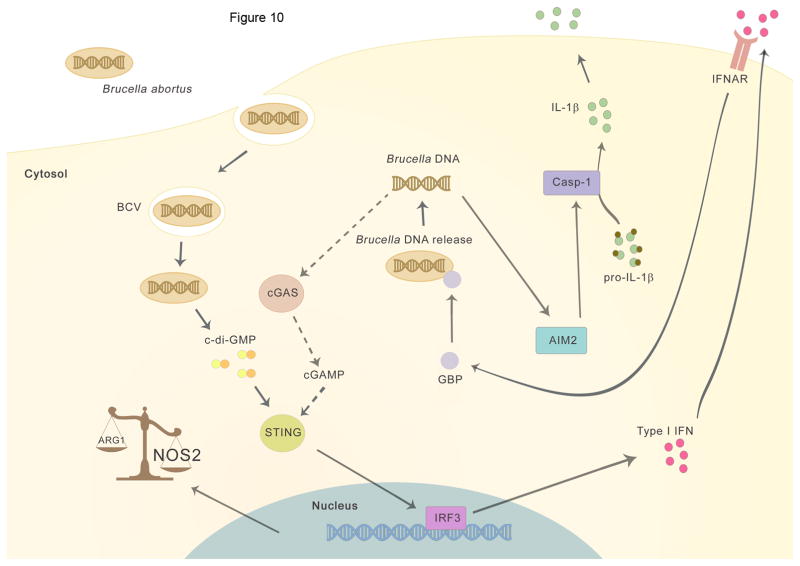
Putting these results together in a pathway, it is possible that Brucella triggers initial direct STING signaling, type I IFN and GBP activation via bacterial c-di-GMP. Our findings suggest that GBPs associate with Brucella-containing vacuole (BCV), and by an as-yet-undefined mechanism, induce lysis of the BCV or maybe direct bacteriolysis which result in bacterial products like DNA being released into the cytosol to be recognized by cytosolic sensors such as AIM2. After DNA is released into the cytosol, the STING signal also could be further amplified by cGAS sensing of genomic DNA generating cGAMP (Figure 10). However, further experiments are necessary to support this hypothesis and define how this DNA signaling hierarchy is governed. Additional questions remain to be answered such as how GBP targeting is regulated and how GBPs act mechanistically to expose Brucella ligands to cytosolic recognition pathways.
Figure 10. Working model.
The intracellular bacteria Brucella abortus enters the host cell and ensures its survival by forming the Brucella-containing vacuole (BCV). Initially, the recognition of bacterial c-di-GMP activates STING and triggers type I interferon response and upregulation of NOS2 and GBPs expression. GBPs promote lysis of the BCV by exposing bacterial components, like bacterial DNA in the cytosol, thus enabling activation of AIM2 and IL-1β secretion. Additionally, release of Brucella genomic DNA in the cytosol may activate cGAS generating cGAMP (dotted line) resulting in further amplification of type I IFN signaling pathway.
In conclusion, our results provide insights into the mechanism by which bacterial-associated ligands are liberated in the cytoplasm to activate inflammasomes and establish a cGAS-independent mechanism of STING-mediated protection against an intracellular bacterial infection.
Supplementary Material
Acknowledgments
This work was supported by grants from the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq, #464711/2014-2, #402527/2013-5, #443662/2014-2 and #302660/2015-1), Fundação de Amparo a Pesquisa do Estado de Minas Gerais (FAPEMIG, APQ #837/15 and Rede Mineira de Imunobiologicos #00140-16), and Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES, #030448/2013-1) and National Institute of Health R01 AI116453.
We thank Dr. Xi Chen, from the Division of Biostatistics, Department of Public Health Sciences, Sylvester Comprehensive Cancer Center and Dr Tianli Xia from the Department of Cell Biology, University of Miami, Florida, USA for doing the bioinformatic analysis of the microarray data and RNA preparation.
