Abstract
Background and Aim
Microscopic colitis (MC) has been associated with increased paracellular permeability. Therefore, we aimed to investigate potential associations between MC and several genes encoding tight junction (TJ) proteins reported to interact with each other.
Methods
The association between MC and single nucleotide polymorphisms (SNP; n = 63) within TJ genes (F11R, MAGI1, MAGI2, MAGI3, PARD3, PTEN, and TJP1) were investigated in a case–control study (n MC patients = 104 and n controls = 423). The genes that exhibited an association with MC were further investigated for gene expression related to genotype, MC phenotype, and gender using colonic biopsies from MC patients (n = 25) and controls (n = 58).
Results
Based on the number of investigated genes and after correction for multiple testing, an association was detected between a SNP marker in PTEN (rs1234224) and both MC overall (OR = 1.70, 95% CI 1.23–2.34, p = 0.001) and collagenous colitis (CC; OR = 1.79, 95% CI 1.22–2.62, p = 0.003). Further, SNP markers in MAGI1 (rs17417230) and F11R (rs790055) were associated with MC overall (OR = 1.58, 95% CI 1.14–2.19, p = 0.006) and with CC (OR = 2.58, 95% CI 1.27–5.25, p = 0.007), respectively. However, none of the associated SNPs contributed markedly to the expression of the respective genes. Nonetheless, decreased MAGI1 (p = 3.47 × 10−4) and PTEN (p = 0.004) expression was associated with lymphocytic colitis (LC) and CC, respectively, compared to controls.
Conclusions
Decreased expression of PTEN and MAGI1 in the colonic mucosa might contribute to the pathogenesis of MC and its sub-phenotypes. Furthermore, our study indicates that genetic variants of TJ components are predisposing factors in the etiology of MC. Finally, F11R, MAGI1, and PTEN are new candidate genes that exhibit an association with MC.
Electronic supplementary material
The online version of this article (10.1007/s10620-017-4857-7) contains supplementary material, which is available to authorized users.
Keywords: Microscopic colitis, Genetic predisposition, Genotype, Gene expression, Single nucleotide polymorphism, Tight junctions
Introduction
Microscopic colitis (MC), including lymphocytic colitis (LC) and collagenous colitis (CC), is a chronic inflammation of the colon, that is characterized by chronic bloodless and watery diarrhea. The mucosal appearance at endoscopy is normal, or near normal, but has typical histopathological abnormalities [1–4]. CC is characterized by a thickened sub-epithelial collagen layer and an increased number of lymphocytes, whereas LC is associated with an increased number of intraepithelial lymphocytes [3–7]. The etiologies of LC and CC are multifactorial and largely unknown, but factors including female gender, increasing age, smoking habits, autoimmune disorders, genetic susceptibility, and environmental factors are implicated [2, 8–10]. Further, MC is associated with increased intestinal permeability as measured using 51CrEDTA and horseradish peroxidase [11] or by performing impedance spectroscopy [12]. Paracellular permeability across the paracellular space is primarily regulated by the tight junction (TJ) structures [13]. In addition, decreased expression of TJ proteins, such as claudin-4 and occludin, has been observed in CC [14].
Genetic variation of the TJ-related gene MAGI2 is associated with Crohn’s disease (CD) [15], ulcerative colitis (UC) [15, 16], and celiac disease [16], and the TJ-related gene PARD3 with celiac disease [16]. In a previous study on IBD [17], a network consisting of seven TJ-related genes [see Fig. 1 in 17] were investigated since we noted that proteins encoded by MAGI2 and PARD3 interacted with proteins encoded by F11R (encoding JAM-A), MAGI1, MAGI3, PTEN, and TJP1 according to the STRING search tool [18], thus indicating that these proteins jointly contribute to a shared function. Here, we extend these observations to encompass MC. Additionally, we investigated the corresponding gene expressions in relation to genotype, MC phenotype, and gender.
Methods
Study Subjects
Swedish patients with an established diagnosis of MC (n total = 104, median age 65 years; 39 LC patients, 22 women and 65 CC patients, 51 women) and controls (n = 423, median age 59 years, 223 women) from an anonymized cohort consisting of randomly selected individuals living in the same geographic region were included in a case–control study. Controls suffering from gastrointestinal symptoms were not included in the investigation.
A second MC cohort (Table 1), from which both RNA from intestinal biopsies and DNA samples were available, was used to further explore significant findings from the case–control study.
Table 1.
