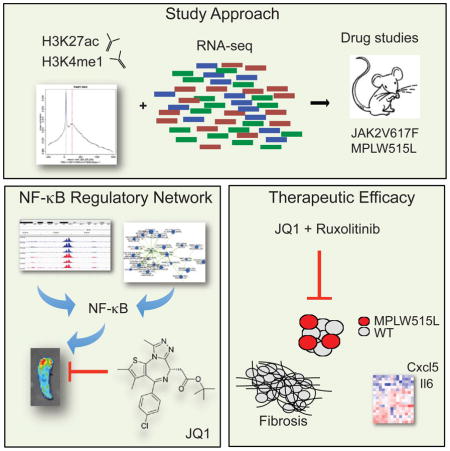
Summary
Genetic and functional studies underscore the central role of JAK/STAT signaling in myeloproliferative neoplasms (MPN). However, the mechanisms that mediate transformation in MPN are not fully delineated and clinically utilized JAK inhibitors have limited ability to reduce disease burden or reverse myelofibrosis. Here we show that MPN progenitor cells are characterized by marked alterations in gene regulation through differential enhancer utilization, and identify NF-κB signaling as a key pathway activated in malignant and non-malignant cells in MPN. Inhibition of BET bromodomain proteins attenuated NF-κB signaling and reduced cytokine production in vivo. Most importantly, combined JAK/BET inhibition resulted in a marked reduction in the serum levels of inflammatory cytokines, reduced disease burden, and reversed bone marrow fibrosis in vivo.
Keywords: Myeloproliferative neoplasms, H3K27ac, chronic inflammation, NF-κB, JQ1
In Brief
Kleppe et al. show that aberrant JAK2 signaling in myeloproliferative neoplasm (MPN) leads to chromatin changes that promote NF-κ B signaling. BET inhibitors reduce NF-κB-induced inflammation and bone marrow fibrosis in MPN models, and combination treatment with BET and JAK inhibitors shows improved efficacy.
Introduction
Myeloproliferative neoplasms (MPNs) are clonal hematopoietic stem cell disorders characterized by dysregulated proliferation of one or more myeloid lineage compartments. The majority of MPNs arise due to somatic mutations that lead to constitutive activation of tyrosine kinase signaling cascades, thus providing the malignant cell with a gain of fitness. The discovery of a single point mutation in the non-receptor tyrosine kinase JAK2 in almost all polycythemia vera (PV) patients and in approximately half of essential thrombocytosis (ET) and primary myelofibrosis (PMF) patients provided critical insight into MPN pathogenesis (Baxter et al., 2005; James et al., 2005; Kralovics et al., 2005; Levine et al., 2005). Subsequent studies have identified mutations that activate JAK2 signaling in JAK2V617F-negative MPN including gain-of function mutations in the thrombopoietin receptor (MPL) (Pikman et al., 2006), loss-of function mutations in the SH2B3 gene (Oh et al., 2010), and recurrent somatic mutations in the calreticulin gene (CALR) (Klampfl et al., 2013; Nangalia et al., 2013).
The discovery of JAK/STAT pathway mutations in the majority of MPN patients provided the rationale for the development of JAK inhibitor therapy, and the JAK1/2 inhibitor ruxolitinib is approved for the treatment of myelofibrosis (MF) (Harrison et al., 2012). JAK kinase inhibitors reduce splenomegaly and alleviate systemic symptom burden, but do not eliminate or markedly attenuate the malignant clone in MPN patients and have little to no impact on bone marrow (BM) fibrosis (Quintas-Cardama et al., 2011). Further, MPN patients exhibit significantly elevated circulating levels of pro-inflammatory cytokines, which contribute to MPN associated symptoms and sequelae; moreover circulating cytokine levels have prognostic relevance in MF (Mondet et al., 2015; Tefferi et al., 2011). Although inflammation is a characteristic feature of MPNs, the underlying mechanisms driving the chronic inflammatory state in MPN patients remain largely elusive. We recently demonstrated that both malignant and non-malignant hematopoietic cells are the source of pro-inflammatory cytokines in MPN and that inhibition of JAK/STAT pathway activation in malignant and non-malignant cells is required to achieve the therapeutic efficacy of JAK kinase inhibition (Kleppe et al., 2015).
Emerging therapeutic strategies targeting epigenetic mechanisms of disease have shown significant promise in various hematological malignancies (Dawson and Kouzarides, 2012; Fong et al., 2014). Notably, it has been reported that different epigenetic mechanisms regulate the expression of inflammatory cytokines in different disease states (Yasmin et al., 2015). Recent studies have suggested an important role for the histone lysine reader BRD4 in mediating pathologic inflammation in different contexts, including sepsis, pulmonary fibrosis and heart failure (Anand et al., 2013; Brown et al., 2014; Nicodeme et al., 2010; Tang et al., 2013). Despite these important insights, the gene regulatory mechanisms that govern aberrant inflammation in MPN and in other malignant contexts have not been delineated. In addition, although mutations in epigenetic regulatory proteins are common in MPN, the role of alterations in transcriptional regulation in MPN pathogenesis is not well elucidated. One seminal study demonstrated a direct link between JAK2 activity to histone phosphorylation (Dawson et al., 2009), however it remains unknown whether constitutive JAK2 signaling induces alterations in the cis-regulatory landscape of MPN cells and how this may lead to cell-autonomous and cell-non-autonomous alterations which mediate transformation in vivo. Given inflammation contributes to constitutional symptoms, BM fibrosis, extramedullary hematopoiesis (EMH), and disease progression, detailed investigation of the mechanisms which regulate inflammatory signaling in MPN is of great importance.
Results
Constitutive JAK2 Activation Leads to Alterations in Enhancer Utilization in MF Progenitors
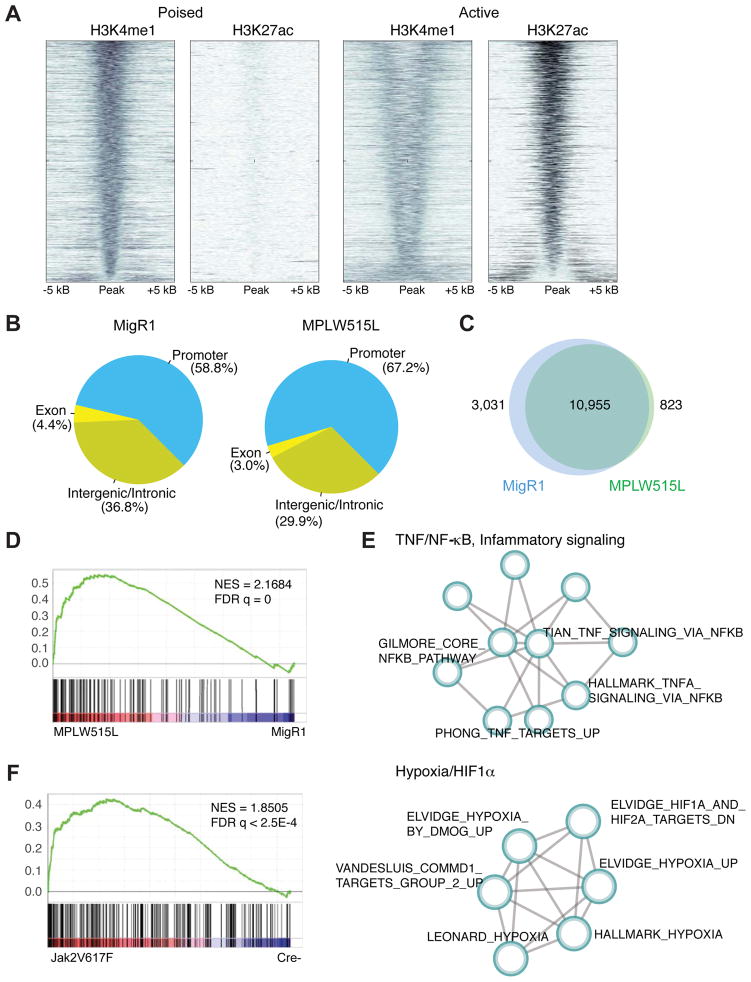
Enhancer function underlies regulatory processes by which cells establish patterns of gene expression. H3K27ac marks demarcate active enhancers, whereas H3K4me1 marks define both active and poised enhancers (Shlyueva et al., 2014). To begin to understand the effect of aberrant JAK2 signaling on the MPN epigenome and to determine underlying regulatory networks in MPN cells, we purified megakaryocyte-erythroid progenitor cells (MEPs) (which we have shown previously mediate aberrant inflammation in MF (Kleppe et al., 2015)) from MPLW515L-diseased mice (MF mice) and controls and performed chromatin immunoprecipitation (ChIP) for H3K4me1 and H3K27ac. This allowed us to reveal differentially active and poised enhancers and promoters in purified MF progenitors (Figure 1A). We identified 11,749 H3K27ac peaks and 28,263 H3K4me1 peaks in MEPs from MF mice. We then classified MF enhancers based on H3K4me1 and H3K27ac deposition as poised (n = 2,465) or active (n = 3,376). Analysis of the genomic binding profile showed altered distribution of H3K27ac peaks across genomic regions in MF progenitors compared to controls. The number of H3K27ac peaks residing in promoter regions of the genome was increased in MF progenitors (67.15% versus 58.81%) while the number of peaks residing in exonic and intronic/intragenic regions was decreased (32.85% versus 41.21%) (Figure 1B). To gain a better understanding of the chromatin landscape of MF progenitors, we next compared the H3K27ac ChIP-seq peak profiles of MPLW515L-positive MEPs and control samples. We identified 3,854 differentially enriched ChIP-seq peaks, with 823 gained and 3,031 lost peaks in MF progenitors in comparison to controls (Figure 1C). We then ranked the differentially enriched ChIP-seq peaks by log2-transformed fold change statistics and applied GSEA to identify functional gene sets. By fitting a Beta-Uniform mixture model to the raw GSEA p values, we selected gene sets that deviated from the random background and subsequently identified optimized sub-networks using a graph partition algorithm. Using this approach, we found that active loci in MF progenitors are significantly associated with signaling pathways linked to the TNF/NF-κB/inflammatory signaling and hypoxia/HIF1 pathways (Figures 1D, 1E, S1A, and Table S1), suggesting that a NF-κB-dependent regulatory network sustains the inflammatory state observed in MF mice. Analysis of the chromatin landscape of JAK2V617F-positive MEPs, but not LSK or GMPs (data not shown), showed similar enrichment of the TNF/NF-κB signaling pathway at both TSS and enhancers compared to control cells (Figures 1F and S1B), further highlighting an important role of an NF-κB-dependent regulatory network in MPN pathogenesis.
Figure 1. Alterations in the cis-regulatory landscape of MPN cells.
A) Density of ChIP-seq reads for H3K4me1 and H3K27ac relative to midpoint at putative poised and active enhancers. Data from sorted MPLW515L-positive MEPs is shown. n=3/mark. B) Distribution (%) of H3K27ac and H3K4me1 peaks over the promoters (5 kb upstream of TSS), coding exons, intronic and distal intergenic regions. Data from GFP-positive MEPs sorted from healthy (MigR1) or MPLW515L-diseased mice are shown. C) Number of differentially enriched ChIP-seq peaks in MF progenitors in comparison to controls. D) GSEA pathways analysis of H3K27ac data (TSS) from MPLW515L-positive MEPs compared to control (MigR1). NES, normalized enrichment score; FDR, false-discovery rate. E) Optimized gene sub-network identified from the analysis of H3K27ac ChIP-seq data from control and MPLW515L-positive MEPs. F) GSEA pathways analysis of H3K27ac data (TSS) from JAK2V617F-positive MEPs compared to control (Cre-). NES, normalized enrichment score; FDR, false-discovery rate. See also Figure S1 and Table S1.
