Abstract
To explore the pathologies of Kashin-Beck disease (KBD) and KBD accompanied with dental fluorosis (DF), we conducted a comparative analysis of gene expression profiles. 12 subjects were recruited, including 4 KBD patients, 4 patients with KBD and DF and 4 healthy subjects. Genome-wide expression profiles from their peripheral blood mononuclear cells were evaluated by customized oligonucleotide microarray. R programming software was used for the microarray data analysis followed by functional enrichment analysis through KOBAS. Several potential biomarkers were identified, and quantitative real-time reverse transcription–polymerase chain reaction (qRT-PCR) was used for their validation. In this study, 28 genes and 8 genes were found to be up- and down-regulated respectively in KBD patients compared with health subjects. In patients with KBD and DF, we obtained 10 up-regulated and 3 down-regulated genes compared with health controls. Strikingly, no differential expression gene (DEG) was identified between the two groups of patients. A total of 10 overlaps (DUSP2, KLRF1, SRP19, KLRC3, CD69, SIK1, ITGA4, ID3, HSPA1A, GPR18) were obtained between DEGs of patients with KBD and patients with KBD and DF. They play important roles in metabolism, differentiation, apoptosis and bone-development. The relative abundance of 8 DEGs, i.e. FCRL6, KLRC3, CXCR4, CD93, CLK1, GPR18, SRP19 and KLRF1, were further confirmed by qRT-PCR analysis.
Introduction
Kashin-Beck disease (KBD) is an endemic and chronic osteoarthropathy which mostly occurs in children between the ages of 3 and 12 in Asia1. The primary pathogenesis of KBD include chondrocyte degeneration, necrosis, progressive loss of articular cartilage and so on, which should result in arthrosis deformities in adult2,3. In China, over 30 million people are at risk of KBD. There have been three etiological hypotheses of KBD, biology-earth-chemistry hypothesis (selenium deficiency), water organic compound poisoning hypothesis, and food fungi toxin poisoning hypothesis, but the accurate etiology of KBD is still unknown4–6.
Li et al. showed that low fluorine in the living environment caused an internal metabolic disorder, leading to pathological change of KBD7. In addition, fluorosis and KBD are endemic diseases, which are closely associated with geology, geochemistry, hydrology and etc8. Dental fluorosis (DF) is an early manifestation of fluorosis, which is caused by the excessive input of fluoride. There are three types of endemic fluorosis, including drinking-water type, coal-burning type and drinking-tea type. It was reported that water type fluorosis is the most common type in China, especially DF in children9,10.
It has been reported that genetic factors play important roles in the pathogenesis of KBD and DF. Extensive genetic studies have been conducted to identify susceptibility genes for KBD and DF, and several valuable biomarkers have been obtained, such as HLA-DRB1, ITPR2, ADAM12, ameloblastin gene, and ER Rsa I. However, the mechanism of KBD and DF remains unclear. The variations of KBD and DF explained by the identified loci were limited, suggesting the existence of undiscovered genetic variants associated with KBD and DF.
The purpose of this study is to explore the common pathogenesis and the underlying molecular functions of KBD and DF. In this study, Affymetrix PrimeView™ Human Gene Expression Array was used for the quantification of genome-wide expression profiles from peripheral blood mononuclear cells of patients with KBD, KBD and DF, as well as healthy controls, for the comparative analysis. Functional enrichment analysis identified important processes related to the progression of KBD and DF. Several biomarkers were further confirmed by qRT-PCR. This should be helpful for the understanding of mechanisms of KBD and DF and the development of novel drugs and therapeutic methods.
Material and Methods
Study Population
Study samples consists of 12 Han Chinese subjects, including 4 patients with KBD, 4 patients with DF and KBD and 4 healthy controls. Patients with KBD were diagnosed as grade II or grade III according to the clinical criteria (diagnostic code GB16395-1996). Donors of dental fluorosis were diagnosed with the modified Dean classification, which is national diagnosis standard and experimental test for fluoride (WS/T 208–2011). The subjects with osteoarthritis (OA), Rheumatoid arthritis (RA) and other skeletal diseases were excluded. All subjects were matched based on age and gender. This study was approved by Qinghai Institute for Endemic Disease Prevention and Control and Medical ethics committee of Qinghai Institute for Endemic Disease Prevention and Control. All participants signed inform-consent documents. This study was carried by Qinghai provincial health and Family Planning Commission of science and Technology Education Department memo (2008) No. 6.