Summary of the study participants and biopsy locations in the second Swedish MC cohort that was used to analyze the significant genetic associations found in the first cohort
| Disease and subgroupsa | Number of individuals/biopsies |
|---|---|
| MC (n = 25) a,b | |
| Ascending colon | 2 |
| Transverse colon | 19 |
| Left-sided colon | 4 |
| CC (n = 10) | |
| Ascending colon | 1 |
| Transverse colon | 6 |
| Left-sided colon | 3 |
| LC (n = 14) | |
| Ascending colon | 1 |
| Transverse colon | 12 |
| Left-sided colon | 1 |
| Non-MC controls (n = 58) c | |
| Ascending colon | 5 |
| Transverse colon | 36 |
| Left-sided colon | 17 |
aIncludes 10 CC patients (median 63, range 47–85, years of age; 10 women), 14 LC patients (median 62, range 49–88, years of age; 10 women), and one female patient with an undefined sub-phenotype (44.6 years of age)
bOne of the CC patients also had biopsy verified celiac disease
cTwo of the non-MC controls (n = 58; median age 51 years, range 18–84) had biopsy verified celiac disease
Colonic biopsies were obtained from 25 MC patients (Table 1) of which 19 (76%) underwent primary investigation for diarrhea; the other six already had an established diagnosis of MC. The patients under primary investigation came to colonoscopy because of ongoing non-bloody diarrhea, while patients with an established diagnosis had colonoscopy either as follow-up after medical treatment or due to an increase in bowel symptoms. There were 14 LC patients, 10 CC patients, and one female patient with an undefined sub-phenotype of MC. For the newly diagnosed cases, study biopsies were sampled at the same occasion as the diagnostic biopsies. In addition, colonic biopsies were obtained from 58 controls (Table 1), who underwent endoscopy in the diagnostic workup of suspected gastrointestinal disorders, mainly suspicion of other inflammatory bowel diseases or cancer. The biopsy specimens were primarily obtained from the transverse colon. When samples from this segment were not available, other colonic segments were used, with a preference for specimens obtained from the ascending colon. One biopsy per individual was selected and categorized as LC, CC, or as a non-inflamed control based on histopathological assessment. The phenotype was based on routine histopathological assessment where the established diagnostic criteria according to Langner et al. [3] were applied. The key histological features are thickened sub-epithelial collagen band in CC (> 10 μm) and an increased number of intraepithelial lymphocytes in LC (> 20 per 100 epithelial cells). All control biopsies were devoid of inflammatory cellular infiltrate and collagen deposition.
Single Nucleotide Polymorphism Selection for Genetic Association Studies
TJ-related genes (F11R, MAGI1, MAGI2, MAGI3, PARD3, PTEN, and TJP1) encoding products interacting with each other [18–20] were investigated. All SNP markers are given in Supplementary Table 1.
SNP markers (minor allele frequency ≥ 10%, pairwise r 2 ≥ 0.8) of F11R, MAGI1, MAGI3, and PTEN were selected using SNPbrowser Software version 4.0 (Applied Biosystems, Foster City, CA, USA), as previously described [17].
Genotyping and Gene Expression Analysis
The DNA isolation and the allelic discrimination (Supplementary Table 1) was performed as previously described [17].
Genes exhibiting an association with MC overall, CC, or LC were further investigated by gene expression analysis in relation to genotype, MC phenotype, and gender. The biopsies were submerged in RNAlater (Qiagen, Düsseldorf, Germany) and stored at either + 4 °C overnight and afterward at − 20 °C, or at room temperature for 1 h and then at − 80 °C until extraction. The different storage temperatures did not affect the expression of genes, neither in biopsies from the MC patients nor the controls (data not shown). RNA purification was performed as previously described [17].
Gene expression of F11R (Hs00170991_m1), MAGI1 Hs00191026_m1, and PTEN Hs02621230_s1) was analyzed using TaqMan Gene Expression Assay (Applied Biosystems), TaqMan Universal Mastermix (Applied Biosystems), and a 7500 Fast Real-Time PCR system (Applied Biosystems). Each individual reaction contained 10 ng cDNA in a total reaction volume of 20 µL. Threshold cycle (C T) values were established (ExpressionSuite Software version 1.0.3; Applied Biosystems) and normalized to the average of selected reference genes, as previously described [17].
Statistical Analysis
Allelic odds ratios (OR) and p values, based on the Chi-squared (χ 2) test, were calculated using the JMP Genomics 6.0 software (JMP Genomics 6.0; SAS Institute Inc., Cary, NC, USA). Deviation from Hardy–Weinberg equilibrium was tested separately in controls and MC patients using the exact test implemented in Haploview version 4.2 (https://www.broad.mit.edu/haploview). For the analysis of genetic associations, a Bonferroni adjusted p value < 0.007 (based on the number of genes analyzed; n = 7) was considered significant.