JAK2 Activation Dependent Alterations in Chromatin State Drive Chronic Inflammation
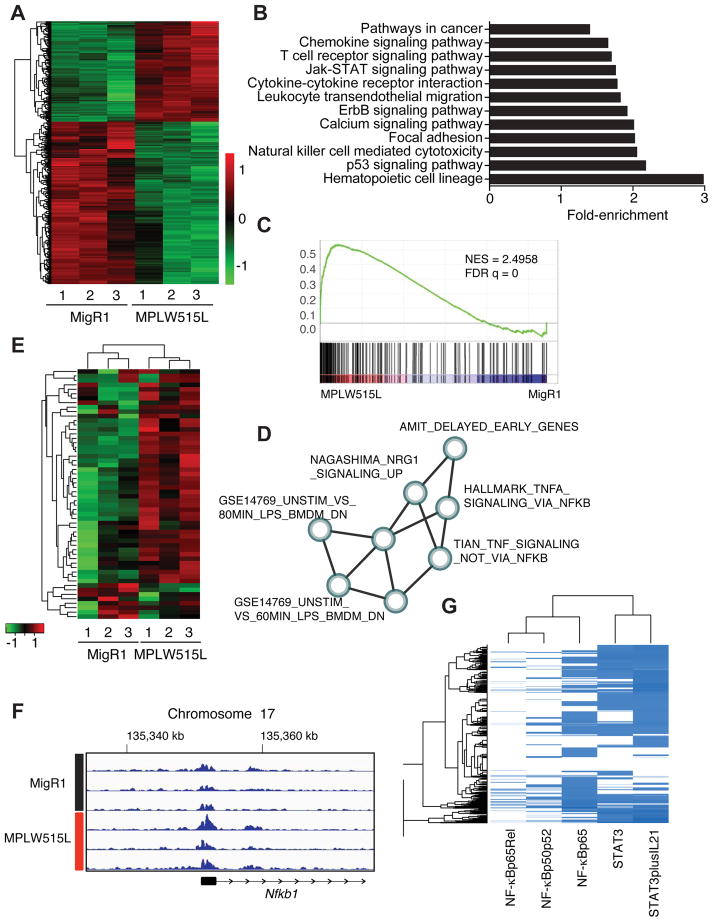
To gain further insight into the transcriptional pathways regulated by JAK/STAT pathway activating mutations, we next performed transcriptional profiling of purified MEPs from MF and control mice. Unsupervised hierarchical clustering robustly partitioned the samples into their respective genotypes (Figure 2A). To elucidate MPLW515L-dependent gene expression changes, we applied DESeq2 to identify differentially expressed genes (DEGs) (adjusted p value <0.01 and absolute value of log2-fold change > 1) of which 850 genes were upregulated and 499 genes were downregulated in MPLW515L-expressing MEPs compared to empty vector control cells (Table S2). KEGG pathway enrichment analysis of DEGs showed enrichment of 28 (p value <0.01) gene ontology terms, including hematopoietic cell lineage, cytokine-cytokine receptor interaction, JAK/STAT signaling pathway, and chemokine signaling pathway, consistent with the known role of JAK2 in cytokine signaling and hematopoietic lineage decision (Figure 2B and Table S3).
Figure 2. TNF/NF-κB signaling axis represents a central signaling node in MF progenitors.
A) Hierachical clustering of differentially expressed genes from control and MPLW515L-positive MEPs. Adjusted p value <0.01 (Wald test) and abs(log2FoldChange)>1. Green: negative values, red: positive values. n=3/group. B) KEGG pathway enrichment analysis of differentially expressed genes. x-axis shows fold enrichment in MPLW515L-positive MEPs compared to control. p value <0.01. C) GSEA pathways analysis of DEGs from MPLW515L-positive MEPs compared to control (MigR1). NES, normalized enrichment score; FDR, false-discovery rate. Kolmogorov-Smirnov test. D) Optimized gene expression sub-network identified from gene expression profiles. Detailed information about the creation of sub-networks and a list of gene sets can be found in STAR Methods, Figure S2 and Table S1. E) Heatmap depicting expression of core genes that accounts for the HALLMARK_TNFA_SIGNALING_VIA_NFKB gene set enrichment signal. MigR1: control MEPs from empty vector transplanted mice, MPLW515L: mutant MEPs isolated from MF mice. n=3. F) ChIP-sequencing tracks for H3K27ac at the Nfkb1 gene locus for MF progenitors (MPLW515L) and control cells (MigR1). Numbers indicate the genes location on chromosome 17. G) Analysis of the co-occurrence between canonical STAT3 (2 sites) and p65/NF-κB (3 sites) transcription factor bindings sites in the regulatory regions of the DEGs in MPLW515L-positive MEPs. See also STAR Methods, Figure S2, and Tables S2 and S3.
We next assessed for pathway enrichment by performing GSEA and subsequently identified key optimal sub-networks (Figures S2A and B) (Bader and Hogue, 2003). Utilizing this network tool, we identified three major clusters related to cytokine signaling, TNF/NF-κB signaling, and STAT signaling (Figure 2C, D and Table S1). The HALLMARK_TNFA_SIGNALING_VIA_NFKB gene set represented the core expression signature of the TNF/NF-κB expression cluster and Tnf itself was found to be highly upregulated in MF progenitors compared to control cells (2.8-fold, q value <0.001, Figures 2C–E, Table S2). Consistent with the finding of dysregulated TNF/NF-κB pathway signaling in MF progenitors by gene expression and chromatin state analysis, integrated analysis of gene expression and H3K27ac occupancy data revealed a significant association between epigenetic and gene transcription data (Figures 2E, 2F, and S2C), suggesting integrated epigenetic and transcriptional networks that regulate inflammatory signaling in MF progenitors. We recently showed that STAT3, a transcription factor known to collaborate and co-regulate key target genes in cis with NF-κB (Grivennikov and Karin, 2010), is required for cytokine production in MPN (Kleppe et al., 2015). We therefore assessed for the presence of STAT3 and NF-κB binding sites (Figure S2D) in the regulatory regions of DEGs in MPLW515L-positive and JAK2V617F-positive MPN cells (Table S2). Notably, we found a strong co-occurrence and enrichment for canonical p65/NF-κB and STAT3 DNA bindings sites in DEGs in both models (Figures 2G and S2E). Taken together, these data underscore the dysregulation of cytokine signaling in MF progenitors and, in addition, suggest an important role for the inflammatory mediator NF-κB in mediating oncogenic effects of JAK2.
Aberrant NF-κB Pathway Activation in MPN Mouse Models
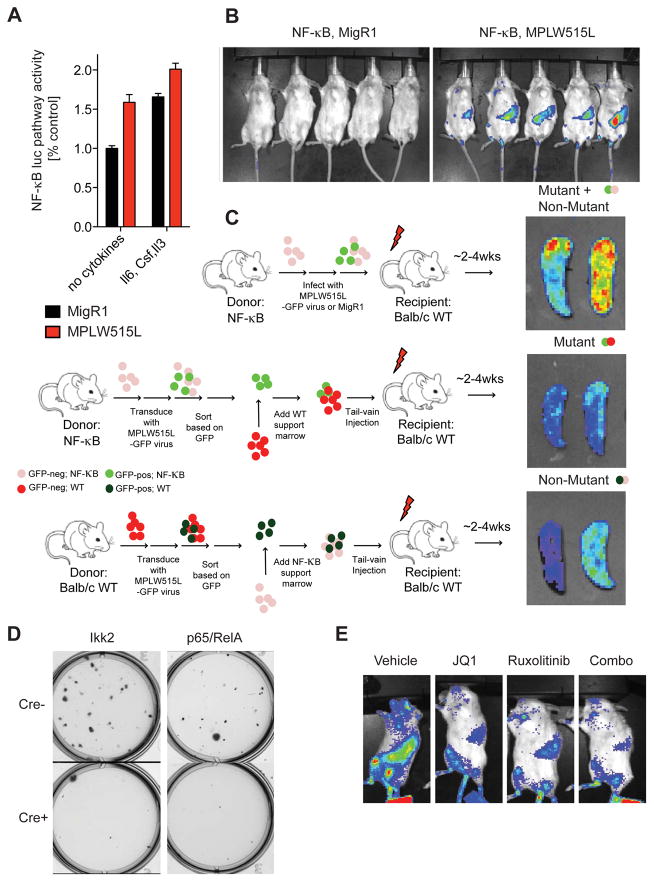
To begin to understand the role of NF-κB signaling in the pathogenesis of MPNs, we first used a reporter mouse expressing firefly luciferase gene under the control of NF-κB DNA binding sites (termed NF-κBluc hereafter, Taconic and (Carlsen et al., 2002)). We measured luciferase expression of sorted GFP-positive, MPLW515L-expressing stem and progenitor cells from NF-κBluc mice upon culture with (Il6, Csf, Il3) or without cytokines for 24 hours. We observed increased NF-κB activity in the presence and absence of cytokines in MPLW515L-positive cells compared to control cells (Figure 3A). By using NF-κBluc as donors in the MPLW515L bone marrow transplantation (BMT) assay, we found that mice transplanted with MPLW515L-positive, reporter-positive cells show marked luciferase activity compared to control mice receiving empty vector, reporter-positive cells consistent with cell autonomous NF-κB activation in vivo (Figure 3B). We next wanted to examine whether NF-κB is active in both mutant and non-mutant cells in MF. We transplanted MPLW515L-mutant, reporter-negative cells with reporter-positive support cells (non-mutant) or mutant, reporter-positive cells with wild-type reporter-negative support cells (mutant) into lethally irradiated wild-type recipient mice (Figure 3C). Bioluminescent imaging showed strong NF-κB pathway activation in each cohort consistent with NF-κB activation in mutant MPN cells and in non-mutant cells through cell non-autonomous mechanisms (Figure 3C). Consistent with these data, genetic deletion of p65 or Ikk2 attenuated cytokine-independent proliferation of MPLW515L-positive progenitor cells suggesting a role for NF-κB signaling in MPN pathogenesis (Figure 3D).
Figure 3. Constitutive NF-κB pathway activation in MPN mouse models.
A) NF-κB luciferase reporter assay of sorted GFP-positive, ckit-positive BM cells. Cells were either cultured for 24 hours in absence of cytokines (no cytokines) or in media supplemented with Il6 (100 ng/ml), Il3 (10 ng/ml), and SCF (10 ng/ml). Luminescent signals were normalized to MigR1-transduced cells cultured without cytokines. n=3. Data represents mean values ± S.E.M. B) In vivo BLI of mice transplanted with NF-κBluc reporter BM cells. BM cells were either infected with viral supernatant containing empty vector (MigR1) or MPLW515L. n=5/group. Images are representative of two independent experiments and were taken ~14 days post transplantation. C) Right: ex vivo BLI of spleens from MPLW515L-diseased mice. Cell type expressing the NF-κB luciferase reporter is indicated above each image. n=4/group. Left: schematic depiction showing BMT design to assess NF-κB activation in mutant and non-mutant cells. D) Representative image of methylcellulose colony plate (6-well plate, 9 cm2 surface area) 7–10 days after plating sorted GFP-positive, ckit-positive BM cells. Cells were harvested from floxed Ikk2 or p65/RelA mice. Images are representative of 2 independent experiments and were performed in triplicate. E) In vivo BLI of MPLW515L-diseased mice treated with ruxolitinib, JQ1, ruxolitinib plus JQ1, or vehicle for three days. See also Figure S3.
