Abstract
Integrating diverse types of prognostic information into accurate, individualized estimates of outcome in colorectal cancer is challenging. Significant heterogeneity in colorectal cancer prognostication tool quality exists. Methodology is incompletely or inadequately reported. Evaluations of the internal or external validity of the prognostic model are rarely performed. Prognostication tools are important devices for patient management, but tool reliability is compromised by poor quality. Guidance for future development of prognostication tools in colorectal cancer is needed.
Keywords: nomogram, colorectal neoplasms, prognosis, clinical prediction tool
Introduction
The Tumour Node Metastasis (TNM) staging classification system is the foundation of prognostication in colorectal cancer; however, variation in survival and optimal clinical management strategies exist within stage groupings.[1–3] The 7th edition of the UICC/AJCC anatomic stage introduced anatomically-based subgroupings within stage II and III disease to account for significant prognostic heterogeneity within these groups.[4] While these additional stratifications were successful, the prognostic power of stage for predicting overall survival in an individual patient could be further enhanced by a number of clinical, disease and patient characteristics.[5] Established prognostic factors include depth of tumor invasion into the intestinal wall and presence of nodal metastases,[5] performance status, co-morbid conditions such as diabetes, the presence of venous or lymphatic invasion, and tumour grade.[6,7] Additional complexity in personalized prognostication lies in newly identified biologic, genetic and other molecular information, which have yet a validated role for colon or rectal cancer.[8–11]
Clinicians and patients are continually challenged as to how to best incorporate established and novel prognostic information alongside anatomic stage into a single, individualized estimate of outcome. Clinical prognostication tools, traditionally based on statistical regression models, are one method of combining prognostic information that avoids further stratification of the TNM staging system, which is based on an inelastic mathematical bin model.[12,13] If appropriately developed and validated, these tools have the potential to integrate and personalize the prognostic information available for individual patients and provide refined risk estimates for application to uncertain clinical management scenarios.[14]
The landscape of prognostication tool quality and clinical relevance is currently unknown in colorectal cancer. The Molecular Modellers Working Group (MMWG) of the American Joint Committee on Cancer (AJCC) was formed to understand how information beyond stage could be used to individualize survival prognostication and personalize patient management. The MMWG chose to review the quality and usability of currently available clinical prognostic tools that predict survival in colorectal and four other cancers as their first task. [15–17] The work of the MMWG established the platform for AJCC for the Precision Medicine Core (PMC) of the 8th Edition of the AJCC Cancer Staging Manual, which is envisioned to continue and expand as a service to the oncology community [18,19]. In this article we provide a detailed catalogue and evaluation of publicly available colorectal cancer prognostication tools.
Materials and Methods
Search Strategy and Selection Criteria
Prognostication tools were identified and documentation on their development and validation gathered using three strategies: 1) A search of the peer-reviewed published literature (including a systematic literature review and cited reference search); 2) A search of the web-based scientific community; and 3) Correspondence with individual tool developers when a web-based tool had no corresponding scientific journal article or technical report.
The search strategy was executed in OVID Medline, OVID Embase and HealthStar from Jan 1, 1996–October 6th, 2015. Medical subject headings (MeSH) did not exist for prognostication tools and so a combination of alternate headings and key words were used following consultation with a scientific librarian. Each set of search terms was modified for the specific search engine. For example, the following search terms were used in Medline: “models, statistical/”, “prognosis”, “predict* model*”, “nomogram/”, “prognos* model*”, and “colorectal neoplasm/”. The searches were limited to English language. Clinically relevant tools originally published prior to 1996 were also included, but these were identified through validation articles found in the systematic literature review. Seemingly eligible studies were excluded if they met any of the following a priori exclusion criteria: 1) assessment of the prognostic impact of a single factor (unless it was updating the accuracy of an existing prognostic tool); 2) inappropriate analytic purpose (e.g. multivariate modeling not aimed at prognostication, application of novel statistical methods); 3) not specific to colorectal cancer patients; 4) not original data/research (e.g. editorial, review) or 5) the outcome was not survival. Studies reporting on genomic classifiers built entirely using gene expression data were not the focus of the review and were excluded.
Prognostication tools in this paper include those developed to estimate the probability of survival at a particular point along the disease trajectory (e.g. at diagnosis, following treatment) or for the purpose of using a survival probability to inform treatment decision-making. Eligible survival end-points included all time-to-death analyses (e.g. overall survival, cause-specific survival, relative survival), as well as vital status analyses (e.g. probability of death at 5-years post-diagnosis). Generally speaking, some form of statistical model underlies most prognostication tools, and we use the terms prognostication tool and prognostication model interchangeably. A single reviewer (AM) assessed the titles and abstracts of citations for inclusion. At the beginning, a second reviewer evaluated a random sample of 20 citations to evaluate reliability. Percent agreement was 85%. The first reviewer was conservative and included more articles at the abstract phase than the second reviewer. These differences were easily resolved, and a discussion of discordant decisions determined that the rules for inclusion and exclusion were being applied consistently. A cited reference search using Web of Science was performed. This was implemented to decrease the probability of missing a relevant article. These peer-reviewed literature search strategies to identify prognostic tools in colorectal cancer were supplemented by a Google web-based search. Search terms included: “clinical prediction tool cancer”, “online calculator cancer”, and “nomogram cancer”. The AJCC contacted tool developers for details on tool development if a tool identified in the Google search did not have a supporting article in the peer-reviewed literature and a technical document was not publically available.
Data Abstraction
A detailed report on data abstraction form development and key definitions was published previously. [16,20,21] The data abstracted allowed an evaluation of tool development and validation methodology and clinical relevance. The final criteria include all key elements described by the Transparent Reporting of a multivariable prediction model for Individual Prognosis Or Diagnosis (TRIPOD) guidelines[20,21] and the Critical Appraisal and Data Extraction for Systematic Reviews of Prediction Modelling Studies (CHARMS) checklist[22]. Clinical relevance was informally assessed by considering the prognostic factors’ relevance to the clinical population and to the question addressed by the tool, and by considering the format of the tool (whether or not the equation was provided, usability in a clinical setting). General descriptive information such as study design, study population characteristics and outcome measurement were abstracted, as well as specific details on tool development (statistical modeling decisions, candidate variable selection) and validation (internal validation methods, measures of model predictive accuracy).
Summary
Key tool development and validation terminology are reported elsewhere.[16,20,21] Descriptive statistics related to tool development and validations are reported in summary tables. The assessment of a tool’s calibration and/or discriminative power was defined as a formal statistical evaluation of the internal or external validity. Model calibration assesses how closely the predicted values of the outcome match the observed outcomes in the study sample. Model discrimination assesses the ability of the model to distinguish between individuals who do and do not have the event, at a particular point in time. These established methods are considered the best means of evaluating a clinical prediction model.[12,20,21] We also described when tools were assessed informally through a comparison of survival time distributions across prognostic groups (Kaplan-Meier survival curves). Note however that although these are the same statistical methods that are often used to evaluate the prognostic ability of TNM stage[2,3] they are not considered statistically robust nor are they considered best practice for clinical prediction tool predictive performance assessment.[20,21]
Results
Literature Search Results
Figure 1 describes the search results. The scientific literature review and web-based search identified 53 tools predicting survival in colon or rectal cancer,[23–73] reported across 63 articles. [23–91] Two articles reported on the development of two tools each.[40,58] Eighteen articles contained external validations only.[74–91] One article updated two tools with additional prognostic information.[76] We did not identify any articles evaluating the effectiveness or implementation of tools in clinical practice. Documentation in the peer-review literature was not available for six prediction tools. Correspondence with tool developers added technical documentation for two of those tools, and we were told that the remaining four were pending publication in the peer-review literature.
Figure 1.