The expressions of genes exhibiting significant associations to MC, CC, or LC were further analyzed. Group differences in gene expression were investigated using Kruskal–Wallis analysis of variance (ANOVA) with Mann–Whitney U post hoc test using Statistica 12.7 (StatSoft Inc., Tulsa, OK, USA). Gene expressions in relation to genotype were investigated using Spearman’s rank correlation test, whereas gene expression in relation to MC phenotype and gender were analyzed using logistic regression (Statistica 12.7; StatSoft Inc.). Significant findings were further explored by multiple logistic regression (Statistica 12.7; StatSoft Inc.), where adjustment for possible confounders (gender and age) was performed one at a time. Correlations between gene expressions were determined using Spearman’s rank correlation (Statistica 12.7; StatSoft Inc.). Scatter plots were created for the relationship between gene expression and phenotype using Statistica 13.1 (StatSoft Inc.). All statistical analyses with respect to gene expression were performed using ΔC T values, and a Bonferroni adjusted p value < 0.017 (based on the number of genes analyzed; n = 3) was considered significant.
Ethical Considerations
The study was conducted under approval by the ethics committees of Linköping University (Dnr M35-07, Dnr 2011/201-31) and Karolinska Institutet (Dnr 2007/791-31/3).
Results
Genetic Associations
Seven out of the 63 selected SNP markers were excluded due to failed genotyping or the absence of Hardy–Weinberg equilibrium (Supplementary Table 1). The strongest association in the case–control study was observed between a PTEN SNP marker (rs1234224; susceptibility allele G) and both MC overall (OR = 1.70, 95% CI 1.23–2.34, p = 0.001) and CC (OR = 1.79, 95% CI 1.22–2.62, p = 0.003) (Supplementary Table 2). Additionally, a MAGI1 SNP marker (rs17417230; susceptibility allele C) was significantly associated with MC overall (OR = 1.58, 95% CI 1.14–2.19, p = 0.006), whereas a F11R marker (rs790055; susceptibility allele G) was significantly associated with CC (OR = 2.58, 95% CI 1.27–5.25, p = 0.007).
Gene Expression
No significant differences in the expression of F11R or MAGI1 were observed between biopsies from the ascending colon, transverse colon, or left-sided colon among controls. PTEN exhibited a slightly increased expression (Kruskal–Wallis ANOVA; p = 0.015) in the proximal colon (ascending colon and transverse colon) compared to the left-sided colon (Mann–Whitney U test; p = 0.006). Since no or only a slight difference in gene expression (1.1 fold on a linear scale) was observed, all biopsies were analyzed together, regardless sampling location.
Gene Expression in Relation to Genotype
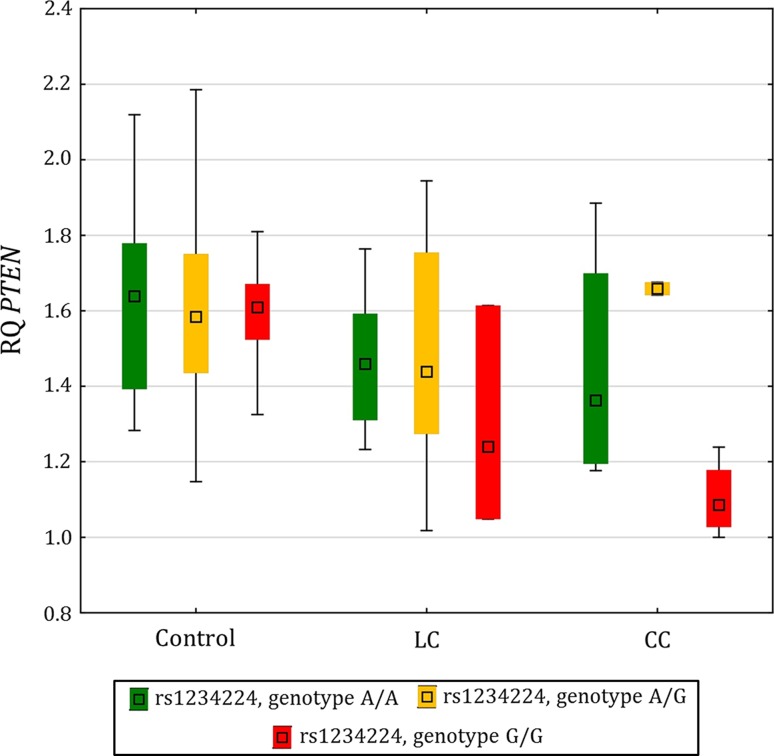
No significant correlations were observed between the susceptibility allele of the identified SNP markers and the expression of the corresponding genes (F11R, MAGI1, and PTEN; Table 2). However, a trend (p = 0.103) toward reduced PTEN expression in relation to the G/G susceptibility genotype was observed among CC patients (Fig. 1).
Table 2.