BET Inhibition Attenuates NF-κB Transactivation in Vivo
Recent data implicating BRD4 function in NF-κB-induced inflammation in atherosclerosis (Brown et al., 2014) suggested to us that BRD4/BET proteins may have an important role in NF-κB-driven MPN-associated inflammation. To investigate whether BET bromodomain inhibitor JQ1 (Filippakopoulos et al., 2010) affects NF-κB pathway activation in vivo, we transplanted lethally irradiated wild-type mice with MPLW515L-expressing, NF-κB reporter positive cells and imaged MPLW515L-diseased mice after 3 days of therapy with vehicle, JQ1 (50 mg/kg, i.p., QD), the JAK1/2 inhibitor ruxolitinib (60 mg/kg, p.o., BID), or JQ1/ruxolitinib combination therapy in vivo. Interestingly, while both mice receiving JQ1 and ruxolitinib alone showed a reduction in NF-κB pathway activation, the effect was significantly more potent when both drugs were administered as combination therapy (Figures 3E and S3). This data suggests that BET protein function and JAK/STAT signaling play a role in aberrant NF-κB activation in MPN, and that these effects may be mitigated using combination targeted therapy against activated signaling and altered epigenetic regulation.
BET Inhibition Attenuates Inflammatory Cytokine Production in MF
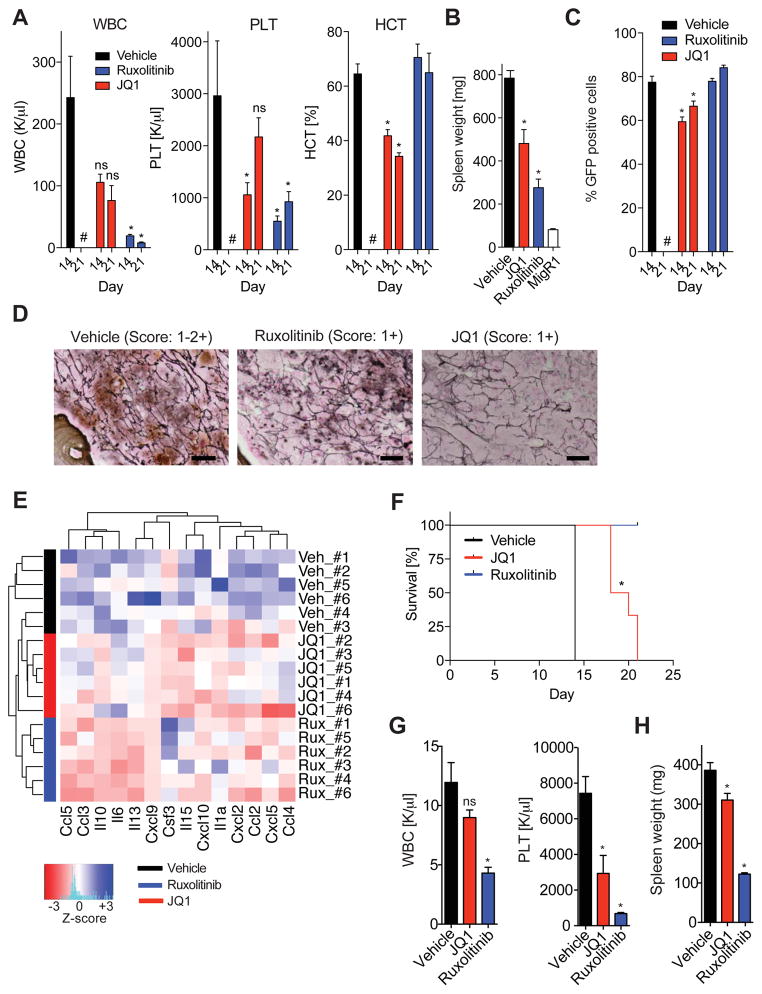
Given the cell autonomous and cell non-autonomous NF-κB activation observed in MF in vivo and the ability of BET inhibition to attenuate NF-κB activation in MPN, we investigated the efficacy of the BET protein inhibitor JQ1 in our adoptive transfer model of MPLW515L-mutant MF (Pikman et al., 2006). After all recipient mice had disease including leukocytosis, inflammatory cytokine production, and BM fibrosis, we began treatment with JQ1 (50mg/kg, QD), ruxolitinib (90mg/kg, BID), or vehicle control. White blood counts (WBC), platelet numbers (PLT), and hematocrit levels (HCT) were significantly reduced in JQ1 treated mice in comparison with control mice (p value <0.05, Figure 4A). Furthermore, JQ1 therapy resulted in lower spleen weights, decreased EMH in liver and spleen, a reduction in reticulin fibrosis, and a decrease of the proportion of GFP-positive mutant cells in the peripheral blood (PB) (Figures 4B–D, and S4). Most importantly, JQ1 therapy reduced the level of pro-inflammatory cytokine levels in the circulation of MF mice (Figure 4E). JQ1 treatment significantly improved survival compared with vehicle-treated mice (19 days versus 14 days, p value <0.001, log-rank test, Figure 4F). Similar to our findings in MF mice, JQ1 also lowered WBC, PLT, and spleen weights in mice engrafted with JAK2V617F-mutant MPN cells (Figures 4G and 4H).
Figure 4. JQ1 monotherapy shows therapeutic efficacy in MPN in vivo.
A) White blood cell counts (WBC, K/μl), platelet counts (PLT, K/μl), and hematocrit levels (HCT, %) of MPLW515L-diseased mice treated with vehicle, ruxolitinib, or JQ1 at 14 days and 21 days. Representative data from two independent experiments are shown with 4–6 mice per treatment condition. #, no data for vehicle-treated mice at 21 days available. *p value <0.05. B) Bar graph showing spleen weights (mg) of MF mice treated with vehicle, ruxolitinib, or JQ1 for 21 days. n=4–6 mice/condition. Data are representative of two independent experiments. *p value <0.05. C) Percentage of GFP-positive cells in the PB of MF mice treated with vehicle, ruxolitinib, or JQ1. Frequencies measured at 14 and 21 days are shown. *p value <0.05. #, no data for vehicle-treated mice at 21 days available. D) Representative images showing reduced reticulin fibrosis in the BM of MF mice treated with ruxolitinib or JQ1 compared to vehicle control mice. n=3 mice per group. Scale bar, 10 μM. E) Serum cytokine levels of MF mice treated with vehicle, ruxolitinib, or JQ1 for 21 days. Color bars indicate treatment group. Heatmap shows z-scores. F) Kaplan-Meier survival analysis (%) of MPLW515L-diseased mice treated with vehicle, ruxolitinib, or JQ1. *p value <0.05 (log-rank test). n=6/condition. G, H) JAK2V617F-diseased mice treated with vehicle, ruxolitinib (60 mg/kg, BID), or JQ1 (50 mg/kg, QD), for 28 days. WBC counts (K/μl) and PTL levels (K/μl). (G) Spleen weights. (H) n=5 mice/condition. *p value <0.05. ns, not significant. Data represent mean values ± S.E.M. The Student’s t-test (unpaired, two-tailed) was used to compare the mean of two groups. See also Figure S4.
Gene Expression Signatures are Unique to each Treatment Group
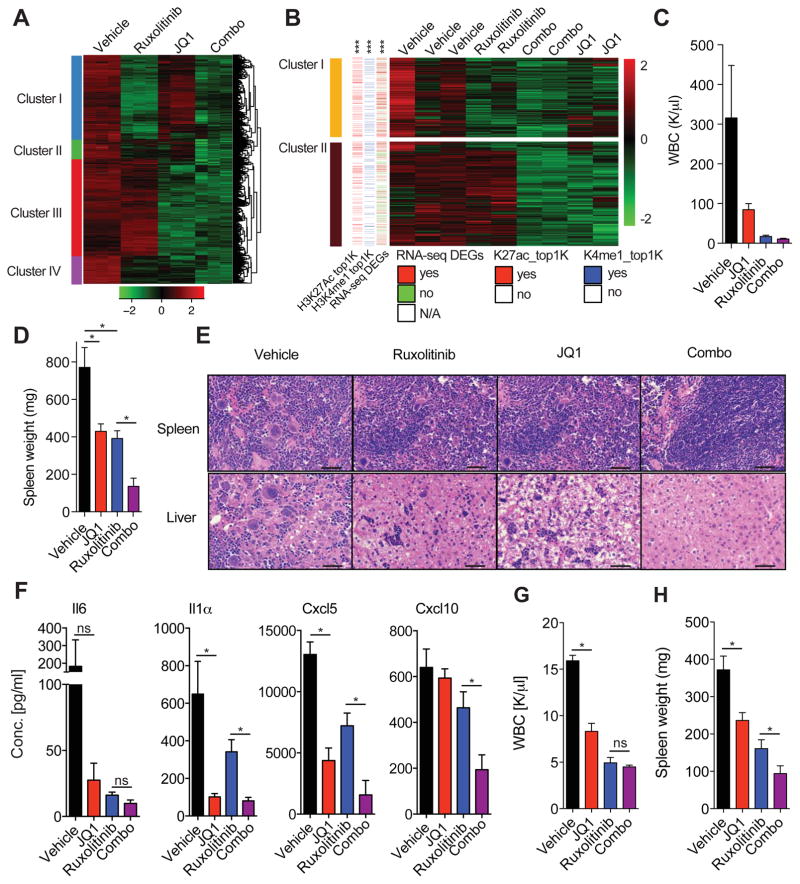
We next studied the effect of vehicle, JQ1 alone, ruxolitinib alone, and combination JQ1/ruxolitinib therapy on transcriptional output of JAK2V617F-mutant SET-2 cells. RNA-sequencing analysis showed clear segregation and clustering of all groups (Figure 5A). Importantly, among the genes, we identified four distinct clusters with differentially expressed genes associated with a specific treatment condition (Figure 5A). Genes in cluster 1 were mostly downregulated in SET-2 cells treated with ruxolitinib alone or with ruxolitinib in combination with JQ1. Cluster 2 contained genes with downregulated expression upon combined JAK/BET inhibition only. Genes downregulated by JQ1 therapy alone or in combination with ruxolitinib were found in cluster 3. Cluster 4 contained genes that were not associated with a specific treatment group, but downregulated in all groups in comparison to vehicle-treated controls (Figure 5A). With the exception of the genes in cluster 2, gene ontology annotations indicated that down-regulated genes were associated with NF-κB signaling (Table S4), in agreement with our data showing reduction of NF-κB activity in response to JQ1 and ruxolitinib therapy which is augmented by combination therapy.
Figure 5. RNA-sequencing analysis reveals inhibitory effect of JQ1 on TNF/NF-κB signaling network.
A) Hierarchical clustering analysis of RNA-sequencing data (z-score of VOOM-normalized read counts, with green negative values and red positive values) from indicated SET-2 samples. Depicted heatmap only shows genes which are downregulated by ruxolitinib/JQ1 combination therapy compared to DMSO control cells (FDR<0.05, log2-fold change greater than 1 or smaller than −1 according to a two class linear model generated following limma standard recommandations). Data from biological triplicates are shown. List of select enriched biological process Gene Ontology (GO) terms for each cluster can be found in Table S4. B) Heatmap depicting RNA-seq data for indicated samples (absolute fold change > 2, FDR-adjusted p-value < 0.01). Color code indicates whether a gene contains a H3K27ac (red: yes, white: no) or H3K4me1 peak (blue: yes, white: no) belonging to the top 1000 significant differential peaks (K27ac_top1K and H3K4me1, respectively) or is differentially expressed (DEGs, red: yes, green: no, white: N/A). ***p value <1e-6 (overlap significance). Clusters 3–5 are presented in Figure S5. C) WBC counts (K/μl) of MPLW515L-diseased mice treated for 14 days. D) Bar graph showing spleen weights (mg) of animals treated for 21 days (vehicle and JQ1) or 28 days (ruxolitinib and combo). n=4–5. *p value <0.05. Data is representative of three independent experiments. E) Representative images showing H&E stain of spleen and liver from MF mice treated for 21 days (vehicle and JQ1) or 28 days (ruxolitinib and combo). Data shown are representative of three independent experiments with 4–5 mice per group. Scale bar, 50 μM. F) Serum cytokine levels of MF mice treated for 21 days (vehicle and JQ1) or 28 days (ruxolitinib and combo). n=4–5/group, *p value <0.05. G, H) JAK2V617F-diseased mice treated with vehicle, JQ1, ruxolitinib, or the combination for 28 days. WBC. (G) Spleen weights. (H) n=3–6 mice per group, *p value <0.05, ns, not-significant. Data represent mean values ± S.E.M. The Student’s t-test (unpaired, two-tailed) was used to compare the mean of two groups. See also Figure S5 and Table S4.
