Abstract
The defective in organelle trafficking/intracellular multiplication (Dot/Icm) Type IVb secretion system (T4SS) is the essential virulence factor for the intracellular life style and pathogenicity of Legionella species. Screens demonstrated that an individual L. pneumophila strain can use the Dot/Icm T4SS to translocate an unprecedented number of more than 300 proteins into host cells, where these, so called Icm/Dot-translocated substrates (IDTS) or effectors, manipulate host cell functions to the benefit of the bacteria. Bioinformatic analysis of the pan-genus genome predicts at least 608 orthologous groups of putative effectors. Deciphering the function of these effectors is key to understanding Legionella pathogenesis; however, the analysis is challenging. Substantial functional redundancy renders classical, phenotypic screening of single gene deletion mutants mostly ineffective. Here, I review experimental approaches that were successfully used to identify, validate and functionally characterize T4SS effectors and highlight new methods, which promise to facilitate unlocking the secrets of Legionella's extraordinary weapons arsenal.
Keywords: Legionella, Type IVb secretion system, Dot/Icm, effectors, toolbox, host targets, infection models, functional genomics
Introduction
Legionella pneumophila was recognized as human pathogen in 1976 after a devastating outbreak of pneumonia, termed Legionnaires' disease, at an American Legion convention (Fraser et al., 1977; McDade et al., 1977). Investigations into the epidemiological and pathological mechanisms soon established that L. pneumophila is a ubiquitous, facultative intracellular pathogen of protozoa (Rowbotham, 1980), which, after inhalation, can also thrive in human alveolar macrophages. Key to exploiting phagocytic hosts is its ability to evade phago-lysosomal degradation (Horwitz, 1983a). Instead the bacteria create the Legionella containing vacuole (LCV) (Horwitz, 1983b), which shelters them from intracellular defenses and intercepts nutrients, supporting replication.
The defective in organelle trafficking/intracellular multiplication (Dot/Icm) Type IVb secretion system (T4SS) is critical for LCV biogenesis and intracellular replication (Berger and Isberg, 1993; Segal et al., 1998). It is located at the bacterial poles and, upon membrane contact, translocates proteins into host cells (Charpentier et al., 2009; Jeong et al., 2017), which manipulate cellular processes and are therefore called effectors. Although for many Icm/Dot-translocated substrates (IDTS) an actual effect on the host awaits demonstration, they will here collectively be referred to as effectors.
Facilitated by the genome sequences of prototype L. pneumophila strains (Cazalet et al., 2004; Chien et al., 2004) screens for T4SS substrates established that each strain translocates more than 300 proteins (Nagai et al., 2002; Luo and Isberg, 2004; De Felipe et al., 2005, 2008; Kubori et al., 2008; Burstein et al., 2009; Huang et al., 2011; Zhu et al., 2011; Lifshitz et al., 2013). Comparative genomics of an increasing number of L. pneumophila isolates and more than 38 Legionella spp. showed that, while sharing the Dot/Icm T4SS, extensive diversity in the effector arsenals exists (Schroeder et al., 2010; Gomez-Valero et al., 2014; Burstein et al., 2016). Only 7 proteins of an estimated 608 orthologous groups of effectors across the genus are conserved in all species (Burstein et al., 2016).
Despite advances in our understanding about some effectors (Finsel and Hilbi, 2015; So et al., 2015; Qiu and Luo, 2017), we still lack knowledge about the functions of the majority. Deciphering their functions is challenging, as effectors are a heterogeneous group with limited homology to characterized proteins. This mini-review summarizes methods that were employed to characterize Dot/Icm T4SS effectors and highlights additional methods that could help uncovering the weapons which Legionella spp. hold in their arsenals.
Characteristics of Dot/Icm T4SS effectors
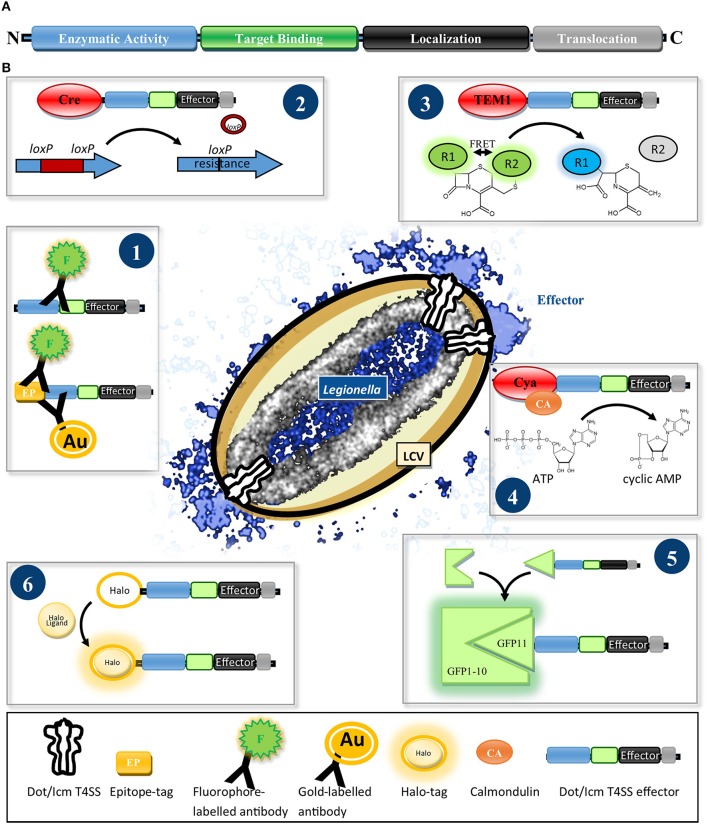
Work over the past 15 years revealed several characteristics of effectors. A translocation signal, which directs them to the T4SS, is commonly found in the C-terminus (Nagai et al., 2005). It consists of a pattern of 20–35 amino acids with specific biophysical properties, e.g., small polar and/or charged residues, and can include a so called E-Block motif encompassing several glutamic acid residues (Nagai et al., 2005; Kubori et al., 2008; Burstein et al., 2009; Huang et al., 2011; Lifshitz et al., 2013). Some effectors comprise an additional internal export signal (Cambronne and Roy, 2007; Jeong et al., 2015). Many effectors are large (>100 kDa), with modular architecture (Figure 1A), consisting of different functional domains, e.g., localization, target binding, and enzymatic activity domains. Most prominent feature, which facilitated the discovery of the first effector RalF, is the occurrence of domains with striking homology to eukaryotic proteins (Nagai et al., 2002; Cazalet et al., 2004; De Felipe et al., 2005; Gomez-Valero et al., 2011).
Figure 1.
(A) Scheme of the typical architecture of Dot/Icm T4SS effectors. Effectors often show a modular structure consisting of a translocation signal and localization, target binding and enzymatic activity domains. (B) Reporter systems for determining the translocation and localization of Dot/Icm T4SS effectors. (1) Fluorophore- or gold particle-conjugated antibodies specific for an effector or an epitope tag are used to detect the effector in host cell lysates by immunoblot or visualize it by immunofluorescence- or electron microscopy. (2) Cre/loxP recombinase system: After delivery of Cre-effector fusions into recipient cells the recombinase removes a loxP-flanked disruptor cassette from a gene reporter conferring antibiotic resistance. (3) β-lactamase (TEM-1) assay: Translocation of TEM-1-effector fusions results in hydrolysis of a green fluorescent β-lactam FRET substrate, separating FRET donor and acceptor, generating a blue fluorescent product. (4) Calmodulin-dependent adenylate-cyclase (Cya) assay: Upon arrival of a Cya-effector fusion in the host, Cya gets activated by binding calmodulin and turns over ATP to cyclic AMP, which can be quantified by ELISA. (5) Split-GFP reporter system: Effectors are fused to the small (GFP11) fragment of a split GFP. Upon delivery into cells expressing the non-fluorescent large (GFP1-10) GFP fragment, spontaneous reassembly of effector-fused GFP11 and GFP1-10 occurs, restoring fluorescence emission which can be visualized by fixed or live imaging fluorescence microscopy. (6) Halo-tag reporter: After translocation into the host the Halo-tagged effector can be ligated with versatile fluorophores for detection by conventional, super-resolution- or electron microscopy.
