Graphical abstract
Keywords: Cassia occidentalis, Anthraquinone, DNA, Molecular docking, Glutathione
Highlights
-
•
Emodin has the maximum binding affinity to calf thymus DNA.
-
•
Anthraquinones form GSH conjugates.
-
•
Anthraquinones oxidizes GSH to GSSG.
-
•
Cytotoxicity of anthraquinones are linked to their DNA binding affinity.
Abstract
Consumption of Cassia occidentalis (CO) seeds has been associated with the hepatomyoencephalopathy (HME) in children. Recently, we have characterized the toxic anthraquinones (AQs) such as Emodin, Rhein, Aloe-emodin, Chrysophanol and Physcion in CO seeds and detected these moieties in the bio fluids of CO poisoning cases. As AQs were detected in the serum of HME patients, their interaction with key biomolecules including protein, DNA and glutathione (GSH) is imperative. In this regard, we have previously reported the interaction of these AQs with serum albumin protein and their subsequent biological effects. However, the interaction of these AQs with DNA and GSH remained unexplored. In the present work, we have studied the binding of these AQs of CO seeds with DNA and GSH by fluorescence spectroscopy, UV–vis spectral analysis, molecular docking, and biochemical studies. Results indicated a higher binding affinity for Emodin (Ka = 3.854 × 104 L mol−1 S−1), Aloe-emodin (Ka = 0.961 × 104 L mol−1 S−1) and Rhein (Ka = 0.034 × 104 L mol−1 S−1) towards calf thymus DNA may be associated with their higher cytotoxicity. Alternatively, Physcion and Chrysophanol which showed less cytotoxicity in our earlier studies exhibited very low DNA binding. The binding pattern of all these AQs is consistent with the in-silico data. Absorption spectroscopy studies indicated the possible formation of GSH conjugate with Aloe-emodin and Physcion. Further biochemical measurement of GSH and GSSG (Glutathione disulfide) following incubation with AQs indicated that Aloe-emodin (28%) and Rhein (30%) oxidizes GSH to GSSG more as compared to other AQs. Taken together, these results suggest that the higher cytotoxicity of Rhein, Emodin and Aloe-emodin may be attributed to their potent DNA and GSH binding affinity.
1. Introduction
In recent past, accidental poisoning of Cassia occidentalis seeds has been known to be the causative factor for children death in several parts of India [[1], [2], [3]]. Our previous studies have established the association of children death with CO poisoning [1] and have identified the toxic anthraquinones (AQs) including Aloe-emodin, Chrysophanol, Emodin, Physcion and Rhein in CO seeds [4]. All these AQs were further detected in the serum of CO seeds exposed patients as well as in the experimental rats, linking their role to the CO toxicity [4]. In another study, we have reported that CO seeds treatment to rats modulate an array of transcripts including oxidative stress and xenobiotic metabolism [5]. Impairment of xenobiotic metabolism due to CO seeds exposure may lead to the accumulation of active ingredients including AQs inside the body that may cause toxic manifestations.
Although the toxicity of some of these AQs is known, the mechanism(s) of toxicity is not fully understood [[6], [7]]. The AQs group of compounds falls under a large group of bioactive Quinones. Quinones primarily exhibit toxicity in two ways; firstly, forming conjugates with cellular macromolecules (Protein, DNA and Glutathione), secondly by generation of semi Quinone radical by one electron transfer leading to oxidative stress [8]. In one of our earlier studies we have examined the binding affinity of AQs of CO seeds including Aloe-emodin, Chrysophanol, Emodin, Physcion and Rhein with bovine serum albumin protein and its association with the cytotoxicity in hepatic cells [9]. However, the interaction of these AQs with DNA and GSH remain unexplored.
Earlier, through the hepatic transcriptional analysis of CO seeds exposed rat, we have demonstrated that the toxic ingredients of CO seeds induce the DNA damage and apoptotic pathways in rat liver [5]. In addition, recently we have seen that Rhein, a toxic anthraquinones in CO seeds causes DNA damage and induce apoptosis in rat primary hepatocytes [10]. Further, we have shown that CO seeds or the anthraquinones therein decreases the free-SH or GSH both in vitro and in vivo [[1], [10]]. Therefore, it is essential to explore the AQs-DNA and AQs-GSH interaction in detail. In this study we have investigated the binding affinity and interactions of AQs with DNA and GSH in a context to their toxicity.
2. Material and methods
2.1. Chemicals and reagents
Calf thymus DNA (ctDNA), Emodin, Rhein, Aloe-emodin, Chrysophanol, Physcion, Glutathione (GSH), oxidized Glutathione (GSSG), Ethidium bromide (EtBr) Orthopthaldehyde (OPT) and N-ethylmaleimide (NEM) were purchased from Sigma Aldrich Co. (St. Louis, MO). All other chemicals used were of the highest purity available from commercial sources.
2.2. DNA stock preparation and quantification
The stock solution of calf thymus DNA was prepared in Tris-EDTA buffer (1 M Tris and 0.5 M EDTA, pH 8.0) as described earlier [11]. DNA solution with UV absorbance ratio at 260 and 280 nm (A260/A280) greater than 1.9 showed that the DNA was free of protein. DNA concentration was determined by Spectrophotometer ND1000 (NanoDrop Technologies Inc., USA) and expressed as ng/μl. Further the concentration was converted to micromolar equivalent using A260 unit of double stranded DNA corresponding to 50 μg/ml, which is equivalent to 0.15 mM. Absorption coefficient of 6600 M−1 cm−1 for DNA was used for quantification.
2.3. Interaction of anthraquinones with DNA-ethidium bromide complex
The fluorescence emission spectra of all the AQs were recorded separately and compared with the emission spectra of DNA, EtBr and DNA-EtBr complex. Since, Emodin emission spectra showed no overlapping with DNA-EtBr, Emodin was further studied by this method as described earlier [12]. In brief, Perkin Elmer Luminescence spectrometer (Waltham, MA) with a quartz cell of 1 cm path length was used to measure the fluorescence. The excitation wavelength was set at 350 nm while the emission spectrum was scanned from 500 to 750 nm. The slit width of the excitation and emission was set at 10 nm. The fluorescence spectra of different concentrations of Emodin (5–50 μM) in a fixed concentration of DNA-EtBr complex (75 μM DNA and 6.3 μM EtBr) were measured in 0.4 M Britton-Robinson buffer (0.04 M acetic acid, 0.04 M boric acid and 0.04 M orthophosphoric acid at pH 7.4). The quenching constants of Emodin to DNA-EtBr complex was quantitatively calculated from the Stern Volmer [13] and modified Stern Volmer plot [14] as described in the results section.