RNA Extraction
The peripheral blood were collected and stored at −80 °C. Blood samples needed to thaw at room temperature for 2 hours before RNA extraction. Total RNA was extracted with Agilent Total RNA Isolation Mini kit (Agilent Technologies, Santa Clara, CA) following the manufacturer’s instructions. The RNA amount was normalized using The Human-Actin Competitive PCR Set (Takara Bio, Kyoto, Japan). To check the integrity of the total RNA, 60 ng (normalized value) denatured total RNA was subjected to 1% agarose gel electrophoresis, and dyed with ethidium bromide. Extracted RNA was stored at −80 °C until cDNA synthesis.
Microarray Hybridization
Total RNA was reverse-transcribed into complementary DNA (cDNA), and then transcribed into cRNA and labled with Cy-Dye using Amino Allyl MessageAmp aRNA Kit (Ambion) following the manufacturer’s instructions. Thereafter, 0.5 μg of each labeled cRNA was purified separately and then mixed with hybridization buffer before being applied on the microarray. The hybridization solution was prepared with the In Situ Hybridization Plus kit (Agilent Technologies), and hybridization was performed in the hybridization chamber (Gene-Machines, San Carlos, CA, USA). Conditions of hybridization and washing were in accordance with the manufacturer’s recommendations (Agilent Technologies).
Analysis of Microarray Data
Affymetrix mRNA microarray were analyzed and transferred into CEL signal files using Affymetrix® GeneChip® Command Console® Software. A possible dye bias in the results was eliminated using an algorithm for the same extraction software (Extraction 9.3 Software, Agilent) that involves normalization factors (global normalization, location normalization). Normalized expression values was used for the identification of DEGs with limma package11 of R based on the thresholds of fold change >2 or <0.5 and FDR adjusted p-value < 0.05. For the exploration of processes involved in the development of KBD and DF, we also conducted functional enrichment analysis for DEGs through KOBAS12 with the thresholds of p-value < 0.05.
Quantitative Real-Time Reverse Transcription PCR
Total RNA was prepared for qRT-PCR. These RNA samples were transformed into complementary DNA (cDNA) using Superscript II reverse transcriptase (Invitrogen, Carlsbad, CA) and random primers. qRT-PCR was operated using the ABI 7500 Real-Time PCR system (Applied Biosys-tems, Foster City, CA) according to the manufacturer’s specification.
To validate the microarray results, 8 significant differentially expressed genes were selected for parallel qRT-PCR analysis, including FCRL6, KLRC3, CXCR4, CD93, CLK1, GPR18, SRP1, KLRF1. Data were analyzed with the 2−ΔΔCt method using GAPDH as internal control.
Statistical analysis
R version3.2.2 was used for all of the statistical analysis. The relative mRNA level in the qPCR analysis was represented by mean ± SD of the three replicates, and p-value < 0.05 was considered as statistical significant.