We next performed RNA-sequencing analysis of mutant MEPs sorted from vehicle-, JQ1-, ruxolitinib-, or JQ1/ruxolitinib-treated mice. We identified five distinct clusters with each cluster associated with a specific therapeutic regimen (Figures 5B and S5A). Genes in cluster 1 were downregulated in all groups in comparison to vehicle-treated controls, genes found in cluster 2 were specifically associated with JQ1 therapy, when given alone and in combination with ruxolitinib (Figure 5B). Notably, integration of ChIP-sequencing and RNA-sequencing data comparing wild-type and mutant MEPs revealed that genes in cluster 1 are significantly enriched for the top 1000 differential H3K27ac peaks (ranked based on DESeq2 statistics), the top 1000 differential H3K4me1 peaks, and genes which were up-regulated in mutant MEPs compared to wild-type MEPs (***p value <1e-6) (Figures 5B and S5A). Genes in cluster 1 were significantly enriched for the JAK2/STAT signaling pathway (p value=1.8e-4), while genes in cluster 2 were significantly enriched for the NF-κB signaling pathway (p value =0.02) (Figure S5B). Taken together, these data demonstrate that BET and JAK inhibition differentially affect the transcriptional output of MPN cells when administered alone and in combination, suggesting that combined JAK/BET inhibition can lead to synergistic alterations in transcriptional output in MPNs.
Combined BET and JAK Inhibition Abrogates Murine MPN
JAK inhibition can reduce cytokine production and attenuate features of MPD in vivo by itself, but JAK inhibitors do not lead to pathologic or molecular responses and have little to no effect on BM fibrosis. We therefore sought to investigate the efficacy of combined JAK/BET inhibition on cytokine production, BM fibrosis, and tumor burden in vivo. Combined JAK/BET inhibition reduced WBC and spleen weights to a degree not observed by either therapy alone (Figures 5C, D and S5C). Furthermore, while EMH in spleen and liver was partially reduced in ruxolitinib-treated mice in comparison with vehicle-treated mice, combined JAK/BET inhibition resulted in near-complete absence of portal and lobular cellular infiltrates and splenic EMH (Figure 5E). In addition, megakaryocyte infiltration was decreased in the spleen of mice receiving combination therapy compared to ruxolitinib alone (Figures 5E). Consistent with these effects, we observed a greater suppression of cytokine production in mice receiving combination therapy, with further attenuation of specific cytokines, including Il1 and Il6 (Figure 5F). Similar to our findings in MF mice, combined JAK/BET inhibition showed increased therapeutic efficacy in JAK2V617F-diseased mice compared to JQ1 and/or ruxolitinib monotherapy (Figures 5G, 5H, and S5D–F). In addition, combined JAK/BET inhibition substantively decreased the number of erythroid progenitors in the bone marrow of primary JAK2V617F mice and led to a significant reduction of red cell blood parameters in JAK2V617F-mutant mice (Figure S5D–F).
Combined BET and JAK inhibition Reduces Mutant Allele Burden and Eliminates Fibrosis
The effect of ruxolitinib on fibrosis and JAK2V617F allele burden in the clinic has only been modest consistent with a lack of long-term disease modification with type I JAK inhibitor therapy. Moreover, there are no agents which have shown an ability to reverse BM fibrosis in MPN. Consistent with previous reports, ruxolitinib treatment of MF mice failed to eliminate fibrosis and did not reduce the proportion of GFP-positive mutant cells in the periphery and target organs (Figures 6A–C). Strikingly, combined JAK/BET inhibitor therapy completely eliminated fibrosis in MF mice (Figures 6A, 6B and S6A). In addition, JQ1 inhibitor therapy alone or in combination with ruxolitinib significantly decreased the proportion of GFP-positive, mutant cells in the peripheral blood and BM of MF mice (Figure 6C). These data suggest that BET inhibition, alone and in combination with JAK kinase inhibition, can attenuate disease burden and reverse myelofibrosis in vivo.
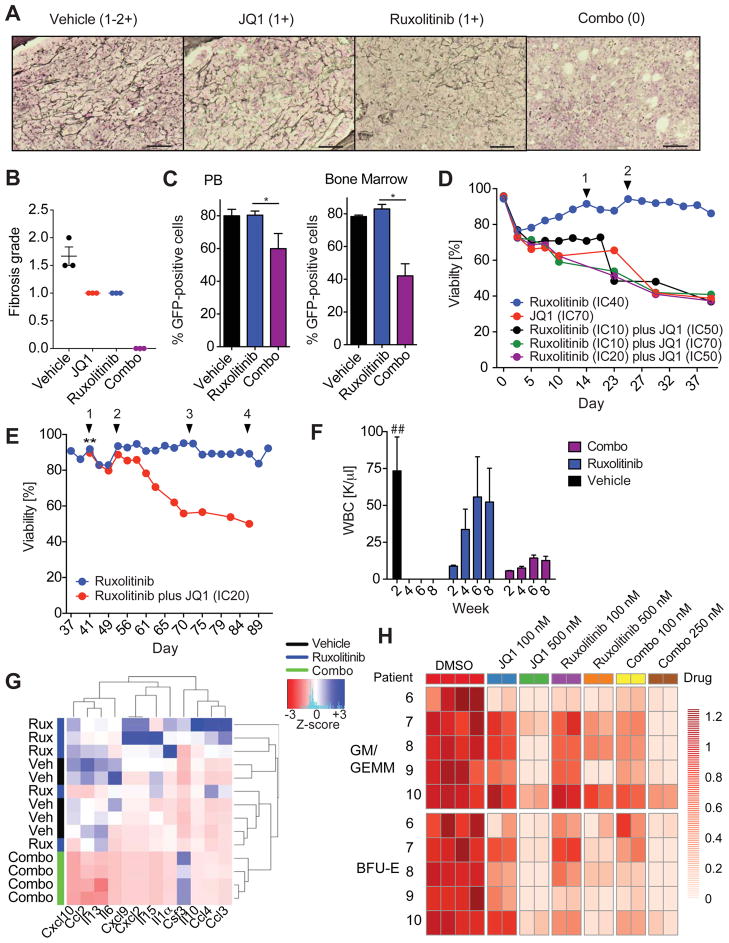
Figure 6. Combined JAK/BET inhibition reverses BM fibrosis and delays JAK inhibitor associated persistence.
A) Representative images showing absence of BM fibrosis in the BM of MF mice treated with vehicle, ruxolitinib, JQ1, or combo for 21 days (vehicle and JQ1) or 28 days (ruxolitinib and combo). Scale bar, 50 μM. B) Fibrosis grading of the bone marrow of MPLW515L-diseased mice treated with vehicle, JQ1, ruxolitinib, or combo (n=3/group). C) Bar graph showing the percentage of viable, GFP-positive cells in the PB and BM of MF mice treated as indicated. n=4–5 mice/condition. *p value <0.05. D) Assessment of viability (%) of SET-2 cells cultured in the presence of increasing concentrations of ruxolitinib and JQ1 alone or in combination. Arrowheads indicate an increase in the ruxolitinib concentration (1: 125 nM, 2: 150 nM). Data are representative of four independent experiments. IC, inhibitor concentration. E) Ruxolitinib-persistent cells (IC40: 125 nM) remain sensitive to treatment with JQ1. ** indicates addition of JQ1 inhibitor (IC20: 100 nM) to SET-2 cells cultured in the presence of increasing concentrations of ruxolitinib. Arrowheads indicate an increase in the concentration of ruxolitinib (1: 150 nM, 2: 200 nM, 3: 300 nM, 4: 500 nM). Data are representative of three independent experiments. F) WBC measured at week 2, 4, 6, and 8 after treatment initiation are shown. Vehicle-treated mice showed signs of morbidity after 2 weeks of treatment and were sacrificed and analyzed at this point. n= 5/treatment group. ##, data for vehicle-treated mice only available for week 2 time point. G) Serum cytokine levels of MF mice treated as indicated. Serum was collect at study endpoint (vehicle: 2 weeks, ruxolitinib and combo: 8 weeks). Color bars indicate treatment group. Hierarchical clustered heatmap shows z-scores. n=4–5/group. H) Colony formation unit (CFU) assays of CD34-positive MF cells. Each row represents the data from one patient with colony counts normalized to DMSO control wells. Combo 100 nM and Combo 250 nM indicates that cells were exposed to 100 nM or 250 nM of each drug. n=2 wells/condition is shown. Results from 5 different patients are shown. Data shown represent mean values ± S.E.M. The Student’s t-test (unpaired, two-tailed) was used to compare the mean of two groups. See also Figure S6, Tables S5 and S6.
BET Inhibition Delays Persistence Associated with JAK Inhibitor Therapy
We previously demonstrated chronic exposure of MPN cells to type-I JAK inhibitors results in a persistence phenotype by which MPN cells survive in the setting of JAK kinase inhibition, highlighting the need for alternative therapies to combat drug resistance/persistence in MPN patients (Koppikar et al., 2012). JAK inhibitor persistence is reversible and is associated with site-specific changes in chromatin state, consistent with an epigenetic mechanism by which MPN cells evade JAK kinase inhibition. We therefore tested the impact of BET protein inhibition on the development and maintenance of JAK inhibitor persistence. JQ1 prevented the development of JAK inhibitor persistence and JAK inhibitor persistent SET-2 cells remained sensitive to BET inhibition (Figures 6D, 6E, and S6B).
To test whether BET inhibition can also delay JAK inhibitor-associated persistence in vivo, we treated MF mice with ruxolitinib alone or in combination with JQ1 for 8 weeks and assessed peripheral counts over time. Mice receiving chronic JAK1/2 inhibitor therapy developed persistence and disease breakthrough as shown by increased WBC counts within 4 weeks of therapy (Figure 6F). In contrast, the WBC of mice treated with JAK/BET combination therapy remained suppressed during the entire course of treatment (Figure 6F). Importantly, clustering analysis of serum cytokine data at study endpoint did not allow segregation of ruxolitnib-treated and vehicle-treated mice (Figure 6G). In contrast, all co-treated mice clustered together and showed a significant reduction of pro-inflammatory cytokine levels in mice receiving combination therapy compared to vehicle and ruxolinitib-treated mice (Figure 6G), consistent with an anti-inflammatory effect of combined JAK/BET inhibition.