2.4. Interaction of DNA with the emission spectra of anthraquinones
The fluorescence emission spectra of all the AQs (20 μM) were recorded. Subsequently, fluorometric titration was carried out by adding ctDNA (10–100 μM). Among all the AQs, only the emission spectra of Rhein and Aloe-emodin were efficiently quenched by the addition of ctDNA. Hence, the binding of these two AQs with DNA was carried out following the method described earlier [15]. In brief, fluorescence measurements were carried out on a Perkin Elmer Luminescence spectrometer (Waltham, MA) by progressive addition of ctDNA (0–1.5 mM) to a fixed concentration (50 μM) of Aloe-emodin or Rhein in BR buffer. The excitation wavelength was set at 400 nm and the emission spectra were recorded from 450 nm to 750 nm. The intrinsic fluorescence of Aloe-emodin and Rhein was obtained at 582 nm when excited at 400 nm. The quantitative analysis of the potential interaction of Aloe-emodin and Rhein with DNA was performed based on the fluorometric titration. The dynamic quenching constant, quenching rate constant and effective quenching constant were calculated using Stern-Volmer equation (SVE) and modified Stern-Volmer equation (MSVE).
2.5. Molecular docking study of anthraquinones with ctDNA
Docking of AQs with DNA were carried out using Discovery Studio version 4.0 (Accelrys, San Diego). The crystal structure of Calf thymus DNA (ctDNA) was obtained from Protein Data Bank (PDBID: 453D) [16]; [17]. In order to prepare the structure for molecular docking, we removed water molecules and added gasteiger charges on the crystal structure. The structures of five anthraquinones (AQs) i.e. Rhein (Pubchem CID:10168), Emodin (Pubchem CID:3220), Aloe-emodin (Pubchem CID:10207), Chrysophanol (Pubchem CID:10208) and Physcion (Pubchem CID:10639) were retrieved from pubchem [18]. The minimum energy conformation of ligand molecules was generated using “Generate Conformation” protocol in Discovery Studio using CHARMm (Chemistry at HARvard Macromolecular Mechanics) force field [[19], [20]]. Molecular docking studies were performed using the CDOCKER module implemented in Discovery Studio. A set of 10 random orientations of each ligand molecule were produced. In order to achieve docking poses with high accuracy and to measure the amelioration of a docking study, CDOCKER score was used as a standard [21]. To further validate our in-silico results, molecular docking studies were also performed using Autodock 4.2 program (version 1.5.6), using the same receptor and ligand set. The detailed methodology for molecular docking using Autodock version 4.2 program has been described in supplementary material.
2.6. Absorption spectroscopy studies of anthraquinones-GSH interaction
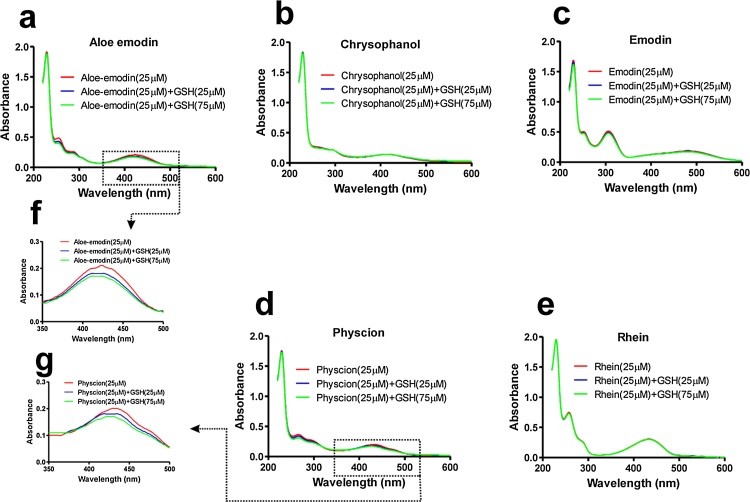
To investigate the interaction of AQs with GSH, UV–vis absorption spectroscopy was employed following the method described earlier [22]. Initially, the UV–vis absorption spectra of all the five AQs (Aloe-emodin, Chrysophanol, Emodin, Physcion, and Rhein) at a maximum concentration of 50 μM were recorded in Shimadzu UV 2550 double beam spectrophotometer (Tokyo, Japan). The AQ-GSH conjugate was studied by mixing the AQ (25 μM) with GSH (25 and 75 μM) in 0.1 M phosphate buffer (pH 7.4) followed by incubation of 15 min at 37 °C. Equal amount of compound was added to the reference and sample cuvettes and the difference spectra of various AQs (350–500 nm for Aloe-emodin, Chrysophanol, Physcion and Rhein; 375–600 nm for Emodin) was recorded.
2.7. Measurement of GSH and GSSG in GSH-anthraquinone incubation mixture
The content of GSH and GSSG was assayed in GSH-AQ incubation mixture using OPT as a fluorescent probe, which binds GSH or GSSG to form highly fluorescent derivative [23]. For GSH assay, the reaction mixture in a final volume of 3.5 mL contained 3.25 mL sodium phosphate-EDTA buffer (0.1 M, pH 8.0), 0.25 mL OPT (1 mg/ml methanol). The reaction was initiated by the addition of AQ (50 μM) in a cuvette containing GSH (100 μM) at room temperature. After 15 min, the formation of fluorescent GSH-OPT adduct was detected and read on the spectrofluorometer at an excitation and emission wavelengths of 350 and 450 nm (slit widths 5 & 10 nm), respectively. For GSSG assay, NEM was used to prevent oxidation of GSH to GSSG during the assay. The above-mentioned reaction mixture (in another set of experiments) along with AQs were incubated with 0.2 mL NEM (0.04 M) for 30 min. Measurement of GSSG was done using the procedure outlined above, except that 0.1 N NaOH was used as diluents instead of phosphate-EDTA buffer. The percentage of GSH and GSSG was calculated by comparing the fluorescence of standard GSH and GSSG.
3. Results
In the present study, two different methods were employed to study the DNA-AQs interactions. The first method involved the ability of AQs to quench the DNA-EtBr complex and second method involved the ability of DNA to quench the emission spectra of the AQs. Among all the AQs, only emodin was suitably studied by the first method whereas, Aloe-emodin and Rhein were effectively studied by the second method. Chrysophanol and Physcion could not studied by any of the above methods.
