Abstract
The present study was conducted to determine the molecular diversity of Staphylococcus aureus strains isolated from human, bovine and food samples based on the polymorphism of the spa gene. A total of 208 S. aureus isolated from human, bovine raw milk and food samples were assessed using polymerase chain reaction–restriction fragment length polymorphism (PCR–RFLP) and single locus sequence typing (SLST) methods, followed by determination of spa types using Ridom SpaServer. Altogether, 15 distinct RFLP patterns were recorded (I–XV). The highest heterogeneity was observed among S. aureus isolated from humans, whereas most of bovine and food S. aureus isolates indicated certain RFLP patterns. Although most of the isolates from patients showed RFLP pattern I, none of the S. aureus isolated from carriers had this spa pattern. Besides, the results of SLST led to the characterization of 16 spa types, and one of them was a novel spa type which has been registered in Ridom SpaServer for the first time and designated as type t16929. Determination of a high number of shared RFLP patterns between human and food S. aureus isolates indicated possible transmission of S. aureus and the source of food contamination. Thus, effective hygiene measures should be taken to break transmission routes. However, it seems that S. aureus isolated from patients, carriers and bovine should be considered in a different way, since some isolates had similar patterns, while the others showed their own specific pattern.
Keywords: Staphylococcus aureus, Typing, PCR–RFLP, spa gene
Introduction
Although Staphylococcus aureus is present on the skin and mucous membrane of warm-blooded animals, it may cause multiple diseases in these hosts (Walther et al. 2008; Persoons et al. 2009). In humans, S. aureus is also an important food-borne pathogen and it is a major causative agent of hospital- and community-associated infections (Wertheim et al. 2004; Lee 2003). Animal infections may result in huge economic losses, in particular, in the case of industrial animal farms (Abebe et al. 2016; Tesfaye et al. 2010).
Hence, typing of S. aureus isolates can be helpful in prevention and control of S. aureus infections, as well as investigation of possible sources of infections. Several phenotypic and genotypic methods are available for this purpose. In the last few years, a variety of molecular methods including pulsed field gel electrophoresis (PFGE) and multilocus sequence typing (MLST) have been used for discrimination of S. aureus isolates (Laplana et al. 2007; Larsen et al. 2012). However, these methods are time consuming, not widely available and expensive (Stranden et al. 2003). PCR-based typing methods such as polymerase chain reaction–restriction fragment length polymorphism (PCR–RFLP) possess a high degree of reproducibility, good discriminatory power and comparable results (Acosta et al. 2014; Omar et al. 2014; Mehndiratta et al. 2009). One of these approaches is RFLP analysis of the spa gene which contains polymorphic repeat regions (21 bp) and can be targeted to type S. aureus strains (Mehndiratta et al. 2009).
In fact, among various molecular typing methods, spa typing (single locus sequence typing: SLST) has been shown to be the most effective and rapid method to differentiate S. aureus isolates. This method is based on the sequence variation and the number of tandem repeats in the region X of the spa gene (Frenay et al. 1996; Sabat et al. 2013). This region is highly polymorphic and consists of up to 12 tandem repeats with a length of 21–27 nucleotides. The result of spa typing is usually in good concordance with the result of PFGE and it has been shown that its discriminatory power is similar to PFGE. However, it is also suggested that simultaneous utilization of both methods may have better results (Hallin et al. 2009).
The aim of the present study was to investigate the molecular diversity and compare the sources of contamination and distribution of S. aureus isolated from different sources based on the PCR–RFLP and SLST of the spa gene.
Materials and methods
Bacterial isolates
A total of 208 S. aureus isolates which had been previously isolated from bovine raw milk samples (83 isolates), humans (40 isolates from patients, 20 isolates from healthy carriers) and food samples (45 isolates from pastries, 20 isolates from cheese) were included in the present study.
DNA extraction
DNA extraction was carried out from 10 ml of overnight cultures of the isolates in tryptic soy broth (TSB, Merck Germany) using phenol–chloroform method as previously described (Wilson 1987). All of these isolates had also been genetically confirmed to be S. aureus using a species-specific PCR which targeted the femA gene (data are not shown).