Combined Ruxolitinib/JQ1 Therapy shows Efficacy against Primary MPN Cells
We next assessed the impact of JQ1 therapy, ruxolitinib therapy, and combination therapy on the proliferation of primary CD34+ progenitor cells from 10 MPN patients in vitro. We found that primary CD34+ progenitor cells from MF patients were sensitive to JQ1 and to ruxolitinib. Of note a subset of patient samples (3 out of 5) which required higher concentrations of ruxolitinib to block colony formation retained sensitivity to BET inhibition (Figure S6C). Further, combined JAK/BET inhibition showed significantly increased efficacy compared to either monotherapy (p value <0.05 (GLM), Figure 6H). Genomic analysis of colonies from patients with multiple mutations, including high-risk MPN disease alleles (ASXL1), showed that combined ruxolitinib/JQ1 therapy showed efficacy against all MPN clones including clones with multiple disease alleles (Tables S5, S6 and Figure S6D).
Discussion
Recent studies in MPN patients and in preclinical MPN models have shown that MPNs, in particular MF, are characterized by a chronic state of inflammation. In addition, increased levels of circulating cytokines are linked to adverse outcome in MF (Tefferi et al., 2011), consistent with a key role for inflammatory signaling in MPN progression and disease maintenance. These observations provide a strong rationale to investigate underlying gene regulatory mechanisms which sustain chronic inflammation in MPN. By integrating RNA-seq and ChIP-seq data, we have uncovered a NF-κB-dependent transcriptional network that fuels the inflammatory state in MPN and is amenable to therapeutic intervention. These data provide insights into MPN pathogenesis and provide a rationale for mechanism-based clinical trials.
The role of NF-κB as a master regulator of inflammation is well understood in many diseases; however, little is known about the role of this key transcriptional pathway in MPN-associated inflammation. We identified an NF-κB-dependent regulatory network by combined analysis of ChIP-seq and gene expression changes in 2 different MPN mouse models, suggesting NF-κB acts as an important inflammatory signaling node in MPN. Using in vivo imaging, we confirmed that NF-κB is activated in both mutant and non-mutant hematopoietic cells in MPL-diseased mice in vivo suggesting that NF-κB activity functions, at least in part, in a non-cell autonomous fashion in MPN. The finding that NF-κB is constitutively active in MPN mouse models raises intriguing questions relating to molecular mechanisms mediating crosstalk between JAK/STAT and NF-κB pathways. We recently reported a role for the JAK2 downstream target STAT3 in MPN pathogenesis and in mediating cytokine production from mutant and non-mutant cells in MF (Kleppe et al., 2015). Growing evidence suggests NF-κB as a key transcriptional co-regulator with activated STAT3 in different pathologic states driven by aberrant inflammation (Grivennikov et al., 2009; Lee et al., 2009). Recent studies in epithelial tumors reported molecular cross-talk between NF-κB and STAT3 in gene regulation (Atkinson et al., 2010; Bollrath and Greten, 2009; Grivennikov and Karin, 2010). Our study suggests that cooperation and co-regulation of key target genes by the two master regulators STAT3 and NF-κB drives the inflammatory state in MPN, similar to what has been described in other pathological states (Grivennikov and Karin, 2010). Future studies will have to determine how precisely these two inflammatory signaling pathways interact in MPN and how cytokine loci are jointly regulated by NF-κB and JAK/STAT mediated alterations in transcriptional control.
The mechanisms by which the NF-κB pathway is regulated by BET proteins remains only particularly understood. BRD4 has been shown to transcriptionally co-activate NF-κB through recognition and direct binding to acetylated p65 (Huang et al., 2009). Our work suggests that BET protein function is required for pathologic transcriptional NF-κB activity in MPN. Future studies will have to delineate whether BRD4 interacts physically with acetylated p65 and/or binds to euchromatin through acetylated histones. Similarly, the therapeutic efficacy of different BET inhibitors and shRNA-mediated silencing of different BET protein family members will have to be tested in order to determine if other bromodomain proteins, including BRD2, play a role in MPN-associated inflammation.
MF patients are characterized by progressive BM fibrosis. Although BM fibrosis is postulated to play an integral role in the pathogenesis of MF and may have prognostic relevance (Gianelli et al., 2012; Lekovic et al., 2014), molecular and mechanisms governing BMF are not well understood. Moreover advanced fibrosis has been associated with worse outcome in the setting of allogeneic stem cell transplantation (allo-SCT) was associated with a better survival (Alchalby et al., 2014; Kroger et al., 2014), underscoring the need for therapies which can attenuate or reverse BM fibrosis. While JAK inhibitors deliver substantial benefits to patients, current JAK kinase inhibitors do not reduce disease burden or reverse pathologic fibrosis. Although previous studies in cell lines have suggested BRD4 as a therapeutic target in MPN (Wyspianska et al., 2014) and that BRD4 inhibition has in vitro/in vivo efficacy in JAK2/EZH2 mutant MPN (Sashida et al., 2016), here we show that BET protein inhibition in combination with JAK kinase inhibition leads to complete reversal of reticulin fibrosis in MF mice. In addition, BET inhibition alone and combined with JAK inhibition reduces inflammatory signaling, reduces disease burden in vivo, and delays persistence associated with JAK inhibitors. These studies suggest that BET inhibition, particularly in combination with JAK kinase inhibition, should be evaluated for the ability to achieve substantive clinical benefit in MPN patients. Our work suggests that therapies blocking simultaneously the JAK/STAT and NF-κB pathways might be more potent in the clinical setting and provide a strong rationale for the clinical evaluation of BET inhibitors in MPN. Most importantly, our studies suggest that targeting inflammatory signaling in tumor and non-tumor cells with epigenetic agents represents a therapeutic approach which should be explored in human cancers where there is inflammatory cross-talk between tumor cells and the microenvironment.
STAR Methods
Contact for Reagent and Resource Sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Ross Levine (leviner@mskcc.org).
Experimental Model and Subject Details
Patient samples
The Institutional Review Boards of Memorial Sloan Kettering Cancer Center (MSKCC) approved sample collection and all experiments (protocol 16-354). Informed consent was obtained from all human subjects prior to study. Information about the age and gender of each patient is listed in Table S5. Mononuclear cells (MNC) were purified using Ficoll-Pacque (GE Healthcare Life Sciences) and CD34-positive cells were isolated from the MNC layer using human CD34 MicroBeads (Miltenyi).
Mouse models
All animal experiments were performed in accordance with our MSKCC Institutional Animal Care and Use Committee-approved animal protocol. Animal care was in strict compliance with institutional guidelines established by the MSKCC, the Guide for the Care and Use of Laboratory Animals (National Academy of Sciences 1996), and the Association for Assessment and Accreditation of Laboratory Animal Care International. Healthy animals with an intact immune system were used for all experiments. All animals were drug and test naïve and not involved in previous procedures. Animals were maintained on a 12 hr light-dark cycle with access to water and standard chow ad libitum. MSKCC animal care staff conducted routine husbandry procedures and provided daily care and monitoring of all animals housed in MSKCC’s animal facilities. Experimental animals were closely monitored by laboratory staff for signs of disease or morbidity, failure to thrive, weight loss > 10 % total body weight, open skin lesions, bleeding, infection, or fatigue. Mice developing any of the above complications were sacrificed immediately. JAK2V617F knock-in mice have been described previously (Mullally et al., 2010). Floxed p65 and Ikk2 mice were kindly provided by Dr. Albert S Baldwin (Lineberger Comprehensive Cancer Center, University of North Carolina) and Dr. Manolis Pasparakis (Institute for Genetics University of Cologne), respectively. Floxed mice were crossed to the interferon-responsive Mx1-Cre and Vav-Cre deleter lines. Mice crossed to the Mx1-Cre line were injected with 2 or 4 doses poly(I:C) to induce gene deletion (~6 weeks of age). 8-week old female CD45.1 (B6.SJL-Ptprca/BoyAiTac) mice were purchased from Taconic and were used for secondary JAK2V617F transplantation experiments. All purchased recipient mice were used at 8–10 weeks and only females were used. Males and females were used as donors for experiments using Balb/c wild-type mice (8 weeks), JAK2V617F transgenic mice (3–6 months), and NFKB luciferase reporter mice (8–12 weeks).
Cell lines
JAK2V617F-positive SET-2 cells were purchased from ATCC. SET-2 cells were originally established from the peripheral blood of a 71-year-old woman with essential thrombocythemia at megakaryoblastic leukemia transformation in 1995. Cells were cultured in RPMI supplemented with 20% fetal bovine serum. A carbon dioxide (CO2) incubator was used to culture the cells at 5% CO2 and 37 degrees celsius.
Methods Details
Fluorescent-activated Cell Sorting
LSKs (Lin-cKIT+Sca1+), GMPs (Lin-cKIT+Sca1−,CD16.32+,CD34+), and MEPs (Lin-cKIT+Sca1−,CD16.32−,CD34−) were purified from control, MPLW515L-diseased, and JAK2V617F-diseased mice using BD FACSAria III cell sorter. MPLW515L-positive and JAK2V617F-positive populations were separated by FACS using GFP or CD45.2, respectively, in conjunction with specific cell-surface markers: PE-conjugated CD117, Alexa Fluor 700-conjugated CD16.CD32, phycoerythrin-Cy7 (PeCy7)-conjugated Sca-1, and e660-conjugated CD34. In addition, lineage-negative cells were identified using a panel of APC-CY7-conjugated antibodies that recognize all mature hematopoietic lineages (CD4, CD19, CD11b, Gr1, TER119, CD3, B220, and NK1.1). 7-Aminoactinomycin-D (7-AAD) or 4,6-Diamidin-2-phenylindol (DAPI) were used as viable cell marker.
ChIP
For chromatin preparation, cells were washed twice in PBS and after the final wash resuspended in PBS at a concentration of ~2 × 106 cells/ml. Cells were then fixed with methanol-free formaldehyde at a final concentration of 1% (8 minutes at room temperature). The crosslinking reaction was quenched by adding 1/10th of the crosslinking volume by adding both 1.25M glycine and 1M Tris-HCl, pH 8.0. The cells were then pelleted and washed twice in 1ml of cold PBS. After washing the pellet, 1 ml of SDS buffer supplemented with protease inhibitors was added to lyse the cytoplasm. Lysed cells were centrifuged at maximum speed for 10 minutes at room temperature. After aspiration of the supernatant, the nuclei pellet was resuspended in 1ml of IP buffer which was then transferred into a Covaris milliTUBE and sheared for 1200 seconds at 140W using 200 bursts per second. After sonication, the sheared chromatin was transferred into a DNA LoBind Tube. Antibodies were added at 1:100 dilution and incubated with sheared chromatin overnight at 4C. After overnight incubation, Protein A/G Dynabeads (mixed 1:1) were added and incubated for an additional 2 hours while rotating at 4C. Using a magnetic stand, the supernatant was discarded and the beads were washed with salt buffers, rotating for 5 minutes at 4C with each wash. After the final salt wash, tubes were spun down and 50 μl of elution buffer was added. Decrosslinking was performed by adding 2 μl of 5M NaCl to the eluent and incubation for at least 4 hours at 65C. Libraries were prepared using the NEBNext® ChIP-seq Library Prep Master Mix Set for Illumina ® and QC’d using Agilent Technologies 2200 TapeStation to determine fragment size. PicoGreen was used to quantify the concentration. Samples were pooled and submitted to New York Genome Center for SE50 sequencing using a HiSeq 2500.