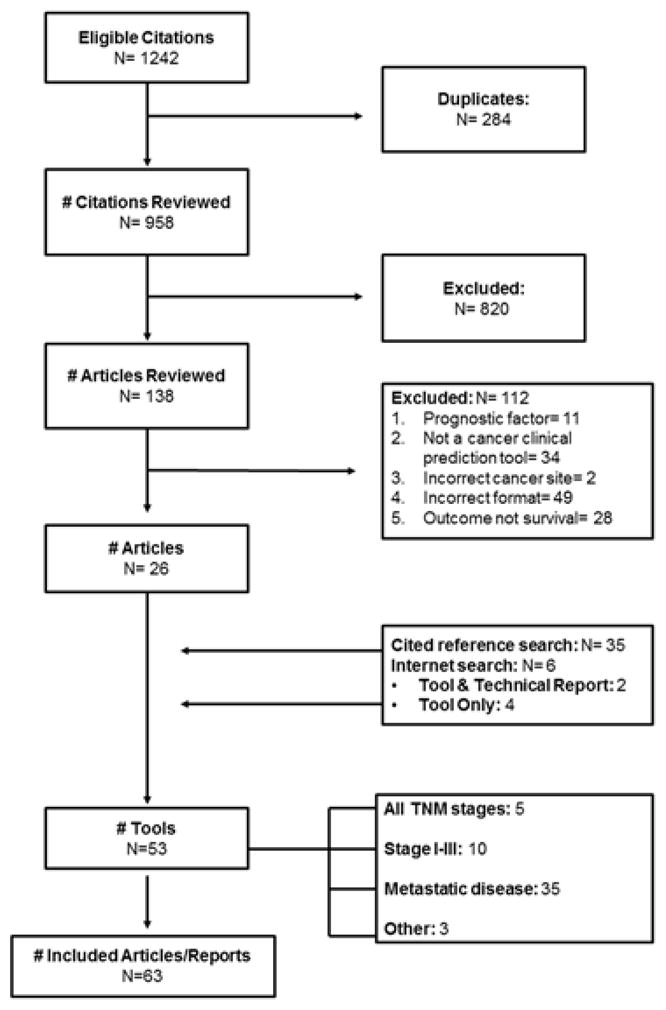
Search results for clinical prognostic tools and their validation
Tool Development Methods
Table 1 describes key information abstracted on the development of each tool. Further supporting aggregate data are reported. Thirty-nine tools were developed for prognosis in colorectal cancer patients; eight tools were targeted to rectal cancer and six targeted to colon cancer patients respectively. Twenty-nine tools (55%) were designed to predict overall survival (defined as the time between an index date and death from any cause), eleven tools predicted disease-specific survival, six tools did not specify the type of survival outcome, two tools predicted both overall and disease-specific survival, and two predicted cumulative survival at a stated time point (e.g. probability of surviving 5 years). Conditional overall survival, conditional diseases-specific survival and cumulative survival (not otherwise specified) were predicted by one tool each. Thirty (57%) of the tools were presented for clinical use as risk scores or risk groupings. Risk groups are not recommended by the TRIPOD guidelines and their validation is only possible if the risk groups are assigned the average outcome value, which is rarely done. Twelve were presented as nomograms, and ten as web-based calculators; one tool was presented as a prognostic tree. Only 6 of 53 tools reported the underlying statistical equation with variable coefficients and the intercept, where appropriate. This information is required for external validation using established and appropriate statistical methods.[20,21]
Table 1.
Details on included prognostic tools for colorectal cancer
| Tool Citation |
Tumour Location |
Population | Dates of Data Collection |
Study Design |
Sample Size |
# Events |
Duration of Follow- Up |
Outcome | Final Variables in Model |
Internal Validation |
External Validation |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Prognostic tools not specific to a particular TNM stage grouping | |||||||||||
| [25] | Colon | Stage I–IV | 1988–1999 | Other | NR | NR | NR | DSS | Age, sex, comorbidity, depth of invasion, # of positive lymph nodes, # of examined lymph nodes, histologic grade | None | Calibration: Y |
| [50] | Colon | Stage I–IV | NR | Other | NR | NR | NR | NOS | Age, sex, tumour diameter, # positive lymph nodes, CEA, histological type, grade, site, farthest tumour extension (including metastasis) | None | None |
| [31] | Colon | Stage I–IV | 1988–2000 | Retrospective Cohort | 83,419 | NR | Median 87 months | CDSS | Age, sex, ethnicity, tumour grade, AJCC stage | Approach: Bootstrap Calibration: Y Discrimination: 0.816 |
None |
| [39] | CRC | Stage I–IV | 1990–1999 | Retrospective Cohort | ** | NR | Mean 43.9 months | OTHER | Tumour depth of invasion, # of metastatic lymph nodes, metastasis, CEA, differentiation, resectability, tumor location, blood transfusion | Approach: Apparent Overall: R2: −0.64, −0.65 |
None |
| [49] | CRC | Locally advanced/Metastatic | 1990–1998 | Retrospective Cohort | 1057 | 161 | ≥ 2 years | OTHER | Performance status, differentiation, primary site, Duke’s stage, number of sites of metastatic disease, CEA, response rate, treatment, number of chemotherapy lines | None | None |
| [56] | CRC | Dukes B | 1988–1996 | Prospective Cohort | 268 | 63 | Median 65 months | DSS | Peritoneal involvement, venous invasion, margin involvement, tumour perforation | Approach: Apparent* | None |
| [65] | Rectal | Locally advanced | 1992–2003 | RCT-PC | 2242 | 850 | Median 55.2 months | OS | pT stage, pN stage, cT stage, age, adjuvant chemotherapy, surgery procedure, radiotherapy dose, sex | Approach: Apparent Discrimination: 0.68 |
Calibration: Y Discrimination: 0.70 |
| [66] | Rectal | Stage I–IV | 1994–2003 | Retrospective Cohort | 42,830 | NR | NR | COS | Age, sex, race, stage | Approach: Bootstrap Calibration: Y Discrimination: 0.75 |
None |
| Stage I–III | |||||||||||
| [33] | CRC | Stage II | 1988–1997 | Retrospective Cohort | 238 | 53 | Median 110 months | DSS | Tumour growth pattern, extent of tumour spread beyond muscularis propria | Type: Apparent* | None |
| [24] | CRC | Stage I–III, PALN | 2001–2011 | Retrospective | 409 | NR | NR | OS | LVI, pN+ status, serum CEA ≥10ng/mL, short axis diameter PALNs ≥10mm | Type: Apparent* | None |
| [73] | Colon | Stage II or III | NR | RCT-PC | 3,302 | NR | Maxiumum 8 years | OS | # of positive lymph nodes, depth of tumour, tumour grade, age | Type: Bootstrap Calibration: Y Discrimination: 0.655 |
Calibration: Y |
| [55] | Rectal | Stage II–III | 1986–2005 | Retrospective | 833 | 263 | Median (in survivors) 51 months | OS | Gender, age, CEA, tumor location, T stage, N stage, Ratio of metastatic lymph nodes, adjuvant chemotherapy, adjuvant chemoradiotherapy | Type: Cross-Validation Discrimination: 0.67 |
Discrimination: 0.76 |
| [29] | Rectal | Stage I–III | NR | NR | NR | NR | NR | NOS | Age, sex, race, grade, stage, extent of surgery | None | None |
| [28] | Rectal | Stage I–III | NR | NR | NR | NR | NR | NOS | Age, sex, race, grade, stage, extent of surgery | None | None |
| [27] | Rectal | Stage I–III | NR | NR | NR | NR | NR | NOS | Age, sex, race, grade, stage, extent of surgery | None | None |
| [59] | Colon | Stage III | RCT-PC | 1989–2002 | 15995 | NR | NR | OS | Age, sex, race, BMI, performance status, tumor grade, tumor stage, ratio of the number of postiive lymph nodes to nodes examines, number and location of primary tumors, systemic treatment class | Type: Bootstrap Calibration: Y Discrimination: 0.66 |
Calibration: Y |
| [72] | Rectal | Stage I–III | RCT-PC | 1987–2002 | 2618 | 1077 | Median 39.5–75.3 months | OS | Age, gender, tumour distance, surgery type, residual disease, p-T, p-N, presence of post-operative complications | Type: Cross-Validation Calibration: Y Discrimination 0.752 |
None |
| [68] | Colon | Stage I–III | Retrospective Cohort | 1994–2005 | 128,853 | NR | NR | OS | T-stage, n-stage, number of positive LN, tumour differentiation, patient age, sex | Type: Split Sample Calibration: Y Discrimination: 0.68 |
None |
| Stage IV | |||||||||||
| [32] | CRC | Stage IV | NR | RCT-PC | 803 | NR | median 35.3–46.3 months | OS | performance status, LDH, ALP, number of metastatic sites, time to metastasis | Approach: Split Sample Calibration: Y Discrimination: 0.60 |
Discrimination: 0.63 |
| [34] | CRC | Stage IV | 1995–2010 | Retrospective | 443 | 385 | Median 62.4 months (range 55.6–77.6) | OS | Number of liver metastases, PCI, type of surgery | Approach: Apparent Calibration: Y Discrimination: 0.61 |
None |
| [71] | CRC | Stage IV | 1997–2007 | Retrospective | 1133 | 278 | NR | OS | Post-operative CEA, depth of tumor invasion, lymph node metastasis, peritoneal dissemination | Approach: Split Sample Calibration: Y Discrimination: 0.64 |
None |
| [44] | CRC | Stage IV | 1984–1999 | Prospective | 9624 | NR | NR | OS | Depth of tumor invasion, regional lymph node metastasis, hitologic grade, liver metastasis, lung metastasis, distant lymph node metastasis, peritoneal metastasis, noncurative resection for metastatic lesions | Approach: Apparent* | None |
| [38] | CRC | Stage IV | 1982–1996 | RCT-PC | 3817 | NR | NR | OS | ECOG, WBC count, number of tumor sites, alkaline phosphatase, platelets, location of primary, hemoglobin, peritoneal metastases | Approach: Apparent* | Calibration: Y Discrimination: 0.52 & 0.54 Overall: Schemper: 1.6% |
| [62] | CRC | Stage IV | 2005–2008 | Retrospective | 124 | 74 | NR | OS | Performance status, pathology, peritoneal metastasis, LDH, PFS interval | Approach: Apparent* | Survival Curves |
| [30] | Rectal | Stage IV | NR | Other | NR | NR | NR | NOS | Age, sex, race, grade, stage, extent of surgery | None | None |
| [26] | CRC | Liver mets | 1988–1999 | Retrospective Cohort | 138 | 99 | mean 48.7 months | OS | location of primary, number of liver metastases, preoperative CA19-9, preoperative tumour size | Approach: Apparent* | Discrimination: 0.64 |
| [23] | CRC | Liver mets | 2003–2010 | Retrospective | 100 | 66 | Mean 60 weeks (range 10–238 weeks) | OS | Prior liver surgery, CEA, transaminase toxicity, CT size of two largest lesions | Approach: Apparent Discrimination: 0.81 |
Discrimination: 0.83 |