PCR amplification of the spa gene
Amplification of the spa gene was done using previously introduced primers (Wichelhaus et al. 2001). The sequences of the primers were as follows: SPA1 (5′-ATCTGGTGGCGTAACACCTG-3′) and SPA2 (5′-CGCTGCACCTAACGCTAATG-3′). PCR reactions (50 μl) contained 30 ng of the extracted DNAs, 5 µl of 10 × PCR buffer (500 mM, KCl and Tris HCl, pH 8.4), 5 µl of 25 mM MgCl2, 200 mM of dNTPs, 1 U of Taq DNA polymerase (all materials were prepared from SinaClon, Iran), and 50 pmol of each primer (BioNeer, Korea). PCR amplifications were performed under the following conditions: initial denaturation at 94 °C for 5 min, followed by denaturation at 94 °C for 45 s, annealing at 55 °C for 90 s and extension at 72 °C for 90 s (32 cycles) and a final extension at 72 °C for 10 min. The PCR products were analyzed using electrophoresis on 1% agarose gel (SinaClon, Iran) containing ethidium bromide (0.5 μg/ml). S. aureus ATCC 25923 and S. epidermidis ATCC 13499 were used as positive and negative controls of the spa gene, respectively. Prior to sequencing of the amplified DNA fragments and RFLP analysis, the PCR products were purified using a commercial gel purification kit (Vivantis, Malaysia) according to the manufacturer’s instructions.
Restriction fragment length polymorphism (RFLP)
The PCR products of the spa gene were digested using two restriction enzymes, HindIII (Jena Bioscience, Germany) and HinfI (Jena Bioscience, Germany). HindIII and HinfI enzymes are type II site-specific deoxyribonuclease restriction enzymes which identify A′AGCTT and G′ANTC sequences, respectively, and cleave them from the specified position (Tang et al. 2000). Briefly, 10.5 µl of distilled water, 2.5 µl of 10 × enzyme buffer, and 7 µl of each of the PCR products were mixed with 2 U of each restriction enzyme and the mixtures were incubated at 37 °C for 1 h. Finally, the digested PCR products were analyzed using electrophoresis on 2% agarose gel containing ethidium bromide (Dallal et al. 2010).
Single locus sequence typing of the spa gene (spa SLST)
To investigate the diversity among the isolates, spa amplicons of each size (representative fragments of the determined RFLP patterns plus one indigestible DNA fragment) were submitted for sequencing (BioNeer, Korea). In the following, the resulting amplicon sequences were analyzed by DNASTAR software (version 14.1) to confirm the results of PCR–RFLP and to determine the spa types (Harmsen et al. 2003).
Statistical analysis
The relationships among S. aureus strains isolated from human, bovine and food samples were compared based on the spa-RFLP patterns using Chi square test and SPSS software (version 16), and P < 0.05 was considered to be statistically significant. Also, the numerical index for the discriminatory power of the spa-RFLP method was calculated using Simpson’s index of diversity equation as follows:
where N is the total number of isolates, S the total number of types, and nj the number of strains belonging to the jth type (Hunter and Gaston 1998).
Results
PCR of the spa gene
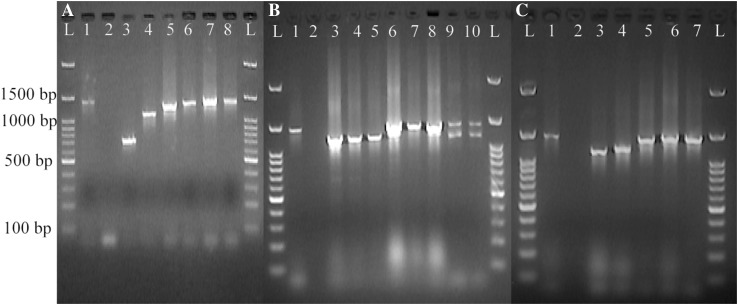
In human isolates, PCR amplification of the spa gene was successfully performed for all of the isolates and resulted in the production of 7 DNA fragments with the sizes ranging from ~ 725 to 1450 bp. The most frequent PCR product among human S. aureus isolates (31.7%) was a DNA band of about 1170 bp (Fig. 1a).
Fig. 1.