For genes with a significant genetic association with investigated phenotypes, gene expression (ΔC T values) was analyzed in relation to genotype in colonic biopsies from the different subgroups
| Gene | SNP marker | Non-MC controls (number of controls) | p value | MC patients (number of patients) | p value | CC patients (number of patients) | p value | LC patients (number of patients) | p value |
|---|---|---|---|---|---|---|---|---|---|
| F11R | rs790055 | AA (2), AG (16), GG (39) | 0.676 | AA (3), AG (8), GG (14) | 0.482 | AA (2), AG (3), GG (5) | 0.428 | AA (0), AG (5), GG (9) | 0.851 |
| MAGI1 | rs17417230 | AA (6), AC (33), CC (19) | 0.480 | AA (6), AC (13), CC (6) | 0.621 | AA (2), AC (4), CC (4) | 0.590 | AA (4), AC (9), CC (1) | 0.825 |
| PTEN | rs1234224 | AA (21), AG (31), GG (6) | 0.978 | AA (10), AG (8), GG (7) | 0.081 | AA (4), AG (2), GG (4) | 0.103 | AA (5), AG (6), GG (3) | 0.535 |
Fig. 1.
Box pot of PTEN expression in the colonic mucosa from non-MC controls, LC patients, and CC patients stratified by the rs1234224 genotype
Gene Expression in Relation to Phenotype
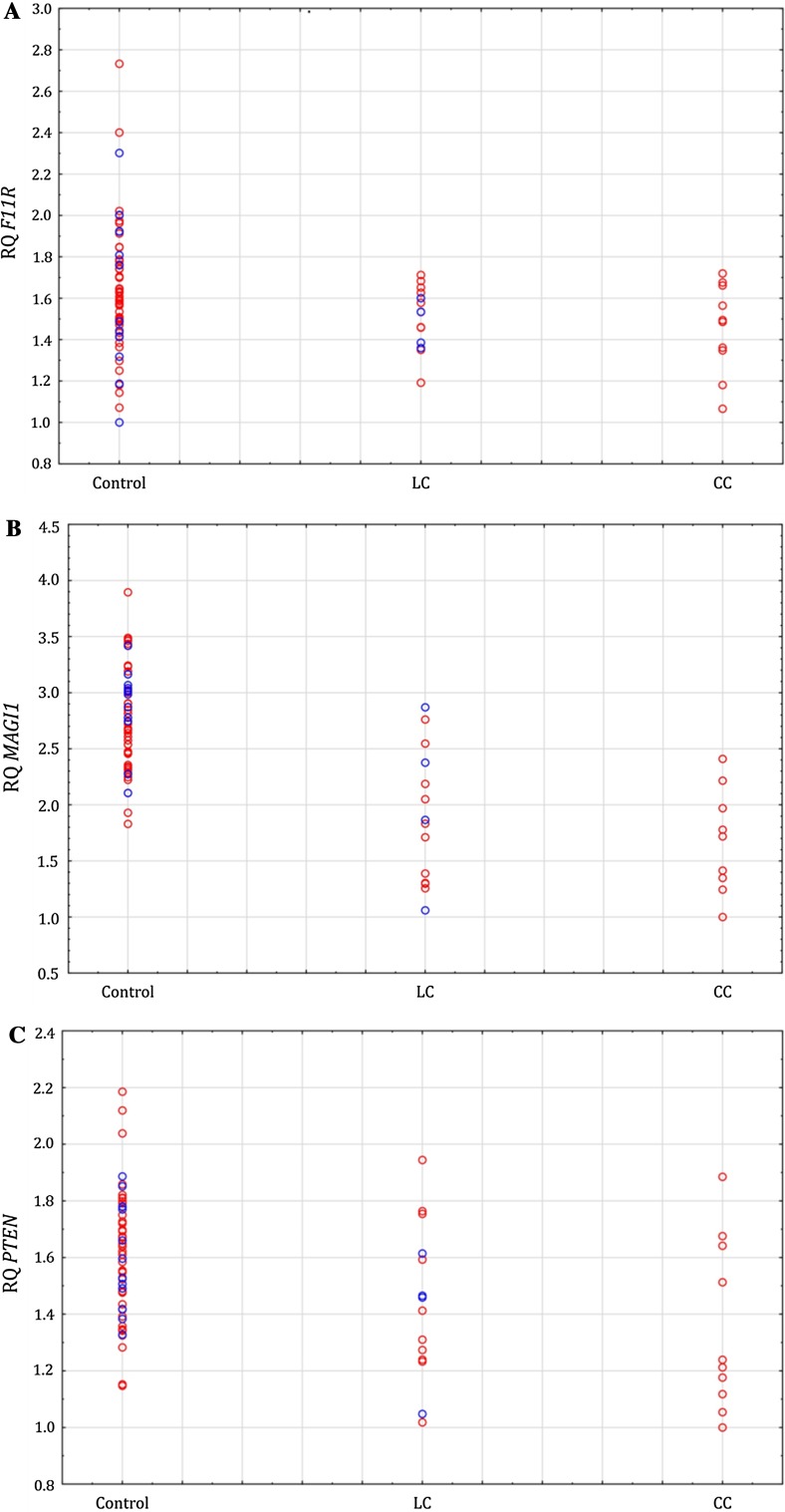
Significant associations were observed between decreased MAGI1 expression and MC overall (p = 2.58 × 10−5) as well as in CC (p = 0.002) and LC (p = 3.47 × 10−4) compared to controls (Table 3, Fig. 2). Furthermore, significant associations were observed between decreased PTEN expression and both MC overall (p = 0.002) and CC (p = 0.004) compared to controls and with a similar trend for LC (p = 0.018). Significant findings, in relation to MC, were confirmed using multiple logistic regression and adjustment, one at a time, for possible confounders (gender and age) (data not shown).
