Abstract
In this paper, the feasibility and advantages of employing high performance liquid chromatographic (HPLC) fingerprints combined with pattern recognition techniques for quality control of Shenmai injection were investigated and demonstrated. The Similarity Evaluation System was employed to evaluate the similarities of samples of Shenmai injection, and the HPLC generated chromatographic data were analyzed using hierarchical clustering analysis (HCA) and soft independent modeling of class analogy (SIMCA). Consistent results were obtained to show that the authentic samples and the blended samples were successfully classified by SIMCA, which could be applied to accurate discrimination and quality control of Shenmai injection. Furthermore, samples could also be grouped in accordance with manufacturers. Our results revealed that the developed method has potential perspective for the original discrimination and quality control of Shenmai injection.
Keywords: Shenmai injection, High performance liquid chromatography, Fingerprint, Pattern recognition
1. Introduction
Fingerprint technique is a powerful tool for the quality control of multi-component herbal medicines and has been accepted as a useful means for the evaluation and quality control of herbal materials and their finished products. Fingerprint analysis has been introduced and accepted by State Food and Drug Administration of China to standardize injections made from traditional Chinese medicines (TCMs) and their raw materials. So far, fingerprint of TCM, especially the chromatographic fingerprint, has gained more and more attention [1], [2], [3], [4]. However, it is obvious that the fingerprint technique, which is based on similarity evaluation, is not efficient enough for distinguishing different origins of the samples because a clear-cut signature could not be given easily even though the calculated cosine values are relatively big enough. Therefore, more effective classification methods, such as pattern recognition, should be employed.
In this study, Shenmai injection, made from Radix Ginseng Rubra and Radix Ophiopogonis, was investigated as a typical drug model to develop HPLC fingerprinting combined with pattern recognition methods for evaluating the quality of herbal medicine. Shenmai injection has been used to treat coronary atherosclerotic cardiopathy and viral myocarditis, and it is also capable of raising tumor patient's immunity. The main constituents in Shenmai injection were found to be ginsenosides and opioponins [5], [6], [7].
Pattern recognition is a primary conceptual activity of the human being. Even without our awareness, clustering on the information that is conveyed to us is constant. However, this clustering activity is frequently based on a few selected properties and is not exempt from personal prejudice. Naturally, when objects are defined by a significant number of properties which are or have been made quantitative, and it is intended to obtain exempt results (natural clusters), the use of mathematical tools is mandatory, and the results can be relatively objective. Pattern recognition techniques are often divided into unsupervised and supervised methodologies. In the former, information stems from the data and there are no pre-classified groups. Hierarchical clustering analysis (HCA) is one of these techniques. In contrast, supervised methodologies, such as soft independent modeling of class analogy (SIMCA), involve classifying samples into predefined structures. Typically, there is a set of labeled objects (training set) that is used to establish decision rules so as to classify new objects.
HCA is a hard modeling method: a provided number of categories should be given. For online analysis of samples, if a new sample, especially the unqualified, is added into HCA, it may be classified into the qualified ones. Moreover, to distinct the qualified and unqualified samples, a batch of former qualified samples should be analyzed, which makes the analysis procedure very tedious. Therefore, HCA is not suitable for online analysis.
SIMCA is a soft modeling method, new samples (belonging or not to one of the studied classes) can be tested for class memberships. If we consider the set of test samples which do not belong to any class, none of them should be assigned to these classes according to SIMCA. In online process, training set can be set up using former qualified samples. So in online analysis, new samples can be imported directly into SIMCA models as test set. Former qualified samples are not required to be analyzed for the second time [8], [9].
In this study, a relatively large volume (1 mL) of qualified sample mixed with a small volume (10 μL) of injection samples produced by the other manufacturer (blended sample) was used to analog the unqualified samples to some extent. High performance liquid chromatographic fingerprints combined with some pattern recognition methods were used to establish an objective pattern recognition system to discriminate blended samples from the authentic ones, and to control the quality of Shenmai injection.
2. Experimental
2.1. Reagents and materials
Acetonitrile of HPLC grade was obtained from Fisher Scientific (New Jersey, USA), and purified water was purchased from Robust Company (Hangzhou, China).
Reference standards of ginsenosides Re, Rg1, Rb1, Rf, Rc, Rb2, and Rd were purchased from National Institute for the Control of Pharmaceutical and Biological Products (Beijing, China). Shenmai injections were purchased from two different manufacturers (Zhejiang and Sichuan Province) in China.