Integration of these characteristics and parameters, such as regulatory motifs, in machine-learning approaches enabled prediction algorithms. Several programs are available (Meyer et al., 2013; Zou et al., 2013; An et al., 2016). Dot/Icm effector-focused algorithms were applied to 38 Legionella spp., revealing not only 608 orthologous groups of effectors, but also 99 frequently-occurring domains, which facilitate the identification of new effectors (Burstein et al., 2009, 2016; Lifshitz et al., 2013).
Probing translocation and localization
Several assays for the validation of T4SS-mediated transport exist (Figure 1B). The visualization of endogenous effectors in host cells using antibodies and immunofluorescence (IF) or electron microscopy (EM) is the gold standard to infer physiologically accurate information (Figure 1B.1) and was achieved for a few effectors, e.g., SdeA, LidA, RidL, SidC, SidM, RalF, (Nagai et al., 2002; Luo and Isberg, 2004; Bardill et al., 2005; Machner and Isberg, 2006; Finsel et al., 2013). An antibody against SidC was used to visualize the reconstitution of translocation of a SidC variant lacking its translocation signal by fusion to putative effectors (Vanrheenen et al., 2006; Huang et al., 2011).
Alternatively, effectors were detected in host cell extracts by immunoblot (Vanrheenen et al., 2006; Lin et al., 2015), which in combination with fractionation steps to isolate organelles or LCVs, also informed about their subcellular localization (Ivanov et al., 2010; Hoffmann et al., 2014a; Lin et al., 2015).
As many effectors seem to be of low abundancy, several assays employ overexpression and exploit that the T4SS tolerates reporter domains fused to the N-terminus of effectors, if these do not fold rapidly into rigid structures (Amyot et al., 2013). One or multiple epitope tags [e.g., M45 (Weber et al., 2006), 4xHemagglutinin (HA) (Dolezal et al., 2012), 13xMyc (Viner et al., 2012) or 3xFlag (Isaac et al., 2015)] were employed to detect translocated effectors.
An early screen using an enzymatic reporter domain measured restoration of an antibiotic resistance gene by the Cre/loxP recombinase after T4SS-mediated translocation of Cre-effector fusions from a Legionella donor into bacterial recipients (Figure 1B.2) (Luo and Isberg, 2004). However, the β-lactamase TEM-1 and the calmodulin-dependent adenylate-cyclase domain of Bordetella pertussis toxin Cya are the most frequently used enzymatic reporters (Figures 1B.3, 4), providing high sensitivity by enzymatic signal amplification (TEM1: cleavage of a β-lactam fluorescence resonance energy transfer (FRET) sensor; Cya; generation of cyclic AMP) (Chen, 2004; Nagai et al., 2005; De Felipe et al., 2008).
Despite localization of several effectors by IF, SidC is the only imaged by super-resolution microscopy (Naujoks et al., 2016) and no live imaging data tracking Legionella effectors during infection exists. Lack of fluorescent protein tags compatible with T4SS-mediated translocation might account for this. Split fluorescent proteins, used for Salmonella T3SS effectors (Figure 1B.5) (Van Engelenburg and Palmer, 2010) and new enzyme-tags (Figure 1B.6) (Halo-, Snap and Clip-tags), which self-label with fluorophores that are suitable for live-, super-resolution- and electron-microscopy (Bottanelli et al., 2016; Liss et al., 2016), are promising tools to reveal the dynamics and distribution of effectors on a nanoscale.
Infection models
Legionella effectors target fundamental processes, conserved between protozoa and mammals, resulting in a range of infection models, which, despite similarities, have each strengths, and weaknesses. Hartmannella vermiformis, Naegleria spp. and in particular Acanthamoeba castellanii and Dictyostelium discoideum are frequently used environmental hosts (Rowbotham, 1980; Newsome et al., 1985; Fields et al., 1990; Moffat and Tompkins, 1992; Solomon and Isberg, 2000; Hoffmann et al., 2014b). Requirements for specific effector subsets in different protozoa vary and are more stringent than in macrophages (O'Connor et al., 2011), making protozoa indispensable to study the evolutionary pressures behind the acquisition of effectors. To analyze the interaction of Legionella with macrophages, various mammalian [e.g., U937 (Pearlman et al., 1988), HL-60 (Marra et al., 1990), THP-1, Raw264.7 (Cirillo et al., 1994), J774 (Husmann and Johnson, 1992), M-HS (Matsunaga et al., 2001)] and insect [S2 and Kc167 (Dorer et al., 2006; Sun et al., 2013)] cell lines served as models. Moreover, non-phagocytic cells [e.g., HEp-2 (Cirillo et al., 1994), A549 (Mousnier et al., 2014), HeLa (Finsel et al., 2013), HEK293 (Losick et al., 2010), CHO (McCusker et al., 1991; Kagan and Roy, 2002)], with optional ectopic-expression of Fcγ-receptor to boost the invasion efficiency of Legionella, were employed. To evaluate the relevance of findings for human disease, differences in patterns of protein family expansion, e.g., Rab GTPases (Klöpper et al., 2012), and innate immune signaling, e.g., inflammasome activation (Krause and Amer, 2016), between cell lines, mice and humans need to be considered. Ultimately, results need validation in primary macrophages and in vivo models that approximate the complexity of the human immune system.
Insects such as Drosophila melanogaster (Kubori et al., 2010) and Galleria mellonella (Harding et al., 2012, 2013b; Aurass et al., 2013) mount innate immune responses and represent straight-forward infection models. Tests in mammals, e.g., guinea pigs, rats, rhesus monkeys, and marmosets, showed that guinea pigs develop disease similar to humans (Baskerville et al., 1983; Davis et al., 1983). Mice, with exception of A/J mice, which are defective in an NAIP5-dependent inflammasome response to flagellin, are resistant to Legionella (Brieland et al., 1994). Nevertheless, because of the wealth of engineered mouse strains, infections of A/J mice with wild-type or non-permissive mice with flagellin-deficient Legionella have become the predominant in vivo models and gave important insight into effector and immune biology (Brown et al., 2016). In the future, humanized mice (Walsh et al., 2017) and ex vivo human lung tissue models (Jäger et al., 2014) will improve our capabilities to define roles of effectors in human infection.
Genetics approaches to determine effector functions
Legionella is amenable for gene deletion by homologous recombination (Merriam et al., 1997; Bryan et al., 2011; O'Connor et al., 2011) and mutagenesis with transposons (Ott, 1994; Pope et al., 1994; Edelstein et al., 1999; O'Connor et al., 2011). Assays recording intracellular growth by colony counting or continuously, using fluorescent or bioluminescent strains, are established (Coers et al., 2007; Tiaden et al., 2013; Schroeder et al., 2015). Mixed infection competition experiments measuring performance of a wild type vs. a mutant strain achieved better resolution of differences in virulence in some cases (Ensminger et al., 2012; Finsel et al., 2013; Harding et al., 2013b). However, attenuation of strains lacking single effectors was rarely observed. Some effectors might be dispensable in a specific host; but Legionella also achieves resilience by deploying families of paralogue effectors, which seem functionally redundant (Cazalet et al., 2004; Chien et al., 2004).
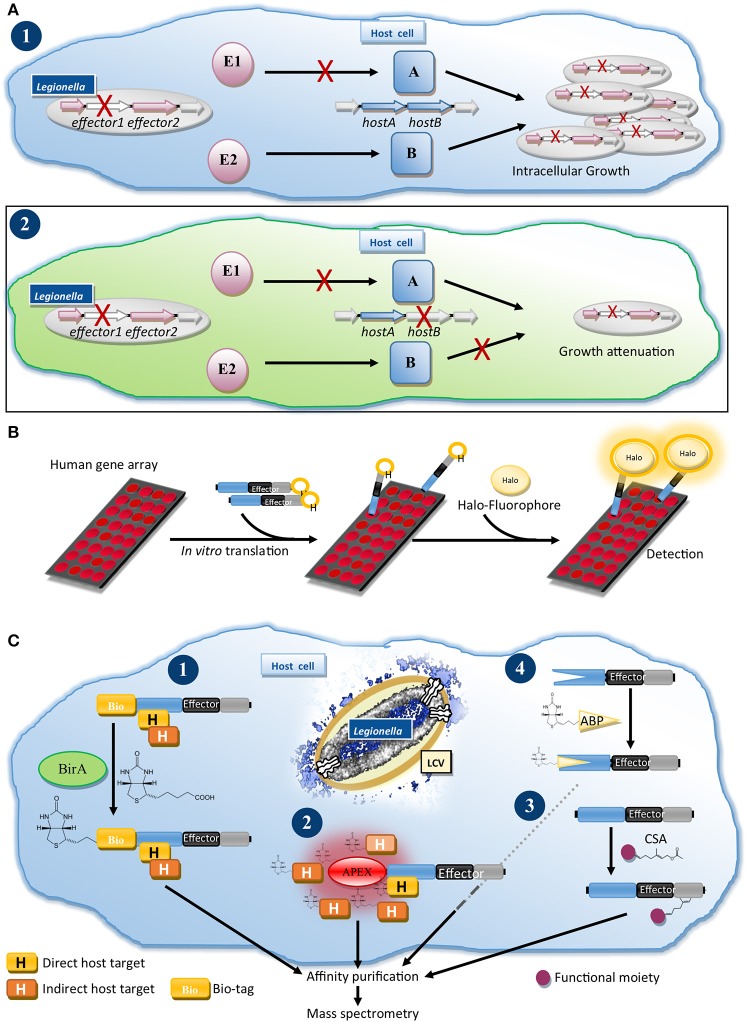
To reduce the complexity of the effector network, O'Connor and Isberg developed two genetic approaches. Insertional mutagenesis and depletion (iMAD, Figure 2A) is based on the combinatorial screening of effector deletion mutants for intracellular growth in hosts, which are also host factor depleted (O'Connor et al., 2012; O'Connor and Isberg, 2014). Additive or compensatory effects of the lack of an effector and a host factor are monitored and interrogated using computational clustering and network analysis, grouping effectors with similar profiles and predicting functional redundancy.