| [35] | CRC | Liver mets | 1985–1998 | Retrospective Cohort | 1001 | 393 | median 32 months | OS | nodal status of primary, disease-free interval before presentation of liver metastases, number of tumours, preoperative CEA level, size of the largest tumour | Approach: Apparent Overall: R2: 0.92 |
Discrimination: 0.533–0.68 |
| [36] | CRC | Liver mets | 1996–2007 | Retrospective Cohort | 280 | NR | median 50.1 months | OS | CEA, number of liver metastases, recurrence pattern, recurrence pattern | Approach: Bootstrap Discrimination: 0.67 |
None |
| [37] | CRC | Liver mets | 1981–1996 | Retrospective Cohort | 243 | 225 | median 32 months | OS | tumour number, tumour size, interval between resection of primary and liver metastases, distribution of liver tumours | None | Discrimination: 0.53–0.64 |
| [40](pre-op) | CRC | Liver mets | 1990–1998 | Retrospective Cohort | 578 | 337 | median 55.2 months | OS, DSS | extrahepatic disease, number of metastatic LN, histology of primary, prehepatectomy CEA level, number of hepatic tumours | Approach: Bootstrap Calibration: Y Discrimination: 0.66 |
Discrimination: 0.69 |
| [40](post-op) | CRC | Liver mets | 1990–1998 | Retrospective Cohort | 578 | 337 | median 55.2 months | OS, DSS | extrahepatic disease, hilar metastatic LN, number of metastatic LN (primary), histology of primary, surgical margin, prehepatectomy CEA | Approach: Bootstrap Calibration: Y Discrimination: 0.68 |
Discrimination: 0.7 |
| [42] | CRC | Liver mets | 1985–1999 | Retrospective Cohort | 148 | NR | NR | OS | extent of liver metastasis, depth of tumour invasion, peritoneal metastasis | Approach: Apparent* | None |
| [43] | CRC | Liver mets | 1986–2004 | Retrospective Cohort | 1477 | NR | NR | DSS | sex, age, primary site, disease-free interval, pre-operative CEA, number of tumours, largest site of metastasis, bilateral resection, number of involved lobes, primary N stage | Approach: Bootstrap Calibration: Y Discrimination: 0.688 |
Discrimination: 0.602 & 0.62 |
| [45] | CRC | Liver mets | 1993–2006 | Retrospective Cohort | 201 | 98 | median 31 months | OS | number of metastases, time of diagnosis of the liver metastases, CEA level | Approach: Apparent* | Discrimination: 0.54–0.58 |
| [46] | CRC | Liver mets | 1994–2005 | Retrospective Cohort | 138 | NR | median 47.2 months | OS | liver resection margin, CEA, number of liver metastasis, lymph node status | Approach: Apparent* | Survival Curves |
| [47] | CRC | Liver mets | 1977–1997 | Retrospective Cohort | 135 | 65 | median 73 months | OS | % liver involvement, colic LN, Duke’s stage, number of metastases, size of metastases, preoperative: -GT, GPT, type of resection, resection margins | Approach: Apparent* | None |
| [48] | CRC | Liver mets | 1993–2006 | Retrospective Cohort | 700 | NR | median 34 months | OS | Number of metastases, inflammatory response to tumour | Approach: Apparent* | None |
| [51] | CRC | Liver mets | 1980–2002 | Retrospective Cohort | 369 | NR | mean 4.11 years | DSS | Hepatic LN metastases, number of LN around primary tumour, CEA at hepatectomy, number of liver metastases | Approach: Split Sample* | None |
| [52] | CRC | Liver mets | 1981–1997 | Retrospective Cohort | 81 | NR | median 36.3 months | DSS | Serosal invasion, LN positivity, number of liver metastases, diameter of largest hepatic metastasis, extrahepatic metastasis | Approach: Apparent* | Discrimination: 0.60–0.65 |
| [53] | CRC | Liver mets | 1990–2005 | Retrospective Cohort | 121 | 52 | median 68 months | OS | Location of hepatic metastases, number of metastatic tumours, LN status of primary | Approach: Apparent* | None |
| [54] | CRC | Liver mets | 1968–1990 | Retrospective Cohort | 1532 | 689 | median 19 months | OS | Age, extension of primary into serosa, lymphatic spread, time interval from primary tumour to metastases, size of largest liver lesion, number of liver lesions, resection margin | Approach: Apparent* | Discrimination: 0.55–0.64 |
| [58](pre-op) | CRC | Liver mets | 1987–2005 | Retrospective Cohort | 929 | 459 | median 26.4 months | DSS | Primary tumour LN status, primary tumour differentiation, CEA, largest tumour diameter, extrahepatic metastatic disease, resection margin | Approach: Split Sample Calibration: Y Discrimination: 0.805 |
Discrimination: 0.59–0.66 |
| [58](post-op) | CRC | Liver mets | 1987–2005 | Retrospective Cohort | 929 | 459 | median 26.4 months | DSS | Primary tumour LN status, primary tumour differentiation, CEA, number of hepatic metastases, largest tumour diameter, extrahepatic metastatic disease | Approach: Split Sample Calibration: Y Discrimination: 0.781 |
Discrimination: 0.63–0.74 |
| [60] | CRC | Liver mets | 1988–2002 | Retrospective Cohort | 337 | NR | median 16.4 months | CS | Duke’s stage, CEA, alkaline phosphatase, number of liver lesions, albumin | Approach: Apparent* | Survival Curves |
| [61] | CRC | Liver mets | 1995–2009 | Retrospective | 382 | 327 | Median (in survivors) 47 months | DSS | Extrahepatic disease, pN category, number of liver lesions | Approach: Apparent* | None |
| [63] | CRC | Liver mets | 2002–2007 | Retrospective | 88 | 76 | Median 99 months | OS | Response to systemic therapy, number of CLM, maximum size of CLM, CEA | Approach: Apparent* | None |
| [64] | CRC | Liver mets | 1995–2005 | Retrospective Cohort | 285 | NR | median 4.4–4.6 years | OS | Tumour grade, nodal status | Approach: Apparent Discrimination: 0.59 |
None |
| [67] | CRC | Liver mets | 1993–2006 | Retrospective Cohort | 252 | NR | NR | DSS | Number of liver metastases, preoperative CEA levels, resection of liver metastases | Approach: Apparent* | None |
| [69] | CRC | Liver mets | 1992–1996 | Prospective Cohort | 478 | NR | NR | OS | Maximum liver metastasis diameter, # of liver metastasis, number of positive lymph nodes, presence of extrahepatic metastases | Approach: Apparent* | Discrimination: 0.54–0.58 |
| [70] | CRC | Liver mets | 1960–1995 | Retrospective Cohort | 662 | 428 | median 3 years | DSS | Diameter of largest liver tumour, interval from primary tumour to metastasis, hepatoduodenal LN positivity, blood transfusions, primary cancer regional LN, number of metastases’ | Approach: Bootstrap Discrimination: 0.61 |
Discrimination: 0.54–0.58 |
| [41] | CRC | Lung mets | 1980–1998 | Retrospective Cohort | 313 | 179 | median 29 months | OS | Primary histology, number of pulmonary tumours, hilar or mediastinal LN, extrathoracic disease, prethoracotomy CEA level | Approach: Bootstrap Calibration: Y Discrimination: 0.72 |
Discrimination: 0.66 & 0.81 |
| [57] | CRC | MSCC | NR | Retrospective | 121 | NR | NR | NOS | ECOG performance status, Ambuulatory status prior to RT, visceral metastases, time of developing motor deficits | Approach: Apparent* | None |
1= 3-year survival (dichotomous); 5-year survival (dichotomous); 2=: more than 2 year survival;
= 3-year survival N=93, 5-year survival N=71;
MSCC=malignant spinal cord compression; OS= overall survival; NOS= not otherwise specified; CRC=colorectal cancer; NR= not reported; DSS= disease specific survival; Schemper=percent of variability in survival explained by the model; LN=lymph nodes
All included prognostication tools were created using data collected for a purpose other than the development of a clinical prediction tool (Table 1). Three tools were developed using data from prospective cohort studies designed with the purpose of investigating prognostic factors, and six others from data collected for one or more randomized controlled trials that were not initially designed to create and/or evaluate a clinical outcome prediction tool. Seventeen tools (32%) were developed using data on cancer populations in the United States, eleven from Japan (21%), five from the United Kingdom (9%) and four each from France and Germany. In the 47 studies that reported colorectal case selection methods, 18 accrued data on patients from multiple institutions and 26 studies used data from a single institution. Data were collected from patients diagnosed or treated for colorectal cancer between 1960 and 2011 and 70% of tools (37/53) were developed on data from patients diagnosed in 2006 or earlier. Sample size for tool development was not reported for six tools, while it ranged from 71 to 128,853 (median = 426) patients when reported. Twenty-eight studies (53%) did not report the number of deaths occurring over the study period. When reported, the number of reported deaths ranged from 52 to 1077 (median = 263).