Data Processing of H3K27ac and H3K4me1
H3K27ac and H3K4me1 FASTQ reads are aligned to mouse reference genome (build mm9) using bowtie2 (Langmead and Salzberg, 2012) after trimming adaptors with trim-galore tool. Duplicated reads are marked with Picard tools and removed from aligned BAM files. Alignment BAMs are sorted and indexed using samtools (Li et al., 2009; Ma et al., 2011). Enriched ChIP-seq peaks genome-wide are called with MACS (v1.4) (Zhang et al., 2008) with local background estimate model using the following MACS parameters: -nolambda --nomodel --bw=300. Nearby peaks within 1 kb window are merged using bedtools. Genome coverage is calculated for individual BAM using bedtools (v2.17) (Quinlan and Hall, 2010) and normalized to 10 million reads. ChIP-seq peaks are annotated either as promoter (promoter or 5′-UTR) or putative enhancer (Distal, Downstream, 3′ UTR) against UCSC mm9 known genes database. ChIP-seq peaks are assigned to the nearest genes. The data processing are done using an in-house shell script. The peak annotation step is done using the R/ChIPseeker packages (Yu et al., 2015).
Analysis of ChIP-Seq Data
P values of differentially enriched ChIP-seq peaks between MPL and control MEPs are calculated using DESeq2 (Love et al., 2014). Peaks with at least 10 reads within the called peak region are ordered by log2-transformed fold change statistics according to DEseq2 (Love et al., 2014) for downstream analysis. GSEA analysis (Subramanian et al., 2005) is applied in weighted mode against the gene sets collection in MSigDB (v5.1). Gene set with size over 5000 genes or smaller than 10 genes are excluded for further analysis. Each gene set is permuted 1000 times to calculate p value and FDR values. We found the raw p values of the GSEA analysis are found to be highly inflated (Figure S2B). Therefore, we fit a Beta-Uniform mixture model (Pounds and Morris, 2003) to the raw GSEA p value to select gene sets deviated from the random background. We follow the Enrichment Map approach (Bader and Hogue, 2003) to build a map of gene sets with nodes representing pre-selected gene sets and edges representing degree of gene overlap using Jaccard index as distance metrics. To further identify common themes among the significant gene sets, we applied a graph partition algorithm (Merico et al., 2010) to the gene set map to identify optimized subnetworks. The graph partition and network visualization are done using Cytoscape(v3.1) (Shannon et al., 2003).
RNA-Sequencing Analysis
We used STAR 2.4.2a (Dobin et al., 2013) to align the RNA-seq samples to the reference genome (mm10) and to count the number of reads mapping to each gene in the ensembl GRCm38.80 gene model. Differential expression between the different groups was performed through the use of DESeq 1.22.1 (Anders and Huber, 2010). We used VOOM (Law et al., 2014) to transform the data into a normalized read count matrix and applied standard hierarchical clustering on the significantly differentially expressed genes in order to separate the gene clusters. We further performed pairwise comparisons of the four experimental conditions (i.e. control, JQ1, Rux, and combo) to identify differentially expressed genes. We selected differential genes from each comparison with at least 1.5-fold change and at least 10 reads mean coverage. We standardized the aggregated differential expression matrix to have mean 0 and standard variation 1. We apply K-Mean algorithm with 100 random restart to cluster the genes into 5 clusters as shown in Figure 5B. Motif signatures were obtained using the ‘de novo’ approach in Homer v4.5 (http://homer.ucsd.edu/homer/index.html) and then matched to the default ‘known TF’ database. To specifically score NF-κB and STAT3 signatures, five candidate motifs were scanned against differentially expressed gene promoters using Homer’s ‘annotatePeaks.pl’ program with a window size of 2 Kb surrounding the transcription start site and the ‘-nmotifs’ option to build the motif matrix. Hierarchical clustering of the resulting matrix was done in R using heatmap2.
DAVID Analysis/Gene Ontology
We used DAVID v6.8 (Huang da et al., 2009) with updated knowledge base to estimate the overrepresented gene ontology and pathways in each of the differentially expressed subgroups from Figure 5A. We considered the Gene Ontology collections GOTERM_BP_DIRECT, GOTERM_CC_DIRECT and GOTERM_MF_DIRECT as well as the KEGG_PATHWAY pathway collection.
Luciferase Assays
Transgenic mice expressing modified firefly luciferase driven by six NF-κB-response-elements were developed by Caliper Life Sciences and obtained from Taconic (BALB/c-Tg(Rela-luc)31Xen). Mice transplanted with luciferase reporter cells were whole-body imaged for evidence of increased luciferase activity on a weekly basis by bioluminescence imaging (BLI). Luciferin (1.5mg) was injected behind the orbit of anaesthetized mice and imaged within 2–5 minutes after that using a Xenogen IVIS system coupled to Live imaging acquisition and analysis software. Photon flux activity was calculated for each mouse taking background and starting BLI signal into account. To assess the effect of different inhibitors on NF-κB pathway activation, mice bearing MPL-induced disease were administrated with INCB018424 (60 mg/kg, BID), JQ1 (50mg/kg, QD), alone or in combination, or vehicle and imaged before first drug administration and again three days later by BLI. To assess NF-κB pathway activation in mutant, female Balb/c mice were transplanted with MPLW515L-positive, NF-κB-REluc (GFP-positive) BM cells along with luciferase-negative, wild-type support marrow. Reversely, to assess NF-κB pathway activation in non-mutant cells, recipient mice were transplanted with MPLW515L-positive, reporter-negative BM cells along with NF-κBRE-luc, wild-type support marrow. For ex vivo imaging of organs, mice were injected with luciferin and mice were sacrificed 5 minutes later. Selected organs were excised and imaged using the IVIS imaging system (Xenogen). Bioluminescence was measured quantitatively by the Living Image® software. Ex vivo imaging of luciferase-reporter positive, cKIT-purified cells expressing MPLW515L-GFP or GFP were measured using the Dual Luciferase Reporter Assay (Promega).
Methylcellulose Assays
Cells were seeded at a density of 10000 cells/replicate into cytokine-supplemented methylcellulose medium (3434; STEMCELL Technologies). Colonies propagated in culture were scored at day 10. For human studies, CD34+ cells isolated from MF patients were plated at a density of 2500 cells/replicate in cytokine supplemented methylcellulose medium (H4435; STEMCELL Technologies) with increasing concentrations of INCB018424 or JQ1. DMSO was added to control wells. Colonies propagated in culture were scored at day 10. All experiments were performed in duplicate using ten different patient samples (Tables S5 and S6). Colony counts were normalized as a ratio to the DMSO control for each patient, with dark red representing an equal ratio or greater for the drug treatment and white representing decreased colony counts with drug treatment. To generate mutational profiles from propagated colonies (pooled), sequencing was performed using a targeted panel covering 156 myeloid genes at an average depth of 600x on Illumina HiSeq 4000 (~100 bp paired-end reads). The raw sequence data was aligned to GRCh37 reference genome using BWA-MEM algorithm (v. 0.7.12-r1039) (Li, 2012). The data quality was assessed using FastQC (v. 0.11.5) (Andrews, 2010). Candidate substitutions and insertions/deletions were called using cgpCaVEMan (v. 1.7.4) and cgpPindel (v. 1.5.4) algorithms as previously described (Breems et al., 2008; Dohner et al., 2010; Papaemmanuil et al., 2016; Papaemmanuil et al., 2013; Rucker et al., 2012; Vardiman et al., 2009). These methods provide post-hoc filters that remove systematic sequencing artifacts as well as artifacts that arise from mapping errors. All candidate mutations were compared to COSMIC (v. 81) (Forbes et al., 2017). ExAC (v. 03.12) (Lek et al., 2016) and 1000 Genomes (phase 3 release) (Genomes Project et al., 2015; Sudmant et al., 2015) databases to provide further annotation that would help to exclude common mutations in normal populations and identify somatic mutations. Following manual annotation of all candidate somatic mutations, each variant was manually visualized using Integrated Genomics Viewer (v. 2.3.92) (Robinson et al., 2011) to ensure of the high quality of the variant at the sequence level. The variants presented here are those that we identified as oncogenic.
Serum Cytokine Analysis
Luminex assays were carried out using the FlexMAP 3D multiplexing platform (Luminex xMAP system). The Millipore Mouse Cytokine 32-plex kit was used to measure the serum concentration of 32 cytokines. xPONENT (Luminex) and Milliplex Analyst Software (Millipore) was applied to convert mean fluorescent intensities (MFI) values into molecular concentrations by the use of a standard curve (5-parameter logistic fitting method). Data were normalized by Z score transformation using the scale() function in R and visualized with the heatmap.2 function of the gplots package.
Transplantation Experiments
For secondary JAK2V617F BMT experiments, 1.5 x106 whole BM cells of CD45.2 heterozygous experimental mice were transplanted via tail vein injection with 1×106 CD45.1 support wild-type bone marrow cells into CD45.1 congenic wild-type recipients. For retroviral MPLW515L BMT experiments, pre-stimulated cKit-enriched BM cells isolated from wild-type Balb/cJ or Balb/c-Tg(Rela-luc)31Xen were subjected to two rounds of cosedimentation with viral supernatant containing MSCV-hMPLW515L-IRES-GFP or empty control vector. Lethally irradiate recipient mice (Balb/cJ or Balb/c-Tg(Rela-luc)31Xen were injected with 1×106 cells (~25–40% GFP+ cells).
In vivo Drug Studies
At first signs of disease (MPLW515L ~14 days, JAK2V617F ~ 28 days), mice were randomized to begin treatment with the JAK1/2 inhibitor INCB108424 (90 mg/kg for monotherapy studies (Figure 4) and 60 mg/kg for combination studies (Figures 5 and 6), p.o., BID), the BET protein inhibitor JQ1 (50 mg/kg, i.p., continuously (Figure 4) or 5 days on 2 days off (Figures 5 and 6), QD), either alone or in combination, or vehicle. Mice were ranked based on baseline white blood cell count (WBC) and assigned to treatment groups to achieve congruent WBC profiles. Primary JAK2V617F transgenic mice used for in vivo drug studies were 2–4 months of age and both males and females were included. Mice were randomized based on HCT and PLT counts and treatment was performed for 28 days as indicated above with the exception that BET inhibitor JQ1 was administered on 7 days per week at 50 mg/kg (Figure S5D–F). At study endpoint, 3–4 hours after last drug administration mice were bled to collect serum for Luminex analysis. WBC profiles were established at different time points during treatments studies. At study endpoint, mice were sacrificed and spleen, sternum, bones (femur, tibia, and hips), and liver were harvested for further processing.
Persistence Studies
JAK2V617F-mutant SET-2 cells were cultured in the presence of increasing concentrations of INCB018424 and JQ1, alone or in combination over a time period of 6–8 weeks. Viability and cell growth were assayed thrice weekly. The concentration was increased when cells showed viability >85% and growth in the presence of the inhibitor. Starting concentrations were chosen based on IC values for each inhibitor. Ruxolitinib: IC10: 25 nM, IC20: 50 nM, IC40: 100 nM. JQ1: IC20: 100 nM, IC50: 250 nM, IC70: 350 nM. To assess cell proliferation, SET-2 cells were seeded out in 96-well plates at a cell density of 10 000 cells/well. Increasing drug concentrations were added to each well and the number of viable cells was determined 48 hours later using the CellTiter-Glo Luminescent Cell Viability Assay (Promega). Each drug dose was measured in triplicate and experiments were performed at least three times.
Immunohistochemistry
Femur, sternum, liver, and spleen samples were fixed in 4% paraformaldehyde overnight and then embedded in paraffin. Paraffin sections were cut on a rotary microtome (Mikrom International AG), mounted on microscope slides (Thermo Scientific) and air-dried in an oven at 37°C overni ght. Tissue section slides were then processed either automatically for H&E staining (COT20 stainer, Medite) or manually for reticulum staining with silver impregnation method kit (Bio-Optica). Pictures were taken at a 20x and 40x (H&E and reticulin) magnification by using Imascope viewer (Aperio).