3.1. Interaction of anthraquinones with DNA: Quenching of DNA-EtBr fluorescence spectra by anthraquinones
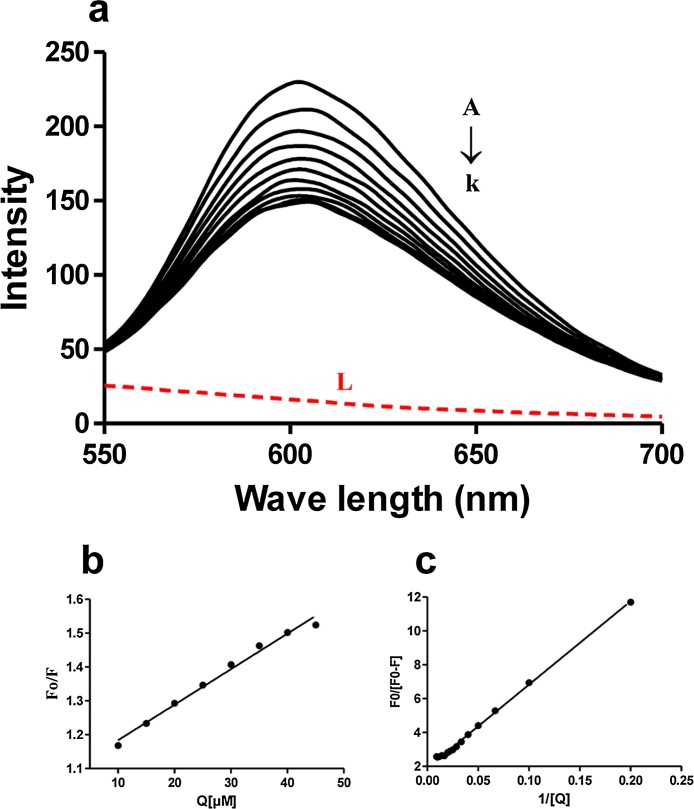
The DNA binding affinities of all the AQs were studied by their ability to quench DNA-EtBr complex. Among all the AQs, Emodin showed no interference with DNA-EtBr emission spectra (Suppl. Fig. 1). Other AQs such as Aloe-emodin, Rhein, Chrysophanol and Physcion could not studied by this method due to spectral overlap (Suppl. Fig. 1). Therefore, the binding of Emodin with DNA was examined by this method which involves the ability of Emodin to quench the DNA-EtBr complex. In brief, Emodin was added into the solution of DNA-EtBr complex and the fluorescence intensity of DNA-EtBr complex decreased with the increasing concentration of Emodin (5–50 μM) (Fig. 1a). The decrease in fluorescence intensity has been used to calculate the dynamic quenching constant (Ksv) from the following Stern-Volmer equation [13].
Where F0 and F are the fluorescence intensities before and after the addition of the quenchers (Emodin), respectively, Ksv is the dynamic/collisional quenching constant (which shows the importance of fluorophore accessibility to the quencher), Kq is the quenching rate constant or bimolecular quenching constant, [Q] is the concentration of compound added; τ0 is the average lifetime of the fluorophore without quencher and its value is considered to be 10−8 s [24]. Stern–Volmer plot for Emodin showed linear relationship in binding with DNA (Fig. 1b). The values of Ksv and Kq for Emodin were found to be 1.05 × 104 (r2 = 0.9873) and 1.05 × 1012 respectively.
Fig. 1.
Fluorescence analysis of Emodin-DNA interactions.
(a) Intrinsic fluorescence emission spectra of DNA: EB (75:6.3 μM) excited at 350 nm in the presence of various concentrations (A–K represent 0, 5, 10, 15, 20, 25, 30, 35, 40, 45 and 50 μM respectively) of Emodin and L (red dotted line) represents emission spectra of 50 μM emodin alone excited at 350 nm, (b) Stern-Volmer plot for quenching of DNA: EB complex by emodin, (c) Modified Stern-Volmer plot for quenching of DNA: EB complex by Emodin.
For a static quenching procedure, the data were analyzed according to the Modified Stern- Volmer equation [14].
ΔF is the difference of fluorescence in the absence and presence of Emodin at concentration [Q], fa is the fraction of accessible fluorescence, and Ka is the effective quenching constant for the accessible fluorophores, which is similar to the binding constant for the quencher-acceptor systems. The dependence of F0/ΔF on the reciprocal value of concentration [Q] was found to be linear with the slope equaling to the value of (faKa)−1 for Emodin. The Modified Stern-Volmer plot [Fo/ΔF vs (faKa)−1] of Emodin is presented in Fig. 1(c). The Ka is calculated to be 3.854 × 104 L mol−1 s−1 (r2 = 0.997).
3.2. Interaction of anthraquinones with DNA: Quenching of anthraquinones fluorescence spectra by ctDNA
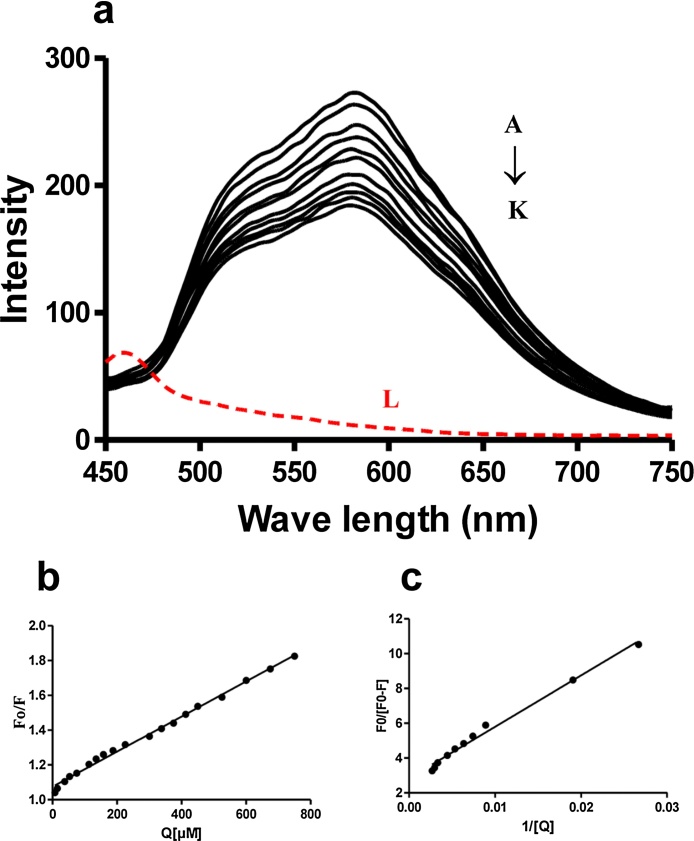
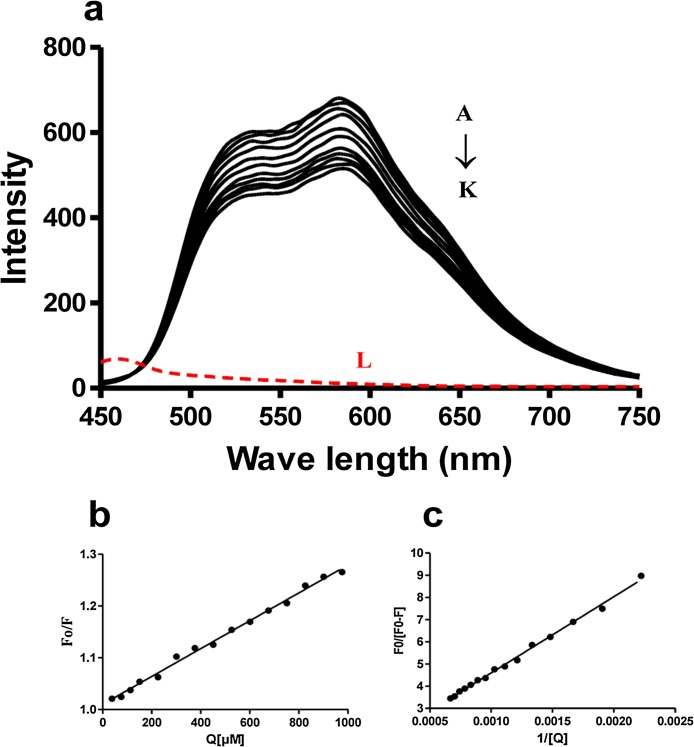
The interaction of AQs with DNA was studied by fluorescence titration experiments, in which ctDNA (0–1.5 mM) was progressively added to quench the fluorescence spectra of AQs. Results indicated that Emodin, Chrysophanol and Physcion did not show any interaction with DNA by this method (Suppl. Fig. 2). However, Aloe-emodin and Rhein showed emission spectra at 582 nm in BR buffer following excitation at 400 nm. The fluorescence intensity of both these AQs was found to be effectively quenched by ctDNA without any change in the position of the peak. The fluorescence emission spectra of Aloe-emodin and Rhein with or without different concentration of DNA are shown in Fig. 2a and 3a, respectively. The Stern Volmer plot and modified Stern Volmer plot for Aloe-emodin are shown in Figs. 2b and 2c respectively, whereas the same for Rhein is presented in Fig. 3b and 3c, respectively. Different quenching constants for Aloe-emodin and Rhein were calculated as mentioned earlier. The values of Ksv, Kq, Ka, r2 (SVE) and r2 (MSVE) for aloe-emodin were calculated to be 0.1008 × 104, 0.1008 × 1012, 0.961 × 104, 0.9946 and 0.9897 respectively; the values of Ksv, Kq, Ka, r2(SVE) and r2(MSVE) for Rhein were found to be 0.02644 × 104, 0.02644 × 1012, 0.034 × 104, 0.9978 and 0.9960 respectively.