2.2. Apparatus and chromatographic conditions
Analysis was performed on a Shimadzu LC-10AT system, equipped with a diode array detector (SPD-10Avp) and CLASS-VP chromatography workstation (Shimadzu, Japan). All the separations were carried out on a Phenomenex Gemini C18 column (250 mm×4.6 mm, 5 μm). The mobile phase consisted of acetonitrile (A)–water (B) pumped at 1.0 mL/min with a gradient elution program of 15–75% (A) in 85 min [10], [11], [12]. The injection volume was 20 μL, the column temperature was 30 °C, and the wavelength was set at 203 nm.
2.3. Preparation of standard solution
Accurately weighted Re, Rg1, Rb1, Rf, Rc, Rb2, and Rd reference standards were dissolved in acetonitrial and diluted quantitatively to obtain working standard solutions, respectively (Table 1).
Table 1.
The preparation of working standard solutions.
| Compound | Preparation | Concentration (mg/mL) |
|---|---|---|
| Re | 2.040 mg→10 mL | 0.2040 |
| Rg1 | 2.033 mg→10 mL | 0.2033 |
| Rb1 | 2.004 mg→10 mL | 0.2004 |
| Rf | 2.054 mg→10 mL | 0.2054 |
| Rc | 1.954 mg→10 mL | 0.1954 |
| Rb2 | 1.970 mg→10 mL | 0.1970 |
| Rd | 2.098 mg→10 mL | 0.2098 |
2.4. Preparation of sample solutions
The authentic sample solution was filtrated through a 0.45 μm filter before HPLC analysis.
The blended sample solution was a mixture of sample (Zhejiang) of 1 mL and another injection sample (Sichuan) of 10 μL, and was filtrated through a 0.45 μm filter before HPLC analysis.
2.5. Data analysis
Data of each chromatogram were exported from the workstation to be further analyzed. All data processing was performed using m-files written for Matlab™ R2010b (The Mathworks, Natick, MA).
Usually, after the procedure of similarity analysis, unsupervised method (HCA) constitutes a second step in data analysis. Without assuming any previous knowledge of sample class, this method enables the visualization of the data in a reduced dimensional space built on the dissimilarities between samples with respect to their chemical composition. Sometimes, the results of the initial unsupervised analysis are confirmed by a third supervised analysis. This employs classification method, such as SIMCA, allowing first to separate samples of different origins, and second to identify more robust samples [8], [13], [14].
2.5.1. Similarity analysis
Similarity analysis was performed by professional software named Similarity Evaluation System for Chromatographic Fingerprint of Traditional Chinese Medicine composed by Chinese Pharmacopoeia Committee (version 2004A). The cosine values in entire chromatographic profiles among samples were calculated and the simulative mean chromatogram as a representative standard chromatogram for a group of chromatograms was calculated and generated. In addition, the relative retention time (RRT) and relative peak area (RPA) of each characteristic peak related to the reference peak were also calculated for quantitative expression of the chemical properties in the chromatographic pattern of Shenmai injection [7], [11].
2.5.2. HCA
HCA is a multivariate analysis technique that is used to sort samples into groups. This technique comprises an unsupervised classification procedure that involves measuring either the distance or the similarity between the samples to be clustered. It is unsupervised because it does not require previous information in the system under study, therefore, is an ideal technique when no previous information is at the scientist's disposal. The similarity or dissimilarity between samples is usually represented in a dendrogram for ease of interpretation. Each sample is similar to the others within a group but different from those in other groups with respect to a predetermined selection criterion. In our study, the HCA of samples was performed using Matlab™ R2010b (The Mathworks, Natick, MA). Ward's method as the amalgamation rule and the Euclidean distance as metric were used to establish clusters [14], [15], [16], [17].
2.5.3. SIMCA
In SIMCA, a separate principal component analysis (PCA) is performed on each class in the training set. Classification in SIMCA is made by comparing the residual variance of a sample with the average residual variance of the samples that make up the class. F-statistic is used to provide a quantitative measure of this comparison (Gemperline, 2006). As a soft modeling method, SIMCA does not forcibly assign a certain sample to a single group if it is an outlier. If the classes in the data set overlapped, a sample might be assigned to more than one group [8], [9], [14], [15], [16], [17].
The model achieved by SIMCA for each category is also evaluated in terms of the recognition rate/sensitivity and rejection rate/specificity. The sensitivity of the model is known as the percentage of samples belonging to the category correctly identified by the mathematical model, and its specificity, as the percentage of samples foreign to the category classified as foreign.