The PCR products of the spa gene in human (a), bovine (b), and food (c) S. aureus isolates. Lane L: a 100 bp DNA ladder; lane 1: a reference S. aureus strain (S. aureus ATCC 25923), lane 2: negative control (S. epidermidis ATCC 13499). The other lanes represent various S. aureus isolates
Amplification of the spa gene in bovine S. aureus isolates resulted in the formation of three different DNA bands with the sizes of ~ 1200 to 1350 bp. However, two isolates were not positive in this PCR. A PCR product of 1350 bp was the most frequently observed DNA band in bovine S. aureus isolates (47%). Moreover, six isolates showed double bands, whereas the rest indicated a single DNA band (Fig. 1b).
As depicted in Fig. 1c, amplification of the spa gene among food S. aureus isolates showed that 98.5% (64 isolates) of them were positive for the spa gene and the PCR assay yielded DNA bands with molecular sizes of ~ 1250, 1300, and 1450 bp. An amplicon of ~ 1450 bp was the mostly observed DNA fragment for these isolates (59.5%).
RFLP
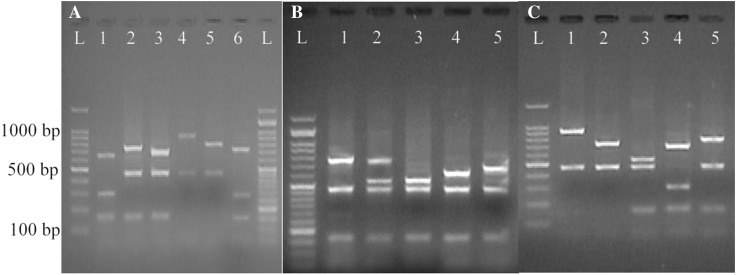
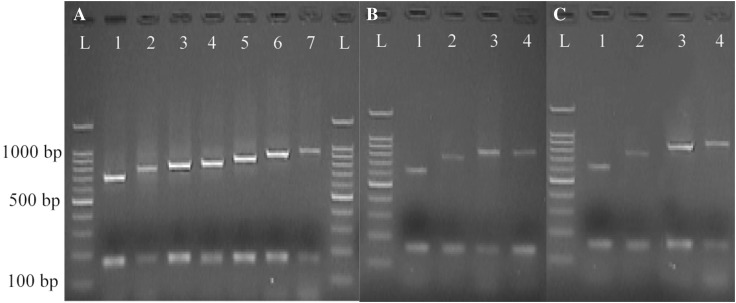
As presented in Table 1, digestion of the purified PCR products with HinfI restriction enzyme yielded 8 distinguishable electrophoretic patterns, whereas 11 patterns were characterized when the same products were digested with HindIII (Figs. 2, 3). However, both enzymes did not digest a DNA fragment of 725 bp. It should be noted that the presence of two DNA bands after digestion of the PCR products with HinfI resulted from the creation of DNA fragments of the same sizes.
Table 1.
RFLP patterns of the spa gene among S. aureus isolates
| Pattern | PCR product (bp) | Digested fragments (HindIII)a | Digested fragments (HinfI)a | Source | Number (%) | ||||
|---|---|---|---|---|---|---|---|---|---|
| C | P | B | Ch | Pa | |||||
| No band | – | – | – | 0 | 0 | 2 | 1 | 0 | 3 (1.4) |
| No digest | 725 | – | – | 0 | 3 | 0 | 0 | 0 | 3 (1.4) |
| I | 1170 | 180-300-680 | 180-180-800 | 0 | 14 | 0 | 2 | 0 | 16 (7.7) |
| II | 1170 | 180-300-680 | 180-180-775 | 2 | 1 | 0 | 0 | 0 | 3 (1.4) |
| III | 1170 | 180-300-680 | 180-180-750 | 2 | 0 | 0 | 0 | 0 | 2 (0.96) |
| IV | 1170 | 450-720 | 170-170-825 | 0 | 0 | 0 | 1 | 0 | 1 (0.48) |
| V | 1200 | 180-470-550 | 180-180-180-650 | 0 | 0 | 35 | 0 | 1 | 36 (17.3) |
| VI | 1270 | 180-300-750 | 180-180-750 | 0 | 1 | 0 | 0 | 0 | 1 (0.48) |
| VII | 1270 | 180-470-630 | 170-180-180-775 | 0 | 0 | 1 | 0 | 0 | 1 (0.48) |
| VIII | 1300 | 180-470-680 | 170-180-180-700 | 0 | 0 | 1 | 0 | 0 | 1 (0.48) |
| IX | 1300 | 470-830 | 170-180-180-700 | 0 | 2 | 0 | 0 | 0 | 2 (0.96) |
| X | 1350 | 180-450-720 | 170-180-180-750 | 7 | 4 | 0 | 0 | 0 | 11 (5.3) |