Results
Microarray Data Analysis
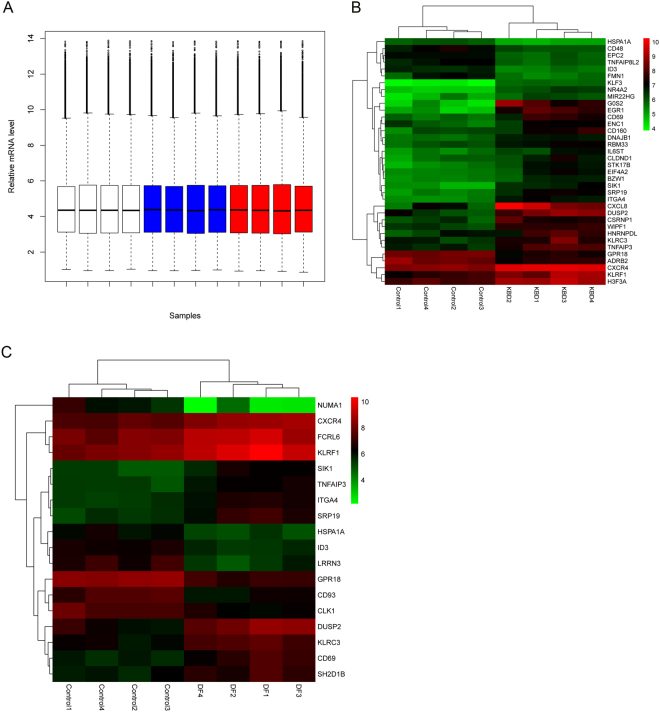
The GeneChip® PrimeView™ Human Gene Expression Array provides comprehensive coverage of the human genome in a cartridge array. Figure 1A illustrates the overall expression profiles in all samples. Comparable expression levels were obtained after normalization, which should be suitable for the following analysis. With the specified thresholds, 28 genes at higher level and 8 genes at lower level in KBD patients compared with controls were obtained. Tables 1 and 2 shows the up- and down-regulated genes and their enriched functions respectively. For patients with KBD and DF, we identified 11 up-regulated and 7 down-regulated genes compared with healthy controls. Tables 3 and 4 is the up- and down-regulated genes and their enriched functions respectively. Figure 1B,C illustrates the heatmap of DEGs in KBD patients and KBD and DF patients respectively in which green and red represents low and high expression level. A total of 10 overlaps, including 7 up-regulated (DUSP2, KLRF1, SRP19, KLRC3, CD69, SIK1 and ITGA4) and 3 down-regulated (ID3, HSPA1A and GPR18) genes, were identified between the two lists of DEGs. Strikingly, no gene was found to be significantly differential expression between patients with KBD and patients with DF.
Figure 1.
Microarray analysis. (A) Relative mRNA level of all samples after normalization. White boxes are healthy controls, blue boxes are KBD samples and red boxes are KBD with DF samples. (B) Two-way hierarchical clustering of expression profiles and samples of healthy controls and KBD samples. (C) Two-way hierarchical clustering of expression profiles and samples of healthy controls and KBD with DF samples.
Table 1.
List of genes differentially expressed in KBD *(up-regulated genes).
| Category | Gene Name | Public ID | Gene Symbol | Fold Change |
|---|---|---|---|---|
| apoptosis | G0/G1 switch 2 | NM_015714 | G0S2 | 10.1038 |
| nuclear receptor subfamily 4, group A, member 2 | NM_006186 | NR4A2 | 3.9593 | |
| salt-inducible kinase 1 | NM_173354 | SIK1 | 3.1203 | |
| early growth response 1 | NM_001964 | EGR1 | 8.6298 | |
| interleukin 6 signal transducer | NM_001190981 | IL6ST | 2.1023 | |
| metabolism | chemokine (C-X-C motif) ligand 8 | NM_000584 | CXCL8 | 7.4122 |
| Kruppel-like factor 3 (basic) | NM_016531 | KLF3 | 3.3914 | |
| signal recognition particle 19 kDa | NM_001204193 | SRP19 | 3.0827 | |
| cysteine-serine-rich nuclear protein 1 | NM_033027 | CSRNP1 | 2.7026 | |
| signal transduction | dual specificity phosphatase 2 | NM_004418 | DUSP2 | 4.3656 |
| CD69 molecule | NM_001781 | CD69 | 3.1153 | |
| killer cell lectin-like receptor subfamily C, member 3 | NM_002261 | KLRC3 | 2.5084 | |
| eukaryotic translation initiation factor 4A2 | NM_001967 | EIF4A2 | 2.3559 | |
| development corrlelated | integrin, alpha 4 | NM_000885 | ITGA4 | 2.8429 |