3. Results and discussion
3.1. Optimization of chromatographic conditions
To give sufficient chemical information and ideal separation in the chromatograms, the detection wavelength, the column types, the mobile phase and the gradient elution procedure, the column temperature and flow rate were investigated in this study. As we know, ginsenosides have relatively strong absorption at 203 nm. So according to the UV absorption maxima of compounds on three-dimensional chromatograms of HPLC-DAD detection, the monitoring wavelength was set at 203 nm with relatively more peaks of ginsenosides.
Four types of chromatographic columns, namely, Agilent Zorbax SB-C18 column, Phenomenex Luna C18 column, Dikma Diamonsil C18 and Phenomenex Gemini C18, were chosen. The injection showed different retention behaviors on these columns. Phenomenex Gemini C18 column was more suitable for separation of the constituents of Shenmai injection.
Acetonitrile (A)–water (B) was investigated as mobile phase based on the chosen wavelength. The gradient elution program was optimized as follows: a linear gradient of 5–100% (A) in 60 min or in 80 min; a linear gradient of 15–100% (A) in 60 min; a linear gradient of 15–75% (A) in 60 min, 85 min, 100 min or in 120 min. Finally, the linear gradient of 15–75% (A) in 85 min was chosen as the optimum gradient with relatively shorter running time, higher resolution and more peaks of ginsenosides and opioponins in chromatograms [5].
From the results of comparative study of column temperature of 25, 30 and 35 °C and flow rate of 0.8, 1.0 and 1.2 mL/min, the flow rate was set at 1.0 mL/min when column temperature was kept at 30 °C considering resolution and running time.
3.2. Method validation
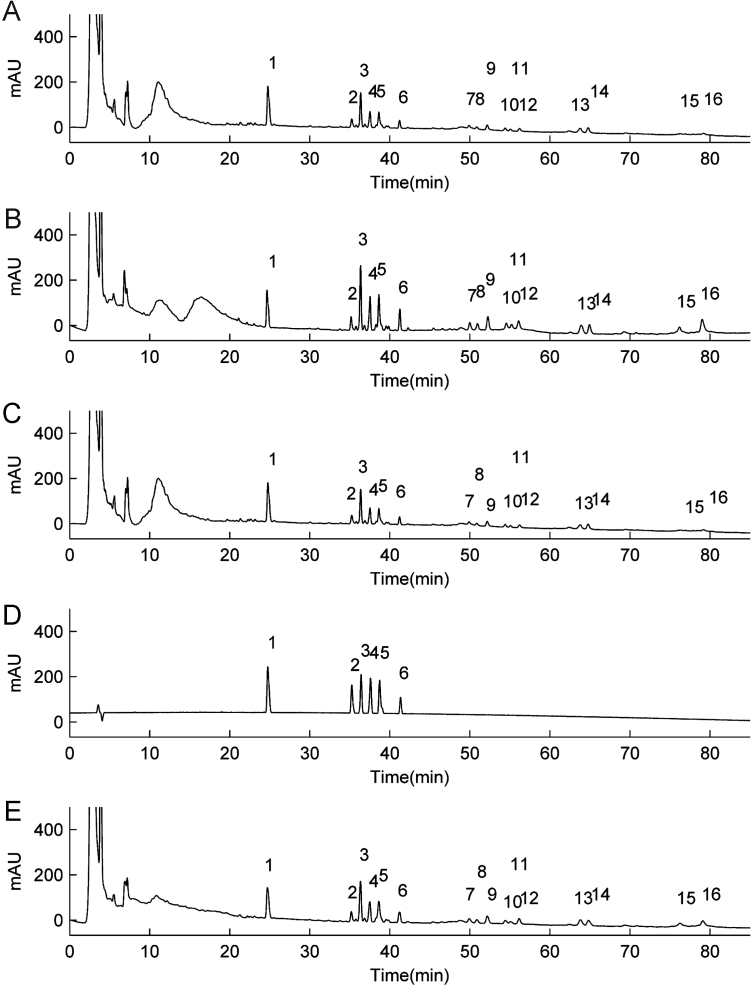
The method was validated for parameters such as precision, repeatability and stability. In order to obtain stable and repeatable chromatographic fingerprint of Shenmai injection for quality control, the method validation of HPLC fingerprint analysis was performed on the basis of the RRT and RPA. The Rb1 peak at retention time 36.1 min (No. 3) was selected as a reference peak because it was a relative strong single peak in the middle of the chromatogram and with a better resolution from the other peaks (Fig. 1A).