| XI | 1350 | 180-450-720 | 170-180-180-700 | 0 | 6 | 38 | 0 | 0 | 44 (21.2) |
| XII | 1400 | 450-950 | 180-180-180-850 | 2 | 6 | 0 | 1 | 37 | 46 (22.1) |
| XIII | 1450 | 180-450-820 | 170-170-170-950 | 5 | 2 | 0 | 15 | 7 | 29 (13.9) |
| XIV | 1450 | 180-450-820 | 170-170-170-170-775 | 2 | 1 | 0 | 0 | 0 | 3 (1.4) |
| XV | 1200, 1350 | 180-470-550-720 | 170-180-180-750 | 0 | 0 | 6 | 0 | 0 | 6 (9.2) |
| Total | 20 | 40 | 83 | 20 | 45 | 208 (100) | |||
C carrier, P patient, b bovine, Ch cheese, Pa pastry
aFragments < 50 bp are not shown
Fig. 2.
spa-RFLP patterns obtained from HindIII digestion of the PCR products. a Human, b bovine and c food S. aureus isolates. Lane L: a 100 bp DNA ladder. The other lanes: RFLP patterns of S. aureus isolates
Fig. 3.
spa-RFLP patterns obtained from HinfI digestion of the PCR products. a Human, b bovine and c food S. aureus isolates. Lane L: a 100 bp DNA ladder. The other lanes: RFLP patterns of S. aureus isolates
Altogether, the isolates were classified into 15 spa-RFLP patterns designated as I–XV. The highest heterogeneity was observed among S. aureus isolated from humans (especially patients), as they showed ten RFLP patterns; whereas bovine and food S. aureus isolates indicated much lower heterogeneity. The results of statistical analysis showed that distribution of S. aureus strains with certain spa-RFLP patterns was different in S. aureus isolated from each source. While spa-RFLP patterns V and XI were the most frequently observed patterns for bovine S. aureus isolates, the majority of pastry and cheese S. aureus isolates showed patterns XII and XIII and this distribution was statistically significant (P < 0.05). However, such concurrent distribution was not detected for human S. aureus isolates (P > 0.05).
The most prevalent patterns in human, bovine, pastry and cheese S. aureus isolates were patterns I, XI, XII and XIII, respectively. Moreover, RFLP patterns I, XII and XIII were common between human and food isolates, while pattern XI was shared between bovine and human isolates.
spa SLST
The sequences of the spa gene were analyzed using DNASTAR and the created restriction maps of HindIII and HinfI enzymes confirmed the results obtained from spa-PCR–RFLP. As summarized in Table 2, sequencing of 16 amplified DNA fragments of the spa gene led to the identification of 16 different spa types. Interestingly, one of these spa sequences was characterized in the world for the first time. This sequence was submitted to Ridom SpaServer (http://www.ridom.de/spaserver), a worldwide database for S. aureus spa types, and designated as a new spa type (t16929). The number of repeats within the spa gene varied between 4 and 11 among these sequences (http://www.ridom.de/spaserver).
Table 2.