Table 3.
Gene expression (ΔC T values) was analyzed in relation to phenotype using logistic regression
| Single logistic regression | ||||
|---|---|---|---|---|
| p value | Estimate | Estimate (95% CI) | Nagelkerke R 2 | |
| MC versus non-MC controls | ||||
| F11R a | 0.042 | 2.14 | 0.08 to 4.21 | 0.08 |
| MAGI1 b | 2.58 × 10 −5 | 6.54 | 3.49 to 9.59 | 0.64 |
| PTEN c | 0.002 | 3.61 | 1.37 to 5.86 | 0.19 |
| CC versus non-MC controls | ||||
| F11R d | 0.084 | 2.34 | − 0.32 to 4.99 | 0.08 |
| MAGI1 e | 0.002 | 9.02 | 3.33 to 14.72 | 0.74 |
| PTEN f | 0.004 | 4.99 | 1.63 to 8.35 | 0.25 |
| LC versus non-MC controls | ||||
| F11R g | 0.181 | 1.66 | − 0.77 to 4.08 | 0.04 |
| MAGI1 h | 3.47 × 10 −4 | 5.62 | 2.54 to 8.70 | 0.53 |
| PTEN i | 0.018 | 3.44 | 0.59 to 6.30 | 0.13 |
| LC versus CC | ||||
| F11R | 0.422 | − 1.90 | − 6.54 to 2.74 | 0.04 |
| MAGI1 | 0.323 | − 1.00 | − 2.98 to 0.98 | 0.06 |
| PTEN | 0.419 | − 1.21 | − 4.14 to 1.72 | 0.04 |
ΔC T values are inversely related to gene expression values. The estimates represent the natural logarithm of the odds ratio with a negative value corresponding to increased odds, while a positive value corresponds to decreased odds. Significant values (p < 0.017) are marked in bold
a n MC = 25; n non-MC = 57
b,c n MC = 25; n non-MC = 58
d n CC = 10; n non-MC = 57
e,f n CC = 10; n non-MC = 58
g,h,i n CC = 14; n non-MC = 58
Fig. 2.
Colonic mucosal expression of F11R (a), MAGI1 (b), and PTEN (c) in relation to phenotype and further stratified by gender (women are illustrated with open red circles and men with open blue circles)
The expression of MAGI1 and PTEN were positively correlated among the MC patients (p = 4.10 × 10−4, r s = 0.65), but not among the controls (p = 0.358, r s = 0.12).
Gene Expression in Relation to Gender
No significant associations were observed between gene expression (F11R, MAGI1, and PTEN) and gender among MC patients or controls (Table 4, Fig. 2).
Table 4.
Gene expression (ΔC T values) was analyzed in relation to gender using single logistic regression
| p value | Estimates | Estimate (95% CI) | Nagelkerke R 2 | |
|---|---|---|---|---|
| Non-MC controls | ||||
| F11R a | 0.751 | − 0.38 | − 2.74 to 1.98 | < 0.01 |
| MAGI1 b | 0.218 | 1.82 | − 1.08 to 4.72 | 0.04 |
| PTEN c | 0.711 | 0.60 | − 2.58 to 3.78 | < 0.01 |
| MC patients | ||||
| F11R d | 0.898 | − 0.39 | − 6.38 to 5.60 | < 0.01 |
| MAGI1 e | 0.453 | 1.02 | − 1.64 to 3.69 | 0.04 |
| PTEN f | 0.979 | − 0.05 | − 3.84 to 3.74 | < 0.01 |
ΔC T values are inversely related to gene expression values. The estimates represent the natural logarithm of the odds ratio with a negative value corresponding to increased odds, while a positive value corresponds to decreased odds
a n female = 45; n male = 12
b,c n female = 45; n male = 13
d,e,f n female = 21; n male = 4
Discussion
Microscopic colitis constitutes multifactorial conditions with mainly watery diarrhea, chronic inflammation of the colon, and an enhanced intestinal permeability [11, 12]. The etiology is largely unknown, but a genetic predisposition might exist [10]. Because of the accompanying increased paracellular permeability, we aimed to investigate the genetic associations between MC and genes that encode tight junction proteins.