Figure 1.
HPLC fingerprint of Shenmai injection from different origins: (A) chromatogram of samples from Zhejiang; (B) chromatogram of blend samples; (C) chromatogram of samples from Sichuan; (D) chromatogram of reference standards: 1 represents ginsenoside Re and Rg1; 2 represents ginsenoside Rf; 3 represents ginsenoside Rb1; 4 represents ginsenoside Rc; 5 represents ginsenoside Rb2, and 6 represents ginsenoside Rd; (E) representative chromatogram of Shenmai injection samples (Average): No. 1–16 represents the common peaks.
The precision was determined by replicate injection of the same sample solution for six times. The RRT and RPA of common peaks were calculated. The relative standard deviations (RSDs) of RRT and RPA of common peaks for precision were not exceeding 3.0% and 5.0%, respectively.
The repeatability was determined by injection of six individual sample solutions from the same sample in the same way. The RRT and RPA of common peaks were calculated. The RSDs of RRT and RPA were within 3.0% and 5.0%, respectively.
Stability was tested with one sample solution stored at room temperature for 0, 1, 3, 6, 12 and 24 h, and the areas of the main peaks were found to be stable within 24 h (RSD<5.0%). The RSDs of RRT and RPA were not more than 3.0% and 5.0%, respectively.
All results indicated that the method was adequate, valid and satisfactory.
3.3. HPLC fingerprints
In this study, 20 authentic samples purchased from two different manufacturers (10 from Sichuan and 10 from Zhejiang) and 10 blended samples were analyzed under the established HPLC conditions. Large amounts of HPLC chromatograms will help us to establish stable and credible standard fingerprints of Shenmai injections. Mean chromatograms of the three groups of samples were generated by the professional software named Similarity Evaluation System for Chromatographic Fingerprint of Traditional Chinese Medicine (Version 2004A), shown in Fig. 1A–C. In this research, the mean chromatograms were regarded as standardized characteristic fingerprints of Shenmai injection. Peaks existed in all chromatograms of the three mean chromatograms were regarded as “common peaks”, which indicated the similarity among various samples. Sixteen common peaks were found from the samples by comparison of their ultraviolet spectra and HPLC retention time. In all 16 peaks, six main peaks were identified. The chromatogram of these six mixed reference standards was marked in Fig. 1D. The mean chromatogram and these 16 common peaks were marked in Fig. 1E.
3.4. Similarities analysis
It is necessary that chromatographic fingerprint should be evaluated by their similarities. The mean chromatograms of samples from different origins had represented their different characters to some extent. Moreover, this software was employed to synchronize the chromatographic peaks and to calculate the cosine values of angles between vectors among different chromatograms, as well as to compute the mean chromatogram as a representative standard chromatogram for a group of chromatograms. The closer the cosine values are to 1, the more similar the two chromatograms are. From Table 2, the results showed that for all those samples, the cosine value of every sample from the same origin was more than 0.929. It is also indicated that the entire chromatograms of samples from the same manufacturer were generally consistent and stable. As to the samples from different manufacturers, the cosine value between mean chromatograms was also more than 0.872 (Table 2). It was obvious that cosine value was not efficient enough for distinguishing different origins of the samples because a clear-cut signature could not be given easily although the calculated cosine values are relatively big enough. The above results have shown the limitation of cosine value for the quality control of TCM.
Table 2.
Similarity comparison (cosine values) of the chromatographic patterns of different samples.
| No.c | Zhejiang |
No.c | Blend |
No.c | Sichuan |
|||
|---|---|---|---|---|---|---|---|---|
| Samea | Totalb | Same | Total | Same | Total | |||
| 1 | 0.971 | 0.959 | 11 | 0.989 | 0.887 | 21 | 0.978 | 0.944 |
| 2 | 0.981 | 0.957 | 12 | 0.987 | 0.886 | 22 | 0.976 | 0.934 |
| 3 | 0.974 | 0.942 | 13 | 0.971 | 0.880 | 23 | 0.987 | 0.936 |
| 4 | 0.932 | 0.895 | 14 | 0.990 | 0.916 | 24 | 0.984 | 0.941 |
| 5 | 0.933 | 0.909 | 15 | 0.995 | 0.882 | 25 | 0.967 | 0.951 |
| 6 | 0.965 | 0.920 | 16 | 0.994 | 0.872 | 26 | 0.954 | 0.951 |
| 7 | 0.973 | 0.944 | 17 | 0.992 | 0.880 | 27 | 0.976 | 0.944 |
| 8 | 0.929 | 0.905 | 18 | 0.992 | 0.879 | 28 | 0.972 | 0.944 |
| 9 | 0.981 | 0.959 | 19 | 0.992 | 0.874 | 29 | 0.987 | 0.945 |
| 10 | 0.978 | 0.947 | 20 | 0.993 | 0.880 | 30 | 0.985 | 0.945 |
The cosine values were calculated using the samples from the same origin.