Figure 2.
Genetics and proteomics methods for the functional characterization of Dot/Icm T4SS effectors. (A) Insertional mutagenesis and depletion (iMAD) disentangles the complex network of effector-host manipulations: (1) Characterization of a Legionella mutant lacking a single effector (E1), which acts through host protein A, does not result in reduced intracellular growth, because a second effector (E2) induces a redundant process through host protein B. (2) Following the iMAD strategy, screening of single effector mutants in host cells, which are also depleted for host factors, eliminates redundant pathways, resulting in attenuation of the strain. (B) Nucleic Acid-Programmable Protein Array (NAPPA) for profiling of host cell targets of effectors: Human genes are printed as array on slides and translated in vitro. Recombinant Halo-tagged effector is added and after washes reacted with a fluorophore-ligand for the Halo-tag, allowing detection of the human proteins, which bound and retained the effector on the array. (C) Proteomics approaches for the characterization of effectors: (1) BirA/Bio-tag system: Bio-tagged effector is translocated into biotin ligase BirA expressing cells leading to biotinlylation of the Bio-tag. After optional cross-linking the bioinylated effector and bound host proteins are isolated by tandem-affinity purification for interactome analysis by MS. (2) Proximity biotinylation: Translocation of an effector fused to e.g., the peroxidase APEX or a promiscuous biotin ligase (BioID) results in biotinylation of host proteins in the proximity of the effector, which can be processed as for (1) to identify potential interactors. (3) Identifying post-translational modifications (PTMs): Infected cells are infused or metabolically-labeled with a chemical substrate analog (CSA) for a PTM-catalyzing enzyme. Host proteins and effectors, which are modified with the CSA, can be isolated after ligation of an affinity handle such as biotin to the CSA and characterized by MS. (4) Profiling the enzymatic activities of effectors with activity-based probes (ABPs): Infected cells or lysates are treated with a chemical ABP, which irreversibly binds to a specific enzyme class and contains or allows addition of an affinity handle. Effectors, which possess such enzymatic activity, react with the probe, are isolated and identified by MS.
In a second approach five genomic regions were deleted to create a minimized genome strain lacking 31% of effectors (O'Connor et al., 2011). This strain grows normally in macrophages; but is attenuated in protozoa, underlining the importance of examining several infection models. Subsequently, the minimized genome strain and intermediates lacking subsets of the genomic regions proved to be valuable tools to link Dot/Icm T4SS dependent phenotypes to a chromosomal region and, through gene-by-gene screening, to individual effectors (Choy et al., 2012; Arasaki et al., 2017; Kotewicz et al., 2017). Deletion of additional effectors could generate even more powerful strains for loss-of-function or gain-of-function experiments, in which the perturbation of host processes by individual effectors can be dissected.
Heterologous expression systems for phenotypic analysis
Alternatives to investigating effectors during infection rely on heterologous expression and delivery. These are often technically and analytically less complex, but do not reflect physiological concentrations, microenvironment of delivery and the effects of other effectors. Microinjection of recombinant effectors, e.g., SetA (Jank et al., 2012), offers excellent control of concentration and timing of injection, enabling the characterization of toxic effectors; however requires protein purification and a microinjector. Relinquishing the tight control over the delivery, but reducing technical requirements, microbial microinjection exploits a Yersinia enterocolitica strain with functional type III secretion system (T3SS), but lacking effectors (Wölke et al., 2011), to deliver individual Dot/Icm effectors (Rothmeier et al., 2013). The suitability of this approach for a wide range of T4SS effectors still needs confirmation.
Ectopic-expression in mammalian cells remains the workhorse to assess effector-induced modulation of host processes and subcellular targeting by co-localization with organelle markers. Numerous studies exist. Libraries containing up to 275 effectors for viral transduction or transfection were used to screen for effectors, which modulate, e.g., caspase activation (Zhu et al., 2013), the cytoskeleton (Liu et al., 2017), translation (Barry et al., 2013), or NF-kB activation (Ge et al., 2009; Losick et al., 2010).
Saccharomyces cerevisiae is an important tool to study effectors (Popa et al., 2016). Phenotypic screens identified Legionella effectors that subvert endosomal trafficking or are cytotoxic for yeast (Campodonico et al., 2005; Shohdy et al., 2005). The availability of yeast gene deletion and overexpression strain collections (Gelperin et al., 2005; Sopko et al., 2006; Giaever et al., 2014) bears particular potential. These have proven useful to test enzymatic activities of effectors, e.g., LegS2 or SidP, in functional complementation assays (Degtyar et al., 2009; Toulabi et al., 2013) and to profile synergistic and antagonistic genetic interactions between yeast and effector genes, allowing to infer affected pathways (Viner et al., 2012). Moreover, screening of overexpressed host proteins or effectors for suppression of effector-induced toxicity toward yeast identified host targets, effectors pairs with antagonistic activities and, so called meta-effectors, which regulate other effectors (Tan and Luo, 2011; Tan et al., 2011; Guo et al., 2014; Urbanus et al., 2016).
Identification of protein targets
Dissecting the molecular mechanisms underlying effector-induced phenotypes often requires the identification of host targets. Yeast two-hybrid screening is a powerful method to identify protein-protein interactions and was used for several effectors (Banga et al., 2007; Lomma et al., 2010; Harding et al., 2013a; Michard et al., 2015). Similarly, pull-down of interactors from host cell lysates using purified effector or co-immunoprecipitation (Co-IP) from cells ectopically expressing an effector bait were frequently used (Machner and Isberg, 2006; Price et al., 2009; Finsel et al., 2013; Urbanus et al., 2016). In a cell-free assay system, the Nucleic Acid-Programmable Protein Array (NAPPA, Figure 2B) (Yu et al., 2015), human bait gene arrays are translated in vitro, exposed to Halo-tagged effector and bound effector detected by ligation of a fluorophore to the Halo-tag. This system, circumventing protein isolation, promises to reveal a global view of interactors.
Despite their proven value, all above-mentioned in vitro and heterologous expression methods struggle with the identification of false-positive and -negative targets, because they do not reflect the unique proteomic landscape which an effector experiences when injected at the LCV membrane into a cell that responds to the infection and is manipulated by hundreds of effectors.
We established a method to determine the interactomes of effectors during infection (Figure 2C.1) (Mousnier et al., 2014). Legionella expressing an effector fused to a tandem-affinity tag including a biotinylation site (Bio-tag), are used to infect cells expressing Escherichia coli biotin ligase BirA. The translocated effector is biotinylated, allowing isolation of effector-host target complexes for analysis by mass spectrometry (MS). Using this approach, we identified new interactors of PieE and profiled the infection-relevant interactions of the promiscuous Rab GTPase-binding effectors SidM and LidA (Mousnier et al., 2014; So et al., 2016).
Exciting prospects for effector target discovery arise from the development of proximity-biotinylation systems (Figure 2C.2). These rely on promiscuous biotinylation of proteins in proximity of engineered BirA (Roux et al., 2012) or the peroxidase APEX2 (Hung et al., 2016) followed by characterization of biotinylated targets by MS. Translocation of T3SS effector-APEX2 fusions by Chlamydia was recently described (Rucks et al., 2017) suggesting that this could be adopted for T4SS effectors.