The strongest associations were observed between a SNP marker in PTEN (rs1234224) and both MC and CC, and a decreased expression of PTEN was apparent in both MC and CC, compared to controls. Recently, we described significantly lower level of PTEN expression in biopsies from inflamed IBD mucosa, compared to non-inflamed IBD mucosa [17]. PTEN is involved in the regulation of fibroblast viability [21], and an overexpression of PTEN inhibits lipopolysaccharide-induced fibroblast proliferation, differentiation, and collagen secretion in mice [22]. Decreased PTEN expression is associated with pulmonary fibrosis that is characterized by collagen secretion and the activation and proliferation of fibroblasts [23]. In parallel, CC is associated with a distinctive, thickened sub-epithelial collagen layer [5], which might be a consequence of the decreased colonic PTEN expression observed in our study.
A SNP marker in MAGI1 (rs17417230) was associated with MC. Compared to controls, decreased MAGI1 expression was associated with MC overall, CC, and LC. The expression of MAGI1 and PTEN was positively correlated to each other, which is in accord with observations of the recruitment of PTEN by MAGI-1b at adherens junctions [24]. Similar to MC, inflamed IBD mucosa expressed lower levels of MAGI1 and PTEN compared to both non-inflamed IBD as well as controls. Compared to the MC mucosa, the expression levels of MAGI1 and PTEN were, however, further reduced in inflamed IBD mucosa (data not shown). Based on these expression levels, there are both similarities and differences between MC and IBD.
A SNP marker in F11R was significantly associated with CC, whereas a marginally significant association was observed between decreased F11R expression and MC overall. Aside from the thickened sub-epithelial collagen layer, CC is also associated with increased mononuclear inflammation in the lamina propria [3]. Further, reduced JAM-A (encoded by F11R) expression is associated with increased infiltration of polymorphonuclear leukocytes (PMN) to the colonic mucosa in patients with colitis [25]. An increased PMN infiltration and lymphoid aggregates are observed in the colonic mucosa from JAM-A−/− mice [26]. Finally, reduced JAM-A expression in the colonic mucosa from patients with inflammatory bowel disease has also been observed [27]. It therefore seems plausible that the regulation of F11R is relevant for the etiology of CC.
None of the associated SNP markers contributed significantly to the expression of their respective genes in MC patients or in controls, i.e, the study was unable to demonstrate an allelic effect on gene expression using the current sample size and/or analyzing whole intestinal mucosa biopsies (representing a heterogeneous collection of cell types). Nevertheless, a nonsignificant decrease in PTEN expression among MC and CC patients was observed in relation to homozygosity for the PTEN SNP marker susceptibility allele (rs1234224, susceptibility allele G; Table 2 and Fig. 1). The majority of hitherto identified risk alleles is noncoding and likely exerts their effects via regulation of gene expression, possibly in a tissue-/cell-type-dependent manner [28, 29]. We have previously established risk allele–gene expression relationships using a disease-relevant sample type (i.e, intestinal mucosa biopsies) and a similar sample size [30]. However, ultimately, success of identifying a risk allele–gene expression relationship using a heterogeneous biological sample will depend on factors such as effect size, disease status, medication, and cell type.
In conclusion, decreased PTEN and MAGI1 expression in the colonic mucosa might contribute to the pathogenesis of MC and its sub-phenotypes. Furthermore, our study indicates that genetic variants of a number of TJ components might be predisposing factors for MC. Similar to the so far identified risk alleles for CD and UC [29, 31–34], we observed small to modest effect sizes of the investigated SNP markers in relation to MC, however, with relatively wide confidence intervals. The identified SNP markers require further investigation in a larger, independent cohort, to both validate and more precisely determine their influence in relation to MC. Nonetheless, F11R, MAGI1, and PTEN are new candidate genes associated with MC.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Table 1: Single nucleotide polymorphism markers included in the study. (XLSX 16 kb)
Supplementary Table 2: Microscopic colitis phenotypes were investigated by using case–control approach in genetic association studies. (XLSX 26 kb)
Funding
This work was supported by FORSS, the Medical Research Council of South-Eastern Sweden [Grant Nos. 236541-2012 and 235131-2012], Futurum—the Academy for Healthcare, Region Jönköping County [Grant No. FUTURUM-338631], Bengt Ihre-Fonden [Grant No. 2012-SLS 254491], and Karolinska Institutets Forskningsfonder [Grant No. 2014fobi42063].