The cosine values were calculated using all the samples from different origins.
Samples 1–10 were the injection samples purchased from Zhengdaqingchunbao Co., Ltd., Zhejiang Province; Samples 11–20 were mixture of 10 batches of Shenmai injection from Zhejiang (1 mL) and one batch of Shenmai injection from Sichuan (10 μL); Samples 21–30 were the injection samples purchased from Sanjingshenghe Co., Ltd., Sichuan province.
3.5. HCA
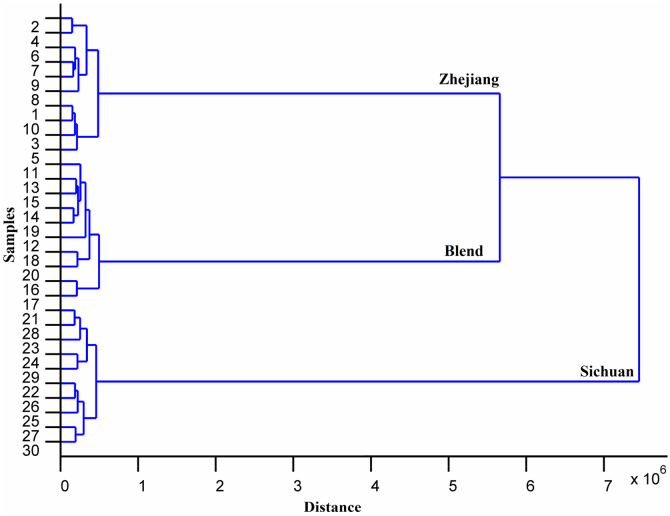
In order to assess this tendency, a hierarchical clustering analysis of samples was performed. This procedure finds natural groupings of the data set. The Euclidean distance and Ward's method were applied. The original data for HCA were exported from Similarity Evaluation System and were normalized by subtracting their arithmetic mean value and then dividing their standard deviation. As is shown in Fig. 2, there were three main clusters corresponding to different origins of samples in the dendrogram, while the subsets gave more information on the samples of different batches.
Figure 2.
Dendrogram of clustering of Shenmai injection samples according to different manufacturers. Nos. 1–10: samples of Zhejiang; Nos. 11–20: blend samples; Nos. 21–30: samples of Sichuan.
However, from the above results, we could find a problem of HCA: how to measure homogeneity or, more exactly, similarity between the individual samples in the same cluster; i.e. how to decide the degree of the similarity among those samples. From this point of view, the conception of class of one kind of injections seems to be more reasonable. Thus, the chemical pattern recognition methods, such as SIMCA, should be taken into consideration for reasonable definition of the class of Shenmai injection.
3.6. SIMCA
After being normalized by subtracting their arithmetic mean value, the data set processed in similarity analysis was considered as the initial data set in SIMCA. F-statistic value was found to be 1.735 (p<0.01). The principal component numbers, which of all three models were found to be 1, were obtained with the use of the cross-validation procedure [8], [9].
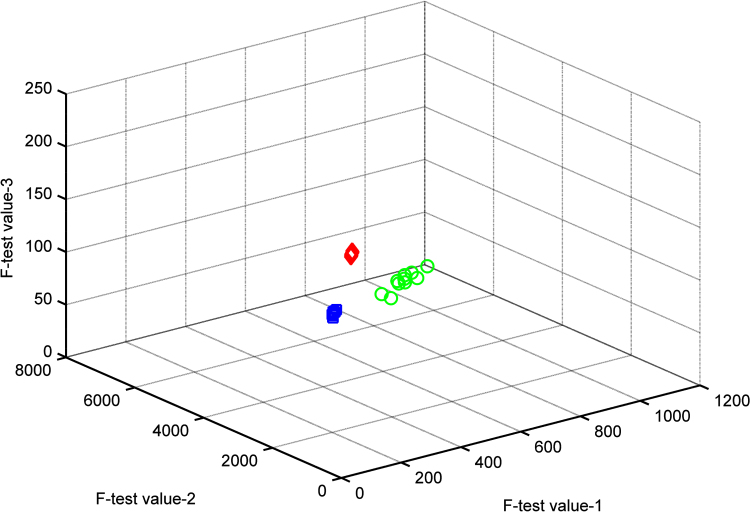
The sensitivity of the three models was high, and only several samples were outside its box. Twenty-eight of all 30 samples were correctly classified, which means that the total sensitivity of the three models was 93% (28/30), whereas the sensitivity of each model was 90% (9/10), 90% (9/10) and 100% (10/10), respectively (Table 3, Table 4). This could also be seen from Cooman's plots of the SIMCA models (Fig. 3).