Profiling post-translational modifications (PTMs) and enzymatic activities
Effectors exploit host proteins as receptors (Gaspar and Machner, 2014) and subvert their functions, which is often achieved by post-translational modification (PTM) (Michard and Doublet, 2015). The discovery of the phosphocholination activity of AnkX and phosphoribosyl-ubiquitin ligase activity in SdeA illustrated that careful analysis of protein targets by MS is key to identify new PTMs (Mukherjee et al., 2011; Bhogaraju et al., 2016; Qiu et al., 2016). PTM specific antibodies were used e.g., to study effector-mediated phosphorylation or histone modifications (Ge et al., 2009; Rolando et al., 2013). This can be complemented by autoradiography assays using radioactive substrates, which excel in sensitivity, and are employed to study, e.g., AMPylation (Neunuebel et al., 2011; Tan and Luo, 2011) or glycosyltransferase effectors (Jank et al., 2012). Non-radioactive chemical substrate analogs (CSAs), which can be functionalized to visualize and isolate modified proteins, were developed for several PTMs (Grammel et al., 2011; Lu et al., 2012; Fischle and Schwarzer, 2016). CSAs can also reveal PTMs on effectors, as demonstrated for the post-translational lipidation of effectors (Figure 2C.3) (Ivanov et al., 2010; Lin et al., 2015; Schroeder et al., 2015). CSAs enable profiling of PTMs on proteome level from cell extracts, living cells or, as shown for SidM-mediated AMPylation, on NAPPA arrays (Yu et al., 2015), promising global overviews of PTMs at a coverage similar to the Legionella-shaped ubiquitinome (Ivanov and Roy, 2013; Bruckert and Abu Kwaik, 2015).
The discovery of new enzymatic activities is challenging as small molecules, e.g., ATP or lipids, can be substrates and/or effectors not necessarily target host proteins. Bioinformatic analysis to identify homologous enzymes and catalytic motifs is critical to find leads (Watson et al., 2005) for focused enzymatic assays, as exemplified by LpdA (lipolysis, Schroeder et al., 2015), SidF (phosphate release, Hsu et al., 2012) or LncP (nucleotide transport, Dolezal et al., 2012). Biophysical methods, e.g., differential scanning fluorimetry, allow screening of ligands (Ciulli, 2013).
For the identification of enzymes-of-interest, e.g., redundant effectors, in the Legionella proteome activity-based probes (ABPs) offer a solution (Figure 2C.4). ABPs are typically small molecules that irreversibly react with a specific enzyme class and functionalized to allow purification of modified enzymes for MS analysis. ABPs are available for many enzymes including the ubiquitin-conjugation and chromatin-modifying machineries (Willems et al., 2014; Fischle and Schwarzer, 2016; Hewings et al., 2017). ABPs will help to disentangle the redundancy problem, probe for eukaryotic-like enzymes and assign functions in new Legionella isolates.
Synthesis
Deciphering the functions of thousands of effectors is a formidable challenge; however new genetics tools and a rapidly growing number of chemical biology and proteomics methods provide a well-suited toolbox to reveal fascinating new mechanisms of host manipulation by Legionella.
Author contributions
The author confirms being the sole contributor of this work and approved it for publication.
Conflict of interest statement
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
I thank Dr. Aurélie Mousnier (QUB) for her critical review of the manuscript.
Footnotes
Funding. This work was supported by institutional funding from Queen's University Belfast for GNS.
References
- Amyot W. M., DeJesus D., Isberg R. R. (2013). Poison domains block transit of translocated substrates via the Legionella pneumophila Icm/Dot system. Infect. Immun. 81, 3239–3252. 10.1128/IAI.00552-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- An Y., Wang J., Li C., Leier A., Marquez-Lago T., Wilksch J., et al. (2016). Comprehensive assessment and performance improvement of effector protein predictors for bacterial secretion systems III, IV and VI. Brief. Bioinform. [Epub ahead of print]. 10.1093/bib/bbw100 [DOI] [PubMed] [Google Scholar]
- Arasaki K., Mikami Y., Shames S. R., Inoue H., Wakana Y., Tagaya M. (2017). Legionella effector Lpg1137 shuts down ER-mitochondria communication through cleavage of syntaxin 17. Nat. Commun. 8:15406. 10.1038/ncomms15406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aurass P., Schlegel M., Metwally O., Harding C. R., Schroeder G. N., Frankel G., et al. (2013). The Legionella pneumophila Dot/Icm-secreted effector PlcC/CegC1 together with PlcA and PlcB promotes virulence and belongs to a novel zinc metallophospholipase C family present in bacteria and fungi. J. Biol. Chem. 288, 11080–11092. 10.1074/jbc.M112.426049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banga S., Gao P., Shen X., Fiscus V., Zong W.-X., Chen L., et al. (2007). Legionella pneumophila inhibits macrophage apoptosis by targeting pro-death members of the Bcl2 protein family. Proc. Natl. Acad. Sci. U.S.A. 104, 5121–5126. 10.1073/pnas.0611030104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardill J. P., Miller J. L., Vogel J. P. (2005). IcmS-dependent translocation of SdeA into macrophages by the Legionella pneumophila type IV secretion system. Mol. Microbiol. 56, 90–103. 10.1111/j.1365-2958.2005.04539.x [DOI] [PubMed] [Google Scholar]
- Barry K. C., Fontana M. F., Portman J. L., Dugan A. S., Vance R. E. (2013). IL-1α signaling initiates the inflammatory response to virulent Legionella pneumophila in vivo. J. Immunol. 190, 6329–6339. 10.4049/jimmunol.1300100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baskerville A., Fitzgeorge R. B., Broster M., Hambleton P. (1983). Histopathology of experimental Legionnaires' disease in guinea pigs, rhesus monkeys and marmosets. J. Pathol. 139, 349–362. 10.1002/path.1711390310 [DOI] [PubMed] [Google Scholar]
- Berger K. H., Isberg R. R. (1993). Two distinct defects in intracellular growth complemented by a single genetic locus in Legionella pneumophila. Mol. Microbiol. 7, 7–19. 10.1111/j.1365-2958.1993.tb01092.x [DOI] [PubMed] [Google Scholar]
- Bhogaraju S., Kalayil S., Liu Y., Bonn F., Colby T., Matic I., et al. (2016). Phosphoribosylation of ubiquitin promotes serine ubiquitination and impairs conventional ubiquitination. Cell 167, 1636–1649.e13. 10.1016/j.cell.2016.11.019 [DOI] [PubMed] [Google Scholar]
- Bottanelli F., Kromann E. B., Allgeyer E. S., Erdmann R. S., Wood Baguley S., Sirinakis G., et al. (2016). Two-colour live-cell nanoscale imaging of intracellular targets. Nat. Commun. 7:10778. 10.1038/ncomms10778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brieland J., Freeman P., Kunkel R., Chrisp C., Hurley M., Fantone J., et al. (1994). Replicative Legionella pneumophila lung infection in intratracheally inoculated A/J mice. A murine model of human Legionnaires' disease. Am. J. Pathol. 145, 1537–1546. [PMC free article] [PubMed] [Google Scholar]
- Brown A. S., Yang C., Hartland E. L., van Driel I. R. (2016). The regulation of acute immune responses to the bacterial lung pathogen Legionella pneumophila. J. Leukoc. Biol. 101, 875–886. 10.1189/jlb.4MR0816-340R [DOI] [PubMed] [Google Scholar]
- Bruckert W. M., Abu Kwaik Y. (2015). Complete and ubiquitinated proteome of the Legionella-containing vacuole within human macrophages. J. Proteome Res. 14, 236–248. 10.1021/pr500765x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryan A., Harada K., Swanson M. S. (2011). Efficient generation of unmarked deletions in Legionella pneumophila. Appl. Environ. Microbiol. 77, 2545–2548. 10.1128/AEM.02904-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burstein D., Amaro F., Zusman T., Lifshitz Z., Cohen O., Gilbert J. A., et al. (2016). Genomic analysis of 38 Legionella species identifies large and diverse effector repertoires. Nat. Genet. 48, 167–175. 10.1038/ng.3481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burstein D., Zusman T., Degtyar E., Viner R., Segal G., Pupko T. (2009). Genome-scale identification of Legionella pneumophila effectors using a machine learning approach. PLoS Pathog. 5:e1000508. 10.1371/journal.ppat.1000508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cambronne E. D., Roy C. R. (2007). The Legionella pneumophila IcmSW complex interacts with multiple Dot/Icm effectors to facilitate type IV translocation. PLoS Pathog. 3, 1837–1848. 10.1371/journal.ppat.0030188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campodonico E. M., Chesnel L., Roy C. R. (2005). A yeast genetic system for the identification and characterization of substrate proteins transferred into host cells by the Legionella pneumophila Dot/Icm system. Mol. Microbiol. 56, 918–933. 10.1111/j.1365-2958.2005.04595.x [DOI] [PubMed] [Google Scholar]