Abbreviations used in this paper
- MOI
multiplicities of infection
- BMDM
bone marrow-derived macrophage
- 2′3′-cGAMP
2′5-3′5′-cyclic GMP-AMP
- GBP
guanylate-binding protein
- cGAS
cyclic GMP-AMP synthase
- STING
stimulator of interferon genes
- c-di-GMP
cyclic dimeric guanosine monophosphate
- CDN
cyclic dinucleotide
- TEM
transmission electron microscopy
References
- 1.Medzhitov R. Recognition of microorganisms and activation of the immune response. Nature. 2007;449:819–826. doi: 10.1038/nature06246. [DOI] [PubMed] [Google Scholar]
- 2.Paludan SR, Bowie AG. Immune sensing of DNA. Immunity. 2013;38:870–880. doi: 10.1016/j.immuni.2013.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chiu YH, Macmillan JB, Chen ZJ. RNA polymerase III detects cytosolic DNA and induces type I interferons through the RIG-I pathway. Cell. 2009;138:576–591. doi: 10.1016/j.cell.2009.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Takaoka A, Wang Z, Choi MK, Yanai H, Negishi H, Ban T, Lu Y, Miyagishi M, Kodama T, Honda K, Ohba Y, Taniguchi T. DAI (DLM-1/ZBP1) is a cytosolic DNA sensor and an activator of innate immune response. Nature. 2007;448:501–505. doi: 10.1038/nature06013. [DOI] [PubMed] [Google Scholar]
- 5.Yang P, An H, Liu X, Wen M, Zheng Y, Rui Y, Cao X. The cytosolic nucleic acid sensor LRRFIP1 mediates the production of type I interferon via a beta-catenin-dependent pathway. Nat Immunol. 2010;11:487–494. doi: 10.1038/ni.1876. [DOI] [PubMed] [Google Scholar]
- 6.Unterholzner L, Keating SE, Baran M, Horan KA, Jensen SB, Sharma S, Sirois CM, Jin T, Latz E, Xiao TS, Fitzgerald KA, Paludan SR, Bowie AG. IFI16 is an innate immune sensor for intracellular DNA. Nat Immunol. 2010;11:997–1004. doi: 10.1038/ni.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kondo T, Kobayashi J, Saitoh T, Maruyama K, Ishii KJ, Barber GN, Komatsu K, Akira S, Kawai T. DNA damage sensor MRE11 recognizes cytosolic double-stranded DNA and induces type I interferon by regulating STING trafficking. Proc Natl Acad Sci U S A. 2013;110:2969–2974. doi: 10.1073/pnas.1222694110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang Z, Yuan B, Bao M, Lu N, Kim T, Liu YJ. The helicase DDX41 senses intracellular DNA mediated by the adaptor STING in dendritic cells. Nat Immunol. 2011;12:959–965. doi: 10.1038/ni.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li Y, Chen R, Zhou Q, Xu Z, Li C, Wang S, Mao A, Zhang X, He W, Shu HB. LSm14A is a processing body-associated sensor of viral nucleic acids that initiates cellular antiviral response in the early phase of viral infection. Proc Natl Acad Sci U S A. 2012;109:11770–11775. doi: 10.1073/pnas.1203405109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hornung V, Ablasser A, Charrel-Dennis M, Bauernfeind F, Horvath G, Caffrey DR, Latz E, Fitzgerald KA. AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature. 2009;458:514–518. doi: 10.1038/nature07725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ishikawa H, Barber GN. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature. 2008;455:674–678. doi: 10.1038/nature07317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ishikawa H, Ma Z, Barber GN. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature. 2009;461:788–792. doi: 10.1038/nature08476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Burdette DL, Monroe KM, Sotelo-Troha K, Iwig JS, Eckert B, Hyodo M, Hayakawa Y, Vance RE. STING is a direct innate immune sensor of cyclic di-GMP. Nature. 