Experimental Design
Mice were randomized based on blood counts and GFP-level (MPLW515L model) or HCT and PLT level (JAK2V617F), respectively. Investigators were not blinded to the identity of mice or samples. No statistical methods were utilized to determine sample size. The experiments described in this study were designed to use the minimum number of animals required. Each experiment was performed at least in duplicate to ensure reproducibility.
Quantification and Statistical Analysis
ChIP-seq and RNA-seq was performed using n=2–3 mice per condition. Blood count analysis and organ weights of vehicle- and drug-treated mice were recorded as indicated in figure legends. Colony formation assays using mouse cells shown in this study were performed in triplicate in two independent experiments. Colony formation assays using human cells from 10 different patients shown in this study were performed in duplicate. For JAK2V617F secondary transplantation experiments, one donor mouse was used for all recipients used in a single drug trial experiment. For retroviral MPLW515L BMT experiments, the donor and recipient mice were used at a ratio of 1:5. In vivo luciferase imaging studies were performed twice to assess the activation status of NF-κB in MPLW515L-diseased mice (n=5/group). A total of n=4 (vehicle) and n=3 (drug-treated) mice were imaged to measure the effect of BET and JAK inhibition of NF-κB signaling in vivo. Blood parameters and organ weights shown from MPLW515L and JAK2V617F drug trials were confirmed in at least two independent experiments. Sternum/femur, spleen, and live from three mice per group from two different experiments were stained with H&E and reticulin and analyzed by a pathologist. Serum levels of pro-inflammatory cytokines in the circulation of vehicle or drug-treated mice were measured in two independent experiments (21–28 days of treatment) or in a single experiment (persistence study). Survival experiments were performed once with n=6 in each arm. In vitro persistence studies were performed at least three times. The number of animals, cells, and experimental replication can be found in the respective figure legend. The Student’s t-test (unpaired, two-tailed) was used to compare the mean of two groups. Normality tests were used to test the assumption of normal distribution. For samples with significantly different variances Welch’s correct was applied. Data were analyzed and plotted using GraphPad Prism 6 software or R. Graphs represent mean values ± S.E.M. Kaplan-Meier survival analysis and long-rank test was used to compare survival outcomes between two groups. To assess effects of combined BET and JAK inhibition (100 nM condition: 100 nM of each inhibitor added), count data were modeled using a generalized model with Poisson family link function and error distribution.
Data and Software Availability
GEO accession number for data generated for this paper: GSE91062. The data set contains the ChIP-sequencing data shown in Figures 1A–G, S1A (MEP, MigR1 versus MPLW515L n=12), and S1B (JAK2fl/WT Cre-negative versus Cre-positive, n=6), Creand RNA-sequencing data shown in Figures 2, (MEP, MigR1 versus MPLW515L, n=6), 5A (SET-2 cell line, n=12), 5B/S4A (GFP-positive MEP, MPLW515L model, n=9), and S2E (JAK2fl/WT Cre-negative versus Cre-positive, n=6).
Supplementary Material
Related to Figure 1. Optimized H3K27ac and gene expression sub-networks
Related to Figure 2. Differentially expressed genes. Provided as an Excel file.
Related to Figure 2. KEGG Pathway enrichment analysis
Significance.
MPN patients have an increased risk of thrombosis and bleeding, and progress to bone marrow fibrosis or acute leukemia. JAK inhibition with current agents in the clinic can reduce cytokine production and attenuate MPN symptoms, but treatment is not curative and chronic therapy does not induce a pathologic nor molecular response in most patients. As such there is a pressing need for therapies for MPN patients. Our results demonstrate that inflammation is associated with alterations in the cis-regulatory landscape of MPN cells and that a BRD4/NF-κB-dependent regulatory network sustains the inflammatory state which can be therapeutically targeted. These data provide insights into the molecular basis of MPN-associated inflammation and to a therapeutic approach to target aberrant inflammatory signaling in MPN.
Highlights.
Inflammation is associated with changes in the chromatin landscape of MPN cells
NF-κB is constitutively active in MPNs and drives MPN-associated inflammation
JQ1 shows potent anti-fibrotic efficacy in MPN mouse models
BET inhibition cooperates with JAK inhibition to ameliorate MPN-driven inflammation
Acknowledgments
The authors thank members of the Levine laboratory for helpful comments and discussion, including Robert Bowman for critical insights and assistance with data presentation. Grant support: This work was supported by NCI R01CA151949, NCI CA173636, and NCI R35197594 to R.L.L; K99 HL122503-01A1 to M.Kl, by the Janus Project to R.L.L and J.E.B, and by a LLS SCOR and Starr Cancer Consortium grant to B.E.B. P.v.G. is supported by a LLS Fellow Award. Studies supported by MSK core facilities were supported in part by MSKCC Support Grant/Core Grant P30 CA008748. R.L.L. was supported by a Leukemia and Lymphoma Society Scholar award. M.Kl is a fellow of the American Society of Hematology and was previously supported by fellowships from the Leukemia and Lymphoma Society and the European Molecular Biology Organization. J.E.B is currently an employee of Novartis. R.L.L is on the Supervisory Board of Qiagen.
Footnotes
Author Contributions
M.Kl and R.L.L. conceived the project. M.Kl., J.E.B., and R.L.L designed experiments. M.Kl, L.Z., J.Re., J.Ro, F.R., R.K., P.V.G., B.E.B., J.M., N.F., and J.T.F. analyzed data. M.Kl, L.D., E.P., C.H., S.D., M.K., M.W., A.V.H.S., P.V.G., and J.Re. performed experiments. R.R. and E.M. provided patient samples for this study. J.Q. and J.E.B. synthesized drugs for this study. M.Kl and R.L.L wrote the manuscript with help from L.Z., R.K. and J.E.B.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alchalby H, Zabelina T, Stubig T, van Biezen A, Bornhauser M, Di Bartolomeo P, Beelen D, Cahn JY, Dreger P, Schroyens W, et al. Allogeneic stem cell transplantation for myelofibrosis with leukemic transformation: a study from the Myeloproliferative Neoplasm Subcommittee of the CMWP of the European Group for Blood and Marrow Transplantation. Biol Blood Marrow Transplant. 2014;20:279–281. doi: 10.1016/j.bbmt.2013.10.027. [DOI] [PubMed] [Google Scholar]
- Anand P, Brown JD, Lin CY, Qi J, Zhang R, Artero PC, Alaiti MA, Bullard J, Alazem K, Margulies KB, et al. BET bromodomains mediate transcriptional pause release in heart failure. Cell. 2013;154:569–582. doi: 10.1016/j.cell.2013.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews S. FastQC: a quality control tool for high throughput sequence data 2010 [Google Scholar]
- Atkinson GP, Nozell SE, Benveniste ET. NF-kappaB and STAT3 signaling in glioma: targets for future therapies. Expert Rev Neurother. 2010;10:575–586. doi: 10.1586/ern.10.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bader GD, Hogue CW. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinformatics. 2003;4:2. doi: 10.1186/1471-2105-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxter EJ, Scott LM, Campbell PJ, East C, Fourouclas N, Swanton S, Vassiliou GS, Bench AJ, Boyd EM, Curtin N, et al. Acquired mutation of the tyrosine kinase JAK2 in human myeloproliferative disorders. Lancet. 2005;365:1054–1061. doi: 10.1016/S0140-6736(05)71142-9. [DOI] [PubMed] [Google Scholar]
- Bollrath J, Greten FR. IKK/NF-kappaB and STAT3 pathways: central signalling hubs in inflammation-mediated tumour promotion and metastasis. EMBO Rep. 2009;10:1314–1319. doi: 10.1038/embor.2009.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breems DA, Van Putten WL, De Greef GE, Van Zelderen-Bhola SL, Gerssen-Schoorl KB, Mellink CH, Nieuwint A, Jotterand M, Hagemeijer A, Beverloo HB, et al. Monosomal karyotype in acute myeloid leukemia: a better indicator of poor prognosis than a complex karyotype. J Clin Oncol. 2008;26:4791–4797. doi: 10.1200/JCO.2008.16.0259. [DOI] [PubMed] [Google Scholar]
- Brown JD, Lin CY, Duan Q, Griffin G, Federation AJ, Paranal RM, Bair S, Newton G, Lichtman AH, Kung AL, et al. NF-kappaB directs dynamic super enhancer formation in inflammation and atherogenesis. Mol Cell. 2014;56:219–231. doi: 10.1016/j.molcel.2014.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlsen H, Moskaug JO, Fromm SH, Blomhoff R. In vivo imaging of NF-kappa B activity. J Immunol. 2002;168:1441–1446. doi: 10.4049/jimmunol.168.3.1441. [DOI] [PubMed] [Google Scholar]
- Dawson MA, Bannister AJ, Gottgens B, Foster SD, Bartke T, Green AR, Kouzarides T. JAK2 phosphorylates histone H3Y41 and excludes HP1alpha from chromatin. Nature. 2009;461:819–822. doi: 10.1038/nature08448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson MA, Kouzarides T. Cancer epigenetics: from mechanism to therapy. Cell. 2012;150:12–27. doi: 10.1016/j.cell.2012.06.013. [DOI] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohner H, Estey EH, Amadori S, Appelbaum FR, Buchner T, Burnett AK, Dombret H, Fenaux P, Grimwade D, Larson RA, et al. Diagnosis and management of acute myeloid leukemia in adults: recommendations from an international expert panel, on behalf of the European LeukemiaNet. Blood. 2010;115:453–474. doi: 10.1182/blood-2009-07-235358. [DOI] [PubMed] [Google Scholar]
- Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, Fedorov O, Morse EM, Keates T, Hickman TT, Felletar I, et al. Selective inhibition of BET bromodomains. Nature. 2010;468:1067–1073. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong CY, Morison J, Dawson MA. Epigenetics in the hematologic malignancies. Haematologica. 2014;99:1772–1783. doi: 10.3324/haematol.2013.092007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forbes SA, Beare D, Boutselakis H, Bamford S, Bindal N, Tate J, Cole CG, Ward S, Dawson E, Ponting L, et al. COSMIC: somatic cancer genetics at high-resolution. Nucleic Acids Res. 2017;45:D777–D783. doi: 10.1093/nar/gkw1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genomes Project C, Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, Korbel JO, Marchini JL, McCarthy S, McVean GA, et al. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gianelli U, Vener C, Bossi A, Cortinovis I, Iurlo A, Fracchiolla NS, Savi F, Moro A, Grifoni F, De Philippis C, et al. The European Consensus on grading of bone marrow fibrosis allows a better prognostication of patients with primary myelofibrosis. Mod Pathol. 2012;25:1193–1202. doi: 10.1038/modpathol.2012.87. [DOI] [PubMed] [Google Scholar]