Populations and Prognostic Factors
The populations addressed by each prognostication tool are described in Table 1. Thirty-five tools (66%) were developed to aid clinical management decisions for patients diagnosed with metastatic colorectal cancer and the majority of these were developed specifically for patients with liver metastasis (26/35 tools). Seven were for all patients with metastatic disease and one each were for patients with lung metastases and patients with malignant spinal cord compression from colorectal cancer. Five were for use with patients diagnosed with all TNM stages of colorectal cancer. Ten tools targeted stage I–III colorectal cancer populations. Two tools were designed for patients with locally advanced and metastatic disease. One tool targeted prognostication in patients with Duke’s B colorectal cancer.
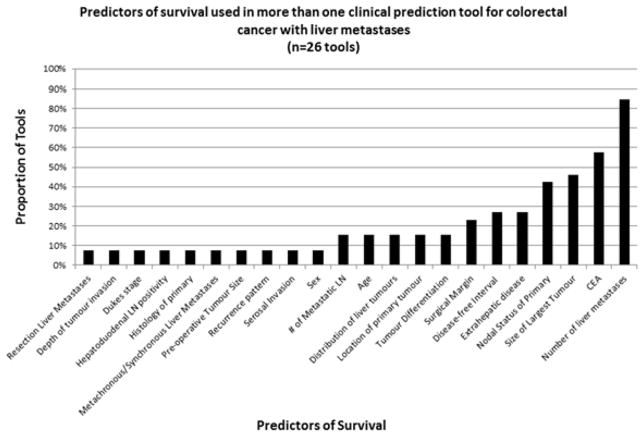
Table 2 outlines information on tool development methodology, including prognostic factor selection methods, underlying statistical model, analytic methods for missing data, and the format of continuous variables. Thirty-two tools (60%) did not provide details on the eligibility criteria used to select prognostic factors for the prediction tool and 11 tools applied p-value cut-points or other statistical rules for variable selection. There was significant heterogeneity in the prognostic factors included in tools addressing the same clinical population. For example, none of the 40 prognostic factors included in the 26 tools for patients with liver metastases were common to all tools (Figure 2). The number of liver metastases was the most commonly included variable (22/26 tools) in tools designed for patients with liver metastases. Eighteen variables were found in one liver metastases prediction tool (Table 3). 44/53 tools reported using Cox Proportional Hazards models for time to event data as the basis for their prediction tool. 23 (43%) studies did not define the index date (e.g. date of diagnosis, date of liver resection), which is critical in order to validate a tool in external populations, as well as for clinical application.
Table 2.
Methodological criteria evaluated for clinical prognostication tools for patients with colorectal cancer (n=53)
| Methodological Criterion | N (%) |
|---|---|
|
| |
| Prognostic Factor Selection Method | |
| Literature-based/clinical reasoning | 7 (13) |
| Screened using univariable analysis | 12 (23) |
| Available in existing dataset | 2 (4) |
| Method not specified | 32 (60) |
|
| |
| Methods for Handling Missing Data | |
| Complete case analysis | 8 (15) |
| Imputation | 9 (17) |
| Unknown variable category used | 1 (2) |
| Method not specified | 35 (66) |
|
| |
| Description of Handling Continuous Predictors | |
| Linear | 2 (4) |
| Cubic spline | 8 (15) |
| Transformation | 2 (4) |
| Dichotomized/categorized | 41 (77) |
|
| |
| Analytic Model Used | |
| Cox proportional hazards regression | 44 (83) |
| Logistic regression | 2 (4) |
| Recursive partition and amalgamation (RECPAM) | 1 (2) |
| Other | 2 (4) |
| Method not specified | 4 (7) |
|
| |
| Statistical Model Assumptions Checked | 7 (13) |
Figure 2.
Prognostic factors used in clinical prediction tools targeted at decision-making and prognosis in patients with colorectal cancer and liver metastases
Table 3.
Prognostic factors included in only one tool predicting survival for patients with colorectal cancer and liver metastases (n=23 tools)
| # lymph nodes around primary |
| % liver involvement |
| Albumin |
| Alkaline Phosphatase |
| Bilateral resection |
| Blood transfusion |
| Colic lymph nodes |
| Hilar metastatic lymph nodes |
| Inflammatory Response to Tumour |
| Peritoneal metastasis |
| Pre-Operative CA 19-9 |
| Preoperative GPT |
| Preoperative GT |
| Recurrence pattern |
| Resection of Liver Mets (yes/no) |
| Response to systemic treatment |
| Transaminase toxicity |
| Type of resection |
Internal Validity
Forty-five tool development studies (85%) included an evaluation of internal validity: an assessment of the predictive accuracy of the model using the same data used for model development. The majority of these evaluations incorrectly used the entire dataset (100% of patients) that the model was initially developed in to perform their assessment, rather than a form of re-sampling (apparent validation). The recommended approach to evaluating model performance is bootstrapping or cross-validation.[20,21] Bootstrapping methods create new training sets to evaluate model performance by drawing the individuals with replacement from the full data. Cross-validation evaluates model performance by repeatedly randomly splitting the original sample into training (model development) and testing (model validation) sets. Twelve studies used bootstrapping or cross-validation methods.
Twenty-three of the 45 internal validity evaluations (50%) were comparisons of the survival distributions using the log-rank statistic among risk scores or groupings, or among particular risk sets determined by values of the prognostic factors. Model calibration was assessed for 16 tools, generally by providing or referencing graphs (11/16); however, calibration slopes or intercepts and the relationship of the lines to the overall line of identity were rarely discussed. Twenty-one tools evaluated the discriminative ability of the prediction model and reported a concordance index. Concordance indices may take on values from 0.5 (model predictions are similar to chance) to 1.0 (perfect prediction). The values reported in the included studies ranged from 0.59 to 0.81.
External Validity
Half of the tools (27/53) did not have an evaluation of external validity (predictive accuracy in an independent sample separate from the one used for tool development). Seventy-nine assessments of external validity were performed on 26 tools by 33 studies, including those studies that both developed and validated a tool in the same publication. Many of the tools were evaluated multiple times, by different authors. For example, the scoring system developed by Fong and colleagues was validated in 19 separate populations,[35] and the risk classification system by Nordlinger and colleagues was validated in 10 separate populations.[54] The predictive accuracy of seven other tools was evaluated in at least three validation populations.[37,38,45,52,56,58,73]
Of the 26 tools with some evaluation of external validity, 22 had at least one assessment of model calibration, discrimination or another measure of overall model fit. Forty assessments of external validity (51%) examined only the statistical significance of separation of survival curves by risk strata. This method is not endorsed by TRIPOD nor does it appropriately assess predictive performance of the model[20,21]. Nine assessments of model calibration in the additional sample population were performed, four of which were accompanied by a calibration plot; five reported sub-group calibration. The discriminative ability of the evaluated prediction tool was reported as a concordance statistic in 33/79 of external validity evaluations; the range of values across all tools was 0.52 to 0.83.