Fig. 2.
Fluorescence analysis of Aloe-emodin-DNA interactions.
(a) Intrinsic fluorescence emission spectra of Aloe-emodin (50 μM) at an excitation of 400 nm in the presence of various concentrations (A–K represent 0, 5, 25, 50, 75, 95, 150, 200, 250, 275 and 300 μM respectively) of DNA and L (red dotted line) represents emission spectra of 100 μM DNA alone at 400 nm, (b) Stern-Volmer plot for quenching of Aloe-emodin by DNA, (c) Modified Stern-Volmer plot for quenching of Aloe-emodin by DNA.
Fig. 3.
Fluorescence analysis of Rhein-DNA interactions.
(a) Intrinsic fluorescence emission spectra of Rhein (50 μM) at an excitation of 400 nm in the presence of various concentrations (A-K represent 0, 25, 75, 150, 200, 350, 500, 550, 650, 700 and 800 μM respectively) of DNA and L (red dotted line) represents emission spectra of 100 μM DNA alone excited at 400 nm, (b) Stern-Volmer plot for quenching of Rhein by DNA, (c) Modified Stern-Volmer plot for quenching of Rhein by DNA
3.3. Molecular docking of anthraquinones with DNA
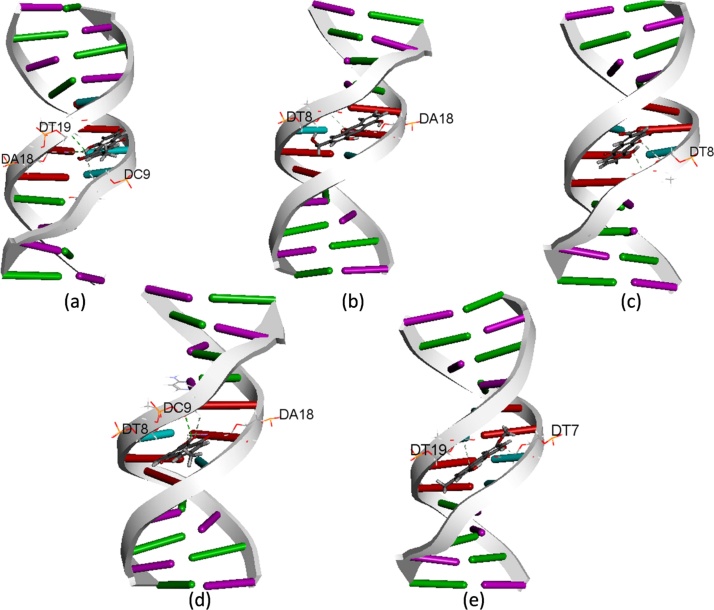
In this study, we examined the probable binding modes of all the five AQs from CO seeds with ctDNA as shown in Fig. 4. Data indicate that all these AQs can enter the minor groove of ctDNA and binds to the A-T rich site. The docking results suggest that AQ possess aromatic rings which allow torsional rotation to fit into the narrower minor groove of ctDNA. The CDOCKER energy and CDOCKER interaction energy for the best binding pose of ligand molecule with the ctDNA was observed in the order of: Emodin > Aloe-emodin > Rhein > Chrysophanol > Physcion (Table 1). Further, we investigated the binding mechanism of ctDNA to all the five AQs. We found that Emodin exhibited the highest binding affinity towards ctDNA with CDOCKER energy of −21.0622 kcal/mol and CDOCKER interaction energy of −25.1604 kcal/mol. The results obtained through Discovery Studio V4.0 were reconfirmed using another docking software, Autodock V4.2 which runs using Amber forcefield whereas CDOCKER runs using CHARMm forcefield [[25], [26]]. The comparative results for both the softwares are presented in Table 1. Interestingly, we found the same hydrogen bonding patterns that occur between backbone atoms of ctDNA and AQs. The docking results showed that the binding pocket of AQs consists mainly of AATT rich sequence i.e. D(A5A6T7T8) and D(A17A18T19T20). The participating nucleotides have been highlighted in Fig. 4. Here, the adenine (A18) and thymine (t8) bases act as both a hydrogen bond donor and hydrogen bond acceptor for ligands. Overall, the in silico studies revealed that the AQs shows preference towards the minor groove of ctDNA and binds in a same orientation in the original structure at same site.
Fig. 4.
In silico analysis of DNA-Anthraquinone interactions.
Molecular docking results of ctDNA–Anthraquinones interaction (a) DNA – Emodin, (b) DNA – Aloe-emodin, (c) DNA – Rhein, (d) DNA – Chrysophanol, (e) DNA – Physcion. Ladder model of ctDNA showed self-complementary duplex sequences and nucleotides in DNA are represented by different color sticks (cytosine (DC) purple, adenine (DA) red, thymine cyan, guanine (DG) green). Anthraquinones entered in the minor groove of DNA and binds with central AATT motifs with intermolecular hydrogen bonds (green color).
Table 1.