Author’s contribution
EN, SA, and JS conceived and designed the study; EN contributed to all laboratory work, performed the statistical analysis, and contributed to data analysis and drafted the manuscript; MRM and SA provided blood samples and patient data; all authors contributed to the context of the manuscript and approved the final manuscript.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no competing interests.
Footnotes
Sven Almer and Jan Söderman have shared senior authorship.
Electronic supplementary material
The online version of this article (10.1007/s10620-017-4857-7) contains supplementary material, which is available to authorized users.
Contributor Information
Elisabeth Norén, Email: elisabeth.noren@rjl.se.
Marie-Rose Mellander, Email: marie-rose.mellander@karolinska.se.
Sven Almer, Email: sven.almer@ki.se.
Jan Söderman, Email: jan.soderman@rjl.se.
References
- 1.Ingle SB, Adgaonkar BD, Ingle CR. Microscopic colitis: common cause of unexplained nonbloody diarrhea. World J Gastrointest Pathophysiol. 2014;5:48–53. doi: 10.4291/wjgp.v5.i1.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ohlsson B. New insights and challenges in microscopic colitis. Ther Adv Gastroenterol. 2015;8:37–47. doi: 10.1177/1756283X14550134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Langner C, Aust D, Ensari A, Villanacci V, Becheanu G, Miehlke S, et al. Histology of microscopic colitis-review with a practical approach for pathologists. Histopathology. 2015;66:613–626. doi: 10.1111/his.12592. [DOI] [PubMed] [Google Scholar]
- 4.Magro F, Langner C, Driessen A, Ensari A, Geboes K, Mantzaris GJ, et al. European consensus on the histopathology of inflammatory bowel disease. J Crohn’s Colitis. 2013;7:827–851. doi: 10.1016/j.crohns.2013.06.001. [DOI] [PubMed] [Google Scholar]
- 5.Brown WR, Tayal S. Microscopic colitis. A review. J Dig Dis. 2013;14:277–281. doi: 10.1111/1751-2980.12046. [DOI] [PubMed] [Google Scholar]
- 6.Pardi DS. Microscopic colitis. Clin Geriatr Med. 2014;30:55–65. doi: 10.1016/j.cger.2013.10.005. [DOI] [PubMed] [Google Scholar]
- 7.Chande N. Microscopic colitis: an approach to treatment. J Can Gastroenterol. 2008;22:686–688. doi: 10.1155/2008/671969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Farrukh A, Mayberry JF. Microscopic colitis: a review. Colorectal Dis Off J Assoc Coloproctol G B Irel. 2014;16:957–964. doi: 10.1111/codi.12716. [DOI] [PubMed] [Google Scholar]
- 9.Tong J, Zheng Q, Zhang C, Lo R, Shen J, Ran Z. Incidence, prevalence, and temporal trends of microscopic colitis: a systematic review and meta-analysis. Am J Gastroenterol. 2015;110:265–276. doi: 10.1038/ajg.2014.431. [DOI] [PubMed] [Google Scholar]
- 10.Westerlind H, Mellander MR, Bresso F, Munch A, Bonfiglio F, Assadi G, et al. Dense genotyping of immune-related loci identifies HLA variants associated with increased risk of collagenous colitis. Gut. 2017;66:421–428. doi: 10.1136/gutjnl-2015-309934. [DOI] [PubMed] [Google Scholar]
- 11.Munch A, Soderholm JD, Wallon C, Ost A, Olaison G, Strom M. Dynamics of mucosal permeability and inflammation in collagenous colitis before, during, and after loop ileostomy. Gut. 2005;54:1126–1128. doi: 10.1136/gut.2004.058750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barmeyer C, Erko I, Fromm A, Bojarski C, Allers K, Moos V, et al. Ion transport and barrier function are disturbed in microscopic colitis. Ann N Y Acad Sci. 2012;1258:143–148. doi: 10.1111/j.1749-6632.2012.06631.x. [DOI] [PubMed] [Google Scholar]
- 13.Vetrano S, Danese S. The role of JAM-A in inflammatory bowel disease: unrevealing the ties that bind. Ann N Y Acad Sci. 2009;1165:308–313. doi: 10.1111/j.1749-6632.2009.04045.x. [DOI] [PubMed] [Google Scholar]
- 14.Burgel N, Bojarski C, Mankertz J, Zeitz M, Fromm M, Schulzke JD. Mechanisms of diarrhea in collagenous colitis. Gastroenterology. 2002;123:433–443. doi: 10.1053/gast.2002.34784. [DOI] [PubMed] [Google Scholar]
- 15.McGovern DP, Taylor KD, Landers C, Derkowski C, Dutridge D, Dubinsky M, et al. MAGI2 genetic variation and inflammatory bowel disease. Inflamm Bowel Dis. 2009;15:75–83. doi: 10.1002/ibd.20611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wapenaar MC, Monsuur AJ, van Bodegraven AA, Weersma RK, Bevova MR, Linskens RK, et al. Associations with tight junction genes PARD3 and MAGI2 in Dutch patients point to a common barrier defect for coeliac disease and ulcerative colitis. Gut. 2008;57:463–467. doi: 10.1136/gut.2007.133132. [DOI] [PubMed] [Google Scholar]
- 17.Noren E, Almer S, Soderman J. Genetic variation and expression levels of tight junction genes identifies association between MAGI3 and inflammatory bowel disease. BMC Gastroenterol. 2017;17:68. doi: 10.1186/s12876-017-0620-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.STRING. http://string-db.org/. Accessed 16 Aug 2016.