Discussion
This study summarized available information on 53 colorectal cancer prognostication tools identified from the peer-reviewed literature and web-based resources. These tools were most commonly intended to help inform clinical management decisions in stage IV patients with liver metastases. There were considerable differences in the prognostic factors included in tools designed to prognosticate in similar clinical sub-populations (e.g. within tools for patients with liver metastases). In many cases, tool development methodology was incompletely reported or inadequate. A large number of internal and external validity assessments were performed; however, the majority did not adhere to recommended guidelines for appropriate statistical methodology[20,21]. It is apparent that a framework for moving the science of prognostic tool development and validation forward, as well as its clinical application in oncology, is still needed in order to address the deficiencies highlighted in this systematic review.
The systematic problems identified in the methods used to develop and validate colorectal cancer prognostication tools support the findings of other authors,[92–95] and call for action in the improvement of prognostic tools in oncology. Over 50% of the tools in this review categorize patients into risk groups rather than providing individual probability estimates of survival, decreasing the accuracy for the individual patient.[21] Only 10% of studies with internal validity assessments used bootstrapping, the recommended method for evaluating internal validity. Although 79 external validation exercises were performed, they evaluated a subset of the prognostic tools developed, and half of tools remained with no assessment of generalizability. In addition, 50% of the internal and external validations performed did not adhere to best practices for evaluating predictive performance and did not include an evaluation of calibration or discrimination. The TRIPOD guidelines, published early in 2015, were designed to assist clinicians and scientists in reporting clinical prediction tool studies. However, it is still too early to measure the impact this reporting guideline will have on the quality of future prognostic tool work in oncology.[20,21]
This study also provided an in depth look at the clinical populations and situations addressed by existing tools and the complement of prognostic factors used to make the survival predictions. Gaps remain in the coverage of clinical populations currently addressed by reliable prognostic tools. The majority of tools (67%) attempted to refine prognosis for patients with metastatic disease, reflecting increased uncertainty in clinical management and the need for better risk assessment to understand the benefit of treatment. A need to refine prognosis to inform decisions around the use of adjuvant chemotherapy and radiation in stage II and III patients was also identified. ACCENT, Numeracy, and Adjuvant Online! have been developed for understanding prognosis and the benefit of adjuvant chemotherapy in stage III disease.[25,59,73,77]
The lack of consistency in which prognostic factors were included in the prognostication tools identified in this review highlights the need for improved understanding of prognosis in colorectal cancer, and cautions authors when developing prognostic tools to think carefully about the inclusion of established prognostic factors and the transferability of their findings. Even when prognosis was being refined in the same clinical population, the prognostic factors included in these tools varied. None of the 40 prognostic factors used in one or more of the 26 tools predicting survival in patients with colorectal liver metastases were common to all of those tools. In the stage IV population with liver metastases, variation in the prognostic factors used across tools may reflect a gap in understanding prognosis for that population or a lack of confidence in the validity of some of those factors. A recent review of prognosis in patients with colorectal liver metastases highlighted 20 different potential prognostic factors, many of which were not included within any of the included tools in our review.[96] In tools designed for non-metastatic patients, the inclusion of clinically significant prognostic factors summarized in the AJCC 7th Edition of the Staging Manual, such as tumour regression grade, serum CEA or tumour deposits were not universal across all prognostic tools reviewed. [4] We have reported similar heterogeneity in prognostic information across prognostic tools in lung cancer[16] and melanoma[17].
The pervasive reliance on retrospective data from single institutions significantly limits what prognostic information may be included in the development of new tools and their widespread clinical usefulness. Only nine prognostication tools for colorectal cancer were developed using prospectively collected data. Designing studies to collect all relevant prognostic information will provide the best individualized estimates of prognosis. New biomarkers and prognostic factors may not be collected in many databases. Half (26/53) relied on data from single institution studies. Prognostication tools developed using data from multi-institutional studies are more likely than single institutional studies to result in relevant, generalizable models. Advances in our ability to understand colorectal cancer will necessitate weighing their added outcome prediction value to existing, affordable, baseline prognostic tools in the future.
This systematic literature review has a number of limitations. Our review may underestimate the number of existing prediction tools designed for survival in colorectal cancer, given the lack of literature search terms at the time to identify relevant studies. However, to account for this both a cited reference search and web-based resources search were performed to widen the net and capture all relevant tools and documentation. The review was also restricted to English language only studies, which may create a language reporting bias. However, the tools included in the study appeared to be developed and validated across a variety of countries and populations. Finally, we did not include tools developed solely using genomic data, as we considered these outside the scope of the review. Therefore, the results of this review many not be representative of the methods and relevance of studies carried out in that area.
The need to refine prognosis for individual patients within TNM stages remains. TNM stage defined the clinical population addressed by the majority of prognostication tools. Within each stage grouping, numerous tools were identified that relied on a multitude of additional prognostic information to individualize predictions. This suggests that clinical prognostic tools may be a viable and clinically relevant option to individualize prognosis without redesigning the TNM stage classification system itself. The AJCC PMC has taken the first step to assuring that existing meritorious tools are made known to the community by establishing the criteria for AJCC endorsement and evaluating tools in major disease areas according to these guidelines.[18,19] The AJCC intends to continue to play a leadership role in the evaluation, development, and promotion of high-quality prognostication tools in coordination with other authoritative groups such as the PROGnosis RESearch Strategy (PROGRESS) Partnership.[13] The evaluation of prognostic models has been incorporated into the 8th Edition of the staging manual.[18]
Overall, prognostication tools are pervasive in colorectal cancer and may be particularly useful in clinical management when the outcome is uncertain. Guidance in the future direction of prognostication tool development and validation in colorectal cancer is needed. Moving forward, many key clinical and methodological issues in the development, validation and clinical usability need to be addressed. However, addressing statistical and methodological concerns alone will not improve this research area, until consideration is given to the practical strengths and limitations of the literature. We need to build capacity and infrastructure to perform optimal prognostic tool research to realize the potential benefit of these tools for the future.[13,20,21] Collaborative, primary research grants with the objective of developing useful prognostic tools using prospectively collected data, and that include an appropriate assessment of internal and external validity, as well as the evaluation of impact on decision-making will be critical to improving the quality of prognostic tools available for use in colorectal cancer.
Table 4.
Details of evaluations of tool internal and external validity (n=53 tools)
| Performance Measure | Internal Validation (n= 45 tools) | External Validation (n= 79 validations) |
|---|---|---|
|
| ||
| Internal Validation Method* | ||
| Apparent | 27 (60) | -- |
| Cross-Validation | 2 (4) | -- |
| Split Sample | 6 (14) | -- |
| Bootstrapping | 10 (22) | -- |
|
| ||
| External Validation Method | ||
| Independent | -- | 61 (77) |
| Geographic | -- | 13 (16) |
| Temporal | -- | 3 (4) |
| Other** | 2 (3) | |
|
| ||
| Overall Model Performance | ||
| R-squared | 2 (4) | 2 (3) |
|
| ||
| Calibration | ||
| Graph (Plot/intercept/slope) | 11 (25) | 4 (5) |
| Hosmer/Lemeshow statistic | 3 (7) | 0 (0) |
| Sub-group calibration*** | 2 (5) | 5 (6) |
|
| ||
| Discrimination | ||
| C-statistic**** | 21 (48) | 33 (42) |
|
| ||
| Survival Analysis Only with Significance Test | 22 (50) | 40 (51) |
One tool applied both split sample and bootstrap methods;
An RCT(s) was used to develop the prognostic tool, and an additional RCT was used for validation;
Tables comparing predicted and observed values for groups of patients were provided;
Concordance index based on the ROC for binary data, Harrell’s C statistic for models using time to event data
Synopsis.
Many prognostication tools have been developed as aids to colorectal cancer patient management, but little is known about their quality. We performed a systematic literature review of colorectal cancer prognostication tools in the peer-reviewed literature and web-based resources. Guidance for future development of prognostication tools in colorectal cancer is needed to assure the quality and clinical utility of these important instruments.
Acknowledgments
Grant Support: P30 CA008748.
Sources of Support: The American Joint Committee on Cancer (AJCC) provided a contract to Patti Groome and Alyson Mahar to support the work of identifying and evaluating existing prognostic tools for lung cancer as a preparatory step in the AJCC’s development of prognostic tools for major cancers.