Binding energies of ctDNA–Anthraquinone interaction and intermolecular hydrogen bonds predicted by CDOCKER.
| LIGANDS (CID No.) | Discovery Studio V4.0 |
Autodock V4.2 |
|||
|---|---|---|---|---|---|
| CDOCKER Energy (Kcal/mole) | CDOCKER Interaction Energy (Kcal/mole) | Intermolecular Interactions | Binding Energy (Kcal/mole) | Intermolecular Interactions | |
| Emodin (CID 3220) | −21.0622 | −25.1604 | 3220:H29- B: DA18:N3; | −8.67 | DT7, DT8, DG10, DA18, DC9, DA17, DT19, DT20 |
| 3220:H29- B: DT19:O4′; | |||||
| A: DC9:H4′- 3220: O2; | |||||
| B: DA18:H2 − 3220: O4 | |||||
| Aloe-emodin (CID 10207) | −20.6581 | −24.1616 | A: DT8:H1′ − 10207: O3; | −8.59 | DT8, DA17, DA18, DT19, DC9, DG10, DT20 |
| B: DA18:H2 − 10207: O3 | |||||
| Rhein (CID 10168) | −18.6288 | −21.256 | A: DT8:H4′ − 10168: O2; A: DT8:H1′ − 10168: O4 | −8.27 | DG10, DA17, DA18, DT8, DC9, DT19, DT20 |
| Chrysophanol (CID 10208) | −18.1836 | −20.5925 | 10208:H29 − A: DT8:O2; A: DC9:H4′ − 10208: O2; B: DA18:H2 − 10208: O4 | −8.07 | DT8, DA17, DA18, DC9, DG10, DT19, DT20 |
| Physcion (CID 10639) | −17.4773 | −20.6081 | A: DT7:H1′ − 10639: O5; B: DT19:H1′ − 10639: O3 | −7.15 | DG10, DA18, DT7, DT8, DC9, DG16, DA17, DT19 |
3.4. Absorption spectroscopy studies on GSH-anthraquinone interaction
The UV–vis spectroscopy was carried out to investigate the formation of GSH-AQ conjugates at physiological pH (7.4) following the method described earlier [22]. The absorption spectra of all the five AQs with or without different concentrations of GSH are presented in Fig. 5. The absorption spectra of Aloe-emodin, Chrysophanol, Emodin, Physcion and Rhein are depicted in Fig. 5(a–e), respectively. It was evident that in case of Chrysophanol, Emodin and Rhein, there was no shift in the spectra upon addition of GSH up to 75 μM; whereas, in case of Physcion and Aloe-emodin there was a hypsochromic shift (blue shift) in the spectra upon addition of GSH, indicating the possibility of formation of GSH conjugate (Fig. 5f and g).
Fig. 5.
UV–vis absorption spectra of GSH-Anthraquinone interactions.
(a) Absorption spectral change resulted by mixing Aloe-emodin (25 μM) with GSH (25 and 75 μM), (b) Absorption spectral change resulted by mixing Chrysophanol (25 μM) with GSH (25 and 75 μM), (c) Absorption spectral change resulted by mixing Emodin (25 μM) with GSH (25 and 75 μM), (d) Absorption spectral change resulted by mixing Physcion (25 μM) with GSH (25 and 75 μM), (e) Absorption spectral change resulted by mixing Rhein (25 μM) with GSH (25 and 75 μM), (f) Hypsochromic shift resulted by mixing aloe Emodin (25 μM) with GSH (25 and 75 μM) in the range of 400–500 nm, (g) Hypsochromic shift resulted by mixing Physcion (25 μM) with GSH (25 and 75 μM) in the range of 400–500 nm.
3.5. Measurement of oxidation of GSH to GSSG by anthraquinones
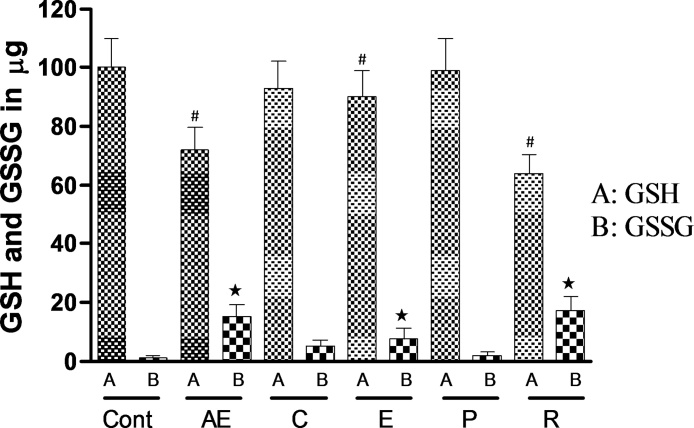
To study the ability of AQs to oxidize GSH to GSSG, fluorometric measurement of GSH and GSSG in AQ-GSH mixtures were carried out. It was observed that all AQs except Chrysophanol and Physcion depleted the GSH concentration significantly. The percentage decrease in GSH contents in the reaction mixture by Aloe-emodin, Emodin and Rhein was 28, 10 and 30%, respectively when compared to the control (tube containing 100 μM GSH in phosphate buffer without test AQs) (Fig. 6). The generation of GSSG following incubation of AQs with GSH is shown in Fig. 6.
Fig. 6.
Depletion of GSH along with generation of GSSG following incubation of anthraquinones (50 μM) with GSH (100 μM).
Cont-Tubes containing 100 μM GSH in 0.1 M phosphate buffer (pH 7.4) without any anthraquinone, AE- Aloe-emodin, C- Chrysophanol, E- Emodin, P- Physcion, R- Rhein. Data represents mean ± SE of 5 replications, #P < 0.05, significant when compared to control GSH. *P < 0.05, significant when compared to control GSSG.
4. Discussion
In addition to the redox cycling, quinones exhibit toxicity by several other mechanisms including protein binding, DNA binding and interaction with GSH. In the present study, we investigated the binding affinity of AQs of CO seed with DNA. Results suggest that the binding affinity of AQs to the DNA is related to their cytotoxic potential. In addition, this study suggests that AQs deplete cellular GSH either by forming conjugates or by oxidizing it to GSSG, which could be a mechanism of toxicity of the AQs.
Among various mechanisms of AQ toxicity, impairment of topoisomerase-IIα activity [[27], [28], [29]], DNA intercalation and nuclear localization are supposed to be the major mode of action of these compounds [[30], [31], [32]]. Earlier biochemical studies have shown that the drugs or toxins inhibit both DNA-directed DNA synthesis and DNA-directed RNA synthesis, presumably by their ability to interact with the DNA [[33], [34], [35], [36]].
The fluorescence quantum yield of DNA is about 10−4 to 10−5 at room temperature [37], hence the intrinsic fluorescence from DNA may not be of relevance in studying DNA interactions. The utility of fluorescent probes may be useful to obtain the biophysical information regarding the interaction of compounds with DNA [12]. The probe for nucleic acids like EtBr is widely used for the DNA interaction studies [38]. Interestingly, in the present study this method was effective to study the interaction of Emodin with DNA. However, due to spectral overlapping of Aloe-emodin and Rhein with DNA-EtBr complex, binding potential of Aloe-emodin and Rhein could not ascertained. Therefore, the binding of Aloe-emodin and Rhein with DNA was investigated by the fluorescence quenching technique using the intrinsic fluorescence of Aloe-emodin and Rhein [15]. Further, DNA binding of Emodin was not studied by this method as the intrinsic fluorescence of Emodin was not quenched by ctDNA. In case of Chrysophanol and Physcion, there was no DNA binding observed in the spectroscopic studies.