2011;478:515–518. doi: 10.1038/nature10429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang X, Shi H, Wu J, Zhang X, Sun L, Chen C, Chen ZJ. Cyclic GMP-AMP containing mixed phosphodiester linkages is an endogenous high-affinity ligand for STING. Mol Cell. 2013;51:226–235. doi: 10.1016/j.molcel.2013.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sun L, Wu J, Du F, Chen X, Chen ZJ. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science. 2013;339:786–791. doi: 10.1126/science.1232458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saitoh T, Fujita N, Hayashi T, Takahara K, Satoh T, Lee H, Matsunaga K, Kageyama S, Omori H, Noda T, Yamamoto N, Kawai T, Ishii K, Takeuchi O, Yoshimori T, Akira S. Atg9a controls dsDNA-driven dynamic translocation of STING and the innate immune response. Proc Natl Acad Sci U S A. 2009;106:20842–20846. doi: 10.1073/pnas.0911267106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Woodward JJ, Iavarone AT, Portnoy DA. c-di-AMP secreted by intracellular Listeria monocytogenes activates a host type I interferon response. Science. 2010;328:1703–1705. doi: 10.1126/science.1189801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Watson RO, Bell SL, MacDuff DA, Kimmey JM, Diner EJ, Olivas J, Vance RE, Stallings CL, Virgin HW, Cox JS. The Cytosolic Sensor cGAS Detects Mycobacterium tuberculosis DNA to Induce Type I Interferons and Activate Autophagy. Cell Host Microbe. 2015;17:811–819. doi: 10.1016/j.chom.2015.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang Y, Yeruva L, Marinov A, Prantner D, Wyrick PB, Lupashin V, Nagarajan UM. The DNA sensor, cyclic GMP-AMP synthase, is essential for induction of IFN-beta during Chlamydia trachomatis infection. J Immunol. 2014;193:2394–2404. doi: 10.4049/jimmunol.1302718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.de Figueiredo P, Ficht TA, Rice-Ficht A, Rossetti CA, Adams LG. Pathogenesis and immunobiology of brucellosis: review of Brucella-host interactions. Am J Pathol. 2015;185:1505–1517. doi: 10.1016/j.ajpath.2015.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.de Almeida LA, Carvalho NB, Oliveira FS, Lacerda TL, Vasconcelos AC, Nogueira L, Bafica A, Silva AM, Oliveira SC. MyD88 and STING signaling pathways are required for IRF3-mediated IFN-beta induction in response to Brucella abortus infection. PLoS One. 2011;6:e23135. doi: 10.1371/journal.pone.0023135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gomes MT, Campos PC, Oliveira FS, Corsetti PP, Bortoluci KR, Cunha LD, Zamboni DS, Oliveira SC. Critical role of ASC inflammasomes and bacterial type IV secretion system in caspase-1 activation and host innate resistance to Brucella abortus infection. J Immunol. 2013;190:3629–3638. doi: 10.4049/jimmunol.1202817. [DOI] [PubMed] [Google Scholar]
- 23.Gao D, Wu J, Wu YT, Du F, Aroh C, Yan N, Sun L, Chen ZJ. Cyclic GMP-AMP synthase is an innate immune sensor of HIV and other retroviruses. Science. 2013;341:903–906. doi: 10.1126/science.1240933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yamamoto M, Okuyama M, Ma JS, Kimura T, Kamiyama N, Saiga H, Ohshima J, Sasai M, Kayama H, Okamoto T, Huang DC, Soldati-Favre D, Horie K, Takeda J, Takeda K. A cluster of interferon-gamma-inducible p65 GTPases plays a critical role in host defense against Toxoplasma gondii. Immunity. 2012;37:302–313. doi: 10.1016/j.immuni.2012.06.009. [DOI] [PubMed] [Google Scholar]
- 25.Man SM, Karki R, Malireddi RK, Neale G, Vogel P, Yamamoto M, Lamkanfi M, Kanneganti TD. The transcription factor IRF1 and guanylate-binding proteins target activation of the AIM2 inflammasome by Francisella infection. Nat Immunol. 2015;16:467–475. doi: 10.1038/ni.3118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sun Q, Sun L, Liu HH, Chen X, Seth RB, Forman J, Chen ZJ. The specific and essential role of MAVS in antiviral innate immune responses. Immunity. 2006;24:633–642. doi: 10.1016/j.immuni.2006.04.004. [DOI] [PubMed] [Google Scholar]