References
- 1.Gunderson LL, Haller DG, Martenson JA, et al. Impact of T and N stage and treatment on survival and relapse in adjuvant rectal cancer: a pooled analysis. J Clin Oncol. 2004;22:1785–1796. doi: 10.1200/JCO.2004.08.173. [DOI] [PubMed] [Google Scholar]
- 2.Gunderson LL, Jessup JM, Sargent DJ, et al. Revised tumor and node categorization for rectal cancer based on surveillance, epidemiology, and end results and rectal pooled analysis outcomes. J Clin Oncol. 2010;28:256–263. doi: 10.1200/JCO.2009.23.9194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gunderson LL, Jessup JM, Sargent DJ, et al. Revised TN categorization for colon cancer based on national survival outcomes data. J Clin Oncol. 2010;28:264–271. doi: 10.1200/JCO.2009.24.0952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Edge SB. AJCC Cancer 7th Edition Staging Manual. New York: Springer; 2010. [Google Scholar]
- 5.Haq AI, Schneeweiss J, Kalsi V, Arya M. The Dukes staging system: a cornerstone in the clinical management of colorectal cancer. Lancet Oncol. 2009;10:1128–2045. doi: 10.1016/S1470-2045(09)70157-3. (1109)70157–70153. [DOI] [PubMed] [Google Scholar]
- 6.Zlobec I, Lugli A. Prognostic and predictive factors in colorectal cancer. J Clin Pathol. 2008;61:561–569. doi: 10.1136/jcp.2007.054858. [DOI] [PubMed] [Google Scholar]
- 7.Compton CC. Colorectal Cancer. In: Gospodarowicz MK, O’Sullivan B, Sobin LH, editors. Prognostic factors in Cancer. Hoboken, New Jersey: John Wiley & Sons, Inc; 2006. pp. 133–137. [Google Scholar]
- 8.Allegra CJ, Jessup JM, Somerfield MR, et al. American Society of Clinical Oncology provisional clinical opinion: testing for KRAS gene mutations in patients with metastatic colorectal carcinoma to predict response to anti-epidermal growth factor receptor monoclonal antibody therapy. J Clin Oncol. 2009;27:2091. doi: 10.1200/JCO.2009.21.9170. [DOI] [PubMed] [Google Scholar]
- 9.De Roock W, De Vriendt V, Normanno N, et al. KRAS, BRAF, PIK3CA, and PTEN mutations: implications for targeted therapies in metastatic colorectal cancer. Lancet Oncol. 2011;12:594–603. doi: 10.1016/S1470-2045(10)70209-6. [DOI] [PubMed] [Google Scholar]
- 10.Febbo PG, Ladanyi M, Aldape KD, et al. NCCN Task Force report: Evaluating the clinical utility of tumor markers in oncology. J Natl Compr Canc Netw. 2011;9(Suppl 5):S1–32. doi: 10.6004/jnccn.2011.0137. quiz S33. [DOI] [PubMed] [Google Scholar]
- 11.Locker GY, Hamilton S, Harris J, et al. ASCO 2006 update of recommendations for the use of tumor markers in gastrointestinal cancer. J Clin Oncol. 2006;24:5313–5327. doi: 10.1200/JCO.2006.08.2644. [DOI] [PubMed] [Google Scholar]
- 12.Steyerberg EW. Clinical prediction models: a practical approach to development, validation, and updating. New York, NY: Springer New York; 2009. [Google Scholar]
- 13.Steyerberg EW, Moons KGM, van der Windt DA, et al. Prognosis Research Strategy (PROGRESS) 3: prognostic model research. PLoS medicine. 2013;10:e1001381. doi: 10.1371/journal.pmed.1001381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vickers AJ. Prediction models in cancer care. CA Cancer J Clin. 2011;61:315–326. doi: 10.3322/caac.20118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mahar AL, Halabi S, McShane L, et al. A survey of clinical prediction tools in colorectal and lung cancers and melanoma. J Clin Oncol. 2013;31:1592. [Google Scholar]
- 16.Mahar AL, Compton C, McShane LM, et al. Refining prognosis in lung cancer: A report on the quality and relevance of clinical prognostic tools. J Thorac Oncol. 2015 doi: 10.1097/JTO.0000000000000652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mahar AL, Compton C, Halabi S, et al. Critical Assessment of Clinical Prognostic Tools in Melanoma. Annals of surgical oncology. 2016;23:2753–2761. doi: 10.1245/s10434-016-5212-5. [DOI] [PubMed] [Google Scholar]
- 18.American Joint Committee on Cancer. AJCC Cancer Staging Manual. Springer International Publishing; 2017. [Google Scholar]
- 19.Kattan MW, Hess KR, Amin MB, et al. American Joint Committee on Cancer acceptance criteria for inclusion of risk models for individualized prognosis in the practice of precision medicine. CA: a cancer journal for clinicians. 2016;66:370–374. doi: 10.3322/caac.21339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Collins GS, Reitsma JB, Altman DG, Moons KGM. Transparent Reporting of a multivariable prediction model for Individual Prognosis Or Diagnosis (TRIPOD): The TRIPOD StatementThe TRIPOD Statement. Ann Intern Med. 2015;162:55–63. doi: 10.7326/M14-0697. [DOI] [PubMed] [Google Scholar]
- 21.Moons KGM, Altman DG, Reitsma JB, et al. Transparent Reporting of a multivariable prediction model for Individual Prognosis Or Diagnosis (TRIPOD): Explanation and ElaborationThe TRIPOD Statement: Explanation and Elaboration. Ann Intern Med. 2015;162:W1–W73. doi: 10.7326/M14-0698. [DOI] [PubMed] [Google Scholar]
- 22.Moons KG, de Groot JA, Bouwmeester W, et al. Critical appraisal and data extraction for systematic reviews of prediction modelling studies: the CHARMS checklist. PLoS Med. 2014:11. doi: 10.1371/journal.pmed.1001744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fendler WP, Ilhan H, Paprottka PM, et al. Nomogram including pretherapeutic parameters for prediction of survival after SIRT of hepatic metastases from colorectal cancer. Eur Radiol. 2015;25:2693–2700. doi: 10.1007/s00330-015-3658-7. [DOI] [PubMed] [Google Scholar]
- 24.Lu HJ, Lin JK, Chen WS, et al. The Prognostic Role of Para-Aortic Lymph Nodes in Patients with Colorectal Cancer: Is It Regional or Distant Disease? PLoS One. 2015;10:e0130345. doi: 10.1371/journal.pone.0130345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Adjuvant! for Colon Cancer. 2011. [Google Scholar]
- 26.Adam R, Delvart V, Pascal G, et al. Rescue Surgery for Unresectable Colorectal Liver Metastases Downstaged by Chemotherapy: A Model to Predict Long-term Survival. Ann Surg. 2004;240:644–658. doi: 10.1097/01.sla.0000141198.92114.f6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.MD Anderson Cancer Center. Rectal Cancer Survival Calculator- No XRT. 2012. [Google Scholar]
- 28.MD Anderson Cancer Center. Rectal Cancer Survival Calculator- Pre-Op XRT. 2012. [Google Scholar]
- 29.MD Anderson Cancer Center. Rectal Cancer Survival Calculator- Post-Op XRT. 2012. [Google Scholar]
- 30.MD Anderson Cancer Center. Rectal Cancer Survival Calculator- Stage IV. 2012. [Google Scholar]
- 31.MD Anderson Cancer Center. Colon Cancer Survival Calculator. 2012. [Google Scholar]
- 32.Chibaudel B, Bonnetain F, Tournigand C, et al. Simplified prognostic model in patients with oxaliplatin-based or irinotecan-based first-line chemotherapy for metastatic colorectal cancer: a GERCOR study. Oncologist. 2011;16:1228. doi: 10.1634/theoncologist.2011-0039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cianchi F, Messerini L, Comin CE, et al. Pathologic Determinants of Survival After Resection of T3N0 (Stage IIA) Colorectal Cancer: Proposal for a New Prognostic Model. Dis Colon Rectum. 2007;50:1332–1341. doi: 10.1007/s10350-007-0222-9. [DOI] [PubMed] [Google Scholar]
- 34.Elias D, Faron M, Goéré D, et al. A Simple Tumor Load-Based Nomogram for Surgery in Patients with Colorectal Liver and Peritoneal Metastases. Ann Surg Oncol. 2014;21:2052–2058. doi: 10.1245/s10434-014-3506-z. [DOI] [PubMed] [Google Scholar]
- 35.Fong Y, Fortner J, Sun RL, et al. Clinical score for predicting recurrence after hepatic resection for metastatic colorectal cancer: analysis of 1001 consecutive cases. Ann Surg. 1999;230:309–309. doi: 10.1097/00000658-199909000-00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hill CRS, Chagpar RB, Callender GG, et al. recurrence following hepatectomy for metastatic colorectal cancer: development of a model that predicts patterns of recurrence and survival. Ann Surg Oncol. 2012;19:139–144. doi: 10.1245/s10434-011-1921-y. [DOI] [PubMed] [Google Scholar]