The quenching of fluorescence intensity of DNA-EtBr complex by increasing concentration of Emodin indicated that Kq value for Emodin is larger than 2.0 × 1010 Lmol−1 s−1, suggesting a static quenching of DNA-EtBr complex by Emodin. There may be three reasons for the decrease in the fluorescence intensity of DNA-EtBr. Firstly, the binding between EtBr and Emodin might occur, that decreases the intensity of DNA-EtBr complex. Secondly, Emodin competes with EtBr in binding with DNA and excludes the intercalated EtBr from the duplex. Thirdly, Emodin binding with DNA-EtBr forms a new non-fluorescence complex DNA-EtBr-Emodin, which causes the fluorescence quenching of DNA-EtBr. The first possibility is not true as Emodin does not react with EtBr when DNA is absent (data not shown). In addition, the binding of EtBr with DNA is strong enough having the binding constant of 2 × 106 L mol−1 [39]. So, there is hardly any chance that Emodin can compete with EtBr to bind DNA; this disregard the second reason. Hence, the third reason indicating Emodin binding with DNA-EtBr to form a new non-fluorescence complex DNA-EtBr-Emodin appears to be more reasonable [12]. Among various mechanisms of non-covalent DNA binding, this has been suggested that Emodin interact with DNA by groove binding [[12], [40]], which has been shown by the molecular docking analysis in our present study.
The quenching efficiency of DNA on the Aloe-Emodin and Rhein emission spectra indicates that either dynamic quenching or static quenching or both the mechanisms may be involved in this interaction. The Kq value for both the AQs (Aloe-emodin and Rhein) are higher than the limiting rate diffusion constant (2.0 × 1010 Lmol−1 s−1), which indicates the possibility of static quenching [15]. The fluorescence quenching of AQs by DNA is considered to be due to the photoelectron transfer from guanine base of DNA to the excited state of both the AQs [41]. Earlier it was observed that the interaction between the groove binders and DNA causes a strong fluorescence quenching [42]. This study indicates that these AQs may interact with DNA by groove binding mechanism as the data related to emission intensity and binding parameters are in concordance with other groove binders [15].
The high affinity of Emodin, Aloe-emodin and Rhein to DNA may be a reason behind their higher cytotoxic potential as we have reported earlier [9]. In the same way, inability of Chrysophanol and Physcion to interact with DNA may be a reason for their minimal toxicity in vitro [9].
Molecular docking studies provide an insight into the interactions between macromolecule and the ligands. The ctDNA has the self-complementary duplex sequences i.e. D(C1G2C3G4A5A6T7T8C9G10C11G12) and D(C13G14C15G16A17A18T19T20C21G22C23G24). The minor groove of ctDNA contains a central AATT sequence which is the site of non-covalent interactions to a large number of antibiotics, drugs and antiviral agents [17]. These DNA binders exert their action by competing with sequence specific transcription factors and thus inhibiting DNA replication process [43].
Further validation of AQs-DNA interactions was done by in silico approach. In consistent with the biophysical data, molecular docking studies both by CDOCKER and Autodock suggest that among the five AQs, Emodin has stronger binding affinity towards ctDNA. Further, the docking results demonstrated the binding of Emodin to ctDNA with the lowest CDOCKER binding energy (−21.0622 kcal/mol) as compared other AQs (Table 1). All the studied AQs were found to be minor groove binders, and the difference in the binding energies is due to the difference in the groups or atoms present in the anthraquinones. Earlier studies highlighted that the hydrogen bonding particularly between N-3 nitrogen of A and minor groove binders play a significant role in the DNA binding [44]. Interestingly, in case of Emodin binding, a total four hydrogen bonds were formed i.e. N-3 nitrogen of DA18, H-2 hydrogen of DA18, O'-4 oxygen of DT19 and H'-4 hydrogen of DC9 nucleotide interaction was observed in the binding process. We believed that this binding might disrupts the normal Watson Crick pairing and may be responsible for producing specific DNA damages.
The reactions between glutathione and quinones are of significant biological importance as these reactions are involved in the toxicity of several xenobiotics. It is generally presumed that most quinones undergo simple electron transfer to form semi quinone radicals and/or hydroquinone [8]. However, several independent studies have shown that the reactions between reduced GSH and quinones in vitro can result in the formation of quinone-glutathione conjugates [[45], [46]] or oxidized glutathione (GSSG) [[47], [48]]. Hence, in our study, interaction of AQs of CO seeds with GSH in physiological condition was investigated. Interestingly, in the absorption spectroscopy experiments no spectral shift was observed in case of Rhein, Emodin and Chrysophanol; while Aloe-emodin and Physcion showed a decrease in the absorption spectra in addition to blue shift upon addition of GSH (25 and 75 μM). These results indicate the possibility of formation of Aloe-emodin-GSH and Physcion-GSH abducts which needs to be investigated further. Though no GSH conjugate was observed for Rhein, Emodin and Chrysophanol, but these AQs may be responsible for oxidation of GSH to GSSG. This prompted us to estimate the levels of GSH and GSSG following incubation with all the AQs with GSH. Depletion of GSH as well as formation of GSSG at different rate was observed in different AQs-GSH incubation mixtures. Rhein, the most cytotoxic AQs was found to oxidize GSH maximally followed by Aloe emodin and Emodin. In case of Physcion and Chrysophanol, the generation of GSSG was relatively less suggesting low electrophilic centers in these moeties that may be a reason for their less toxicity.
5. Conclusion
Overall, this study highlighted that among five AQs detected in CO seeds, the maximal cytotoxicity of Rhein, Emodin and Aloe-emodin may be due to their higher DNA binding affinity and higher GSH oxidizing potential.
Conflicts of interest
All authors declare they have no Conflicts of Interest.
Acknowledgements
We are grateful to the Director of CSIR-IITR for his keen interest in the study. GKP is thankful to Indian Council of Medical Research, New Delhi for the award of Senior Research Fellowship during the study period. We acknowledge Dr. Asit Mridha at All India Institute of Medical Science for editing the manuscript.
Footnotes
Supplementary data associated with this article can be found, in the online version, at https://doi.org/10.1016/j.toxrep.2017.12.024.
Contributor Information
Gati Krushna Panigrahi, Email: gpanigra@wakehealth.edu, panigrahigk@gmail.com.
Mukul Das, Email: mditrc@rediffmail.com.