- 27.Petersen E, Chaudhuri P, Gourley C, Harms J, Splitter G. Brucella melitensis cyclic di-GMP phosphodiesterase BpdA controls expression of flagellar genes. J Bacteriol. 2011;193:5683–5691. doi: 10.1128/JB.00428-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Khan M, Harms JS, Marim FM, Armon L, Hall CL, Liu YP, Banai M, Oliveira SC, Splitter GA, Smith JA. The Bacterial Second Messenger Cyclic di-GMP Regulates Brucella Pathogenesis and Leads to Altered Host Immune Response. Infect Immun. 2016;84:3458–3470. doi: 10.1128/IAI.00531-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Macedo GC, Magnani DM, Carvalho NB, Bruna-Romero O, Gazzinelli RT, Oliveira SC. Central role of MyD88-dependent dendritic cell maturation and proinflammatory cytokine production to control Brucella abortus infection. J Immunol. 2008;180:1080–1087. doi: 10.4049/jimmunol.180.2.1080. [DOI] [PubMed] [Google Scholar]
- 30.Kim BH, Shenoy AR, Kumar P, Das R, Tiwari S, MacMicking JD. A family of IFN-gamma-inducible 65-kD GTPases protects against bacterial infection. Science. 2011;332:717–721. doi: 10.1126/science.1201711. [DOI] [PubMed] [Google Scholar]
- 31.Meunier E, Dick MS, Dreier RF, Schurmann N, Kenzelmann Broz D, Warming S, Roose-Girma M, Bumann D, Kayagaki N, Takeda K, Yamamoto M, Broz P. Caspase-11 activation requires lysis of pathogen-containing vacuoles by IFN-induced GTPases. Nature. 2014;509:366–370. doi: 10.1038/nature13157. [DOI] [PubMed] [Google Scholar]
- 32.Ahn J, Barber GN. Self-DNA, STING-dependent signaling and the origins of autoinflammatory disease. Curr Opin Immunol. 2014;31:121–126. doi: 10.1016/j.coi.2014.10.009. [DOI] [PubMed] [Google Scholar]
- 33.Campos PC, Gomes MT, Guimaraes ES, Guimaraes G, Oliveira SC. TLR7 and TLR3 Sense Brucella abortus RNA to Induce Proinflammatory Cytokine Production but They Are Dispensable for Host Control of Infection. Front Immunol. 2017;8:28. doi: 10.3389/fimmu.2017.00028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fernandes-Alnemri T, Yu JW, Juliana C, Solorzano L, Kang S, Wu J, Datta P, McCormick M, Huang L, McDermott E, Eisenlohr L, Landel CP, Alnemri ES. The AIM2 inflammasome is critical for innate immunity to Francisella tularensis. Nat Immunol. 2010;11:385–393. doi: 10.1038/ni.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Barron L, Smith AM, El Kasmi KC, Qualls JE, Huang X, Cheever A, Borthwick LA, Wilson MS, Murray PJ, Wynn TA. Role of arginase 1 from myeloid cells in th2-dominated lung inflammation. PLoS One. 2013;8:e61961. doi: 10.1371/journal.pone.0061961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lieberman OJ, Orr MW, Wang Y, Lee VT. High-throughput screening using the differential radial capillary action of ligand assay identifies ebselen as an inhibitor of diguanylate cyclases. ACS Chem Biol. 2014;9:183–192. doi: 10.1021/cb400485k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.White LC, Wright KL, Felix NJ, Ruffner H, Reis LF, Pine R, Ting JP. Regulation of LMP2 and TAP1 genes by IRF-1 explains the paucity of CD8+ T cells in IRF-1−/− mice. Immunity. 1996;5:365–376. doi: 10.1016/s1074-7613(00)80262-9. [DOI] [PubMed] [Google Scholar]
- 38.Harada H, Fujita T, Miyamoto M, Kimura Y, Maruyama M, Furia A, Miyata T, Taniguchi T. Structurally similar but functionally distinct factors, IRF-1 and IRF-2, bind to the same regulatory elements of IFN and IFN-inducible genes. Cell. 1989;58:729–739. doi: 10.1016/0092-8674(89)90107-4. [DOI] [PubMed] [Google Scholar]