| chemokine (C-X-C motif) receptor 4 | NM_001008540 | CXCR4 | 2.6099 | |
| cystoskeleton and cell movement | serine/threonine kinase 17b | NM_004226 | STK17B | 2.5863 |
| WAS/WASL interacting protein family, member 1 | NM_001077269 | WIPF1 | 2.5435 | |
| ectodermal-neural cortex 1 (with BTB domain) | NM_001256574 | ENC1 | 2.0278 | |
| bone resorption | tumor necrosis factor, alpha-induced protein 3 | NM_001270507 | TNFAIP3 | 2.5231 |
| membrane protein | killer cell lectin-like receptor subfamily F, member 1 | NM_001291822 | KLRF1 | 2.4904 |
| CD160 molecule | NM_007053 | CD160 | 2.2795 | |
| claudin domain containing 1 | NM_001040181 | CLDND1 | 2.1154 | |
| DNA modification | H3 histone, family 3A | NM_002107 | H3F3A | 2.3112 |
| RNA process | heterogeneous nuclear ribonucleoprotein D-like | NM_001207000 | HNRNPDL | 2.2607 |
| basic leucine zipper and W2 domains 1 | NM_001207067 | BZW1 | 2.039 | |
| miscellaneous | MIR22 host gene (non-protein coding) | NM_032895 | MIR22HG | 2.2257 |
| DnaJ (Hsp40) homolog, subfamily B, member 1 | NM_006145 | DNAJB1 | 2.0591 | |
| RNA binding motif protein 33 | NM_001008408 | RBM33 | 2.0549 |
Table 2.
List of genes differentially expressed in KBD *(down-regulated genes).
| Category | Gene Name | Public ID | Gene Symbol | Fold Change |
|---|---|---|---|---|
| metabolism | enhancer of polycomb homolog 2 | NM_015630 | EPC2 | 0.4687 |
| inhibitor of DNA binding 3 | NM_002167 | ID3 | 0.4964 | |
| signal transduction | G protein-coupled receptor 18 | NM_001098200 | GPR18 | 0.4933 |
| cystoskeleton and cell | heat shock 70 kDa protein 1 A | NM_005345 | HSPA1A | 0.4949 |
| movement | formin 1 | NM_001103184 | FMN1 | 0.4882 |
| membrane protein | CD48 molecule | NM_001256030 | CD48 | 0.4984 |
| adrenoceptor beta 2 | NM_000024 | ADRB2 | 0.4836 | |
| miscellaneous | tumor necrosis factor, alpha-induced protein 8-like 2 | NM_024575 | TNFAIP8L2 | 0.4928 |
Table 3.
List of genes differentially expressed in KBD with DF (up-regulated genes).
| Category | Gene Name | Public ID | Gene Symbol | Fold Change |
|---|---|---|---|---|
| development correlated | dual specificity phosphatase 2 | NM_004418 | DUSP2 | 3.4894 |
| integrin, alpha 4 | NM_000885 | ITGA4 | 2.261 | |
| menbrane protein | killer cell lectin-like receptor subfamily F, member 1 | NM_001291822 | KLRF1 | 2.6744 |
| signal recognition particle 19 kDa | NM_001204193 | SRP19 | 2.6216 | |
| Fc receptor-like 6 | NM_001004310 | FCRL6 | 2.098 | |
| signal transduction | killer cell lectin-like receptor subfamily C, member 3 | NM_002261 | KLRC3 | 2.639 |
| CD69 molecule | NM_001781 | CD69 | 2.4183 | |
| salt-inducible kinase 1 | NM_173354 | SIK1 | 2.3002 | |
| apoptosis | tumor necrosis factor, alpha-induced protein 3 | NM_001270507 | TNFAIP3 | 2.1203 |
| cystoskeleton and cell movement | chemokine (C-X-C motif) receptor 4 | NM_001008540 | CXCR4 | 2.0905 |
| miscellaneous | SH2 domain containing 1B | NM_053282 | SH2D1B | 2.182 |
Table 4.