Table 3.
The test results of SIMCA.
| No.a | Model 1 |
Model 2 |
Model 3 |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Residue variance | F-test value | Category | Residue variance | F-test value | Category | Residue variance | F-test value | Category | |
| 1 | 0.1144 | 0.7677 | 1b | 6.4610 | 248.90 | 1 | 125.00 | 148.50 | 1 |
| 2 | 0.0270 | 0.1814 | 1 | 5.6240 | 216.70 | 1 | 130.40 | 154.90 | 1 |
| 3 | 0.2501 | 1.6780 | 1 | 5.4200 | 208.80 | 1 | 130.00 | 154.40 | 1 |
| 4 | 0.0910 | 0.6105 | 1 | 5.7790 | 222.60 | 1 | 130.00 | 154.50 | 1 |
| 5 | 0.0827 | 0.5549 | 1 | 6.1120 | 235.50 | 1 | 129.80 | 154.30 | 1 |
| 6 | 0.0307 | 0.2066 | 1 | 6.3700 | 245.40 | 1 | 126.90 | 150.80 | 1 |
| 7 | 0.3600 | 2.4160 | 0 | 4.3200 | 166.40 | 0 | 132.20 | 157.10 | 0 |
| 8 | 0.1665 | 1.1180 | 1 | 6.7540 | 260.20 | 1 | 127.70 | 151.70 | 1 |
| 9 | 0.0390 | 0.2620 | 1 | 5.9110 | 227.70 | 1 | 129.60 | 154.00 | 1 |
| 10 | 0.1050 | 0.7047 | 1 | 6.9300 | 267.00 | 1 | 127.30 | 151.20 | 1 |
| 11 | 5.0060 | 33.600 | 2 | 0.0091 | 0.3487 | 2 | 175.70 | 208.80 | 2 |
| 12 | 4.9570 | 33.270 | 2 | 0.0112 | 0.4304 | 2 | 175.10 | 208.00 | 2 |
| 13 | 4.8710 | 32.690 | 2 | 0.0186 | 0.7149 | 2 | 176.10 | 209.20 | 2 |
| 14 | 4.7860 | 32.120 | 2 | 0.0244 | 0.9388 | 2 | 175.00 | 207.90 | 2 |
| 15 | 5.6990 | 38.250 | 0 | 0.0612 | 2.3593 | 0 | 178.00 | 211.50 | 0 |
| 16 | 5.3610 | 35.980 | 2 | 0.0237 | 0.9142 | 2 | 177.90 | 211.40 | 2 |
| 17 | 5.4140 | 36.340 | 2 | 0.0200 | 0.7723 | 2 | 178.90 | 212.60 | 2 |
| 18 | 5.0160 | 33.660 | 2 | 0.0253 | 0.9737 | 2 | 176.80 | 210.00 | 2 |
| 19 | 4.9860 | 33.460 | 2 | 0.0163 | 0.6281 | 2 | 175.60 | 208.60 | 2 |
| 20 | 5.1140 | 34.320 | 2 | 0.0109 | 0.4196 | 2 | 176.00 | 209.10 | 2 |
| 21 | 158.10 | 1061.0 | 3 | 197.00 | 7591.0 | 3 | 0.5104 | 0.6065 | 3 |
| 22 | 159.20 | 1069.0 | 3 | 193.10 | 7438.0 | 3 | 0.8133 | 0.9663 | 3 |
| 23 | 166.20 | 1116.0 | 3 | 194.30 | 7486.0 | 3 | 0.8093 | 0.9615 | 3 |
| 24 | 177.60 | 1192.0 | 3 | 203.90 | 7854.0 | 3 | 1.0620 | 1.2620 | 3 |
| 25 | 143.60 | 963.60 | 3 | 179.90 | 6929.0 | 3 | 0.7950 | 0.9445 | 3 |
| 26 | 162.00 | 1087.0 | 3 | 197.70 | 7616.0 | 3 | 0.4053 | 0.4815 | 3 |
| 27 | 164.40 | 1103.0 | 3 | 201.00 | 7744.0 | 3 | 0.8024 | 0.9533 | 3 |
| 28 | 156.90 | 1053.0 | 3 | 194.20 | 7481.0 | 3 | 0.4183 | 0.4970 | 3 |
| 29 | 168.50 | 1131.0 | 3 | 202.00 | 7780.0 | 3 | 0.5632 | 0.6691 | 3 |
| 30 | 143.40 | 962.70 | 3 | 186.70 | 7193.0 | 3 | 0.9752 | 1.1590 | 3 |
Samples 1–10 were the injection samples purchased from Zhengdaqingchunbao Co., Ltd., Zhejiang province; Samples 11–20 were mixture of 10 batches of Shenmai injection from Zhejiang (1 mL) and one batch of Shenmai injection from Sichuan (10 μL); Samples 21–30 were the injection samples purchased from Sanjingshenghe Co., Ltd., Sichuan province.