- Cazalet C., Rusniok C., Brüggemann H., Zidane N., Magnier A., Ma L., et al. (2004). Evidence in the Legionella pneumophila genome for exploitation of host cell functions and high genome plasticity. Nat. Genet. 36, 1165–1173. 10.1038/ng1447 [DOI] [PubMed] [Google Scholar]
- Charpentier X., Gabay J. E., Reyes M., Zhu J. W., Weiss A., Shuman H. A. (2009). Chemical genetics reveals bacterial and host cell functions critical for type IV effector translocation by Legionella pneumophila. PLoS Pathog. 5:e1000501. 10.1371/journal.ppat.1000501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J. (2004). Legionella effectors that promote nonlytic release from protozoa. Science 303, 1358–1361. 10.1126/science.1094226 [DOI] [PubMed] [Google Scholar]
- Chien M., Morozova I., Shi S., Sheng H., Chen J., Gomez S. M., et al. (2004). The genomic sequence of the accidental pathogen Legionella pneumophila. Science 305, 1966–1968. 10.1126/science.1099776 [DOI] [PubMed] [Google Scholar]
- Choy A., Dancourt J., Mugo B., O'Connor T. J., Isberg R. R., Melia T. J., et al. (2012). The Legionella effector RavZ inhibits host autophagy through irreversible Atg8 deconjugation. Science 338, 1072–1076. 10.1126/science.1227026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirillo J. D., Falkow S., Tompkins L. S. (1994). Growth of Legionella pneumophila in Acanthamoeba castellanii enhances invasion. Infect. Immun. 62, 3254–3261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciulli A. (2013). Biophysical screening for the discovery of small-molecule ligands. Methods Mol. Biol. 1008, 357–388. 10.1007/978-1-62703-398-5_13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coers J., Vance R. E., Fontana M. F., Dietrich W. F. (2007). Restriction of Legionella pneumophila growth in macrophages requires the concerted action of cytokine and Naip5/Ipaf signalling pathways. Cell. Microbiol. 9, 2344–2357. 10.1111/j.1462-5822.2007.00963.x [DOI] [PubMed] [Google Scholar]
- Davis G. S., Winn W. C., Gump D. W., Craighead J. M., Beaty H. N. (1983). Legionnaires' pneumonia in guinea pigs and rats produced by aerosol exposure. Chest 83(5 Suppl.), 15S−16S. [DOI] [PubMed] [Google Scholar]
- De Felipe K. S., Glover R. T., Charpentier X., Anderson O. R., Reyes M., Pericone C. D., et al. (2008). Legionella eukaryotic-like type IV substrates interfere with organelle trafficking. PLoS Pathog. 4:e1000117. 10.1371/journal.ppat.1000117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Felipe K. S., Pampou S., Jovanovic O. S., Pericone C. D., Ye S. F., Kalachikov S., et al. (2005). Evidence for acquisition of Legionella type IV secretion substrates via interdomain horizontal gene transfer. J. Bacteriol. 187, 7716–7726. 10.1128/JB.187.22.7716-7726.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degtyar E., Zusman T., Ehrlich M., Segal G. (2009). A Legionella effector acquired from protozoa is involved in sphingolipids metabolism and is targeted to the host cell mitochondria. Cell. Microbiol. 11, 1219–1235. 10.1111/j.1462-5822.2009.01328.x [DOI] [PubMed] [Google Scholar]
- Dolezal P., Aili M., Tong J., Jiang J.-H., Marobbio C. M., Lee S. F., et al. (2012). Legionella pneumophila secretes a mitochondrial carrier protein during infection. PLoS Pathog. 8:e1002459. 10.1371/journal.ppat.1002459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorer M. S., Kirton D., Bader J. S., Isberg R. R. (2006). RNA interference analysis of Legionella in Drosophila cells: exploitation of early secretory apparatus dynamics. PLoS Pathog. 2, 315–327. 10.1371/journal.ppat.0020034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelstein P. H., Edelstein M. A., Higa F., Falkow S. (1999). Discovery of virulence genes of Legionella pneumophila by using signature tagged mutagenesis in a guinea pig pneumonia model. Proc. Natl. Acad. Sci. U.S.A. 96, 8190–8195. 10.1073/pnas.96.14.8190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ensminger A. W., Yassin Y., Miron A., Isberg R. R. (2012). Experimental evolution of Legionella pneumophila in mouse macrophages leads to strains with altered determinants of environmental survival. PLoS Pathog. 8:e1002731. 10.1371/journal.ppat.1002731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields B. S., Nerad T. A., Sawyer T. K., King C. H., Barbaree J. M., Martin W. T., et al. (1990). Characterization of an axenic strain of Hartmannella vermiformis obtained from an investigation of nosocomial Legionellosis. J. Protozool. 37, 581–583. 10.1111/j.1550-7408.1990.tb01269.x [DOI] [PubMed] [Google Scholar]
- Finsel I., Hilbi H. (2015). Formation of a pathogen vacuole according to Legionella pneumophila: how to kill one bird with many stones. Cell. Microbiol. 17, 935–950. 10.1111/cmi.12450 [DOI] [PubMed] [Google Scholar]
- Finsel I., Ragaz C., Hoffmann C., Harrison C. F., Weber S., Van Rahden V. A., et al. (2013). The Legionella effector RidL inhibits retrograde trafficking to promote intracellular replication. Cell Host Microbe 14, 38–50. 10.1016/j.chom.2013.06.001 [DOI] [PubMed] [Google Scholar]
- Fischle W., Schwarzer D. (2016). Probing Chromatin-modifying enzymes with chemical tools. ACS Chem. Biol. 11, 689–705. 10.1021/acschembio.5b01023 [DOI] [PubMed] [Google Scholar]
- Fraser D. W., Tsai T. R., Orenstein W., Parkin W. E., Beecham H. J., Sharrar R. G., et al. (1977). Legionnaires' disease: description of an epidemic of pneumonia. N. Engl. J. Med. 297, 1189–1197. 10.1056/NEJM197712012972201 [DOI] [PubMed] [Google Scholar]
- Gaspar A. H., Machner M. P. (2014). VipD is a Rab5-activated phospholipase A1 that protects Legionella pneumophila from endosomal fusion. Proc. Natl. Acad. Sci. U.S.A. 111, 4560–4565. 10.1073/pnas.1316376111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge J., Xu H., Li T., Zhou Y., Zhang Z., Li S., et al. (2009). A Legionella type IV effector activates the NF-κB pathway by phosphorylating the IκB family of inhibitors. Proc. Natl. Acad. Sci. U.S.A. 106, 13725–13730. 10.1073/pnas.0907200106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelperin D. M., White M. A., Wilkinson M. L., Kon Y., Kung L. A., Wise K. J., et al. (2005). Biochemical and genetic analysis of the yeast proteome with a movable ORF collection. Genes Dev. 19, 2816–2826. 10.1101/gad.1362105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giaever G., Nislow C., Alonso J. M., Stepanova A. N., Leisse T. J., Kim C. J., et al. (2014). The yeast deletion collection: a decade of functional genomics. Genetics 197, 451–465. 10.1534/genetics.114.161620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Valero L., Rusniok C., Cazalet C., Buchrieser C. (2011). Comparative and functional genomics of Legionella identified eukaryotic like proteins as key players in host-pathogen interactions. Front. Microbiol. 2, 1–20. 10.3389/fmicb.2011.00208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Valero L., Rusniok C., Rolando M., Neou M., Dervins-Ravault D., Demirtas J., et al. (2014). Comparative analyses of Legionella species identifies genetic features of strains causing Legionnaires' disease. Genome Biol. 15:505. 10.1186/PREACCEPT-1086350395137407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grammel M., Luong P., Orth K., Hang H. C. (2011). A chemical reporter for protein AMPylation. J. Am. Chem. Soc. 133, 17103–17105. 