List of genes differentially expressed in KBD with DF (down-regulated genes).
| Category | Gene Name | Public ID | Gene Symbol | Fold Change |
|---|---|---|---|---|
| development correlated | inhibitor of DNA binding 3 | NM_002167 | ID3 | 0.4673 |
| G protein-coupled receptor 18 | NM_001098200 | GPR18 | 0.3675 | |
| menbrane protein | CD93 molecule | NM_012072 | CD93 | 0.4206 |
| cystoskeleton and cell movement | nuclear mitotic apparatus protein 1 | NM_001286561 | NUMA1 | 0.1127 |
| metabolism | heat shock 70 kDa protein 1 A | NM_005345 | HSPA1A | 0.4804 |
| leucine rich repeat neuronal 3 | NM_001099658 | LRRN3 | 0.3146 | |
| miscellaneous | CDC-like kinase 1 | NM_001024646 | CLK1 | 0.4413 |
qRT-PCR Validation of Microarray Data
8 genes were further verified by qRT-PCR. The results of qRT-PCR experiment were consistent with microarray analysis. According to qRT-PCR results, the expression levels of KLRC3, KLRF1, SRP19 and CXCR4 were higher in KBD and KBD with DF than controls, while expression levels of CLK1 and GPR18 were lower in both KBD and KBD with DF samples compared with healthy controls (shown in Fig. 2). Besides, FCRL6 was at higher level and CD93 was at lower level only in patients with KBD and DF.
Figure 2.

Histogram showing the ratio of expression levels of the 8 selected genes in three groups, as analyzed by quantitative real-time reverse transcription–polymerase chain reaction (PCR) (n = 12). Bars show the standard deviations. (A), (B) and (C) is KBD samples, KBD with DF samples and healthy subjects respectively.
Discussion
KBD and DF are complex diseases that determined by genetic to a large extent. In this study, we conducted a comparative analysis of gene expression profiles for patients with KBD, KBD and DF, and healthy controls using Affymetrix PrimeView™ Human Gene Expression Array. qRT-PCR was used to validate the oligonucleotide array data. Based on the gene ontology enrichment analysis, we divided these genes into different categories, including metabolism, apoptosis, cytoskeleton, signal transduction and bone development-related genes. Cartilage damage is the main characteristics of pathological changes in KBD, including necrosis in deeper articular cartilage, excessive apoptosis of chondrocytes, extracellular matrix degradation and so on13–15. Endemic fluorosis is a chronic systemic diseases, characterized with lumbocrural pain, anchylosis, bone-deformity. It is necessary to study bone development-related genes of KBD and DF. Significant differences in gene expression pattern were observed between patients with KBD and healthy controls, as well as between patients with KBD and DF and healthy controls. Ten genes (7 up-regulated and 3 down-regulated) were found to be consistently differentially expressed in KBD and KBD with DF samples, which contains several bone development-related genes, such as DUSP2, ITGA4, ID3, GPR18, and they should provide valuable information for further understanding of KBD and DF.
In mammalian cells, the dual-specificity phosphatase (DUSP) family is responsible for the dephosphorylation of threonine and tyrosine residues. Hamamura et al. showed that Dusp2 could suppress inflammation in antibody-induced arthritis in a mouse model through down-regulating inflammatory signs16. Besides, DUSP2 is involved in response to oxidative stress and apoptotic signaling, which play important roles in the development of KBD. Yin et al. reported that DUSP2 transcription was induced in response to oxidative stress, causing p53-dependent apoptosis17. Moreover, DUSP2 involved in the process of salvianolic acid a (SAA) effects rat cardiomyocytes by mediating regulation of the ERK1/2/JNK pathway18.
It was reported that the expression of integrin was associated with the osteoarthritis severity, especially ITGA4 (integrinα4). Becerril M et al. declared that ITGA4 played an important role in the loss of proteoglycans and clusters formation at OA early stages19. Proteoglycans, the main component of extracellular matrix of cartilage, were associated with articular cartilage metabolism in patients with KBD1,20. Consistent with the previous studies, we also identified ITGA4 as an important biomarker in the pathology of KBD.