1 represents that the sample was classified as category 1 (Zhejiang), 2 represents that the sample was classified as category 2 (Blend), and 3 represents that the sample was classified as category 3 (Sichuan), 0 represents that the sample was classified to none of these categories.
Table 4.
Sensitivity and specificity using SIMCA.
| REC and REJ* | Sample |
|||
|---|---|---|---|---|
| Zhejiang | Blend | Sichuan | Total | |
| Sensitivity | 90% (9/10) | 90% (9/10) | 100% (10/10) | 93% (28/30) |
| Specificity | 100% (20/20) | 100% (20/20) | 100% (20/20) | – |
* represents the recognition rate and rejection rate using SIMCA.
Figure 3.
Cooman's plot of SIMCA for the samples. F-test value-1 represents the F-test value based on model 1. F-test value-2 represents the F-test value based on model 2. F-test value-3 represents the F-test value based on model 3.  : data points of model 1 (Zhejiang).
: data points of model 1 (Zhejiang).  : data points of model 2 (blend).
: data points of model 2 (blend). : data points of model 3 (Sichuan).
: data points of model 3 (Sichuan).
None of the models built admitted samples from another category, and thus the specificity was 100% in all cases (20/20) (Table 4). Cooman's plot showed that a large distance separated the three classes, especially the samples from Sichuan and the other samples. The distance between samples from Zhejiang and the blend was small, which was in accordance with their origins.
However, it should be noted that we do not have a sufficiently large data set in the current research, and the obtained clustering tendency may have an over-fitting problem and its prediction ability would be limited. If many new samples are available in the future, they will be pretreated in the same way like the old ones. Then one can do supervised discriminate analysis with testing its prediction performance.
4. Conclusions
The proposed HPLC fingerprint combined with pattern recognition methods is an effective and comprehensive tool for quality consistency evaluations of Shenmai injection. The HPLC method showed good precision and repeatability. Using fingerprint technique combined with pattern recognition methods, such as HCA and SIMCA, the authentic samples and the blended samples were successfully classified. Furthermore, samples could also be grouped in accordance with manufacturers. Attributed to statistical method (F-test) and the aid of computer technique, the method of SIMCA showed more objective and accurate results, and the analyzing procedure was more simplified. Our results revealed that the developed method has potential perspective for the original discrimination and quality control of Shenmai injection.
Acknowledgments
This work was financially supported by National Key Scientific Project for New Drug Discovery and Development of China (Grant no. 2009ZX09301-012).
Footnotes
Peer review under responsibility of Xi'an Jiaotong University.