10.1021/ja205137d [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Z., Stephenson R., Qiu J., Zheng S., Luo Z. Q. (2014). A Legionella effector modulates host cytoskeletal structure by inhibiting actin polymerization. Microbes Infect. 16, 225–236. 10.1016/j.micinf.2013.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding C. R., Mattheis C., Mousnier A., Oates C. V., Hartland E. L., Frankel G., et al. (2013a). LtpD is a novel Legionella pneumophila effector that binds phosphatidylinositol 3-phosphate and inositol monophosphatase IMPA1. Infect. Immun. 81, 4261–4270. 10.1128/IAI.01054-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding C. R., Schroeder G. N., Reynolds S., Kosta A., Collins J. W., Mousnier A., et al. (2012). Legionella pneumophila pathogenesis in the Galleria mellonella infection model. Infect. Immun. 80, 2780–2790. 10.1128/IAI.00510-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding C. R., Stoneham C. A., Schuelein R., Newton H., Oates C. V., Hartland E. L., et al. (2013b). The Dot/Icm effector SdhA is necessary for virulence of Legionella pneumophila in Galleria mellonella and A/J mice. Infect. Immun. 81, 2598–2605. 10.1128/IAI.00296-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewings D. S., Flygare J. A., Bogyo M., Wertz I. E. (2017). Activity-based probes for the ubiquitin conjugation–deconjugation machinery: new chemistries, new tools, and new insights. FEBS J. 284, 1555–1576. 10.1111/febs.14039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann C., Finsel I., Otto A., Pfaffinger G., Rothmeier E., Hecker M., et al. (2014a). Functional analysis of novel Rab GTPases identified in the proteome of purified Legionella-containing vacuoles from macrophages. Cell. Microbiol. 16, 1034–1052. 10.1111/cmi.12256 [DOI] [PubMed] [Google Scholar]
- Hoffmann C., Harrison C. F., Hilbi H. (2014b). The natural alternative: protozoa as cellular models for Legionella infection. Cell. Microbiol. 16, 15–26. 10.1111/cmi.12235 [DOI] [PubMed] [Google Scholar]
- Horwitz M. A. (1983a). The Legionnaires' disease bacterium (Legionella pneumophila) inhibits phagosome-lysosome fusion in human monocytes. J. Exp. Med. 158, 2108–2126. 10.1084/jem.158.6.2108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horwitz M. A. (1983b). Formation of a novel phagosome by the Legionnaires' disease bacterium (Legionella pneumophila) in human monocytes. J. Exp. Med. 158, 1391–1331. 10.1084/jem.158.4.1319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu F., Zhu W., Brennan L., Tao L., Luo Z.-Q., Mao Y. (2012). Structural basis for substrate recognition by a unique Legionella phosphoinositide phosphatase. Proc. Natl. Acad. Sci. U.S.A. 109, 13567–13572. 10.1073/pnas.1207903109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L., Boyd D., Amyot W. M., Hempstead A. D., Luo Z. Q., O'Connor T. J., et al. (2011). The E Block motif is associated with Legionella pneumophila translocated substrates. Cell. Microbiol. 13, 227–245. 10.1111/j.1462-5822.2010.01531.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung V., Udeshi N. D., Lam S. S., Loh K. H., Cox K. J., Pedram K., et al. (2016). Spatially resolved proteomic mapping in living cells with the engineered peroxidase APEX2. Nat. Protoc. 11, 456–475. 10.1038/nprot.2016.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husmann L. K., Johnson W. (1992). Adherence of Legionella pneumophila to guinea pig peritoneal macrophages, J774 mouse macrophages, and undifferentiated U937 human monocytes: role of Fc and complement receptors. Infect. Immun. 60, 5212–5218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaac D. T., Laguna R. K., Valtz N., Isberg R. R. (2015). MavN is a Legionella pneumophila vacuole-associated protein required for efficient iron acquisition during intracellular growth. Proc. Natl. Acad. Sci. U.S.A. 112, E5208–E5217. 10.1073/pnas.1511389112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov S. S., Charron G., Hang H. C., Roy C. R. (2010). Lipidation by the host prenyltransferase machinery facilitates membrane localization of Legionella pneumophila effector proteins. J. Biol. Chem. 285, 34686–34698. 10.1074/jbc.M110.170746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov S. S., Roy C. R. (2013). Pathogen signatures activate a ubiquitination pathway that modulates the function of the metabolic checkpoint kinase mTOR. Nat. Immunol. 14, 1219–1228. 10.1038/ni.2740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jäger J., Marwitz S., Tiefenau J., Rasch J., Shevchuk O., Kugler C., et al. (2014). Human lung tissue explants reveal novel interactions during Legionella pneumophila infections. Infect. Immun. 82, 275–285. 10.1128/IAI.00703-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jank T., Böhmer K. E., Tzivelekidis T., Schwan C., Belyi Y., Aktories K. (2012). Domain organization of Legionella effector SetA. Cell. Microbiol. 14, 852–868. 10.1111/j.1462-5822.2012.01761.x [DOI] [PubMed] [Google Scholar]
- Jeong K. C., Ghosal D., Chang Y.-W., Jensen G. J., Vogel J. P. (2017). Polar delivery of Legionella type IV secretion system substrates is essential for virulence. Proc. Natl. Acad. Sci. U.S.A. 114, 8077–8082. 10.1073/pnas.1621438114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong K. C., Sutherland M. C., Vogel J. P. (2015). Novel export control of a Legionella Dot/Icm substrate is mediated by dual, independent signal sequences. Mol. Microbiol. 96, 175–188. 10.1111/mmi.12928 [DOI] [PubMed] [Google Scholar]
- Kagan J. C., Roy C. R. (2002). Legionella phagosomes intercept vesicular traffic from endoplasmic reticulum exit sites. Nat. Cell Biol. 4, 945–954. 10.1038/ncb883 [DOI] [PubMed] [Google Scholar]
- Klöpper T. H., Kienle N., Fasshauer D., Munro S. (2012). Untangling the evolution of Rab G proteins: implications of a comprehensive genomic analysis. BMC Biol. 10:71. 10.1186/1741-7007-10-71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotewicz K. M., Ramabhadran V., Sjoblom N., Behringer J., Scheck R. A., Isberg R. R., et al. (2017). A single Legionella effector catalyzes a multistep ubiquitination pathway to rearrange tubular endoplasmic reticulum for replication. Cell Host Microbe 21, 169–181. 10.1016/j.chom.2016.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause K., Amer A. O. (2016). Caspase exploitation by Legionella pneumophila. Front. Microbiol. 7, 1–10. 10.3389/fmicb.2016.00515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubori T., Hyakutake A., Nagai H. (2008). Legionella translocates an E3 ubiquitin ligase that has multiple U-boxes with distinct functions. Mol. Microbiol. 67, 1307–1319. 10.1111/j.1365-2958.2008.06124.x [DOI] [PubMed] [Google Scholar]
- Kubori T., Shinzawa N., Kanuka H., Nagai H. (2010). Legionella metaeffector exploits host proteasome to temporally regulate cognate effector. PLoS Pathog. 6, 1–8. 10.1371/journal.ppat.1001216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lifshitz Z., Burstein D., Peeri M., Zusman T., Schwartz K., Shuman H. A., et al. (2013). Computational modeling and experimental validation of the Legionella and Coxiella virulence-related type-IVB secretion signal. Proc. Natl. Acad. Sci. U.S.A. 110, E707–E715. 10.1073/pnas.1215278110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y. H., Doms A. G., Cheng E., Kim B., Evans T. R., Machner M. P. (2015). Host cell-catalyzed S-palmitoylation mediates Golgi targeting of the Legionella ubiquitin ligase GobX. J. Biol. Chem. 290, 25766–25781. 10.1074/jbc.M115.637397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liss V., Barlag B., Nietschke M., Hensel M. (2016). Self-labelling enzymes as universal tags for fluorescence microscopy, super-resolution microscopy and electron microscopy. Sci. Rep. 5:17740. 10.1038/srep17740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Zhu W., Tan Y., Nakayasu E. S., Staiger C. J., Luo Z.-Q. (2017). A Legionella effector disrupts host cytoskeletal structure by cleaving actin. PLOS Pathog. 13:e1006186. 10.1371/journal.ppat.1006186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomma M., Dervins-Ravault D., Rolando M., Nora T., Newton H. J., Sansom F. M., et al. (2010). The Legionella pneumophila F-box protein Lpp2082 (AnkB) modulates ubiquitination of the host protein parvin B and promotes intracellular replication. Cell. Microbiol. 12, 1272–1291. 10.1111/j.1462-5822.2010.01467.x [DOI] [PubMed] [Google Scholar]