Representative spa type for each spa-RFLP pattern
| spa type | spa-RFLP pattern | Repeat succession | Source |
|---|---|---|---|
| t701 | No pattern | 11-10-21-17-34-24-34-22-25-25 | Human |
| t037 | I | 15-12-16-02-25-17-24 | Cheese |
| t030 | II | 15-12-16-02-24-24 | Human |
| t1149 | III | 08-16-34-24-34-17-17 | Human |
| t1155 | IV | 15-12-16-02-25-17-16 | Cheese |
| t162 | V | 14-44-12-17-17-23-18-17 | Bovine |
| t5584 | VI | 14-44-13-12-17-17-23-17 | Human |
| t1534 | VII | 04-31-17-25-17-17 | Bovine |
| t16929a | VIII | 26-13-33-34 | Bovine |
| t4348 | IX | 26-16-25-16-28 | Human |
| t189 | X | 07-23-12-21-17-34 | Human |
| t359 | XI | 07-23-12-21-17-34-34-33-34 | Bovine |
| t2062 | XII | 26-23-13-23-31-29-17-25-16-28 | Human |
| t521 | XIII | 07-23-12-21-17-34-34-34-34-33-34 | Cheese |
| t14870 | XIV | 11-10-13-25 | Human |
| t13981 | XV | 07-12-21-17-34-13-34-13-33-34 | Bovine |
aNew type
Statistical analysis
The discrimination index (DI) for the spa-RFLP method using HindIII enzyme (0.8316) was higher than that of HinfI enzyme (0.8027). Generally, the DI of spa-RFLP was found to be 0.8506 in this analysis.
Discussion
As the spa gene is highly polymorphic at the 3′ end region, PCR amplification of this gene may yield amplicons of different sizes and this can help in the differentiation of S. aureus isolates to determine possible sources of contamination and control infections caused by this bacterium (Strommenger et al. 2008).
In this study, the target sequence of the spa gene was successfully amplified from most of the S. aureus isolates (98.5%). However, some researchers have reported lower prevalence of S. aureus without the spa gene (Adesida et al. 2006; Shakeri et al. 2010). We also found a number of isolates (2.9%) with dual-band spa. This may have resulted from hybridization of the primers with more than one specific site, indicating that the sequence of the spa gene for such isolates may contain repeated annealing sites for the used primers. This phenomenon was also documented by Shakeri and Ghaemi (2014) who reported 10.6% frequency for isolates with double-band spa in PCR (Shakeri and Ghaemi 2014).
Thereafter, RFLP analysis of the spa gene was used to investigate the distribution and clonal relatedness of the examined S. aureus isolates. Although HindIII enzyme has been used in previous studies (Rohinishree and Negi 2011; Sato’o et al. 2013), here we also used HinfI enzyme to type S. aureus for the first time.
Our findings indicated that the discriminatory power of HindIII enzyme was generally higher than that of HinfI enzyme for all isolates and the RFLP patterns obtained from digestion with this enzyme were quite distinctive between human and bovine S. aureus isolates. However, HinfI enzyme created more RFLP patterns among the human isolates, suggesting that combination of both enzymes is beneficial in typing of human S. aureus isolates.
According to the results of digestion, 15 different patterns were created by the restriction enzymes. As presented in Table 1, the majority of bovine and food S. aureus isolates showed certain spa-RFLP patterns (V and XI for bovine and XII and XIII for food isolates), indicating that isolates of each type may originate from the same source of contamination, or some strains may be dominant because of their compatibility with the environment (Oliver et al. 2005). Nevertheless, a high diversity was observed among human isolates. In addition, there was little similarity between the RFLP patterns of human and bovine S. aureus strains, and only 6 out of 60 human isolates shared the same RFLP pattern with bovine isolates (RFLP pattern XI). As previously reported; a certain host specificity is usually seen between bovine and human isolates (Zadoks et al. 2000; Alni et al. 2017).
On the other hand, some strains of different origins showed similar RFLP pattern including I, V, XI, XII and XIII. This may be due to the circulation of these strains among various sources. These findings are in agreement with the results obtained by Lee (2003) who showed that S. aureus can be transmitted from food of animal origin to humans.
The results of RFLP also revealed that most of the food S. aureus isolates (62 out of 65 strains) shared similar RFLP patterns with human isolates (patterns I, XII and XIV). This possibly came from the fact that S. aureus may be present as normal flora on the skin and this can lead to contamination of food by workers handling them.