- 37.Iwatsuki S, Dvorchik I, Madariaga JR, et al. Hepatic resection for metastatic colorectal adenocarcinoma: a proposal of a prognostic scoring system. J Am Coll Surg. 1999;189:291. doi: 10.1016/s1072-7515(99)00089-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kohne CH, Cunningham D, Di Costanzo F, et al. Clinical determinants of survival in patients with 5-fluorouracil-based treatment for metastatic colorectal cancer: results of a multivariate analysis of 3825 patients. Ann Oncol. 2002;13:308–317. doi: 10.1093/annonc/mdf034. [DOI] [PubMed] [Google Scholar]
- 39.Kama NA, Kologlu M, Reis E, et al. A prognostic score for colorectal cancer. Hepatogastroenterology. 2003;50:1356–1361. [PubMed] [Google Scholar]
- 40.Kanemitsu Y, Kato T. Prognostic models for predicting death after hepatectomy in individuals with hepatic metastases from colorectal cancer. World J Surg. 2008;32:1097–1107. doi: 10.1007/s00268-007-9348-0. [DOI] [PubMed] [Google Scholar]
- 41.Kanemitsu Y, Kato T, Hirai T, Yasui K. Preoperative probability model for predicting overall survival after resection of pulmonary metastases from colorectal cancer. Br J Surg. 2004;91:112–120. doi: 10.1002/bjs.4370. [DOI] [PubMed] [Google Scholar]
- 42.Kato H, Yoshimatsu K, Ishibashi K, et al. A New Staging System for Colorectal Carcinoma with Liver Metastasis. Anticancer Res. 2005;25:1251. [PubMed] [Google Scholar]
- 43.Kattan MW, Gonen M, Jarnagin WR, et al. A nomogram for predicting disease-specific survival after hepatic resection for metastatic colorectal cancer. Ann Surg. 2008;247:282. doi: 10.1097/SLA.0b013e31815ed67b. [DOI] [PubMed] [Google Scholar]
- 44.Kobayashi H, Kotake K, Sugihara K. Prognostic scoring system for stage IV colorectal cancer: is the AJCC sub-classification of stage IV colorectal cancer appropriate? Int J Clin Oncol. 2013;18:696–703. doi: 10.1007/s10147-012-0433-5. [DOI] [PubMed] [Google Scholar]
- 45.Konopke R, Kersting S, Distler M, et al. Prognostic factors and evaluation of a clinical score for predicting survival after resection of colorectal liver metastases. Liver International. 2009;29:89–102. doi: 10.1111/j.1478-3231.2008.01845.x. [DOI] [PubMed] [Google Scholar]
- 46.Lee W-S, Kim MJ, Yun SH, et al. Risk factor stratification after simultaneous liver and colorectal resection for synchronous colorectal metastasis. Arch Surg. 2008;393:13–19. doi: 10.1007/s00423-007-0231-0. [DOI] [PubMed] [Google Scholar]
- 47.Lise M, Bacchetti S, Da Pian P, et al. Patterns of recurrence after resection of colorectal liver metastases: prediction by models of outcome analysis. World J Surg. 2001;25:638–644. doi: 10.1007/s002680020138. [DOI] [PubMed] [Google Scholar]
- 48.Malik HZ, Prasad KR, Halazun KJ, et al. Preoperative prognostic score for predicting survival after hepatic resection for colorectal liver metastases. Ann Surg. 2007;246:806–814. doi: 10.1097/SLA.0b013e318142d964. [DOI] [PubMed] [Google Scholar]
- 49.Massacesi C, Norman A, Price T, et al. A clinical nomogram for predicting long-term survival in advanced colorectal cancer. Eur J Cancer. 2000;36:2044–2052. doi: 10.1016/s0959-8049(00)00286-0. [DOI] [PubMed] [Google Scholar]
- 50.Michaelson JS. Colon Cancer Outcome Calculator. 2011. [Google Scholar]
- 51.Minagawa M, Yamamoto J, Kosuge T, et al. Simplified staging system for predicting the prognosis of patients with resectable liver metastasis: development and validation. Arch Surg. 2007;142:269–276. doi: 10.1001/archsurg.142.3.269. [DOI] [PubMed] [Google Scholar]
- 52.Nagashima I, Takada T, Matsuda K, et al. A new scoring system to classify patients with colorectal liver metastases: proposal of criteria to select candidates for hepatic resection. J Hepatobiliary Pancreat Surg. 2004;11:79–83. doi: 10.1007/s00534-002-0778-7. [DOI] [PubMed] [Google Scholar]
- 53.Nanashima A, Sumida Y, Abo T, et al. A modified grading system for post-hepatectomy metastatic liver cancer originating from colorectal carcinoma. J Surg Oncol. 2008;98:363–370. doi: 10.1002/jso.21114. [DOI] [PubMed] [Google Scholar]
- 54.Nordlinger B, Guiguet M, Vaillant J-C, et al. Surgical resection of colorectal carcinoma metastases to the liver: A prognostic scoring system to improve case selection, based on 1568 patients. Cancer. 1996;77:1254–1262. [PubMed] [Google Scholar]
- 55.Peng J, Ding Y, Tu S, et al. Prognostic nomograms for predicting survival and distant metastases in locally advanced rectal cancers. PLoS One. 2014;9:e106344. doi: 10.1371/journal.pone.0106344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Petersen VC, Baxter KJ, Love SB, Shepherd NA. Identification of objective pathological prognostic determinants and models of prognosis in Dukes’ B colon cancer. Gut. 2002;51:65–69. doi: 10.1136/gut.51.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rades D, Douglas S, Huttenlocher S, et al. Prognostic factors and a survival score for patients with metastatic spinal cord compression from colorectal cancer. Strahlenther Onkol. 2012;188:1114–1118. doi: 10.1007/s00066-012-0141-0. [DOI] [PubMed] [Google Scholar]
- 58.Rees M, Tekkis PP, Welsh FKS, et al. Evaluation of long-term survival after hepatic resection for metastatic colorectal cancer: a multifactorial model of 929 patients. Ann Surg. 2008;247:125. doi: 10.1097/SLA.0b013e31815aa2c2. [DOI] [PubMed] [Google Scholar]
- 59.Renfro LA, Loprinzi CL, Yothers G, et al. ACCENT-based web calculators to predict recurrence and overall survival in stage III colon cancer. J Natl Cancer Inst. 2014;106:dju333–dju333. doi: 10.1093/jnci/dju333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schindl M, Wigmore SJ, Currie EJ, et al. Prognostic scoring in colorectal cancer liver metastases: development and validation. Arch Surg. 2005;140:183–189. doi: 10.1001/archsurg.140.2.183. [DOI] [PubMed] [Google Scholar]
- 61.Settmacher U, Dittmar Y, Knösel T, et al. Predictors of long-term survival in patients with colorectal liver metastases: a single center study and review of the literature. Int J Colorectal Dis. 2011;26:967–981. doi: 10.1007/s00384-011-1195-7. [DOI] [PubMed] [Google Scholar]
- 62.Shitara K, Najima M, Muro K, et al. Prognostic factors for metastatic colorectal cancer patients undergoing irinotecan-based second-line chemotherapy. Gastrointest Cancer Res. 2011;4:168–172. [PMC free article] [PubMed] [Google Scholar]
- 63.Stang A, Oldhafer K, Jr, Weilert H, et al. Selection criteria for radiofrequency ablation for colorectal liver metastases in the era of effective systemic therapy: a clinical score based proposal. BMC Cancer. 2014;14:500–500. doi: 10.1186/1471-2407-14-500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tan MCB, Castaldo ET, Gao F, et al. A Prognostic System Applicable to Patients with Resectable Liver Metastasis from Colorectal Carcinoma Staged by Positron Emission Tomography with [ 18F]Fluoro-2-Deoxy-D-Glucose: Role of Primary Tumor Variables. J Am Coll Surg. 2008;206:857–868. doi: 10.1016/j.jamcollsurg.2007.12.023. [DOI] [PubMed] [Google Scholar]
- 65.Valentini V, van Stiphout RGPM, Lammering G, et al. Nomograms for predicting local recurrence, distant metastases, and overall survival for patients with locally advanced rectal cancer on the basis of European randomized clinical trials. J Clin Oncol. 2011;29:3163. doi: 10.1200/JCO.2010.33.1595. [DOI] [PubMed] [Google Scholar]