- Losick V. P., Haenssler E., Moy M., Isberg R. R. (2010). LnaB: a Legionella pneumophila activator of NF-kB. Cell. Microbiol. 12, 1083–1097. 10.1111/j.1462-5822.2010.01452.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C., Liu K., Tan L. P., Yao S. Q. (2012). Current chemical biology tools for studying protein phosphorylation and dephosphorylation. Chemistry 18, 28–39. 10.1002/chem.201103206 [DOI] [PubMed] [Google Scholar]
- Luo Z.-Q., Isberg R. R. (2004). Multiple substrates of the Legionella pneumophila Dot/Icm system identified by interbacterial protein transfer. Proc. Natl. Acad. Sci. U.S.A. 101, 841–846. 10.1073/pnas.0304916101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machner M. P., Isberg R. R. (2006). Targeting of host Rab GTPase function by the intravacuolar pathogen Legionella pneumophila. Dev. Cell 11, 47–56. 10.1016/j.devcel.2006.05.013 [DOI] [PubMed] [Google Scholar]
- Marra A., Horwitz M. A., Shuman H. A. (1990). The HL-60 model for the interaction of human macrophages with the Legionnaires' disease bacterium. J. Immunol. 144, 2738–2744. [PubMed] [Google Scholar]
- Matsunaga K., Klein T. W., Friedman H., Yamamoto Y. (2001). Alveolar macrophage cell line MH-S is valuable as an in vitro model for Legionella pneumophila infection. Am. J. Respir. Cell Mol. Biol. 24, 326–331. 10.1165/ajrcmb.24.3.4359 [DOI] [PubMed] [Google Scholar]
- McCusker K. T., Braaten B. A., Cho M. W., Low D. A. (1991). Legionella pneumophila inhibits protein synthesis in Chinese hamster ovary cells. Infect. Immun. 59, 240–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDade J. E., Shepard C. C., Fraser D. W., Tsai T. R., Redus M. A., Dowdle W. R. (1977). Legionnaires' disease: isolation of a bacterium and demonstration of its role in other respiratory disease. N. Engl. J. Med. 297, 1197–1203. 10.1056/NEJM197712012972202 [DOI] [PubMed] [Google Scholar]
- Merriam J. J., Mathur R., Maxfield-Boumil R., Isberg R. R. (1997). Analysis of the Legionella pneumophila fliI gene: intracellular growth of a defined mutant defective for flagellum biosynthesis. Infect. Immun. 65, 2497–2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer D. F., Noroy C., Moumène A., Raffaele S., Albina E., Vachiéry N. (2013). Searching algorithm for type IV secretion system effectors 1.0: a tool for predicting type IV effectors and exploring their genomic context. Nucleic Acids Res. 41, 9218–9229. 10.1093/nar/gkt718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michard C., Doublet P. (2015). Post-translational modifications are key players of the Legionella pneumophila infection strategy. Front. Microbiol. 6:00087. 10.3389/fmicb.2015.00087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michard C., Sperandio D., Baïlo N., Pizarro-Cerdá J., LeClaire L., Chadeau-Argaud E., et al. (2015). The Legionella kinase Legk2 targets the Arp2/3 complex to inhibit actin nucleation on phagosomes and allow bacterial evasion of the late endocytic pathway. MBio 6, 1–14. 10.1128/mBio.00354-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffat J. F., Tompkins L. S. (1992). A quantitative model of intracellular growth of Legionella pneumophila in Acanthamoeba castellanii. Infect. Immun. 60, 296–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mousnier A., Schroeder G. N., Stoneham C. A., So E. C., Garnett J. A., Yu L., et al. (2014). A new method to determine in vivo interactomes reveals binding of the Legionella pneumophila effector PieE to multiple Rab GTPases. MBio 5:e01148–14. 10.1128/mBio.01148-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee S., Liu X., Arasaki K., McDonough J., Galan J. E., Roy C. R. (2011). Modulation of Rab GTPase function by a protein phosphocholine transferase. Nature 477, 103–106. 10.1038/nature10335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai H., Cambronne E. D., Kagan J. C., Amor J. C., Kahn R. A., Roy C. R. (2005). A C-terminal translocation signal required for Dot/Icm-dependent delivery of the Legionella RalF protein to host cells. Proc. Natl. Acad. Sci. U.S.A. 102, 826–831. 10.1073/pnas.0406239101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai H., Kagan J. C., Zhu X., Kahn R. A., Roy C. R. (2002). A bacterial guanine nucleotide exchange factor activates ARF on Legionella phagosomes. Science 295, 679. 10.1126/science.1067025 [DOI] [PubMed] [Google Scholar]
- Naujoks J., Tabeling C., Dill B. D., Hoffmann C., Brown A. S., Kunze M., et al. (2016). IFNs modify the proteome of Legionella-containing vacuoles and restrict infection via IRG1-derived itaconic acid. PLoS Pathog. 12:e1005408. 10.1371/journal.ppat.1005408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neunuebel M. R., Chen Y., Gaspar A. H., Backlund P. S., Yergey A., Machner M. P., et al. (2011). De-AMPylation of the small GTPase Rab1 by the pathogen Legionella pneumophila. Science 333, 453–456. 10.1126/science.1207193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newsome A. L., Baker R. L., Miller R. D., Arnold R. R. (1985). Interactions between Naegleria fowleri and Legionella pneumophila. Infect. Immun. 50, 449–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connor T. J., Adepoju Y., Boyd D., Isberg R. R. (2011). Minimization of the Legionella pneumophila genome reveals chromosomal regions involved in host range expansion. Proc. Natl. Acad. Sci. U.S.A. 108, 14733–14740. 10.1073/pnas.1111678108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connor T. J., Boyd D., Dorer M. S., Isberg R. R. (2012). Aggravating genetic interactions allow a solution to redundancy in a bacterial pathogen. Science 338, 1440–1444. 10.1126/science.1229556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connor T. J., Isberg R. R. (2014). iMAD, a genetic screening strategy for dissecting complex interactions between a pathogen and its host. Nat. Protoc. 9, 1916–1930. 10.1038/nprot.2014.133 [DOI] [PubMed] [Google Scholar]
- Ott M. (1994). Genetic approaches to study Legionella pneumophila pathogenicity. FEMS Microbiol. Rev. 14, 161–176. 10.1111/j.1574-6976.1994.tb00085.x [DOI] [PubMed] [Google Scholar]
- Pearlman E., Jiwa A. H., Engleberg N. C., Eisenstein B. I. (1988). Growth of Legionella pneumophila in a human macrophage-like (U937) cell line. Microb. Pathog. 5, 87–95. 10.1016/0882-4010(88)90011-3 [DOI] [PubMed] [Google Scholar]
- Popa C., Coll N. S., Valls M., Sessa G. (2016). Yeast as a heterologous model system to uncover Type III effector function. PLoS Pathog. 12:e1005360. 10.1371/journal.ppat.1005360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pope C. D., Dhand L., Cianciotto N. P. (1994). Random mutagenesis of Legionella pneumophila with mini-Tn10. FEMS Microbiol. Lett. 124, 107–111. 10.1111/j.1574-6968.1994.tb07269.x [DOI] [PubMed] [Google Scholar]
- Price C. T., Al-Khodor S., Al-Quadan T., Santic M., Habyarimana F., Kalia A., et al. (2009). Molecular mimicry by an F-box effector of Legionella pneumophila hijacks a conserved polyubiquitination machinery within macrophages and protozoa. PLoS Pathog. 5:e1000704. 10.1371/journal.ppat.1000704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu J., Luo Z. (2017). Legionella and Coxiella effectors: strength in diversity and activity. Nat. Rev. Micro. 15, 591–605. 10.1038/nrmicro.2017.67 [DOI] [PubMed] [Google Scholar]
- Qiu J., Sheedlo M. J., Yu K., Tan Y., Nakayasu E. S., Das C., et al. (2016). Ubiquitination independent of E1 and E2 enzymes by bacterial effectors. Nature 533, 120–124. 10.1038/nature17657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolando M., Sanulli S., Rusniok C., Gomez-Valero L., Bertholet C., Sahr T., et al. (2013). Legionella pneumophila effector RomA uniquely modifies host chromatin to repress gene expression and promote intracellular bacterial replication. Cell Host Microbe 13, 395–405. 10.1016/j.chom.2013.03.004 [DOI] [PubMed] [Google Scholar]