Meanwhile, 18 (90%) out of 20 strains isolated from carriers had similar RFLP patterns with isolates from patients, and only two isolates (10%) indicated a different spa-RFLP pattern (III). This high similarity reflected the fact that carriers of S. aureus are at increased risk to infect individuals. In this case, some researchers reported that nosocomial S. aureus bacteremia in carriers is indicated as an endogenous source and S. aureus nasal carriers are more susceptible to nosocomial infections (Wertheim et al. 2004). In another study, Toshkova et al. (2001) also indicated the importance of nasal carriage of S. aureus in causing skin disease in humans. Therefore, elimination of S. aureus from nasal carriers is a great help in controlling of such staphylococcal diseases. Conversely, although most of the isolates from patients showed RFLP pattern I, none of the S. aureus isolated from carriers had this spa pattern suggesting that more studies are needed to clarify the potentials of various S. aureus strains as causative agents of different diseases.
In the present study, to global comparison of S. aureus spa types, the corresponding DNA fragment of each spa-RFLP pattern was sequenced and the spa types were determined using the Ridom SpaServer. Among the examined isolates, some identified spa types such as t13981, t037, t030, t1149, t14870 and t701 have previously been reported from Iran (http://www.ridom.de/spaserver). However, here, we reported a panel of new spa types including t1155, t162, t1534, t189, t4348, t5584, t359, t521 and t2062 in the country. Besides, a novel S. aureus spa type, type t16929, was identified and has been registered in Ridom SpaServer in the present study for the first time.
According to Ridom SpaServer, spa type t037 has the highest frequency (23.1%) in the world and has been registered in 40 countries so far (http://www.ridom.de/spaserver); whereas spa type t16929 is the lowest observed (0%). The results of our study were also in agreement with these data, although not all of the spa-PCR products were sequenced. spa type t037 (spa-RFLP pattern I) was also a dominant RFLP type among the human isolates, and spa type t16929 which belonged to an S. aureus strain isolated from a bovine sample (spa-RFLP pattern VIII) had the lowest frequency.
Conclusion
In the present study, spa-PCR–RFLP with HindIII and HinfI restriction enzymes was successfully used to assess S. aureus isolated from human, bovine and food samples. The results of the RFLP analysis confirmed the possibility of transmission of pathogenic S. aureus strains between humans and foodstuffs. However, S. aureus isolates were predominantly associated with a host species in case of human and bovine samples. Given that spa-PCR–RFLP method is an available and cost-effective technique with high discriminatory power and repeatable results, the method can be considered as an appropriate procedure to genotypically type S. aureus isolates and to determine their clonal relationships.
Acknowledgements
This project was a part of Hakimi’s PhD thesis and was financially supported by research grants from Bu-Ali Sina University of Hamedan.
Compliance with ethical standards
Conflict of interest
The authors declare no conflicts of interest.
References
- Abebe R, Hatiya H, Abera M, Megersa B, Asmare K. Bovine mastitis: prevalence, risk factors and isolation of Staphylococcus aureus in dairy herds at Hawassa milk shed, South Ethiopia. BMC Vet Res. 2016;12:270–281. doi: 10.1186/s12917-016-0905-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acosta A, Guelfer E, Ruiz A, Mota RA, Uffo O, Gomes-Filho MA, Barbosa SB. Molecular subtyping of Staphylococcus aureus isolated from bovine mastitis in Pernambuco State, Brazil. Comp Clin Pathol. 2014;23:1037–1041. doi: 10.1007/s00580-013-1739-z. [DOI] [Google Scholar]