Appendix A. Supplementary data
The following are Supplementary data to this article:
References
- 1.Panigrahi G., Tiwari S., Ansari K.M., Chaturvedi R.K., Khanna V.K., Chaudhari B.P., Vashistha V.M., Raisuddin S., Das M. Association between children death and consumption of Cassia occidentalis seeds: clinical and experimental investigations. Food Chem. Toxicol. 2014;67:236–248. doi: 10.1016/j.fct.2014.03.005. [DOI] [PubMed] [Google Scholar]
- 2.Vashishtha V.M., Kumar A., John T.J., Nayak N.C. Cassia occidentalis poisoning as the probable cause of hepatomyoencephalopathy in children in western Uttar Pradesh. Indian J. Med. Res. 2007;125:756–762. [PubMed] [Google Scholar]
- 3.Vashishtha V.M., Nayak N.C., John T.J., Kumar A. Recurrent annual outbreaks of a hepato-myo-encephalopathy syndrome in children in western Uttar Pradesh, India. Indian J. Med. Res. 2007;125:523–533. [PubMed] [Google Scholar]
- 4.Panigrahi G.K., Ch R., Mudiam M.K., Vashishtha V.M., Raisuddin S., Das M. Activity-guided chemo toxic profiling of Cassia occidentalis (CO) seeds: detection of toxic compounds in body fluids of CO-exposed patients and experimental rats. Chem. Res. Toxicol. 2015;28:1120–1132. doi: 10.1021/acs.chemrestox.5b00056. [DOI] [PubMed] [Google Scholar]
- 5.Panigrahi G.K., Yadav A., Yadav A., Ansari K.M., Chaturvedi R.K., Vashistha V.M., Raisuddin S., Das M. Hepatic transcriptional analysis in rats treated with Cassia occidentalis seed: involvement of oxidative stress and impairment in xenobiotic metabolism as a putative mechanism of toxicity. Toxicol. Lett. 2014;229:273–283. doi: 10.1016/j.toxlet.2014.06.037. [DOI] [PubMed] [Google Scholar]
- 6.Bironaite D., Ollinger K. The hepatotoxicity of rhein involves impairment of mitochondrial functions. Chem. Biol. Interact. 1997;103:35–50. doi: 10.1016/s0009-2797(96)03747-7. [DOI] [PubMed] [Google Scholar]
- 7.Sendelbach L.E. A review of the toxicity and carcinogenicity of anthraquinone derivatives. Toxicology. 1989;57:227–240. doi: 10.1016/0300-483x(89)90113-3. [DOI] [PubMed] [Google Scholar]
- 8.O'Brien P.J. Molecular mechanisms of quinone cytotoxicity. Chem. Biol. Interact. 1991;80:1–41. doi: 10.1016/0009-2797(91)90029-7. [DOI] [PubMed] [Google Scholar]
- 9.Panigrahi G.K., Suthar M.K., Verma N., Asthana S., Tripathi A., Gupta S.K., Saxena J.K., Raisuddin S., Das M. Investigation of the interaction of anthraquinones of Cassia occidentalis seeds with bovine serum albumin by molecular docking and spectroscopic analysis: correlation to their in vitro cytotoxic potential. Food Res. Int. 2015;77:368–377. [Google Scholar]
- 10.Panigrahi G.K., Yadav A., Srivastava A., Tripathi A., Raisuddin S., Das M. Mechanism of rhein-induced apoptosis in rat primary hepatocytes: beneficial effect of cyclosporine A. Chem. Res. Toxicol. 2015;28:1133–1143. doi: 10.1021/acs.chemrestox.5b00063. [DOI] [PubMed] [Google Scholar]
- 11.Gholivand M.B., Kashanian S., Peyman H., Roshanfekr H. DNA-binding study of anthraquinone derivatives using Chemometrics methods. Eur. J. Med. Chem. 2011;46:2630–2638. doi: 10.1016/j.ejmech.2011.03.034. [DOI] [PubMed] [Google Scholar]
- 12.Bi S.Y., Zhang H.Q., Qiao C.Y., Sun Y., Liu C.M. Studies of interaction of emodin and DNA in the presence of ethidium bromide by spectroscopic method. Spectrochim. Acta A. 2008;69:123–129. doi: 10.1016/j.saa.2007.03.017. [DOI] [PubMed] [Google Scholar]
- 13.Lakowicz J.R., Weber G. Quenching of fluorescence by oxygen: a probe for structural fluctuations in macromolecules. Biochemistry. 1973;12:4161–4170. doi: 10.1021/bi00745a020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sharma M., Chauhan K., Shivahare R., Vishwakarma P., Suthar M.K., Sharma A., Gupta S., Saxena J.K., Lal J., Chandra P., Kumar B., Chauhan P.M. Discovery of a new class of natural product-inspired quinazolinone hybrid as potent antileishmanial agents. J. Med. Chem. 2013;56:4374–4392. doi: 10.1021/jm400053v. [DOI] [PubMed] [Google Scholar]
- 15.Gholivand M.B., Kashanian S., Peyman H. DNA-binding, DNA cleavage and cytotoxicity studies of two anthraquinone derivatives. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2012;87:232–240. doi: 10.1016/j.saa.2011.11.045. [DOI] [PubMed] [Google Scholar]
- 16.Berman H.M., Westbrook J., Feng Z., Gilliland G., Bhat T.N., Weissig H., Shindyalov I.N., Bourne P.E. The protein data bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Neidle S., Mann J., Rayner E.L., Baron A., Opoku-Boahen Y., Simpson I.J., Smith N.J., Fox K.R., Hartley J.A., Kelland L.R. Symmetric bis-benzimidazoles: new sequence-selective DNA-binding molecules. Chem. Commun. 1999:929–930. [Google Scholar]
- 18.Li Q.L., Chen T.J., Wang Y.L., Bryant S.H. PubChem as a public resource for drug discovery. Drug Discov. Today. 2010;15:1052–1057. doi: 10.1016/j.drudis.2010.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Momany F.A., Rone R. Validation of the general-purpose Quanta(R)3.2/Charmm(R) force-field. J. Comput. Chem. 1992;13:888–900. [Google Scholar]
- 20.Singh N., Freiesleben S., Gupta S.K., Shukla Y., Wolkenhauer O. Identification of antineoplastic targets with systems approaches, using resveratrol as an in-depth case study. Curr. Pharm. Des. 2017 doi: 10.2174/1381612823666170710152918. [DOI] [PubMed] [Google Scholar]
- 21.Miller B.T., Singh R.P., Klauda J.B., Hodoscek M., Brooks B.R., Woodcock H.L., 3rd. CHARMMing: a new, flexible web portal for CHARMM. J. Chem. Inf. Model. 2008;48:1920–1929. doi: 10.1021/ci800133b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Butler J., Hoey B.M. Reactions of glutathione and glutathione radicals with benzoquinones. Free Radic. Bio. Med. 1992;12:337–345. doi: 10.1016/0891-5849(92)90082-r. [DOI] [PubMed] [Google Scholar]
- 23.Hissin P.J., Hilf R. A fluorometric method for determination of oxidized and reduced glutathione in tissues. Anal. Biochem. 1976;74:214–226. doi: 10.1016/0003-2697(76)90326-2. [DOI] [PubMed] [Google Scholar]