- 39.Ko J, Gendron-Fitzpatrick A, Splitter GA. Susceptibility of IFN regulatory factor-1 and IFN consensus sequence binding protein-deficient mice to brucellosis. J Immunol. 2002;168:2433–2440. doi: 10.4049/jimmunol.168.5.2433. [DOI] [PubMed] [Google Scholar]
- 40.Warren SE, Armstrong A, Hamilton MK, Mao DP, Leaf IA, Miao EA, Aderem A. Cutting edge: Cytosolic bacterial DNA activates the inflammasome via Aim2. J Immunol. 2010;185:818–821. doi: 10.4049/jimmunol.1000724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mills CD, Ley K. M1 and M2 macrophages: the chicken and the egg of immunity. J Innate Immun. 2014;6:716–726. doi: 10.1159/000364945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xavier MN, Winter MG, Spees AM, den Hartigh AB, Nguyen K, Roux CM, Silva TM, Atluri VL, Kerrinnes T, Keestra AM, Monack DM, Luciw PA, Eigenheer RA, Baumler AJ, Santos RL, Tsolis RM. PPARgamma-mediated increase in glucose availability sustains chronic Brucella abortus infection in alternatively activated macrophages. Cell Host Microbe. 2013;14:159–170. doi: 10.1016/j.chom.2013.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kawai T, Akira S. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat Immunol. 2010;11:373–384. doi: 10.1038/ni.1863. [DOI] [PubMed] [Google Scholar]
- 44.Man SM, Hopkins LJ, Nugent E, Cox S, Gluck IM, Tourlomousis P, Wright JA, Cicuta P, Monie TP, Bryant CE. Inflammasome activation causes dual recruitment of NLRC4 and NLRP3 to the same macromolecular complex. Proc Natl Acad Sci U S A. 2014;111:7403–7408. doi: 10.1073/pnas.1402911111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Man SM, Place DE, Kuriakose T, Kanneganti TD. Interferon-inducible guanylate-binding proteins at the interface of cell-autonomous immunity and inflammasome activation. J Leukoc Biol. 2017;101:143–150. doi: 10.1189/jlb.4MR0516-223R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim BH, Chee JD, Bradfield CJ, Park ES, Kumar P, MacMicking JD. Interferon-induced guanylate-binding proteins in inflammasome activation and host defense. Nat Immunol. 2016;17:481–489. doi: 10.1038/ni.3440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Meunier E, Wallet P, Dreier RF, Costanzo S, Anton L, Ruhl S, Dussurgey S, Dick MS, Kistner A, Rigard M, Degrandi D, Pfeffer K, Yamamoto M, Henry T, Broz P. Guanylate-binding proteins promote activation of the AIM2 inflammasome during infection with Francisella novicida. Nat Immunol. 2015;16:476–484. doi: 10.1038/ni.3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sauer JD, Witte CE, Zemansky J, Hanson B, Lauer P, Portnoy DA. Listeria monocytogenes triggers AIM2-mediated pyroptosis upon infrequent bacteriolysis in the macrophage cytosol. Cell Host Microbe. 2010;7:412–419. doi: 10.1016/j.chom.2010.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Peng K, Broz P, Jones J, Joubert LM, Monack D. Elevated AIM2-mediated pyroptosis triggered by hypercytotoxic Francisella mutant strains is attributed to increased intracellular bacteriolysis. Cell Microbiol. 2011;13:1586–1600. doi: 10.1111/j.1462-5822.2011.01643.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Man SM, Karki R, Sasai M, Place DE, Kesavardhana S, Temirov J, Frase S, Zhu Q, Malireddi RK, Kuriakose T, Peters JL, Neale G, Brown SA, Yamamoto M, Kanneganti TD. IRGB10 Liberates Bacterial Ligands for Sensing by the AIM2 and Caspase-11-NLRP3 Inflammasomes. Cell. 2016;167:382–396. e317. doi: 10.1016/j.cell.2016.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.