- Adesida SAY, Likhoshvay W, Eisner AO, Coker OA, Abioye F, Ogunsola T. Repeats in the 3’region of the protein A gene is unique in a strain of Staphylococcus aureus recovered from wound infections in Lagos, Nigeria. Afr J Biotechnol. 2006;16:1858–1863. [Google Scholar]
- Alni RH, Mohammadzadeh A, Mahmoodi P, Alikhani MY. RAPD-PCR analysis of Staphylococcus aureus strains isolated from different sources. Comp Clin Pathol. 2017;4:1–8. [Google Scholar]
- Dallal MM, Salehipour Z, Eshraghi S, Mehrabadi JF, Bakhtiari R. Occurrence and molecular characterization of Staphylococcus aureus strains isolated from meat and dairy products by PCR–RFLP. Ann Microbiol. 2010;60:189–196. doi: 10.1007/s13213-010-0025-4. [DOI] [Google Scholar]
- Frenay HM, Bunschoten AE, Schouls LM, Van Leeuwen WJ, Vandenbroucke-Grauls CM, Verhoef J, Mooi FR. Molecular typing of methicillin-resistant Staphylococcus aureus on the basis of protein A gene polymorphism. Eur J Clin Microbiol Infect. 1996;15:60–64. doi: 10.1007/BF01586186. [DOI] [PubMed] [Google Scholar]
- Hallin M, Friedrich AW, Struelens MJ. spa typing for epidemiological surveillance of Staphylococcus aureus. Mol Epidemiol Microorg Methods Protoc. 2009;551:189–202. doi: 10.1007/978-1-60327-999-4_15. [DOI] [PubMed] [Google Scholar]
- Harmsen D, Claus H, Witte W, Rothgänger J, Claus H, Turnwald D, Vogel U. Typing of methicillin-resistant Staphylococcus aureus in a university hospital setting by using novel software for spa repeat determination and database management. J Clin Microbiol. 2003;41:5442–5448. doi: 10.1128/JCM.41.12.5442-5448.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter PR, Gaston MA. Numerical index of the discriminatory ability of typing systems: an application of Simpson’s index of diversity. J Clin Microbiol. 1998;26:2465–2466. doi: 10.1128/jcm.26.11.2465-2466.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laplana LM, Cepero MP, Ruiz J, Zolezzi PC, Calvo MC, Erazo MC, Gómez-Lus R. Molecular typing of Staphylococcus aureus clinical isolates by pulsed-field gel electrophoresis, staphylococcal cassette chromosome mec type determination and dissemination of antibiotic resistance genes. Int J Antimicrob Agents. 2007;30:505–513. doi: 10.1016/j.ijantimicag.2007.06.020. [DOI] [PubMed] [Google Scholar]
- Larsen J, Enright MC, Godoy D, Spratt BG, Larsen AR, Skov RL. Multilocus sequence typing scheme for Staphylococcus aureus: revision of the gmk locus. J Clin Microbiol. 2012;5:2538–2539. doi: 10.1128/JCM.00290-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH. Methicillin (oxacillin)-resistant Staphylococcus aureus strains isolated from major food animals and their potential transmission to humans. Appl Environ Microbiol. 2003;69:6489–6494. doi: 10.1128/AEM.69.11.6489-6494.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehndiratta PL, Bhalla P, Ahmed A, Sharma YD. Molecular typing of methicillin-resistant Staphylococcus aureus strains by PCR–RFLP of SPA gene: a reference laboratory perspective. Indian J Med Microbiol. 2009;27:116–122. doi: 10.4103/0255-0857.45363. [DOI] [PubMed] [Google Scholar]
- Oliver SP, Jayarao BM, Almeida RA. Foodborne pathogens in milk and the dairy farm environment: food safety and public health implications. Foodborne Pathog Dis. 2005;2:115–129. doi: 10.1089/fpd.2005.2.115. [DOI] [PubMed] [Google Scholar]
- Omar NY, Ali HA, Harfoush RA, El Khayat EH. Molecular typing of methicillin resistant Staphylococcus aureus clinical isolates on the basis of protein A and coagulase gene polymorphisms. Int J Microbiol. 2014;15:1–11. doi: 10.1155/2014/650328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persoons D, Van Hoorebeke S, Hermans K, Butaye P, de Kruif A, Haesebrouck F, Dewulf J. Methicillin-resistant Staphylococcus aureus in poultry. Emerg Infect Dis. 2009;15:452–453. doi: 10.3201/eid1503.080696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohinishree YS, Negi PS. Detection, identification and characterization of staphylococci in street vend foods. Food Sci Nutr. 2011;2:304–313. doi: 10.4236/fns.2011.24044. [DOI] [Google Scholar]