- 24.Eftink M.R., Ghiron C.A. Fluorescence quenching studies with proteins. Anal. Biochem. 1981;114:199–227. doi: 10.1016/0003-2697(81)90474-7. [DOI] [PubMed] [Google Scholar]
- 25.Morris G.M., Goodsell D.S., Halliday R.S., Huey R., Hart W.E., Belew R.K., Olson A.J. Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J. Comput. Chem. 1998;19:1639–1662. [Google Scholar]
- 26.Wu G., Robertson D.H., Brooks C.L., Jr., Vieth M. Detailed analysis of grid-based molecular docking: a case study of CDOCKER-A CHARMm-based MD docking algorithm. J. Comput. Chem. 2003;24:1549–1562. doi: 10.1002/jcc.10306. [DOI] [PubMed] [Google Scholar]
- 27.Coldwell K.E., Cutts S.M., Ognibene T.J., Henderson P.T., Phillips D.R. Detection of Adriamycin-DNA adducts by accelerator mass spectrometry at clinically relevant Adriamycin concentrations. Nucleic Acids Res. 2008;36:e100. doi: 10.1093/nar/gkn439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gewirtz D.A. A critical evaluation of the mechanisms of action proposed for the antitumor effects of the anthracycline antibiotics adriamycin and daunorubicin. Biochem. Pharmacol. 1999;57:727–741. doi: 10.1016/s0006-2952(98)00307-4. [DOI] [PubMed] [Google Scholar]
- 29.Tewey K.M., Rowe T.C., Yang L., Halligan B.D., Liu L.F. Adriamycin-induced DNA damage mediated by mammalian DNA topoisomerase II. Science. 1984;226:466–468. doi: 10.1126/science.6093249. [DOI] [PubMed] [Google Scholar]
- 30.Cummings J., McArdle C.S. Studies on the in vivo disposition of adriamycin in human tumours which exhibit different responses to the drug. Br. J. Cancer. 1986;53:835–838. doi: 10.1038/bjc.1986.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gigli M., Doglia S.M., Millot J.M., Valentini L., Manfait M. Quantitative study of doxorubicin in living cell nuclei by microspectrofluorometry. Biochim. Biophys. Acta. 1988;950:13–20. doi: 10.1016/0167-4781(88)90068-1. [DOI] [PubMed] [Google Scholar]
- 32.Terasaki T., Iga T., Sugiyama Y., Sawada Y., Hanano M. Nuclear binding as a determinant of tissue distribution of adriamycin, daunomycin, adriamycinol, daunorubicinol and actinomycin D. J. Pharmacobiodyn. 1984;7:269–277. doi: 10.1248/bpb1978.7.269. [DOI] [PubMed] [Google Scholar]
- 33.Calendi E., Dimarco A., Reggiani M., Scarpinato B., Valentini L. On physico-chemical interactions between daunomycin and nucleic acids. Biochim. Biophys. Acta. 1965;103:25–49. doi: 10.1016/0005-2787(65)90539-3. [DOI] [PubMed] [Google Scholar]
- 34.Fialkoff H., Goodman M.F., Seraydarian M.W. Differential effect of adriamycin on DNA replicative and repair synthesis in cultured neonatal rat cardiac cells. Cancer Res. 1979;39:1321–1327. [PubMed] [Google Scholar]
- 35.Pigram W.J., Fuller W., Hamilton L.D. Stereochemistry of intercalation: interaction of daunomycin with DNA. Nat. New Biol. 1972;235:17–19. doi: 10.1038/newbio235017a0. [DOI] [PubMed] [Google Scholar]
- 36.Ward D.C., Reich E., Goldberg I.H. Base specificity in the interaction of polynucleotides with antibiotic drugs. Science. 1965;149:1259–1263. doi: 10.1126/science.149.3689.1259. [DOI] [PubMed] [Google Scholar]
- 37.Vigny P., Favre A. Fluorescence and photochemistry of oligocytidylic and polycytidylic acids in aqueous solution. Photochem. Photobiol. 1974;20:345–349. doi: 10.1111/j.1751-1097.1974.tb06586.x. [DOI] [PubMed] [Google Scholar]
- 38.Reinhardt C.G., Krugh T.R. A comparative study of ethidium bromide complexes with dinucleotides and DNA: direct evidence for intercalation and nucleic acid sequence preferences. Biochemistry. 1978;17:4845–4854. doi: 10.1021/bi00616a001. [DOI] [PubMed] [Google Scholar]
- 39.Sarker M., Chen F.M. Binding of mithramycin to DNA in the presence of second drugs. Biochemistry. 1989;28:6651–6657. doi: 10.1021/bi00442a018. [DOI] [PubMed] [Google Scholar]
- 40.Kumar C.V., Asuncion E.H. DNA-binding studies and site-selective fluorescence sensitization of an anthryl probe. J. Am. Chem. Soc. 1993;115:8547–8553. [Google Scholar]
- 41.Vaidyanathan V.G., Nair B.U. Photooxidation of DNA by a cobalt(II) tridentate complex. J. Inorg. Biochem. 2003;94:121–126. doi: 10.1016/s0162-0134(02)00620-7. [DOI] [PubMed] [Google Scholar]
- 42.Ye B.F., Ju H.X. Determination of yeast DNA based on its quenching the fluorescence emission of norfloxacin. Anal. Lett. 2003;36:1351–1364. [Google Scholar]
- 43.Zimmer C., Wahnert U. Nonintercalating DNA-binding ligands: specificity of the interaction and their use as tools in biophysical, biochemical and biological investigations of the genetic material. Prog. Biophys. Mol. Biol. 1986;47:31–112. doi: 10.1016/0079-6107(86)90005-2. [DOI] [PubMed] [Google Scholar]
- 44.Nguyen B., Hamelberg D., Bailly C., Colson P., Stanek J., Brun R., Neidle S., Wilson W.D. Characterization of a novel DNA minor-groove complex. Biophys. J . 2004;86:1028–1041. doi: 10.1016/s0006-3495(04)74178-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Koob M., Dekant W. Bioactivation of xenobiotics by formation of toxic glutathione conjugates. Chem. Biol. Interact. 1991;77:107–136. doi: 10.1016/0009-2797(91)90068-i. [DOI] [PubMed] [Google Scholar]
- 46.Rossi L., Moore G.A., Orrenius S., O'Brien P.J. Quinone toxicity in hepatocytes without oxidative stress. Arch. Biochem. Biophys. 1986;251:25–35. doi: 10.1016/0003-9861(86)90047-0. [DOI] [PubMed] [Google Scholar]
- 47.Gant T.W., Rao D.N., Mason R.P., Cohen G.M. Redox cycling and sulphydryl arylation; their relative importance in the mechanism of quinone cytotoxicity to isolated hepatocytes. Chem. Biol. Interact. 1988;65:157–173. doi: 10.1016/0009-2797(88)90052-x. [DOI] [PubMed] [Google Scholar]
- 48.Ross D., Thor H., Orrenius S., Moldeus P. Interaction of menadione (2-methyl-1,4-naphthoquinone) with glutathione. Chem. Biol. Interact. 1985;55:177–184. doi: 10.1016/s0009-2797(85)80126-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.