- Grivennikov S, Karin E, Terzic J, Mucida D, Yu GY, Vallabhapurapu S, Scheller J, Rose-John S, Cheroutre H, Eckmann L, et al. IL-6 and Stat3 are required for survival of intestinal epithelial cells and development of colitis-associated cancer. Cancer Cell. 2009;15:103–113. doi: 10.1016/j.ccr.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grivennikov SI, Karin M. Dangerous liaisons: STAT3 and NF-kappaB collaboration and crosstalk in cancer. Cytokine Growth Factor Rev. 2010;21:11–19. doi: 10.1016/j.cytogfr.2009.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison C, Kiladjian JJ, Al-Ali HK, Gisslinger H, Waltzman R, Stalbovskaya V, McQuitty M, Hunter DS, Levy R, Knoops L, et al. JAK inhibition with ruxolitinib versus best available therapy for myelofibrosis. N Engl J Med. 2012;366:787–798. doi: 10.1056/NEJMoa1110556. [DOI] [PubMed] [Google Scholar]
- Huang B, Yang XD, Zhou MM, Ozato K, Chen LF. Brd4 coactivates transcriptional activation of NF-kappaB via specific binding to acetylated RelA. Mol Cell Biol. 2009;29:1375–1387. doi: 10.1128/MCB.01365-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- James C, Ugo V, Casadevall N, Constantinescu SN, Vainchenker W. A JAK2 mutation in myeloproliferative disorders: pathogenesis and therapeutic and scientific prospects. Trends Mol Med. 2005;11:546–554. doi: 10.1016/j.molmed.2005.10.003. [DOI] [PubMed] [Google Scholar]
- Klampfl T, Gisslinger H, Harutyunyan AS, Nivarthi H, Rumi E, Milosevic JD, Them NC, Berg T, Gisslinger B, Pietra D, et al. Somatic mutations of calreticulin in myeloproliferative neoplasms. N Engl J Med. 2013;369:2379–2390. doi: 10.1056/NEJMoa1311347. [DOI] [PubMed] [Google Scholar]
- Kleppe M, Kwak M, Koppikar P, Riester M, Keller M, Bastian L, Hricik T, Bhagwat N, McKenney AS, Papalexi E, et al. JAK-STAT pathway activation in malignant and nonmalignant cells contributes to MPN pathogenesis and therapeutic response. Cancer discovery. 2015;5:316–331. doi: 10.1158/2159-8290.CD-14-0736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koppikar P, Bhagwat N, Kilpivaara O, Manshouri T, Adli M, Hricik T, Liu F, Saunders LM, Mullally A, Abdel-Wahab O, et al. Heterodimeric JAK-STAT activation as a mechanism of persistence to JAK2 inhibitor therapy. Nature. 2012;489:155–159. doi: 10.1038/nature11303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kralovics R, Passamonti F, Buser AS, Teo SS, Tiedt R, Passweg JR, Tichelli A, Cazzola M, Skoda RC. A gain-of-function mutation of JAK2 in myeloproliferative disorders. N Engl J Med. 2005;352:1779–1790. doi: 10.1056/NEJMoa051113. [DOI] [PubMed] [Google Scholar]
- Kroger N, Zabelina T, Alchalby H, Stubig T, Wolschke C, Ayuk F, von Hunerbein N, Kvasnicka HM, Thiele J, Kreipe HH, et al. Dynamic of bone marrow fibrosis regression predicts survival after allogeneic stem cell transplantation for myelofibrosis. Biol Blood Marrow Transplant. 2014;20:812–815. doi: 10.1016/j.bbmt.2014.02.019. [DOI] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law CW, Chen Y, Shi W, Smyth GK. voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 2014;15:R29. doi: 10.1186/gb-2014-15-2-r29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Herrmann A, Deng JH, Kujawski M, Niu G, Li Z, Forman S, Jove R, Pardoll DM, Yu H. Persistently activated Stat3 maintains constitutive NF-kappaB activity in tumors. Cancer Cell. 2009;15:283–293. doi: 10.1016/j.ccr.2009.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, O’Donnell-Luria AH, Ware JS, Hill AJ, Cummings BB, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lekovic D, Gotic M, Perunicic-Jovanovic M, Vidovic A, Bogdanovic A, Jankovic G, Cokic V, Milic N. Contribution of comorbidities and grade of bone marrow fibrosis to the prognosis of survival in patients with primary myelofibrosis. Med Oncol. 2014;31:869. doi: 10.1007/s12032-014-0869-8. [DOI] [PubMed] [Google Scholar]
- Levine RL, Wadleigh M, Cools J, Ebert BL, Wernig G, Huntly BJ, Boggon TJ, Wlodarska I, Clark JJ, Moore S, et al. Activating mutation in the tyrosine kinase JAK2 in polycythemia vera, essential thrombocythemia, and myeloid metaplasia with myelofibrosis. Cancer Cell. 2005;7:387–397. doi: 10.1016/j.ccr.2005.03.023. [DOI] [PubMed] [Google Scholar]
- Li H. Exploring single-sample SNP and INDEL calling with whole-genome de novo assembly. Bioinformatics. 2012;28:1838–1844. doi: 10.1093/bioinformatics/bts280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R Genome Project Data Processing S. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma C, Fan R, Ahmad H, Shi Q, Comin-Anduix B, Chodon T, Koya RC, Liu CC, Kwong GA, Radu CG, et al. A clinical microchip for evaluation of single immune cells reveals high functional heterogeneity in phenotypically similar T cells. Nature medicine. 2011;17:738–743. doi: 10.1038/nm.2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merico D, Isserlin R, Stueker O, Emili A, Bader GD. Enrichment map: a network-based method for gene-set enrichment visualization and interpretation. PLoS One. 2010;5:e13984. doi: 10.1371/journal.pone.0013984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mondet J, Hussein K, Mossuz P. Circulating Cytokine Levels as Markers of Inflammation in Philadelphia Negative Myeloproliferative Neoplasms: Diagnostic and Prognostic Interest. Mediators Inflamm. 2015;2015:670580. doi: 10.1155/2015/670580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullally A, Lane SW, Ball B, Megerdichian C, Okabe R, Al-Shahrour F, Paktinat M, Haydu JE, Housman E, Lord AM, et al. Physiological Jak2V617F expression causes a lethal myeloproliferative neoplasm with differential effects on hematopoietic stem and progenitor cells. Cancer cell. 2010;17:584–596. doi: 10.1016/j.ccr.2010.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nangalia J, Massie CE, Baxter EJ, Nice FL, Gundem G, Wedge DC, Avezov E, Li J, Kollmann K, Kent DG, et al. Somatic CALR mutations in myeloproliferative neoplasms with nonmutated JAK2. N Engl J Med. 2013;369:2391–2405. doi: 10.1056/NEJMoa1312542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicodeme E, Jeffrey KL, Schaefer U, Beinke S, Dewell S, Chung CW, Chandwani R, Marazzi I, Wilson P, Coste H, et al. Suppression of inflammation by a synthetic histone mimic. Nature. 2010;468:1119–1123. doi: 10.1038/nature09589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh ST, Simonds EF, Jones C, Hale MB, Goltsev Y, Gibbs KD, Jr, Merker JD, Zehnder JL, Nolan GP, Gotlib J. Novel mutations in the inhibitory adaptor protein LNK drive JAK-STAT signaling in patients with myeloproliferative neoplasms. Blood. 2010;116:988–992. doi: 10.1182/blood-2010-02-270108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papaemmanuil E, Gerstung M, Bullinger L, Gaidzik VI, Paschka P, Roberts ND, Potter NE, Heuser M, Thol F, Bolli N, et al. Genomic Classification and Prognosis in Acute Myeloid Leukemia. N Engl J Med. 2016;374:2209–2221. doi: 10.1056/NEJMoa1516192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papaemmanuil E, Gerstung M, Malcovati L, Tauro S, Gundem G, Van Loo P, Yoon CJ, Ellis P, Wedge DC, Pellagatti A, et al. Clinical and biological implications of driver mutations in myelodysplastic syndromes. Blood. 2013;122:3616–3627. doi: 10.1182/blood-2013-08-518886. quiz 3699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pikman Y, Lee BH, Mercher T, McDowell E, Ebert BL, Gozo M, Cuker A, Wernig G, Moore S, Galinsky I, et al. MPLW515L is a novel somatic activating mutation in myelofibrosis with myeloid metaplasia. PLoS Med. 2006;3:e270. doi: 10.1371/journal.pmed.0030270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pounds S, Morris SW. Estimating the occurrence of false positives and false negatives in microarray studies by approximating and partitioning the empirical distribution of p-values. Bioinformatics. 2003;19:1236–1242. doi: 10.1093/bioinformatics/btg148. [DOI] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintas-Cardama A, Kantarjian H, Cortes J, Verstovsek S. Janus kinase inhibitors for the treatment of myeloproliferative neoplasias and beyond. Nat Rev Drug Discov. 2011;10:127–140. doi: 10.1038/nrd3264. [DOI] [PubMed] [Google Scholar]
- Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rucker FG, Schlenk RF, Bullinger L, Kayser S, Teleanu V, Kett H, Habdank M, Kugler CM, Holzmann K, Gaidzik VI, et al. TP53 alterations in acute myeloid leukemia with complex karyotype correlate with specific copy number alterations, monosomal karyotype, and dismal outcome. Blood. 2012;119:2114–2121. doi: 10.1182/blood-2011-08-375758. [DOI] [PubMed] [Google Scholar]
- Sashida G, Wang C, Tomioka T, Oshima M, Aoyama K, Kanai A, Mochizuki-Kashio M, Harada H, Shimoda K, Iwama A. The loss of Ezh2 drives the pathogenesis of myelofibrosis and sensitizes tumor-initiating cells to bromodomain inhibition. J Exp Med. 2016;213:1459–1477. doi: 10.1084/jem.20151121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shlyueva D, Stampfel G, Stark A. Transcriptional enhancers: from properties to genome-wide predictions. Nat Rev Genet. 2014;15:272–286. doi: 10.1038/nrg3682. [DOI] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudmant PH, Rausch T, Gardner EJ, Handsaker RE, Abyzov A, Huddleston J, Zhang Y, Ye K, Jun G, Hsi-Yang Fritz M, et al. An integrated map of structural variation in 2,504 human genomes. Nature. 2015;526:75–81. doi: 10.1038/nature15394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X, Peng R, Phillips JE, Deguzman J, Ren Y, Apparsundaram S, Luo Q, Bauer CM, Fuentes ME, DeMartino JA, et al. Assessment of Brd4 inhibition in idiopathic pulmonary fibrosis lung fibroblasts and in vivo models of lung fibrosis. Am J Pathol. 2013;183:470–479. doi: 10.1016/j.ajpath.2013.04.020. [DOI] [PubMed] [Google Scholar]
- Tefferi A, Vaidya R, Caramazza D, Finke C, Lasho T, Pardanani A. Circulating interleukin (IL)-8, IL-2R, IL-12, and IL-15 levels are independently prognostic in primary myelofibrosis: a comprehensive cytokine profiling study. J Clin Oncol. 2011;29:1356–1363. doi: 10.1200/JCO.2010.32.9490. [DOI] [PubMed] [Google Scholar]
- Vardiman JW, Thiele J, Arber DA, Brunning RD, Borowitz MJ, Porwit A, Harris NL, Le Beau MM, Hellstrom-Lindberg E, Tefferi A, et al. The 2008 revision of the World Health Organization (WHO) classification of myeloid neoplasms and acute leukemia: rationale and important changes. Blood. 2009;114:937–951. doi: 10.1182/blood-2009-03-209262. [DOI] [PubMed] [Google Scholar]
- Wyspianska BS, Bannister AJ, Barbieri I, Nangalia J, Godfrey A, Calero-Nieto FJ, Robson S, Rioja I, Li J, Wiese M, et al. BET protein inhibition shows efficacy against JAK2V617F-driven neoplasms. Leukemia. 2014;28:88–97. doi: 10.1038/leu.2013.234. [DOI] [PubMed] [Google Scholar]
- Yasmin R, Siraj S, Hassan A, Khan AR, Abbasi R, Ahmad N. Epigenetic regulation of inflammatory cytokines and associated genes in human malignancies. Mediators Inflamm. 2015;2015:201703. doi: 10.1155/2015/201703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G, Wang LG, He QY. ChIPseeker: an R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics. 2015;31:2382–2383. doi: 10.1093/bioinformatics/btv145. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Related to Figure 1. Optimized H3K27ac and gene expression sub-networks
Related to Figure 2. Differentially expressed genes. Provided as an Excel file.
Related to Figure 2. KEGG Pathway enrichment analysis