- Sabat AJ, Budimir A, Nashev D, Sá-Leão R, Van Dijl JM, Laurent F, Grundmann H, Friedrich AW, ESCMID Study Group of Epidemiological Markers (ESGEM) Overview of molecular typing methods for outbreak detection and epidemiological surveillance. Euro Surveill. 2013;18:20380. doi: 10.2807/ese.18.04.20380-en. [DOI] [PubMed] [Google Scholar]
- Shakeri F, Ghaemi EA. New Spa types among MRSA and MSSA isolates in north of Iran. Adv Microbiol. 2014;4:899–995. doi: 10.4236/aim.2014.413100. [DOI] [Google Scholar]
- Shakeri F, Shojai A, Golalipour M, Rahimi Alang S, Vaez H, Ghaemi EA. Spa diversity among MRSA and MSSA Strains of Staphylococcus aureus in North of Iran. Int J Microbiol. 2010;31:1–5. doi: 10.1155/2010/351397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stranden A, Frei R, Widmer AF. Molecular typing of methicillin-resistant Staphylococcus aureus: can PCR replace pulsed-field gel electrophoresis? J Clin Microbiol. 2003;41:3181–3186. doi: 10.1128/JCM.41.7.3181-3186.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strommenger B, Braulke C, Heuck D, Schmidt C, Pasemann B, Nübel U, Witte W. spa typing of Staphylococcus aureus as a frontline tool in epidemiological typing. J Clin Microbiol. 2008;46:574–581. doi: 10.1128/JCM.01599-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang D, Ando S, Takasaki Y, Tadano J. Mutational analyses of restriction endonuclease-HindIII mutant E86K with higher activity and altered specificity. Protein Eng. 2000;13:283–289. doi: 10.1093/protein/13.4.283. [DOI] [PubMed] [Google Scholar]
- Tesfaye GY, Regassa FG, Kelay B. Milk yield and associated economic losses in quarters with subclinical mastitis due to Staphylococcus aureus in Ethiopian crossbred dairy cows. Trop Anim Health Prod. 2010;42:925–931. doi: 10.1007/s11250-009-9509-2. [DOI] [PubMed] [Google Scholar]
- Toshkova K, Annemüller C, Akineden Ö, Lämmler C. The significance of nasal carriage of Staphylococcus aureus as risk factor for human skin infections. FEMS Microbiol Lett. 2001;202:17–24. doi: 10.1111/j.1574-6968.2001.tb10774.x. [DOI] [PubMed] [Google Scholar]
- Walther B, Wieler LH, Friedrich AW, Hanssen A, Kohn B, Brunnberg L, Lübke-Becker A. Methicillin-resistant Staphylococcus aureus (MRSA) isolated from small and exotic animals at a university hospital during routine microbiological examinations. Vet Microbiol. 2008;127:171–178. doi: 10.1016/j.vetmic.2007.07.018. [DOI] [PubMed] [Google Scholar]
- Wertheim HF, Vos MC, Ott A, van Belkum A, Voss A, Kluytmans JA, van Keulen PH, Vandenbroucke-Grauls CM, Meester MH, Verbrugh HA. Risk and outcome of nosocomial Staphylococcus aureus bacteraemia in nasal carriers versus non-carriers. Lancet. 2004;27:703–705. doi: 10.1016/S0140-6736(04)16897-9. [DOI] [PubMed] [Google Scholar]
- Wichelhaus TA, Hunfeld KP, Böddinghaus B, Kraiczy P, Schafer V, Brade V. Rapid molecular typing of methicillin-resistant Staphylococcus aureus by PCR–RFLP. Infect Control Hosp Epidemiol. 2001;22:294–298. doi: 10.1086/501903. [DOI] [PubMed] [Google Scholar]
- Wilson K. Preparation of genomic DNA from bacteria. Curr Protoc Mol Biol. 1987;2:2–4. doi: 10.1002/0471142727.mb0204s56. [DOI] [PubMed] [Google Scholar]
- Sato’o Y, Omoe K, Ono HK, Nakane A, Hu DL. A novel comprehensive analysis method for Staphylococcus aureus pathogenicity islands. Microbiol Immunology. 2013;57:91–99. doi: 10.1111/1348-0421.12007. [DOI] [PubMed] [Google Scholar]
- Zadoks R, Van Leeuwen W, Barkema H, Sampimon O, Verbrugh H, Schukken YH, Van Belkum A. Application of pulsed-field gel electrophoresis and binary typing as tools in veterinary clinical microbiology and molecular epidemiologic analysis of bovine and human Staphylococcus aureus isolates. J Clin Microbiol. 2000;38:1931–1939. doi: 10.1128/jcm.38.5.1931-1939.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]