- 19.KEGG PATHWAY Database. http://www.genome.jp/kegg/pathway.html. Accessed 19 Nov 2015.
- 20.Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, et al. STRING v10: protein–protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43:D447–D452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nho RS, Xia H, Diebold D, Kahm J, Kleidon J, White E, et al. PTEN regulates fibroblast elimination during collagen matrix contraction. J Biol Chem. 2006;281:33291–33301. doi: 10.1074/jbc.M606450200. [DOI] [PubMed] [Google Scholar]
- 22.He Z, Deng Y, Li W, Chen Y, Xing S, Zhao X, et al. Overexpression of PTEN suppresses lipopolysaccharide-induced lung fibroblast proliferation, differentiation and collagen secretion through inhibition of the PI3-K-Akt-GSK3beta pathway. Cell Biosci. 2014;4:2. doi: 10.1186/2045-3701-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.He Z, Gao Y, Deng Y, Li W, Chen Y, Xing S, et al. Lipopolysaccharide induces lung fibroblast proliferation through Toll-like receptor 4 signaling and the phosphoinositide3-kinase-Akt pathway. PLoS ONE. 2012;7:e35926. doi: 10.1371/journal.pone.0035926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kotelevets L, van Hengel J, Bruyneel E, Mareel M, van Roy F, Chastre E. Implication of the MAGI-1b/PTEN signalosome in stabilization of adherens junctions and suppression of invasiveness. FASEB J Off Publ Fed Am Soc Exp Biol. 2005;19:115–117. doi: 10.1096/fj.04-1942fje. [DOI] [PubMed] [Google Scholar]
- 25.Kucharzik T, Walsh SV, Chen J, Parkos CA, Nusrat A. Neutrophil transmigration in inflammatory bowel disease is associated with differential expression of epithelial intercellular junction proteins. Am J Pathol. 2001;159:2001–2009. doi: 10.1016/S0002-9440(10)63051-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Laukoetter MG, Nava P, Lee WY, Severson EA, Capaldo CT, Babbin BA, et al. JAM-A regulates permeability and inflammation in the intestine in vivo. J Exp Med. 2007;204:3067–3076. doi: 10.1084/jem.20071416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vetrano S, Rescigno M, Cera MR, Correale C, Rumio C, Doni A, et al. Unique role of junctional adhesion molecule—a in maintaining mucosal homeostasis in inflammatory bowel disease. Gastroenterology. 2008;135:173–184. doi: 10.1053/j.gastro.2008.04.002. [DOI] [PubMed] [Google Scholar]
- 28.Consortium GT Human genomics. The genotype-tissue expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348:648–660. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jostins L, Ripke S, Weersma RK, Duerr RH, McGovern DP, Hui KY, et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491:119–124. doi: 10.1038/nature11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Söderman J, Berglind L, Almer S. Gene expression-genotype analysis implicates GSDMA, GSDMB, and LRRC3C as contributors to inflammatory bowel disease susceptibility. Biomed Res Int. 2015;2015:834805. doi: 10.1155/2015/834805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Anderson CA, Boucher G, Lees CW, Franke A, D’Amato M, Taylor KD, et al. Meta-analysis identifies 29 additional ulcerative colitis risk loci, increasing the number of confirmed associations to 47. Nat Genet. 2011;43:246–252. doi: 10.1038/ng.764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Barrett JC, Hansoul S, Nicolae DL, Cho JH, Duerr RH, Rioux JD, et al. Genome-wide association defines more than 30 distinct susceptibility loci for Crohn’s disease. Nat Genet. 2008;40:955–962. doi: 10.1038/ng.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Franke A, McGovern DP, Barrett JC, Wang K, Radford-Smith GL, Ahmad T, et al. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn’s disease susceptibility loci. Nat Genet. 2010;42:1118–1125. doi: 10.1038/ng.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Control Wellcome Trust Case. C. Genome-wide association study of 14,000 cases of seven common diseases and 3000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1: Single nucleotide polymorphism markers included in the study. (XLSX 16 kb)
Supplementary Table 2: Microscopic colitis phenotypes were investigated by using case–control approach in genetic association studies. (XLSX 26 kb)