- 66.Wang SJ, Wissel AR, Luh JY, et al. An interactive tool for individualized estimation of conditional survival in rectal cancer. Ann Surg Oncol. 2011;18:1547–1552. doi: 10.1245/s10434-010-1512-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang Y, Liu YF, Cheng Y, et al. Prognosis of colorectal cancer with liver metastasis: value of a prognostic index. Braz J Med Biol Res. 2010;43:1116–1122. doi: 10.1590/s0100-879x2010007500103. [DOI] [PubMed] [Google Scholar]
- 68.Weiser MR, Gönen M, Chou JF, et al. Predicting survival after curative colectomy for cancer: individualizing colon cancer staging. J Clin Oncol. 2011;29:4796–4802. doi: 10.1200/JCO.2011.36.5080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yamaguchi T, Mori T, Takahashi K, et al. A new classification system for liver metastases from colorectal cancer in Japanese multicenter analysis. Hepatogastroenterology. 2008;55:173–178. [PubMed] [Google Scholar]
- 70.Zakaria S, Donohue JH, Que FG, et al. Hepatic resection for colorectal metastases: value for risk scoring systems? Ann Surg. 2007;246:183–191. doi: 10.1097/SLA.0b013e3180603039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kawai K, Ishihara S, Yamaguchi H, et al. Nomograms for predicting the prognosis of stage IV colorectal cancer after curative resection: a multicenter retrospective study. Eur J Surg Oncol. 2015;41:457–465. doi: 10.1016/j.ejso.2015.01.026. [DOI] [PubMed] [Google Scholar]
- 72.van Gijn W, van Stiphout RG, van de Velde CJ, et al. Nomograms to predict survival and the risk for developing local or distant recurrence in patients with rectal cancer treated with optional short-term radiotherapy. Ann Oncol. 2015;26:928–935. doi: 10.1093/annonc/mdv023. [DOI] [PubMed] [Google Scholar]
- 73.Adjuvant systemic therapy for resected colon cancer. 2012. Numeracy. [Google Scholar]
- 74.Ayez N, Lalmahomed ZS, van der Pool AEM, et al. Is the clinical risk score for patients with colorectal liver metastases still useable in the era of effective neoadjuvant chemotherapy? Ann Surg Oncol. 2011;18:2757–2763. doi: 10.1245/s10434-011-1819-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Desot E, Marquis E, Bouché O, et al. Prognostic factors in patients with non resectable metastatic colorectal cancer in the era of targeted biotherapies: relevance of Köhne’s risk classification. Dig Liver Dis. 2013;45:330. doi: 10.1016/j.dld.2012.10.016. [DOI] [PubMed] [Google Scholar]
- 76.Diouf M, Hadengue A, de Gramont A, et al. Could baseline health-related quality of life (QoL) predict overall survival in metastatic colorectal cancer? The results of the GERCOR OPTIMOX 1 study. Health and quality of life outcomes. 2014;12:69–69. doi: 10.1186/1477-7525-12-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gill S, Loprinzi C, Kennecke H, et al. Prognostic web-based models for stage II and III colon cancer: A population and clinical trials-based validation of Numeracy and Adjuvant! online. Cancer. 2011;117:4155. doi: 10.1002/cncr.26003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ivanecz A, Potrc S, Horvat M, et al. The validity of clinical risk score for patients undergoing liver resection for colorectal metastases. Hepatogastroenterology. 2009;56:1452–1458. [PubMed] [Google Scholar]
- 79.Kanemitsu Y, Kato T, Komori K, et al. Validation of a nomogram for predicting overall survival after resection of pulmonary metastases from colorectal cancer at a single center. World J Surg. 2010;34:2973–2978. doi: 10.1007/s00268-010-0745-4. [DOI] [PubMed] [Google Scholar]
- 80.Mala T, Bohler G, Mathisen O, et al. Hepatic resection for colorectal metastases: can preoperative scoring predict patient outcome? World J Surg. 2002;26:1348–1353. doi: 10.1007/s00268-002-6231-x. [DOI] [PubMed] [Google Scholar]
- 81.Mann CD, Metcalfe MS, Leopardi LN, Maddern GJ. The clinical risk score: emerging as a reliable preoperative prognostic index in hepatectomy for colorectal metastases. Arch Surg. 2004;139:1168–1172. doi: 10.1001/archsurg.139.11.1168. [DOI] [PubMed] [Google Scholar]
- 82.Mavros MN, Hyder O, Pulitano C, et al. Survival of patients operated for colorectal liver metastases and concomitant extra-hepatic disease: External validation of a prognostic model. J Surg Oncol. 2013;107:481–485. doi: 10.1002/jso.23260. [DOI] [PubMed] [Google Scholar]
- 83.Merkel S, Bialecki D, Meyer T, et al. Comparison of clinical risk scores predicting prognosis after resection of colorectal liver metastases. J Surg Oncol. 2009;100:349–357. doi: 10.1002/jso.21346. [DOI] [PubMed] [Google Scholar]
- 84.Morris EJA, Maughan NJ, Forman D, Quirke P. Who to treat with adjuvant therapy in Dukes B/stage II colorectal cancer? The need for high quality pathology. Gut. 2007;56:1419–1425. doi: 10.1136/gut.2006.116830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Reddy SK, Kattan MW, Yu C, et al. Evaluation of peri-operative chemotherapy using a prognostic nomogram for survival after resection of colorectal liver metastases. HPB (Oxford) 2009;11:592. doi: 10.1111/j.1477-2574.2009.00106.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Reissfelder C, Rahbari NN, Koch M, et al. Validation of prognostic scoring systems for patients undergoing resection of colorectal cancer liver metastases. Ann Surg Oncol. 2009;16:3279–3288. doi: 10.1245/s10434-009-0654-7. [DOI] [PubMed] [Google Scholar]
- 87.Roberts KJ, White A, Cockbain A, et al. Performance of prognostic scores in predicting long-term outcome following resection of colorectal liver metastases. Br J Surg. 2014;101:856–866. doi: 10.1002/bjs.9471. [DOI] [PubMed] [Google Scholar]
- 88.Roxburgh CS, Crozier JE, Maxwell F, et al. Comparison of tumour-based (Petersen Index) and inflammation-based (Glasgow Prognostic Score) scoring systems in patients undergoing curative resection for colon cancer. Br J Cancer. 2009;100:701–706. doi: 10.1038/sj.bjc.6604926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Shin SJ, Kim NK, Ahn JB, et al. Implications of clinical risk score to predict outcomes of liver-confined metastasis of colorectal cancer. Surg Oncol. 2012;21:e125. doi: 10.1016/j.suronc.2012.04.002. [DOI] [PubMed] [Google Scholar]
- 90.Shitara K, Yuki S, Yamazaki K, et al. Validation study of a prognostic classification in patients with metastatic colorectal cancer who received irinotecan-based second-line chemotherapy. J Cancer Res Clin Oncol. 2013;139:595–603. doi: 10.1007/s00432-012-1349-1. [DOI] [PubMed] [Google Scholar]
- 91.Takakura Y, Okajima M, Kanemitsu Y, et al. External Validation of Two Nomograms for Predicting Patient Survival After Hepatic Resection for Metastatic Colorectal Cancer. World J Surg. 2011;35:2275–2282. doi: 10.1007/s00268-011-1194-4. [DOI] [PubMed] [Google Scholar]
- 92.Bouwmeester W, Zuithoff NPA, Mallett S, et al. Reporting and methods in clinical prediction research: a systematic review. PLoS Med. 2012;9:1. doi: 10.1371/journal.pmed.1001221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Mallett S, Royston P, Dutton S, et al. Reporting methods in studies developing prognostic models in cancer: a review. BMC Med. 2010;8:20. doi: 10.1186/1741-7015-8-20. 7015-7018-7020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Mallett S, Royston P, Waters R, et al. Reporting performance of prognostic models in cancer: a review. BMC Med. 2010;8:21. doi: 10.1186/1741-7015-8-21. 7015-7018-7021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Isariyawongse BK, Kattan MW. Prediction tools in surgical oncology. Surg Oncol Clin N Am. 2012;21:439–447. viii–ix. doi: 10.1016/j.soc.2012.03.007. [DOI] [PubMed] [Google Scholar]
- 96.Spolverato G, Ejaz A, Azad N, Pawlik TM. Surgery for colorectal liver metastases: The evolution of determining prognosis. World J Gastrointest Oncol. 2013;5:207–221. doi: 10.4251/wjgo.v5.i12.207. [DOI] [PMC free article] [PubMed] [Google Scholar]