References
- 1.Yang D.Z., An Y.Q., Jiang X.L. Development of a novel method combining HPLC fingerprint and multi-ingredients quantitative analysis for quality evaluation of traditional Chinese medicine preparation. Talanta. 2011;85:885–890. doi: 10.1016/j.talanta.2011.04.059. [DOI] [PubMed] [Google Scholar]
- 2.Zhao Y.L., Fan R.H., Yuan H.X. Development of the fingerprints for the quality evaluation of Viscum coloratum by high performance liquid chromatography. J. Pharm. Anal. 2011;1:113–118. doi: 10.1016/S2095-1779(11)70020-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yan S.K., Xin W.F., Luo G.A. An approach to develop two-dimensional fingerprint for the quality control of Qingkailing injection by high-performance liquid chromatography with diode array detection. J. Chromatogr. A. 2005;1090(1–2):90–97. doi: 10.1016/j.chroma.2005.07.066. [DOI] [PubMed] [Google Scholar]
- 4.Kong W.J., Wang J.B., Zang Q.C. Fingerprint-efficacy study of artificial Calculus bovis in quality control of Chinese materia medica. Food Chem. 2011;127:1342–1347. doi: 10.1016/j.foodchem.2011.01.095. [DOI] [PubMed] [Google Scholar]
- 5.Zhang H.J., Wu Y.J., Cheng Y.Y. Analysis of ‘SHENMAI’ injection by HPLC/MS/MS. J. Pharm. Biomed. Anal. 2003;31:175–183. doi: 10.1016/s0731-7085(02)00565-4. [DOI] [PubMed] [Google Scholar]
- 6.Fan X.H., Wang Y., Cheng Y.Y. LC/MS fingerprinting of Shenmai injection: a novel approach to quality control of herbal medicines. J. Pharm. Biomed. Anal. 2006;40:591–597. doi: 10.1016/j.jpba.2005.10.036. [DOI] [PubMed] [Google Scholar]
- 7.Huang L., Wu X.M., Ji Y. Fingerprint analysis of placenta polypeptide injection by high performance liquid chromatography. J. Pharm. Anal. 2012;2:71–75. doi: 10.1016/j.jpha.2011.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Branden K.V., Hubert M. Robust classification in high dimensions based on the SIMCA method. Chem. Intell. Lab. Syst. 2005;79:10–21. [Google Scholar]
- 9.Flumignan D.L., Tininis A.G., Ferreira F.D.O. Screening Brazilian C gasoline quality: application of the SIMCA chemometric method to gas chromatographic data. Anal. Chim. Acta. 2007;595:128–135. doi: 10.1016/j.aca.2007.02.049. [DOI] [PubMed] [Google Scholar]
- 10.Wang Y.H., Qiu C., Wang D.W. Identification of multiple constituents in the traditional Chinese medicine formula Sheng-Mai San and rat plasma after oral administration by HPLC–DAD–MS/MS. J. Pharm. Biomed. Anal. 2011;54:1110–1127. doi: 10.1016/j.jpba.2010.11.034. [DOI] [PubMed] [Google Scholar]
- 11.Zhao A.J., Shen P.Q., Qi D.Q. Reserch of the HPLC fingerprint of Shenmai injection. Qilu Pharm. Aff. 2010;29:525–527. [Google Scholar]
- 12.Zhang J.L., Cui M., He Y. Chemical fingerprint and metabolic fingerprint analysis of Danshen injection by HPLC-UV and HPLC-MS methods. J. Pharm. Biomed. Anal. 2005;36:1029–1035. doi: 10.1016/j.jpba.2004.09.009. [DOI] [PubMed] [Google Scholar]
- 13.Zhou J.D. Improvement of determination method for the content of Shenmai injection. J. Chin. Med. 2011;26:196–197. in Chinese. [Google Scholar]
- 14.Chen Y., Zhu S.B., Xie M.Y. Quality control and original discrimination of Ganoderma lucidum based on high-performance liquid chromatographic fingerprints and combined chemometrics methods. Anal. Chim. Acta. 2008;623:146–156. doi: 10.1016/j.aca.2008.06.018. [DOI] [PubMed] [Google Scholar]
- 15.Ni Y.N., Lai Y.H., Brandes S. Multi-wavelength HPLC fingerprints from complex substances: an exploratory chemometrics study of the Cassia seed example. Anal. Chim. Acta. 2009;647:149–158. doi: 10.1016/j.aca.2009.06.021. [DOI] [PubMed] [Google Scholar]
- 16.Lucio-Gutiérrez J.R., Coello J., Maspoch S. Enhanced chromatographic fingerprinting of herb materials by multi-wavelength selection and chemometrics. Anal. Chim. Acta. 2012;710:40–49. doi: 10.1016/j.aca.2011.10.010. [DOI] [PubMed] [Google Scholar]
- 17.Kong W.J., Zhao Y.L., Xiao X.H. Quantitative and chemical fingerprint analysis for quality control of Rhizoma Coptidischinensis based on UPLC-PAD combined with chemometrics methods. Phytomedicine. 2009;16:950–959. doi: 10.1016/j.phymed.2009.03.016. [DOI] [PubMed] [Google Scholar]