Inhibitor of DNA Binding 3 (ID3), a transcription factor involved in the development of T cell and growth inhibition of a B cell progenitors, plays an important role in controlling cell cycle progression21–23. Thornemo et al. reported that ID3 is important for chondrocyte differentiation and ID proteins are expressed in a lot of cell types and decrease in various cell lines during differentiation24. Here, ID3 was also proved to be down-regulated in KBD, as well as KBD and DF samples, which should indicate its roles in the progression of KBD and DF.GPR18, one of the orphan G protein-coupled receptors, has been found to be a receptor for endogenous lipid neurotransmitters. Ramos et al. reported that GPR18 was differentially expressed in osteoarthritis patients25. Takenouchi et al. declared that GPR18 involved in the regulation of apoptosis26, and apoptosis plays an important role on pathological process of KBD, so GPR18 might contribute the development of KBD.
In summary, we conducted a comparative analysis of gene expression profiles to explore the common pathogenesis and the underlying molecular functions between KBD and DF. Significant differences in gene expression pattern were found between KBD, KBD with DF samples and healthy controls. Our results should provide novel insights for further study of the molecular mechanism of KBD and DF. While, further studies should be conducted to confirm our findings.
Acknowledgements
This study was supported by grants from the Science and Technology project of Qinghai Province (2014-ZJ-7 754) and Qinghai provincial Health and Family planning research project (2016-wjzdx-37) .
Author Contributions
Z.Q. takes responsibility for the integrity of the work as a whole, from inception to the finished manuscript. Conception & design: Z.Q., L.H.Q., H.D.L. Collection & Asembly of Data: M.J., L.H.Q., C.L.L., W.H.K. Analysis & Interpretation of the data: W.H.K., J.H., L.Q., B.S.L. Drafting & final approval of the manuscript: H.D.L., C.L.L., W.H.K., J.H., L.Q., B.S.L., Z.Q., M.J., L.H.Q.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Cao J, et al. Articular cartilage metabolism in patients with Kashin-Beck Disease: an endemic osteoarthropathy in China. Osteoarthritis and cartilage. 2008;16:680–688. doi: 10.1016/j.joca.2007.09.002. [DOI] [PubMed] [Google Scholar]
- 2.Xiong G. Diagnostic, clinical and radiological characteristics of Kashin-Beck disease in Shaanxi Province, PR China. International orthopaedics. 2001;25:147–150. doi: 10.1007/s002640100248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zeng Y, et al. X-ray image characteristics and related measurements in the ankles of 118 adult patients with Kashin-Beck disease. Chinese medical journal. 2014;127:2479–2483. [PubMed] [Google Scholar]
- 4.Yang J. Etiology of Kashin-Beck disease. Chinese Journal of Epidemiology. 1995;14:201–204. [Google Scholar]
- 5.Guo XZS, Mo DX. A role of low selenium in the occurrence of Kashin-Beck disease. Journal of Xi’an Jiaotong University(Medical Sciences) 1992;4:99–108. [Google Scholar]
- 6.Ping ZGGX, Wang FQ, Wang ZW. Epidemiological charac-ters of Kashin-Beck disease in nuclear families. Zhonghua liu xing bing xue za zhi = Zhonghua liuxingbingxue zazhi. 2004;25:848–851. [PubMed] [Google Scholar]
- 7.Li D. Pathogenetic cause of fluorine deficiency in Kaschin-Beck disease. Jilin Geology. 2007;4:20. [Google Scholar]
- 8.Liu HLH, Li F. Epidemiological survey of cases suffering Kaschin-eck disease and skeletal fluorosis. Chinese Journal of Control of Endemic Diseases. 2014;3:4. [Google Scholar]
- 9.Zhao LJ, et al. Surveillance on drinking-water-born endemic fluorosis in China, 2013. Zhonghua liu xing bing xue za zhi = Zhonghua liuxingbingxue zazhi. 2016;37:816–820. doi: 10.3760/cma.j.issn.0254-6450.2016.06.015. [DOI] [PubMed] [Google Scholar]
- 10.Sun DJ, G. Y. Progress and perspective of the prevention, control, and research of endemic fluorosis in China. Chinese Journal of Epidemiology. 2013;32:119–120. [Google Scholar]