- Rothmeier E., Pfaffinger G., Hoffmann C., Harrison C. F., Grabmayr H., Repnik U., et al. (2013). Activation of Ran GTPase by a Legionella effector promotes microtubule polymerization, pathogen vacuole motility and infection. PLoS Pathog. 9:e1003598. 10.1371/journal.ppat.1003598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux K. J., Kim D. I., Raida M., Burke B. (2012). A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. J. Cell Biol. 196, 801–810. 10.1083/jcb.201112098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowbotham T. J. (1980). Preliminary report on the pathogenicity of Legionella pneumophila for freshwater and soil amoebae. J. Clin. Pathol. 33, 1179–1183. 10.1136/jcp.33.12.1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rucks E. A., Olson M. G., Jorgenson L. M., Srinivasan R. R., Ouellette S. P. (2017). Development of a proximity labeling system to map the Chlamydia trachomatis inclusion membrane. Front. Cell. Infect. Microbiol. 7:40. 10.3389/fcimb.2017.00040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder G. N., Aurass P., Oates C. V., Tate E. W., Hartland E. L., Flieger A., et al. (2015). Legionella pneumophila effector LpdA Is a palmitoylated phospholipase D virulence factor. Infect. Immun. 83, 3989–4002. 10.1128/IAI.00785-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder G. N., Petty N. K., Mousnier A., Harding C. R., Vogrin A. J., Wee B., et al. (2010). Legionella pneumophila strain 130b possesses a unique combination of type IV secretion systems and novel Dot/Icm secretion system effector proteins. J. Bacteriol. 192, 6001–6016. 10.1128/JB.00778-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal G., Purcell M., Shuman H. A. (1998). Host cell killing and bacterial conjugation require overlapping sets of genes within a 22-kb region of the Legionella pneumophila genome. Proc. Natl. Acad. Sci. U.S.A. 95, 1669–1674. 10.1073/pnas.95.4.1669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shohdy N., Efe J. A., Emr S. D., Shuman H. a. (2005). Pathogen effector protein screening in yeast identifies Legionella factors that interfere with membrane trafficking. Proc. Natl. Acad. Sci. U.S.A. 102, 4866–4871. 10.1073/pnas.0501315102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- So E. C., Mattheis C., Tate E. W., Frankel G., Schroeder G. N. (2015). Creating a customized intracellular niche: subversion of host cell signaling by Legionella type IV secretion system effectors. Can. J. Microbiol. 635, 617–635. 10.1139/cjm-2015-0166 [DOI] [PubMed] [Google Scholar]
- So E. C., Schroeder G. N., Carson D., Mattheis C., Mousnier A., Broncel M., et al. (2016). The Rab-binding profiles of bacterial virulence factors during infection. J. Biol. Chem. 291, 5832–5843. 10.1074/jbc.M115.700930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon J. M., Isberg R. R. (2000). Growth of Legionella pneumophila in Dictyostelium discoideum: a novel system for genetic analysis of host-pathogen interactions. Trends Microbiol. 8, 478–480. 10.1016/S0966-842X(00)01852-7 [DOI] [PubMed] [Google Scholar]
- Sopko R., Huang D., Preston N., Chua G., Papp B., Kafadar K., et al. (2006). Mapping pathways and phenotypes by systematic gene overexpression. Mol. Cell 21, 319–330. 10.1016/j.molcel.2005.12.011 [DOI] [PubMed] [Google Scholar]
- Sun E. W., Wagner M. L., Maize A., Kemler D., Garland-Kuntz E., Xu L., et al. (2013). Legionella pneumophila infection of Drosophila S2 cells induces only minor changes in mitochondrial dynamics. PLoS ONE 8:e62972. 10.1371/journal.pone.0062972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Y., Arnold R. J., Luo Z.-Q. (2011). Legionella pneumophila regulates the small GTPase Rab1 activity by reversible phosphorylcholination. Proc. Natl. Acad. Sci. U.S.A. 108, 21212–21217. 10.1073/pnas.1114023109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Y., Luo Z.-Q. (2011). Legionella pneumophila SidD is a deAMPylase that modifies Rab1. Nature 475, 506–509. 10.1038/nature10307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiaden A. N., Kessler A., Hilbi H. (2013). Analysis of Legionella infection by flow cytometry. Methods Mol. Biol. 954, 233–249. 10.1007/978-1-62703-161-5_14 [DOI] [PubMed] [Google Scholar]
- Toulabi L., Wu X., Cheng Y., Mao Y. (2013). Identification and structural characterization of a Legionella phosphoinositide phosphatase. J. Biol. Chem. 288, 24518–24527. 10.1074/jbc.M113.474239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbanus M. L., Quaile A. T., Stogios P. J., Morar M., Rao C., Di R., et al. (2016). Diverse mechanisms of metaeffector activity in an intracellular bacterial pathogen, Legionella pneumophila. Mol. Syst. Biol. 12:893. 10.15252/msb.20167381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Engelenburg S. B., Palmer A. E. (2010). Imaging type-III secretion reveals dynamics and spatial segregation of Salmonella effectors. Nat. Methods 7, 325–330. 10.1038/nmeth.1437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanrheenen S. M., Luo Z. Q., O'Connor T., Isberg R. R. (2006). Members of a Legionella pneumophila family of proteins with ExoU (Phospholipase A) active sites are translocated to target cells. Infect. Immun. 74, 3597–3606. 10.1128/IAI.02060-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viner R., Chetrit D., Ehrlich M., Segal G. (2012). Identification of two Legionella pneumophila effectors that manipulate host phospholipids biosynthesis. PLoS Pathog. 8:e1002988. 10.1371/journal.ppat.1002988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh N. C., Kenney L. L., Jangalwe S., Aryee K.-E., Greiner D. L., Brehm M. A., et al. (2017). Humanized mouse models of clinical disease. Annu. Rev. Pathol. Mech. Dis. 12, 187–215. 10.1146/annurev-pathol-052016-100332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson J. D., Laskowski R. A., Thornton J. M. (2005). Predicting protein function from sequence and structural data. Curr. Opin. Struct. Biol. 15, 275–284. 10.1016/j.sbi.2005.04.003 [DOI] [PubMed] [Google Scholar]
- Weber S. S., Ragaz C., Reus K., Nyfeler Y., Hilbi H. (2006). Legionella pneumophila exploits PI(4)P to anchor secreted effector proteins to the replicative vacuole. PLoS Pathog. 2:e46. 10.1371/journal.ppat.0020046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willems L. I., Overkleeft H. S., Van Kasteren S. I. (2014). Current developments in activity-based protein profiling. Bioconjug. Chem. 25, 1181–1191. 10.1021/bc500208y [DOI] [PubMed] [Google Scholar]
- Wölke S., Ackermann N., Heesemann J. (2011). The Yersinia enterocolitica type 3 secretion system (T3SS) as toolbox for studying the cell biological effects of bacterial Rho GTPase modulating T3SS effector proteins. Cell. Microbiol. 13, 1339–1357. 10.1111/j.1462-5822.2011.01623.x [DOI] [PubMed] [Google Scholar]
- Yu X., Decker K. B., Barker K., Neunuebel M. R., Saul J., Westcott N., et al. (2015). Host-Pathogen interaction profiling using self-assembling human protein arrays. J. Proteome Res. 14, 1920–1936. 10.1021/pr5013015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W., Banga S., Tan Y., Zheng C., Stephenson R., Gately J., et al. (2011). Comprehensive identification of protein substrates of the Dot/Icm type IV transporter of Legionella pneumophila. PLoS ONE 6:e17638. 10.1371/journal.pone.0017638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W., Hammad L. A., Hsu F., Mao Y., Luo Z.-Q. (2013). Induction of caspase 3 activation by multiple Legionella pneumophila Dot/Icm substrates. Cell. Microbiol. 15, 1783–1795. 10.1111/cmi.12157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou L., Nan C., Hu F., Hancock J. (2013). Accurate prediction of bacterial type IV secreted effectors using amino acid composition and PSSM profiles. Bioinformatics 29, 3135–3142. 10.1093/bioinformatics/btt554 [DOI] [PMC free article] [PubMed] [Google Scholar]