- 11.Diboun I, Wernisch L, Orengo CA, Koltzenburg M. Microarray analysis after RNA amplification can detect pronounced differences in gene expression using limma. BMC genomics. 2006;7:252. doi: 10.1186/1471-2164-7-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xie C, et al. KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases. Nucleic acids research. 2011;39:W316–322. doi: 10.1093/nar/gkr483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu SX, et al. X. Difference in apoptosis-associated genes expression profiling and immunohistology analysis between Kashin-Beck disease and primary osteoarthritis. Chinese Science Bulletin. 2014;59:833–839. doi: 10.1007/s11434-014-0130-z. [DOI] [Google Scholar]
- 14.Chen J, et al. Altered proteolytic activity and expression of MMPs and aggrecanases and their inhibitors in Kashin-Beck disease. Journal of orthopaedic research: official publication of the Orthopaedic Research Society. 2015;33:47–55. doi: 10.1002/jor.22708. [DOI] [PubMed] [Google Scholar]
- 15.Wang W, et al. Oxidative stress and status of antioxidant enzymes in children with Kashin-Beck disease. Osteoarthritis and cartilage. 2013;21:1781–1789. doi: 10.1016/j.joca.2013.08.002. [DOI] [PubMed] [Google Scholar]
- 16.Hamamura K, et al. Salubrinal acts as a Dusp2 inhibitor and suppresses inflammation in anti-collagen antibody-induced arthritis. Cellular signalling. 2015;27:828–835. doi: 10.1016/j.cellsig.2015.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yin Y, Liu YX, Jin YJ, Hall EJ, Barrett JC. PAC1 phosphatase is a transcription target of p53 in signalling apoptosis and growth suppression. Nature. 2003;422:527–531. doi: 10.1038/nature01519. [DOI] [PubMed] [Google Scholar]
- 18.Xu T, et al. The anti-apoptotic and cardioprotective effects of salvianolic acid a on rat cardiomyocytes following ischemia/reperfusion by DUSP-mediated regulation of the ERK1/2/JNK pathway. PloS one. 2014;9:e102292. doi: 10.1371/journal.pone.0102292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Almonte-Becerril M, Costell M, Kouri JB. Changes in the integrins expression are related with the osteoarthritis severity in an experimental animal model in rats. Journal of orthopaedic research: official publication of the Orthopaedic Research Society. 2014;32:1161–1166. doi: 10.1002/jor.22649. [DOI] [PubMed] [Google Scholar]
- 20.Li S, Cao J, Caterson B, Hughes CE. Proteoglycan metabolism, cell death and Kashin-Beck disease. Glycoconjugate journal. 2012;29:241–248. doi: 10.1007/s10719-012-9421-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Szczepanowski M, et al. Immunohistochemical detection of inhibitor of DNA binding 3 mutational variants in mature aggressive B-cell lymphoma. Haematologica. 2016;101:e259–261. doi: 10.3324/haematol.2015.138701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maruyama T, et al. Control of the differentiation of regulatory T cells and T(H)17 cells by the DNA-binding inhibitor Id3. Nature immunology. 2011;12:86–95. doi: 10.1038/ni.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang Y, Zhang B, Fang J, Cao X. Hypomethylation of DNA-binding inhibitor 4 serves as a potential biomarker in distinguishing acquired tamoxifen-refractory breast cancer. International journal of clinical and experimental pathology. 2015;8:9500–9505. [PMC free article] [PubMed] [Google Scholar]
- 24.Thornemo M, Jansson ES, Lindahl A. Expression of the ID1 and ID3 genes during chondrocyte differentiation. Annals of the New York Academy of Sciences. 1996;785:337–339. doi: 10.1111/j.1749-6632.1996.tb56302.x. [DOI] [PubMed] [Google Scholar]
- 25.Ramos YF, et al. Genes expressed in blood link osteoarthritis with apoptotic pathways. Annals of the rheumatic diseases. 2014;73:1844–1853. doi: 10.1136/annrheumdis-2013-203405. [DOI] [PubMed] [Google Scholar]
- 26.Takenouchi R, Inoue K, Kambe Y, Miyata A. N-arachidonoyl glycine induces macrophage apoptosis via GPR18. Biochemical and biophysical research communications. 2012;418:366–371. doi: 10.1016/j.bbrc.2012.01.027. [DOI] [PubMed] [Google Scholar]